Abstract
Aminoacyl-tRNA synthetases are multidomain proteins responsible for the attachment of specific amino acids to their tRNA substrates. Prolyl-tRNA synthetases (ProRSs) are notable due to their particularly diverse architectures through evolution. For example, Saccharomycese cerevisiae ProRS possesses an N-terminal extension with weak homology to a bacterial-specific domain typically present as an insertion (INS) within the aminoacylation active site. The INS domain has been shown to contain a “posttransfer” editing active site responsible for cleaving the aminoacyl-ester bond of misacylated Ala-tRNAPro species. However, wild-type S. cerevisiae ProRS does not perform posttransfer editing in vitro. Here, we show that replacement of the N-terminal domain of S. cerevisiae ProRS with the Escherichia coli INS domain confers posttransfer editing function to this chimeric enzyme, with specificity for yeast Ala-tRNAPro. In contrast, the isolated INS domain displays only weak editing activity and lacks tRNA sequence specificity. These results emphasize the modular nature of synthetase editing active sites and demonstrate how in evolution, a weak editing activity can be converted to a more robust state through fusion to the body of a synthetase. In this manner, a single editing module can be distributed to different synthetases, and simultaneously acquire specificity and enhanced activity.
Keywords: Prolyl-tRNA synthetase, Saccharomyces cerevisiae
Protein synthesis is a dynamic process that occurs within the complex machinery of the ribosome. Aminoacylated or “charged” tRNAs are delivered to the ribosome following amino acid attachment by a family of multidomain enzymes known as aminoacyl-tRNA synthetases (aaRS). Each aaRS possesses an ancient catalytic core domain, which functions to activate a specific amino acid and attach it to either the 2′ or 3′ hydroxyl of the terminal ribose associated with its cognate tRNA molecules (1). In some cases, the inability to discriminate between noncognate amino acids that are isosteric to or smaller than the desired substrate results in misactivated amino acids and mischarged tRNAs. To ensure high fidelity of genetic code translation, some synthetases have evolved error-correcting mechanisms known as “pretransfer” editing (hydrolysis of misactivated adenylates) and “posttransfer” editing (hydrolysis of mischarged tRNAs). Whereas posttransfer editing has been shown to occur in an active site that is distinct from the site of aminoacylation (2–7), the site of pretransfer editing is less clear. In the case of some synthetases such as isoleucyl-tRNA synthetase, the pretransfer editing reaction has also been suggested to take place in the posttransfer editing domain (8). In contrast, in the case of prolyl-tRNA synthetase (ProRS), pretransfer editing appears to occur in the aminoacylation active site (9).
Posttransfer editing domains are found in both classes of aminoacyl-tRNA synthetase. The class I synthetases specific for isoleucine, leucine, and valine share a highly conserved editing motif known as connective polypeptide 1 (CP1), which is inserted into the Rossmann-fold catalytic domain (3, 10–13). CP1 is present in all extant species and is believed to be of ancient origin (14). The class II synthetases, ProRS (15, 16), alanyl-tRNA synthetase (AlaRS) (17, 18), threonyl-tRNA synthetase (ThrRS) (19, 20), and phenylalanyl-tRNA synthetase (21), contain a variety of editing domains that are unrelated to CP1. In the case of AlaRS, phylogenetic analysis suggested that, as for class I enzymes, the editing domain is present throughout evolution and co-evolved with the aminoacylation active site (18); it is thus believed to be of ancient origin. Bacterial and eukaryotic ThrRSs, contain an editing domain with weak homology to the AlaRS editing domain, whereas the archaeal editing domain is distinct (19, 22, 23). The latter was recently shown to be structurally similar to d-amino acid deacylases found in eubacteria and eukaryotes (24), and is found as a free-standing editing module in some species (23).
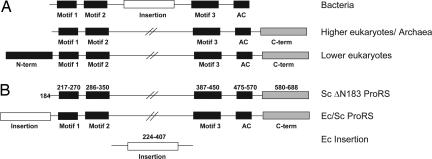
The unique ProRS editing domain is found in at least three different structural contexts: as an insertion (INS) between motifs 2 and 3 of the aminoacylation active site of bacterial enzymes (16, 25, 26), as an N-terminal extension in ProRSs of lower eukaryotes (27, 28), and as a free-standing editing module (25, 28). A highly conserved lysine (K279 in Ec ProRS) found in most INS domain homologs is critical for posttransfer editing function (16, 29). Archaeal and higher eukaryotic ProRSs lack an INS-like editing domain (Fig. 1A). Based on phylogenetic and structural arguments, the bacterial ProRS editing domain has been hypothesized to be a late addition in evolution (14, 18). However, the possibility exists that the domain is also ancient and was lost in some cases.
