Abstract
The recent report of the synthesis of glycoproteins by the abundant intestinal symbionts Bacteroides showed that these organisms use a novel bacterial enzyme to decorate their surfaces with a sugar residue derived from their environment. As a first step in understanding the importance of these glycoproteins to the bacteria and to the bacterial–host symbiosis, we identified and characterized the abundant glycoproteins of Bacteroides distasonis (proposed reclassification as Parabacteroides distasonis) [Sakamoto M, Benno Y (2006) Int J Syst Evol Microbiol 56:1599–1605]. Using lectin-affinity purification followed by tandem mass spectrometry, we identified a family of at least nine glycoproteins, similar only to the S-layer glycoproteins of Tannerella forsythia. Analysis of one of these purified glycoproteins demonstrated that the glycan is primarily a polymer of xylose, a monosaccharide rarely found in bacterial glycans. Even more unexpected was the finding that seven of nine of the glycoprotein promoters undergo DNA inversion, a process that we show is active in their endogenous human environment. Using cross-species functional assays, we show that a single serine family site-specific recombinase globally mediates the inversions of these glycoprotein promoters. This regulatory mechanism is similar to that of the Bacteroides fragilis capsular polysaccharides and establishes DNA inversion as a general and ancient means of regulation of glycan-containing surface molecules of these important human intestinal symbionts.
Keywords: Bacteroides, DNA inversion, microbiota, xylose
Humans are colonized with trillions of microorganisms with which they have coevolved to form mutually beneficial relationships. The human colon, in particular, is a complex ecosystem dense with hundreds of species of organisms, most of which are just beginning to be appreciated. Studies over the last 5 years have yielded important insights into the composition, genomic content, and genetic features of the human colonic microbiota. Scientific interest in members of this complex environment has increased significantly in part because of studies demonstrating important health benefits provided by certain members of the microbiota. Few reports, however, have linked particular bacterial molecules to specific beneficial effects. Elucidation of these symbiotic factors will greatly benefit from a comprehensive analysis of the biology of the intestinal bacteria themselves.
Analyses of the numerically abundant Bacteroides spp. have revealed that a large portion of their genomes is dedicated to the acquisition and degradation of polysaccharides derived from their ecosystem. In addition, a large amount of their genetic material is also devoted to the synthesis and regulation of their own surface glycans. For example, Bacteroides fragilis and Bacteroides thetaiotaomicron synthesize at least eight and seven capsular polysaccharides, respectively (1, 2). The expression of these surface molecules is subject to phase variation, a reversible ON–OFF phenotype, dictated by DNA inversions of the polysaccharide biosynthesis loci promoters (1, 2). In B. fragilis, a single serine family site-specific recombinase, designated Mpi, mediates inversion of all of the invertible polysaccharide promoters (3). These abilities allow the bacteria to elaborate an extremely flexible and adaptive surface architecture that is likely pivotal for their long-term survival and predominance in the human colon.
The importance of bacterial surface molecules in mediating crucial interactions with the host has been well established for pathogenic bacteria. The surfaces of these commensal organisms will likely have equally important roles in the establishment of mutually beneficial relationships with the host. In fact, capsular polysaccharide A of B. fragilis is one of the few molecules produced by the intestinal microbiota that has been shown to have a symbiotic function, in this case in modulating the host immune system (4).
We recently showed that a novel bacterial enzyme, Fkp, allows B. fragilis to incorporate exogenous fucose, acquired from its ecosystem by the breakdown of plant polysaccharides and host glycans, into its capsular polysaccharides and glycoproteins (5). The importance of these fucosylated molecules to the Bacteroides is demonstrated by the defect in intestinal colonization by a B. fragilis mutant that cannot add fucose to capsular polysaccharides and glycoproteins (5). An fkp-specific probe hybridized with the genomic DNA of four other related species and each of these organisms incorporates exogenous fucose into glycoproteins (5). The amount of exogenous fucose incorporated into the glycoproteins of the numerically abundant colonic species Bacteroides distasonis far exceeded that of the other species tested, making it an interesting subject for further study.
Because of a lack of knowledge of the glycoproteins of these intestinal organisms, we do not yet understand their importance to the biology of the bacteria, their role in intestinal colonization, or their possible contribution to a symbiotic relationship with the host. The present study was undertaken to identify and characterize the abundant glycoproteins of B. distasonis and to analyze their expression potential in their native human intestinal ecosystem.
Results
Identification and Characterization of a B. distasonis Glycoprotein Family.