Fig. 1.
Schematic representation of the domain architecture of ProRS enzymes. (A) ProRS enzymes from all three domains of life. The class II consensus motifs and anticodon domains are shown in black and the bacterial INS domain is indicated as a white box. The C-terminal extension domains present in eukaryotic and archaeal ProRSs, which are not homologous to the INS domain, are shown in gray. The N-terminal extension unique to lower eukaryotic ProRSs is indicated as a white box with black stripes to indicate weak homology with the bacterial INS domain. (B) ProRS constructs used in this work: Sc ProRS with a deletion of the N-terminal 183 residues (Sc ΔN183 ProRS, Top); chimera with the Ec INS domain substituted for the first 183 aa of Sc ProRS (Ec/Sc ProRS, Middle); the Ec ProRS INS domain (Ec Insertion, Bottom).
In this work, we investigate the posttransfer editing activity of Saccharomyces cerevisiae (Sc) ProRS, a lower eukaryotic enzyme containing an N-terminal extension with weak homology to the bacterial editing domain. Our data support the notion that the N-terminal domain of Sc ProRS is a defunct editing module that was likely inactivated during evolution, but nevertheless retained due to its role in optimizing tRNA aminoacylation efficiency. The lack of posttransfer editing by wild type Sc ProRS allowed investigation of the effect of fusing an active posttransfer editing module derived from the Escherichia coli (Ec) INS domain to the N terminus of a nonediting synthetase. Taken together, this work emphasizes the modular nature of editing domains, as well as the important contribution of synthetase domains outside the editing active site to enhancing editing efficiency and tRNA specificity.
Results
Sc ProRS N Terminus, Haemophilus influenzae (Hi) YbaK Protein, and Ec INS Domain Share Strong Structural Homology.
The known posttransfer editing domain (INS) of Ec ProRS does not appear to share homology to any of the other known synthetase-editing domains. Sequence alignments have revealed that yeast and other lower eukaryotic ProRSs contain an “extra” N-terminal domain with weak homology to the bacterial ProRS INS domain (Fig. 1A) (15, 27, 28). Although the extra N-terminal domain common to lower eukaryotes is absent from higher eukaryotes, both share a C-terminal extension as a common feature (in addition to the class-defining active-site core and anticodon binding domain).
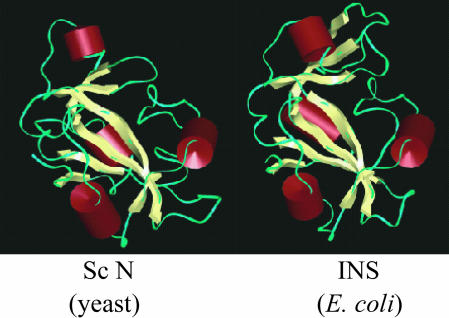
The N terminus of Sc ProRS also shares weak sequence similarity to the free-standing Hi YbaK protein (27). Although sequence identity is relatively low (13%), to explore structural similarity, we prepared a homology model of the Sc ProRS N-terminal extension based on the known structure of Hi YbaK (27). A model of the Ec INS domain was generated in a similar manner. The secondary structures of the proteins were determined based on Kabsch–Sander secondary structure definitions (30). The three-dimensional models show overall similarity in secondary structure, with the same number and arrangement of α-helices and β-sheets (Fig. 2). Based on this analysis, we conclude that, despite the relatively low sequence identity, the N terminus of Sc ProRS and the INS domain of Ec ProRS are likely to exhibit significant structural homology with the Hi YbaK protein.
Fig. 2.
Homology models. Models of the Ec INS domain (Right) and the Sc ProRS N terminus (Left) generated by using the known x-ray structure of the Hi YbaK protein (27).
Probing Sc ProRS Posttransfer Editing Activity.