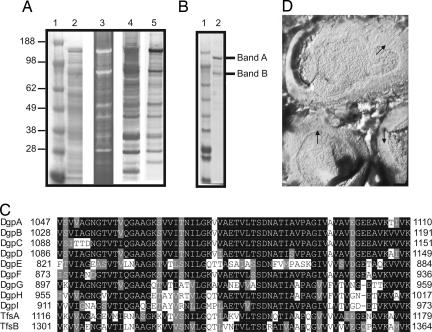
We previously showed that exogenous 3H-fucose is incorporated into multiple B. distasonis glycoproteins (5). Two glyco-proteins in particular, of ≈140 kDa and 90 kDa, were abundantly labeled with 3H-fucose (Fig. 1A, lane 5). The strong signal observed from these proteins might indicate their relative abundance in the cell or could arise from a large number of fucose residues per molecule. A Coomassie-stained SDS/PAGE gel of B. distasonis whole-cell lysate demonstrates that molecules that migrate consistent with the 140-kDa and 90-kDa glycoproteins are very abundant in the cell (Fig. 1A, lane 2). Because 3H-fucose labeling identifies only glycoproteins that contain fucose, we stained a gel containing B. distasonis whole-cell lysate with a general glycoprotein stain and found that the glycoproteins that stain most strongly with the glycostain also label most intensely with 3H-fucose (Fig. 1A, lane 3). Therefore, under the in vitro conditions in which these bacteria were grown, the majority of the glycoproteins produced by this organism contain fucose. Aleuria aurantia lectin (AAL) binds to fucose-linked α-1,3 or α-1,6. Using biotinylated AAL, we determined that this lectin binds the majority of B. distasonis glycoproteins (Fig. 1A, lane 4).
Fig. 1.
Analysis of B. distasonis glycoproteins. (A) Whole-cell lysates were subjected to SDS/PAGE and stained with Coomassie (lane 2), stained with a glycoprotein reagent (lane 3), blotted and probed with A. aurantia lectin (AAL) (lane 4), or metabolically labeled with 3H-fucose before lysis and analyzed with a phosphoimager (lane 5). (B) Coomassie-stained SDS/PAGE of the AAL-purified fraction. Two bands excised from the gel for further analyses are marked. (C) Box shade alignment demonstrating the similarity of the C-terminal domains of the Dgp and Tfs proteins. (D) Freeze–fracture EM of B. distasonis showing a 2D array (arrows) consistent with the appearance of an S-layer.
An AAL-agarose column was used to affinity-purify the glycoproteins bound by the lectin. The AAL purification resulted in enrichment of the 140-kDa and 90-kDa glycoproteins (Fig. 1B, lane 2). To determine the total glycan composition of the AAL-purified fraction, this material was analyzed at the Complex Carbohydrate Research Center at the University of Georgia (Athens, GA), which revealed that 2% of the sample was carbohydrate by weight. These data, combined with those shown in Fig. 1A, demonstrate that these glycoproteins are abundant but not heavily glycosylated. Glycosyl composition analysis revealed that the most abundant monosaccharide in this fraction was xylose (Table 1), a residue rarely found in bacterial glycans. Common monosaccharides such as glucose and rhamnose were also present, as was fucose.
Table 1.
Glycan composition of the AAL-purified fraction
| Glycosyl residue | Mol% |
|---|---|
| Xylose | 38.0 |
| Glucose | 24.9 |
| Rhamnose | 19.0 |
| Fucose | 9.5 |
| Mannose | 8.6 |
To identify the 140-kDa and 90-kDa glycoproteins, the AAL-purified fraction was subjected to SDS/PAGE (Fig. 1B, lane 2). Bands A and B, corresponding to the 140-kDa and 90-kDa glycoproteins, respectively, were excised from the gel, trypsin-digested, and analyzed by tandem MS (MS/MS). To generate a protein database to which we could compare the experimental spectra, we used the most complete B. distasonis sequence released by the Genome Sequencing Center at Washington University School of Medicine (St. Louis, MO). This database comprised 13 contigs containing 4,462,845 bp, or 92.8% of the 4,811,369-bp genome.
The spectral output from band A matched two proteins in the database; these were tentatively designated ORF1144 and ORF1097. ORF1144 has a calculated molecular mass of 121.89 kDa, of which 78.7 kDa were experimentally observed (64.5%). In contrast, three peptides of ORF1097 were identified corresponding to 3.7 kDa of the calculated 128.0-kDa protein (2.9%). The experimental spectra from band B also matched two proteins, ORF889 and ORF1144. For ORF889, 69 of 106 tryptic peptides (34.2 kDa of the calculated 93.5-kDa protein, or 36.6%) were identified. In addition, 25 of the 249 tryptic peptides from ORF1144 were identified, all localizing to the N-terminal two-thirds of the protein.
Analysis of these proteins demonstrated that all three are similar to each other, especially at the C termini (Fig. 1C). When we searched the B. distasonis protein database for additional products with C-terminal similarity, a total of nine proteins were detected that are very similar over their last 63 aa (Fig. 1C) and some with full-length similarity [supporting information (SI) Tables 3 and 4]. The proteins ranged in size from 884 aa to 1191 aa and were encoded by genes on noncontiguous regions of the genome. Based on the previous data showing that bands A and B were glycoproteins, we predict that these nine products comprise a family of related glycoproteins and therefore designate them DgpA–DgpI for distasonis glycoprotein. Based on the abundance of some of these glycoproteins, we predict that they may be S-layer proteins, which are abundant proteins forming 2D crystalline arrays around the surface of some bacteria. Two S-layer proteins, TfsA and TfsB, have recently been described for the related oral organism Tannerella forsythia (6). Comparison of the protein sequences of TfsA and TfsB with the Dgp proteins revealed that they also have the same C-terminal motif, and each shares full-length similarity with various Dgp products (SI Tables 3 and 4). No other products currently in the publicly available databases are similar to the Dgp proteins.
B. fragilis and B. thetaiotaomicron, the two most studied Bacteroides species, do not make S-layers and do not synthesize products similar to the Dgp glycoproteins. To determine whether B. distasonis synthesizes an S-layer, freeze-etch electron microscopy, which is the best method to visualize S-layers, was used. This analysis revealed 2D arrays on the surface of B. distasonis consistent with the appearance of S-layers (Fig. 1D), but not on B. fragilis (data not shown). In an attempt to directly correlate the Dgp products with S-layer formation, we expressed DgpA (ORF1144) and DgpC (ORF1097) individually in B. fragilis 9343. Although both of these products were expressed in B. fragilis, DgpA was not expressed full-length (SI Fig. 5). Freeze–fracture EM did not reveal an S-layer in either of these B. fragilis transconjugants likely because of incorrect size of the products, because of missing or incorrect glycosylation, and/or because B. fragilis does not contain other machinery necessary for S-layer formation. Treatment of B. distasonis at 55°C for 2 min, a relatively gentle treatment that does not lyse the cells, liberated a subset of cellular proteins including DgpA and DgpC, supporting their localization on the cell surface (data not shown). The combined data indicating the abundance of these glycoproteins, their similarity to known S-layer proteins, their extraction from the cell surface, and the visualization of an S-layer lattice on B. distasonis strongly suggest that the Dgp products are S-layer glycoproteins.
Purification and Glycan Analysis of DgpC.
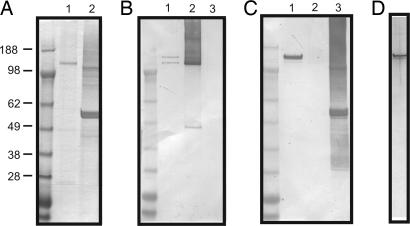
Glycan analysis of the AAL-purified fraction of B. distasonis demonstrated the presence of xylose, glucose, rhamnose, fucose, and mannose. To determine the glycan component of individual glycoproteins, we attempted to purify DgpA and DgpC from B. distasonis. His-tagged fusions of both DgpA and DgpC were purified from Escherichia coli. The DgpC-His tagged product remained full-length in E. coli, but the majority of the DgpA-His-tagged protein was degraded (Fig. 2A). These purified His-tagged proteins were used to generate antisera in rabbits, which was tested for reactivity with B. distasonis cell lysates by immunoblot. The anti-DgpC serum reacted with two bands from B. distasonis, possibly corresponding to glycosylated and unglycosylated forms of the molecule (Fig. 2B). This blot also shows that there is no cross-reactivity between this antiserum and the His-tagged DgpA product (Fig. 2B, lane 3). The antiserum to DgpA-His reacted specifically with a single molecule of B. distasonis and did not react with His-tagged DgpC (Fig. 2C). The DgpC-His antiserum was used successfully to purify the native form of this molecule from a B. distasonis cell lysate (Fig. 2D). Attempts to purify DgpA in the same manner were unsuccessful presumably because the avidity of the DgpA-His antiserum for native DgpA was too low. Consistent with this assumption, DgpA-His antiserum did not deplete DgpA from a B. distasonis cell lysate in an immunoprecipitation assay (data not shown).