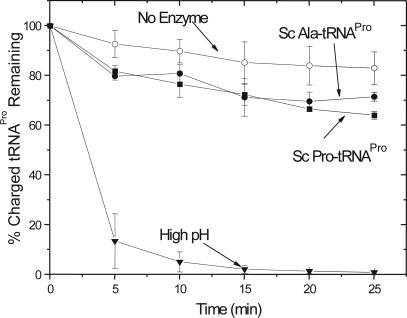
To investigate WT Sc ProRS posttransfer editing activity, a Sc tRNAPro U70/A73 double mutant was generated to facilitate high levels of alanine mischarging by Ec AlaRS. As has been reported for Ec Ala-tRNAPro (28), Sc U70/A73-Ala-tRNAPro was not a substrate for deacylation by Sc ProRS (Fig. 3). As expected, cognate Sc Pro-tRNAPro was also not a substrate for the yeast enzyme (Fig. 3). Thus, despite the fact that the N terminus of yeast ProRS displays significant structural homology to the known editing modules, Ec ProRS INS and Hi YbaK, WT Sc ProRS lacks the capacity to hydrolyze mischarged Ala-tRNAPro. Consistent with this result, weak misacylation with alanine of WT Sc tRNAPro by yeast ProRS is observed (Fig. 4). The extent of misacylation by the yeast enzyme is low, which indicates that pretransfer editing may be the predominant mode of clearing misactivated alanine by Sc ProRS (9).
Fig. 3.
Deacylation by WT Sc ProRS. Deacylation of Sc U70/A73-[3H]Ala-tRNAPro (filled circle) or Sc [3H]Pro-tRNAPro (filled square) by 4 μM Sc ProRS. A background (no protein) reaction was performed for all tRNAs tested and all were within 11% of the representative background data shown (open circle). An additional reaction (filled inverted triangle) was performed by incubating Sc U70/A73-[3H]Ala-tRNAPro with 0.2 M NaOH (pH 13). The plots represent the average of at least three assays.
Fig. 4.
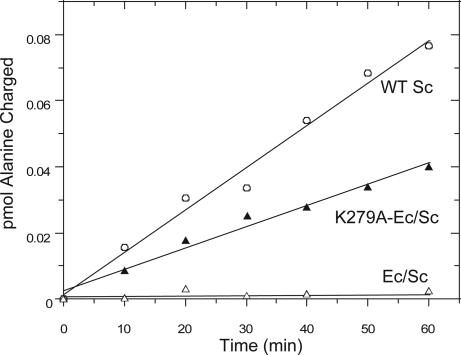
Aminoacylation assays. Reaction time courses showing charging of alanine onto Sc tRNAPro by WT Sc ProRS (open circle), Ec/Sc chimera (open triangle), and K279A-Ec/Sc chimera (filled triangle). Assays were performed at 30°C using 12 μM enzyme and 8 μM WT Sc tRNAPro. The plots represent the average of two assays with the values differing by <15%.
Ec/Sc Chimera Aminoacylates Sc tRNAPro and Gains Posttransfer Editing Function.
Although Sc ProRS does not appear to possess posttransfer editing activity in vitro, the high structural homology between the Ec INS domain and the Sc N terminus suggests that the latter may be the remnant of a once functional editing domain. To probe this idea further, a chimeric Ec/Sc ProRS enzyme was constructed, wherein the first 183 aa of Sc ProRS were replaced with residues 224–407 of the Ec INS domain (Fig. 1B, Ec/Sc chimera). An N-terminal deletion mutant lacking the first 183 residues of Sc ProRS was also prepared (Fig. 1B, Sc ΔN183 ProRS).To establish whether the N terminus of yeast ProRS contributes to aminoacylation activity, the ΔN183 variant was tested for its ability to charge a WT Sc tRNAPro transcript in vitro. Interestingly, the kcat/KM for cognate proline charging was only 3-fold reduced relative to charging by WT Sc ProRS (9). Similar reductions were observed in the kcat/KM for proline (5-fold) and alanine (2-fold) activation upon deletion of the N-terminal domain (Table 1). These results suggest that the N terminus of yeast ProRS contributes modestly to amino acid activation and aminoacylation in vitro. The N-terminal truncation mutant was also tested for alanine mischarging activity. Very weak mischarging was detected, but no increase in this activity was observed relative to the WT enzyme (Fig. 4 and data not shown). Taken together, these results suggest that the N terminus of yeast ProRS contributes modestly to amino acid activation and aminoacylation in vitro, but plays no role in posttransfer editing.
Table 1.