Fig. 2.
Purification and glycan analysis of DgpC. (A) Coomassie-stained SDS/PAGE showing purification of His-tagged DgpC (lane 1) and DgpA (lane 2) from E. coli. (B) Immunoblot analysis of B. distasonis whole-cell lysate (lane 1), DgpC-His from E. coli (lane 2), and DgpA-His from E. coli (lane 3) probed with antiserum generated to DgpC-His. (C) Immunoblot analysis of B. distasonis whole-cell lysate (lane 1), DgpC-His from E. coli (lane 2), and DgpA-His from E. coli (lane 3) probed with DgpA-His antiserum. (D) Silver-stained SDS/PAGE analysis of native DgpC purified from B. distasonis.
The purified native DgpC glycoprotein was analyzed for glycan composition and glycosyl linkages. Similar to the AAL-purified fraction, the glycan comprised 2.1% of the molecule by weight. Unlike the AAL fraction, the DgpC glycan was composed almost entirely of xylose (Table 2). Glucose, mannose, and rhamnose were also present in low amounts, as were trace amounts of C16:0 and C18:0 fatty acids, suggesting a possible anchoring mechanism for these glycoproteins. Although a small fraction of DgpC was identified by MS/MS analysis of band A from the AAL-purified fraction, no fucose was detected. It is possible that the cell lysis preparation was not stringent enough to disaggregate the S-layer, and, if DgpC was a minor component of an S-layer complex with DgpA, these molecules could have been copurified by AAL. Based on the percentage of the molecule that is glycan and the molar percentage of each monosaccharide, we calculated that there are on average 16 xylose residues and one or two glucose or other six-carbon sugar per DgpC molecule. Glycosyl linkage analysis revealed that the xylose molecules are largely linked 1,2 and 1,4 (SI Table 5).
Table 2.
Glycan composition of native DgpC
| Glycosyl residue | Mol% |
|---|---|
| Xylose | 89.6 |
| Glucose | 6.9 |
| Mannose | 2.2 |
| Rhamnose | 1.3 |
dgp Promoters Undergo DNA Inversion.
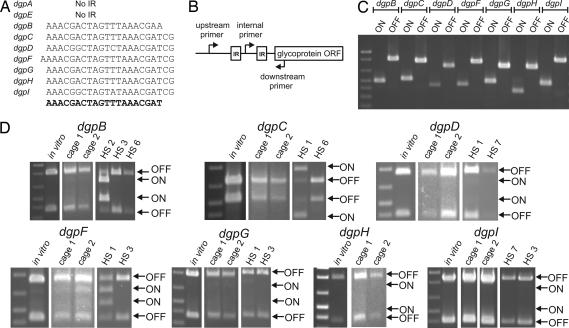
Analysis of the DNA immediately surrounding the Dgp coding regions revealed the presence of 20- to 23-bp inverted repeats (IRs) upstream of seven of nine of the dgp genes (Fig. 3A). DgpA and DgpE, which were the major proteins identified in bands A and B, did not have upstream IR regions. These seven sets of IRs are remarkably similar to each other, and their presence is reminiscent of the invertible regions upstream of seven of the capsular polysaccharide biosynthesis loci of B. fragilis. Between the IRs, there is 173–177 bp of intervening DNA, each containing a nearly exact match with the consensus promoter sequence described for B. fragilis (7) (SI Fig. 6). The database contains each of these seven promoter sequences only in the OFF orientation for transcription of the corresponding dgp. Therefore, PCR experiments were performed and confirmed that each of these seven promoter regions inverts (Fig. 3C). We sequenced the PCR products corresponding to the ON orientation and found that, for each region, the result of the inversion is the flipping of DNA exactly between the IRs (SI Fig. 6).
Fig. 3.
dgp promoter inversions. (A) The upstream dgp IR sequences showing a consensus at the bottom in bold. (B) Schematic of the procedure used to demonstrate DNA inversions. (C) EtBr-stained agarose gel of PCR products amplified from B. distasonis chromosome when the internal primer is used with either the upstream or downstream primer. (D) EtBr-stained agarose gels of PCR digestion experiments showing the percentage of organisms with ON and OFF orientations for the invertible dgp promoters from in vitro cultures, from the feces of mice, or from human fecal samples.
Our analyses of bands A and B identified both of the glycoproteins whose promoters do not undergo inversion and may be constitutively expressed and only one phase-variable glycoprotein. We therefore predicted that the majority of in vitro grown bacteria have the seven invertible promoters oriented OFF. To quantify these promoter orientations, PCR digestion was used and demonstrated that, as expected, almost none of the in vitro grown bacteria have any of the invertible Dgp promoters oriented ON (Fig. 3D). The most abundant ON promoter is DgpF, but still <10% of the bacterial population has this promoter oriented ON.
Because in vitro culture is a poor mimic of the conditions the organisms encounter in the mammalian intestine, we performed this quantitative PCR digestion assay using DNA isolated from the feces of gnotobiotic mice that had been colonized with B. distasonis for 1 week. The results of these analyses were similar to those of in vitro grown organisms: nearly all of the bacteria had the seven invertible dgp promoters oriented OFF. Although the gnotobiotic mouse colonization model more closely represents the environment that B. distasonis encounters in the intestine, it is still an artificial system. B. distasonis are symbionts of humans rather than of mice, and, because we are using gnotobiotic animals, other members of the microbiota are not present. Therefore, we chose to analyze the orientations of the dgp promoters directly from human fecal samples. DNA was isolated from the feces of eight healthy adult humans, and PCR digestion was performed for each of the invertible promoters. We obtained PCR products for all of the regions, except for the dgpH IR region, from at least two of the human samples. Human fecal sample 1 (HS1) and HS3 were the most successful, yielding products for four dgp IR regions, whereas HS4, HS5, and HS8 yielded no PCR products. Digestion of the resulting PCR products demonstrated ON orientations for some of the dgp promoters from some human samples. In particular, the dgpB promoter is oriented ON in ≈90% of HS2, but almost completely OFF in the other two samples. Also, HS1 contained a proportion of bacteria with ON promoters for both the dgpC and dgpF regions. Therefore, in the bacteria's normal environment, expression of Dgp glycoproteins is phase-variable.
Identification of the dgp Promoter Invertase.
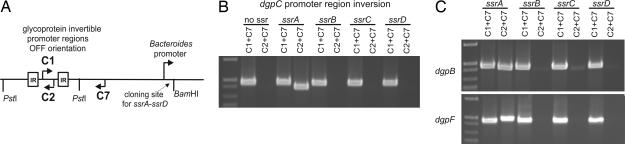
The invertible promoter regions upstream of the seven capsular polysaccharide biosynthesis loci of B. fragilis have a common core IR sequence (3) with half sites (8), and inversion of all seven is mediated by a single serine family site-specific recombinase (Ssr) designated Mpi (3). Our analysis of the dgp IR regions shows that they also have half sites (SI Fig. 6), indicating that their inversions are mediated by Ssrs rather than tyrosine family site-specific recombinases. Based on the similarity of the seven dgp IR regions, we predicted that their inversions would be mediated by a single global DNA invertase, similar to Mpi. Analysis of the B. distasonis genome revealed four products, which we designated SsrA–SsrD, that are 66–43% similar to Mpi. Unlike B. fragilis, there is currently no described means of introducing DNA into B. distasonis; therefore, creation of site-directed mutants is currently not possible. Thus, to identify which of the four Ssrs inverts the dgp promoters, we performed cross-species functional assays where both an invertible promoter region and a candidate ssr gene were cloned on a plasmid (Fig. 4A), conjugally transferred into the heterologous host B. fragilis, and monitored for inversion. Using the dgpC promoter region as our model, we first confirmed that this promoter does not invert when present in B. fragilis without a B. distasonis ssr gene (Fig. 4B). Addition of ssrA to this vector, but not the other three ssr genes, resulted in inversion of the dgpC promoter in B. fragilis (Fig. 4B). We then selected two other promoters, dgpB and dgpF, and performed the same analysis. There was extensive inversion of both of these regions in B. fragilis when ssrA was added, compared with the other ssr genes for which little or no inversion was detected (Fig. 4C). Based on these results, we have redesignated SsrA as Gfi for glycoprotein family invertase.
Fig. 4.
Identification of the dgp promoter DNA invertase. (A) Scheme used to clone the invertible dgp promoter regions and site-specific recombinase genes on the same vector for functional inversion assays in B. fragilis. (B) EtBr-stained agarose gel of PCR products demonstrating that ssrA mediates inversion of the dgpC promoter in B. fragilis. (C) EtBr-stained agarose gel of PCR products demonstrating that ssrA also mediates inversion of the dgpB and dgpF promoters in B. fragilis.