Kinetic parameters for activation of proline and alanine by WT and truncated Sc ProRS and WT Ec ProRS
| ProRS | Amino acid | kcat, s−1 | KM, mM | kcat/KM, s−1 mM−1 | kcat/KM, relative* |
|---|---|---|---|---|---|
| Sc-WT† | Proline | 8.83 | 2.32 | 3.73 | 1 |
| Sc-WT† | Alanine | 0.114 | 5.0 × 103‡ | 2.3 × 10−5 | 6 × 10−6 |
| Sc-ΔN† | Proline | 2.85 | 3.77 | 0.74 | 1 |
| Sc-ΔN† | Alanine | 0.071 | 5.9 × 103‡ | 1.2 × 10−5 | 1.6 × 10−5 |
| Ec WT§ | Proline | 70 | 0.25 | 280 | 1 |
| Ec WT§ | Alanine | 1.7 | 140 | 0.013 | 5 × 10−5 |
*kcat/KM is relative to proline in each set, which was set to 1.0.
†Each assay was performed at least twice with the values differing by <30%.
‡Values given are estimates using 200–800 mM alanine; more accurate determinations require prohibitively high amino acid concentrations.
§As reported in ref. 17.
The Ec/Sc chimera was able to charge Sc tRNAPro transcripts with proline. Similar to the ΔN183 variant, substitution of the N-terminal domain of Sc ProRS with the INS domain of Ec ProRS resulted in an ≈3-fold reduction in aminoacylation activity (kcat/KM = 0.0116 ± 0.003 s−1 μM−1) relative to WT Sc ProRS (kcat/KM = 0.0308 ± 0.005 s−1 μM−1).
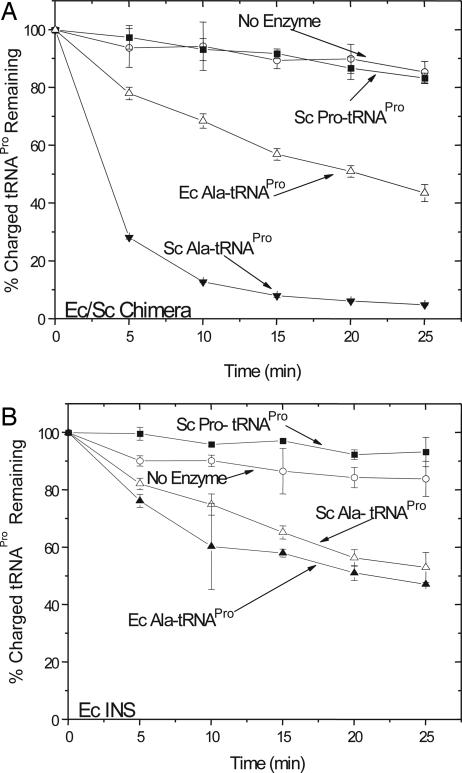
The Ec/Sc chimera was next tested for posttransfer editing activity in vitro. The chimera (1 μM) rapidly deacylated mischarged Sc U70/A73-Ala-tRNAPro, while preserving the cognate Pro-tRNAPro species (Fig. 5A). The chimera demonstrated species-specificity, as much weaker deacylation activity was observed in the presence of mischarged Ec Ala-tRNAPro. We estimate that the rate of deacylation of Ec tRNAPro was ≈8-fold lower than that of Sc tRNAPro. The separately cloned Ec INS domain (Fig. 1B) was previously shown to weakly deacylate a mischarged Ala-microhelixPro variant (25). High concentrations (20 μM) of purified Ec INS were also required to deacylate Ec Ala-tRNAPro, and very similar levels of Sc Ala-tRNAPro deacylation by the INS module were observed (Fig. 5B). Thus, whereas the cloned INS domain displays little, if any, sequence preference, the Ec/Sc chimera represents a gain of function mutant with species specificity for Sc tRNAPro. Additionally, this result shows that the activity of the INS domain fused to ProRS in cis, is significantly greater than its activity as a trans editing protein.
Fig. 5.
Deacylation by the Ec/Sc chimera and the Ec INS domain. (A) Deacylation of Sc U70/A73-[3H]Ala-tRNAPro (filled inverted triangle), Sc [3H]Pro-tRNAPro (filled square), and Ec [3H]Ala-tRNAPro (open triangle) in the presence of Ec/Sc chimeric ProRS (1 μM). (B) Deacylation of Sc U70/A73-[3H]Ala-tRNAPro (open triangle), Sc [3H]Pro-tRNAPro (filled square), and Ec [3H]Ala-tRNAPro (filled triangle) in the presence of the Ec INS domain (20 μM). A background (no protein, open circle) reaction was performed for all tRNAs tested, and all of the plots present the average of at least three assays.
Deacylation by the Ec/Sc Chimera Depends on the K279 Residue.
To establish whether the observed posttransfer editing activity of the Ec/Sc chimera depends on the strictly conserved K279 residue shown to be critical for editing by Ec ProRS (16), we mutated this residue to alanine in the context of the chimera and tested the alanine mischarging efficiency. As described earlier, under conditions of high enzyme and tRNA concentration, WT Sc ProRS can mischarge alanine onto Sc-tRNAPro transcripts. As expected, the Ec/Sc chimera prevents any detectable mischarging (Fig. 4). In contrast, in the presence of the K279A Ec/Sc chimera, significant mischarging is observed (Fig. 4). Thus, in the context of the chimera, the INS domain activity depends on K279, just as it does when present as an internal domain of Ec ProRS. This result suggests that the mechanism of posttransfer editing by the N-terminal INS domain is similar to that of the internal editing module.
Role of the N-Terminal Domain of Sc ProRS in Vivo.
Although deletion of the N-terminal domain of Sc ProRS had only minor effects on the in vitro kinetic parameters for tRNA aminoacylation and amino acid activation, the N terminus may still contribute to an additional function in vivo. To probe this hypothesis, the ability of ΔN183 Sc ProRS to complement a Sc ProRS null strain was examined. Under minimal growth conditions, the truncation mutant was fully functional [see supporting information (SI) Fig. 6]. Furthermore, the Ec/Sc chimera also appeared fully functional in vivo and maintained growth as effectively as WT and ΔN183 Sc ProRS at 30°C (SI Fig. 6). Complementation assays were also performed in the presence of 1 M alanine in rich media. Under these conditions, cell growth was significantly reduced. However, these experiments also revealed no difference between WT Sc ProRS and the chimeric enzyme (data not shown).
Discussion
Unlike the CP1 editing module found in class I synthetases or the class II AlaRS editing domain, the bacterial ProRS editing domain is not conserved through evolution. However, free-standing proteins with homology to the INS domain are found in the genomes of representative species from all three kingdoms of life (25, 28). Interestingly, there appear to be at least two distinct classes of ProRS-like editing modules with different specificities. Whereas the Ec ProRS INS domain (16, 25) and the Clostridium sticklandii PrdX protein (28) edit Ala-tRNAPro, the Hi YbaK protein functions primarily as a Cys-tRNAPro deacylase (29), with only weak activity toward Ala-tRNAPro (25, 31).
In addition to these free-standing ProRS-like editing proteins, a module with homology to the INS domain is appended to the N terminus of yeast and other lower eukaryotic ProRSs (27, 28). Previously, Plasmodium falciparum ProRS, a lower eukaryotic enzyme with an N-terminal INS-like domain, was shown to catalyze editing of Ec Ala-tRNAPro, whereas Sc ProRS lacked this activity (28). The latter result is not surprising, given the low cross-species aminoacylation demonstrated by Sc ProRS (J.S., P.S., and K.M.-F., unpublished observations). Here, we use mischarged Sc Ala-tRNAPro to confirm that Sc ProRS appears to lack posttransfer editing function. We also show that the presence of the N-terminal domain plays a minor role in enhancing in vitro adenylate formation (Table 1) and tRNA aminoacylation (9). The specificity of amino acid activation (i.e., relative kcat/KM for cognate proline vs. alanine) is also enhanced ≈3-fold in vitro (Table 1). However, because the truncated Sc ΔN183 ProRS complements yeast ProRS null strain as effectively as the WT enzyme, the role of the N-terminal domain in vivo is still unclear. The possibility exists that the N-terminal domain plays a nonessential or redundant function that is not detected in our in vivo complementation assay, such as facilitating interactions with other synthetases. We have shown that this domain is not associated with Sc ProRS editing function and based on a blast (blastp and CDART) search, there does not appear to be a free-standing INS-like editing domain homolog in S. cerevisiae. Thus, posttransfer editing does not appear to be required in this organism, and the N-terminal domain may have been retained based on its role in optimizing the synthetic step.