Discussion
More than any bacteria studied to date, members of the predominant intestinal Bacteroidetes have evolved a very complex biology that depends heavily on both the acquisition and synthesis of polysaccharides. This fact is illustrated by the large amount of genetic material, up to 10% of some genomes, dedicated to these two processes. In fact, the expression of structurally distinct, phase-variable, glycan-containing surface molecules seems to be a hallmark of these organisms. The interplay between the acquisition and surface expression of these glycans is not completely understood but likely has important consequences for both the bacteria and the host.
The accumulating data demonstrate that these intestinal organisms decorate their surfaces with monosaccharides that are abundant in their ecosystem, such as fucose and xylose. Xylose is prevalent in plant polymers and therefore abundant in the human colon because of the host's inability to digest plant polysaccharides. Xylose is also present on host proteoglycans but is rarely found as a component of bacterial glycans. Whether the surface expression of these particular monosaccharides by endogenous bacteria helps to mask the organisms so they appear more like their surroundings is a possible explanation for the evolution of this mechanism.
An unexpected finding of this study is that the glycoprotein promoters undergo inversion. These inversions are mediated by a global Ssr, designated Gfi, which is, of the four Ssrs of B. distasonis, the most similar to Mpi. We found that gfi is adjacent to the fkp homolog in a head-to-head orientation on the B. distasonis genome, a genomic pairing not present in B. fragilis. Therefore, Fkp and Mpi, which are very important to the synthesis and regulation of capsular polysaccharides of B. fragilis, have homologs in B. distasonis with equally important roles in the synthesis and regulation of its glycoproteins.
This study, coupled with those of previous studies, demonstrates that DNA inversions are ubiquitous in this family of intestinal symbionts, especially in regulating glycan-containing molecules. Our analysis of the dgp promoters from human fecal samples shows that expression of these glycoproteins is phase-variable in their normal habitat. The strong evidence suggesting that the Dgp products are S-layer glycoproteins reveals a very significant role for these molecules to the bacteria. S-layers are believed to have evolved as a consequence of different and changing ecological conditions, likely providing a selective advantage (9). Indeed, the ability to synthesize a vast assortment of S-layer glycoproteins, coupled with the ability to rapidly and reversibly turn their synthesis ON and OFF, likely provides a survival advantage to these organisms during their lifelong association with the host. This study provides a foundation for future work to determine whether this family of glycoproteins also confers beneficial properties to the host.
Materials and Methods
Bacterial Strains and Growth Conditions.
B. distasonis strain 8503 was used for all studies and was grown as previously described (10). Labeling with 3H-l-fucose was achieved as previously described (5). E. coli strains DH5α and BL21(DE3) containing recombinant plasmids were grown in L broth or on L agar plates containing 100 μg/ml ampicillin.
Analysis of Products Separated by SDS/PAGE.
SDS/PAGE and Western blotting were performed essentially as described (10). Analyses of bacteria metabolically labeled with 3H-fucose and AAL probing of blots were performed as described (5). Glycoproteins were stained in gel with the Pro-Q Emerald 300 Glycoprotein Stain (Invitrogen, Carlsbad, CA).
AAL Affinity Chromatography.
B. distasonis (100 ml) was grown to stationary phase, harvested by centrifugation, resuspended in 10 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 0.1% Tween 20, and lysed by freeze–thaw and French press. The cleared lysate was mixed with AAL-agarose (Vector Laboratories, Burlingame, CA) and washed extensively with the binding buffer, and bound molecules were eluted with 100 mM l-fucose in the same buffer.
MS/MS Analysis.
AAL-purified molecules were separated by SDS/PAGE by using a 40-cm vertical gel and visualized with Simply Blue SafeStain (Invitrogen). Proteins contained in gel slices were digested with trypsin, and peptides were extracted with CH3CN and subjected to nano-LC/MS/MS using a LCQ Deca Thermo-Finnegan instrument. Using the partially completed B. distasonis 8503 genome sequence, we built a database from all possible ORFs encoding 100 aa or more in the forward and reverse orientations. The Turbo-SEQUEST algorithm was used to match the MS/MS spectra with tryptic fragments of the indexed database. Data were then filtered with BioWorks 3.2 by using stringent filters for charge state and cross-correlation coefficient as follows: MH1+ = 1.8, MH2+ = 2.5, and MH3+ = 3.5.
Freeze–Fracture EM.