The structure of a prokaryotic ProRS possessing an INS domain has recently been reported (32). The posttransfer editing active site is poised over the aminoacylation catalytic domain, with ≈35-Å separation between the two active sites. The results reported here suggest that the N-terminal domain of Sc ProRS, an enzyme of unknown structure, may be similarly oriented even though its function has diverged.
Recently, it was shown that human mitochondrial leucyl-tRNA synthetase appears to possess a defunct CP1 editing domain (33). In addition, it was reported that a “single sieve” editing mechanism efficiently discriminates against noncognate isoleucine based on the enzyme's enhanced specificity for activation of cognate leucine relative to Ec leucyl-tRNA synthetase (33). Similarly, in the case of Sc ProRS, we estimate that the KM for alanine is ≈2,000-fold greater than that of proline (Table 1). This, together with an ≈80-fold more favorable kcat value for proline activation, likely precludes the need for posttransfer editing in this system. Moreover, the overall amino acid discrimination factor (i.e., kcat/KMPro/kcat/KMAla) measured for Sc ProRS is enhanced at least 10-fold relative to the Ec enzyme (Table 1), and is comparable to the level of discrimination achieved by M. jannaschii and human ProRS, which lack an INS-like domain (34).
A sequence alignment of the N-terminal domain of lower eukaryotic ProRSs was performed along with the Ec INS domain (see SI Fig. 7). The highly conserved nature of the lysine residue that aligns with K279 of Ec ProRS is striking. This residue was previously shown to be critical for posttransfer editing of both Ec ProRS (16) and Hi YbaK (29), and we confirmed in this work that it also plays a critical role in editing by the EC/SC chimera. Two other residues are highly conserved among these domains: K284 and D350 (Ec ProRS numbering). Although the function of K284 has not yet been probed, D350 was also shown to be a key determinant for editing by the Ec enzyme (16). Although Sc ProRS contains these three residues, a comparison of the sequences shows that it also lacks several residues previously shown to be important for posttransfer editing by the bacterial INS domain, including H369 (16) and G336 (C. Silvers and K.M.-F., unpublished data). The latter residue, which is part of a GXXXP motif, was shown to be part of an oxyanion hole proposed to form a putative ligand binding crevice in Hi YbaK (27). The lack of H369 and the GXXXP motif may explain the lack of detectable posttranfer editing activity in the case of yeast ProRS. In support of this hypothesis, P. falciparum ProRS, which possesses editing activity (28), contains both of these elements. The fact that substitution of the N-terminal domain of Sc ProRS with the Ec INS domain restores posttransfer editing function of the Ec/Sc chimera also supports the requirement for an intact GXXXP motif.
As more genome sequence information becomes available, the widespread presence of synthetase-like proteins is becoming more evident (35). In particular, the functional and evolutionary importance of these free standing synthetase-like modules is of great interest. Although some of these single-domain homologs have been shown to carry out functions unrelated to tRNA charging (35, 36) others, such as YbaK and PrdX, have clearly been shown to possess aminoacylation or editing activity (22, 23, 25, 28, 29, 31, 37). Free-standing editing proteins found in nature appear to have evolved alternate mechanisms to function as efficient and specific editing modules in trans (23, 28, 29, 31). Precisely how they accomplish this in vivo is an open question, but plausible mechanisms include noncovalent interactions with synthetases (38) or other factors.
In contrast, cloned editing modules derived from both classes of synthetases and expressed as free-standing proteins show significantly lower activity (3, 25) or lack editing activity altogether (11). It has been speculated that some editing domains have been incorporated into synthetases to increase editing efficiency and specificity (28). In support of this hypothesis, the free-standing INS domain displays very weak tRNA binding affinity (Kd > 5 μM, data not shown) and lacks tRNA specificity. However, in the case of the chimera, the domain is at least operationally acting in cis, through association with a core synthetase domain that serves to enhance the local concentration of tRNA around the editing module. The fact that this domain can be readily transplanted into the heterologous Sc ProRS framework and confer efficient and specific editing function on a non-posttransfer editing synthetase is also a remarkable demonstration of the modular nature of editing domains. These results suggest how a generalized editing module can be distributed to different synthetases, and simultaneously acquire specificity and activity. The enhanced activity and newly acquired tRNA specificity is a strong evolutionary driving force for trans to cis conversion of the editing domain, with adaptation to different amino acids occurring through accumulation of mutations.
Materials and Methods
All chemicals and amino acids were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. [3H]Alanine (54 Ci/mmol) was from Amersham Biosciences (Piscataway, NJ). All restriction enzymes were purchased from New England Biolabs (Ipswich, MA) unless otherwise noted.
Plasmid Construction for in Vitro Expression.
WT yeast ProRS and an N-terminal deletion allele lacking 183 residues (ΔN183 Sc ProRS) were cloned into protein expression vector pQE30 (Qiagen, Valencia, CA). The Sc ProRS PCR product was amplified from yeast genomic DNA using primers yPRS SphI/P1 (5′-ACACATGCAGCGCCTGTTTCGGAAGCGTTTGCC-3′) and MBP/P2 (5′-CCCAAGCTTCTAATAAGAACGACCGAACAT-3′). Both the pQE30 vector and the Sc ProRS PCR product were digested with SphI and HindIII. After digestion, the products were ligated together to generate plasmid pJS550. The ΔN183 yeast ProRS gene was PCR amplified via primers ΔN-term183 SphI (5′-ACACATGCATGCCTTATT GGTATCACCGTAGAC-3′) and MBP/P2, and cloned into pQE30 to construct pJS371. All constructs were confirmed through sequencing.
The recombinant Ec/Sc chimera construct for protein purification was generated via a stepwise process. This construct contains an N-terminal INS domain derived from Ec ProRS (residues 224–407), appended to residues 184–688 of Sc ProRS. PCR was first performed using plasmid pCS-M1S as a template (39) and primers cc-oJS1-INS 670 (5′-CGCGGATCCTCCGACACCTCTGACTATGCA-3′) and cc-oJS2-INS 1221 (5′-CATGCATGCACGTTTGAT CAGCTTACC-3′) to generate the Ec INS domain bearing BamHI and SphI restriction sites. Plasmid pJS371 and the PCR product were digested with BamHI and SphI and ligated together to create plasmid pJS1058. All constructs were confirmed through sequencing. The K279A mutant of the Ec/Sc chimera was created using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutant plasmid was generated via PCR using WT plasmid pJS1058 as template and primers 5′-GAAAACGGTTGCGACTCTGCTGG-3′ and 5′-CCAGCAGAGTCGCAACCGTTTTC-3′.
Plasmid Construction for in Vivo Expression.
The gene encoding the Ec/Sc was PCR amplified using plasmid pJS1058 as a template and primers oJS-pG-1 chimera PRS (5′-CGCGGATCCGCCACCATGTCCGACACCTCTGACTATGCA-3′) and oJS3-pG-1 yPRS C-terminal (5′-ACGGTCGACCTAATAAGAACGACCGAACAT-3′). The PCR products were digested with BamHI and SalI, and cloned into the high copy yeast expression vector, pG-1 (TRP1 2 μM) (40), to construct plasmid pJS1085.
The ΔN183 Sc PROS gene was PCR amplified using template pJS371 and primers ΔN183 pG-1 (5′-CGCGGATCCGCCACCATGCTTATTGGTATCACCGTAGAC-3′) and oJS3-pG-1 yPRS C-terminal. The PCR product was digested with BamHI and SalI and cloned into pG-1 to construct plasmid pJS380.
The WT Sc PROS gene was amplified via PCR using plasmid pJS550 and primers oJS10-pG-1 N-terminal (5′-CGCGTCGACGCCACCATGCCTGTTTCGGAAGCGTTTG-3′) and oJS11-Sc-pG-1 C-terminal (5′-CGCGTCGACCTAATAAGAACGACCGAACAT-3′). The PCR product and the yeast expression vector pG-1 were digested with SalI and ligated together to create plasmid pJS505. All constructs were confirmed through sequencing.
Yeast Strains and Growth Conditions.
All yeast strains in this study (see SI Table 2), with the exception of SSL212, are isogenic with TR2 (41). Yeast transformations were performed either by electroporation or using the lithium acetate transformation method (42). Media, mating, sporulation, and tetrad dissection were carried out according to standard methods (40). The deletion of the yeast PROS chromosomal copy is described in SI Text.
Protein Preparation.