Samples for freeze etching were rapidly frozen between two planchets by using a propane jet freezer (RMC MF7200) and transferred to a Balzers model 360 freeze etch apparatus as a sandwich. After the system stabilized at −100°C, the planchets were separated, allowed to etch for 4.5 min, then shadowed with ≈2–4 nm of carbon platinum then 20 nm of carbon. After raising to room temperature, the replicas were floated onto water, cleaned in 50% sulfuric acid/water, washed, and picked up onto 200 mesh copper grids. The replicas were observed in a Japan Electron Optical Laboratories JEM 100 CX II at 60 kV and photographed.
Cloning dgpA, dgpC, and dgpE for Expression in B. fragilis.
These three dgp genes were PCR-amplified from B. distasonis by using the primers listed in SI Table 6. The PCR products were digested with BamHI and cloned into the BamHI site of pFD340 (11), and transformants were screened for proper orientation in relation to the vector-borne promoter. Recombinant plasmids were transferred to B. fragilis 9343 by conjugation as described (3).
Purification of DgpC.
Native DgpC glycoprotein was purified from B. distasonis by affinity chromatography by using polyclonal antibodies raised to His-tagged DgpC. dgpA and dgpC were PCR-amplified from the B. distasonis chromosome by using primers listed in SI Table 5 and cloned in the correct orientation into the BamHI site of pET16b. His-tagged DgpA and DgpC were overexpressed in E. coli BL21(DE3), purified under denaturing conditions by using the ProBond Purification System (Invitrogen), and used to raise polyclonal antisera in rabbits (Lampire Biological Laboratories, Pipersville, CA).
Antibodies to DgpC were purified from the crude antiserum by affinity to His-tagged DgpC as described (12) using a precolumn containing His-tagged DgpA to remove nonspecific antibodies. His-tagged DgpA and DgpC were purified from E. coli by using the hybrid denaturing/nondenaturing procedure of the Ni-NTA Purification System (Invitrogen), leaving the proteins bound noncovalently to the columns. DgpC antiserum was mixed with the His-tagged DgpA precolumn, and then the flow-through was mixed with the His-tagged DgpC column. This column was washed with 50 mM Tris·HCl (pH 7.4)/150 mM NaCl and 50 mM Tris·HCl (pH 7.4)/2 M NaCl, and then antibodies were eluted with 4 M MgCl2, dialyzed immediately against PBS, and concentrated.
The purified antibodies were bound and covalently cross-linked to a protein A column by using the ImmunoPure rProtein A IgG Plus Orientation Kit (Pierce, Rockford, IL). A precolumn was prepared by using crude antiserum raised to an irrelevant His-tagged protein. B. distasonis was grown to an OD600 of 0.9–1.0, harvested by centrifugation, resuspended in PBS (pH 7.4), 1 mM EDTA, 0.1% Triton X-100, and one Complete Protease Inhibitor Tablet (Roche Diagnostics, Mannheim, Germany) per 25 ml, and lysed by freeze–thaw and sonication. The cleared lysate was added to the precolumn, and the flow-through was added to the DgpC antibody column. This was washed with PBS (pH 7.4)/0.1% Triton X-100, and bound protein was eluted with 4 M MgCl2, adjusting the pH immediately to 8.0. Fractions containing pure DgpC were identified by SDS/PAGE stained with the SilverQuest Silver Staining Kit (Invitrogen) and Western blots probed with DgpC antiserum; these fractions were pooled, concentrated, dialyzed against deionized water, and lyophilized.
Carbohydrate Composition and Glycosyl Linkage Analyses.
Carbohydrate compositions and glycosyl linkage analyses were performed at the Complex Carbohydrate Research Center at the University of Georgia. For composition analysis, dry samples were used to generate methyl glycosides, which were then per-O-trimethylsilylated and analyzed by GC/MS as described (13). For linkage analysis, the sample was permethylated, depolymerized, reduced, and acetylated, and the resulting partially methylated alditol acetates were analyzed by GC/MS as described (14).
Demonstration of dgp Promoter Inversions.
Inversion of the promoter regions that are flanked by IRs was demonstrated by PCR (Fig. 3). For each region, PCR was performed by using B. distasonis 8503 chromosomal DNA with a central primer between the two IRs and either a forward primer upstream of the first IR or a reverse primer downstream of the second IR. Primer sequences are listed in SI Table 6. To obtain a sequence across the upstream IR junction in the ON orientation, PCR was performed by using the forward primer with the central primer for each of these seven regions, and the products were sequenced at the Brigham and Women's Hospital DNA Core Sequencing Facility.
Extraction of DNA from Mouse and Human Feces.
Mouse studies were approved by the Harvard Medical Area standing committee on animals. Male Swiss–Webster (3-week-old) germ-free mice were purchased from Taconic (Germantown, NY). The mice were housed in gnotobiotic isolators (two mice per cage) and fed an autoclaved rodent chow diet (Zeigler Bros., Gardners, PA). Mice were monoassociated with B. distasonis by spreading organisms on their fur. Fresh fecal samples were collected and pooled from both mice in each cage 1 week after colonization. Eight human fecal samples were obtained as secondary use material from a study of healthy pregnant women where informed consent was obtained from all subjects. The samples were renumbered so that there are no identifiers linking them to specific individuals. Approval for experimentation with these samples was obtained from the Partners Human Research Committee. Mouse and human fecal samples were suspended in PBS, particulate material was allowed to sediment for 5 min, and 200 μl of the nonsedimented material was pelleted and used for chromosomal extraction with the ExtractMaster Fecal DNA Extraction Kit (Epicentre, Madison, WI).
Quantitation of dgp Promoter Orientations.
The proportions of bacteria with dgp promoters in the ON and OFF orientations from in vitro grown and from mouse and human fecal samples were measured by PCR amplification and restriction digestion by using a protocol similar to that described previously (1). For each DNA sample and each dgp region, a DNA segment spanning the IRs and promoter region was PCR-amplified with the same forward and reverse primers used to demonstrate inversion (see above). The amplified DNA segments were digested with MnlI for the dgpC region and MluI for the six other dgp regions. The DNA fragments were separated by electrophoresis and visualized by EtBr staining.
Ability of B. distasonis Ssrs to Invert Glycoprotein Promoters in Heterologous Host.
ssrA, ssrB, ssrC, and ssrD were PCR-amplified from the B. distasonis chromosome by using the primers listed in SI Table 6. The PCR products were digested with BamHI and cloned into the BamHI site of pFD340. E. coli transformants were screened for those plasmids having the ssr gene in the correct orientation for transcription from the vector-borne promoter. Segments of ≈1 kb containing both IRs and additional flanking DNA of the dgpB, dgpC, and dgpF promoter regions were PCR-amplified by using the primers listed in SI Table 5. These three products were digested with PstI and cloned into the PstI site of each of the four pFD340-ssr plasmids, resulting in a total of 12 plasmid constructs containing each of the three invertible promoters with each of the four ssr genes. These plasmids were conjugally transferred to B. fragilis 9343, and inversion was tested by PCR using the primers listed in SI Table 6.
Supplementary Material
Acknowledgments
The B. distasonis 8503 genome sequence data were produced by the Genome Sequencing Center at Washington University School of Medicine. We thank A. Onderdonk and A. Dubois for human samples; P. Azadi at the Complex Carbohydrate Research Center (Channing Laboratory); B. Castor for PCR digestion; and Y. Lin, L. Hu, and P. Messner for helpful discussions. This work was supported in part by National Institutes of Health/National Center for Research Resources Grant RR018502 to the Complex Carbohydrate Research Center and largely by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant AI067711.
Abbreviations
- IR
inverted repeat
- AAL
Aleuria aurantia lectin
- MS/MS
tandem MS
- HSn
human fecal sample n.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608797104/DC1.
References
- 1.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 3.Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. Proc Natl Acad Sci USA. 2003;100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Coyne MJ, Reinap B, Lee M, Comstock LE. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 6.Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. Gene. 2006;371:102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Bayley DP, Rocha ER, Smith CJ. FEMS Microbiol Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 8.Patrick S, Parkhill J, McCoy LJ, Lennard N, Larkin MJ, Collins M, Sczaniecka M, Blakely G. Microbiology. 2003;149:915–924. doi: 10.1099/mic.0.26166-0. [DOI] [PubMed] [Google Scholar]
- 9.Sleytr UB, Messner P, Pum D, Sára M. Angew Chem Int Ed. 1999;38:1034–1054. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1034::AID-ANIE1034>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 10.Coyne MJ, Kalka-Moll W, Tzianabos AO, Kasper DL, Comstock LE. Infect Immun. 2000;68:6176–6181. doi: 10.1128/iai.68.11.6176-6181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CJ, Rogers MB, McKee ML. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Stephenson CG, Iadarola MJ. BioTechniques. 1994;17:257, 260, 262. [PubMed] [Google Scholar]
- 13.Merkle RK, Poppe I. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 14.York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Methods Enzymol. 1985;118:3–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.