WT Sc ProRS, ΔN183 Sc ProRS, WT Deinococcus radiodurans ProRS, WT Ec AlaRS, and the Ec ProRS INS domain were purified using a Talon cobalt affinity resin (Clontech, Mountain View, CA) as described (9, 43). Plasmids encoding the Ec/Sc chimera and the K279A Ec/Sc chimera were transformed into Ec SG13009 [pREP4] (Qiagen) competent cells and protein expression was induced with 0.1 mM isopropyl β-d-thiogalactoside for 22 h at room temperature. Histidine-tagged proteins were purified using a Talon resin according to the manufacturer's protocol. The desired proteins were eluted with 50–200 mM imidazole, concentrated, and stored as described (30). Total protein concentrations were determined by the Bradford assay (Bio-Rad, Hercules, CA) (44). The concentrations of synthetases were determined using the active site titration assay (45). In the case of the INS protein, the concentration was obtained by using the Bradford assay.
Preparation of tRNAPro.
All tRNAs used in this study were prepared by in vitro transcription as described (15, 46, 47). Point mutations in Sc tRNAPro to generate U70/A73-tRNAPro were introduced by using the QuikChange Site-Directed Mutagenesis kit (Stratagene).
Preparation of Aminoacyl-tRNAs.
Aminoacyl-tRNAs for use in deacylation assays were prepared as described (15) using WT Ec AlaRS (5.0 μM) to aminoacylate Sc U70/A73-tRNAPro (10 μM), and WT D. radiodurans ProRS (10 μM) to aminoacylate WT Ec tRNAPro (8 μM) with [3H]alanine in buffer containing 50 mM Hepes (pH 7.5), 4 mM ATP, 20 mM KCl, 20 mM 2-mercaptoethanol, 25 mM MgCl2, and 0.1 mg/ml BSA. The misacylated tRNAs were purified by repeated phenol extractions, followed by ethanol precipitation. The purified aminoacyl-tRNAs were quantified by scintillation counting and stored at −80°C in 50 mM KPO4 (pH 5.0). Pro-tRNAPro species were prepared by using WT Sc ProRS (4 μM) to aminoacylate both Sc WT tRNAPro (12 μM) and U70/A73-tRNAPro (12 μM) with [3H]proline as described above. Preparation of Ec Pro-tRNAPro was performed similarly using Ec ProRS (4 μM).
ATP–PPi Exchange Assays.
To test for activation of proline and alanine, the ATP–PPi exchange assay was performed as described (43) at 37°C in 144 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 10 mM KF, 10 mM 2-mercaptoethanol, 1 mM PPi, and 0.2 mg/ml BSA. The following concentrations were used: proline (2–25 mM), alanine (200–800 mM), WT Sc ProRS (10 nM), and ΔN183 Sc ProRS (500 nM).
Cognate Aminoacylation and Mischarging Assays.
Aminoacylation assays were performed at 30°C as described (18, 47, 48) using purified cognate Sc tRNAPro transcripts (0.5 μM), 100 nM Sc WT ProRS, Ec/Sc chimera, or K279A chimera, and 22.7 μM [3H]proline. Mischarging assays in the presence of alanine were also carried out at 30°C using standard conditions (15) with 8 μM Sc tRNAPro, 12 μM enzyme (Sc WT ProRS, Ec/Sc chimera, or K279A chimera), and 22.7 μM [3H]alanine.
Deacylation Assays.
Deacylation assays were carried out at room temperature according to published conditions (15). Reactions contained ≈0.2 μM Ec [3H]Ala-tRNAPro or 0.6 μM Sc U70/A73 [3H]Ala-tRNAPro, and were initiated with either 20 μM Ec ProRS INS domain, 1 μM Ec/Sc chimera, or 4 μM WT Sc ProRS. A background reaction was carried out in each case in which buffer (0.15 M KPO4, pH 7.0) was used to initiate the reaction. Results are the average of at least three trials.
Sc ProRS Structure Modeling.
A homology model of the Sc N-terminal domain (residues 9–147) was generated based on the known x-ray structure (residues 31–158) of the Hi YbaK protein (27). The sequence alignment of the target and template proteins was first achieved using CLUSTALW (49), followed by manual editing. Using the MODELLER-4 program (50), 40 models were generated, and the model with the lowest value of the objective function (derived from template protein spatial restraints and the CHARMM force field) was taken as the final comparative model. The stereo-chemical quality of the models was further checked using internal routines in the MODELLER and by PROCHECK (51, 52). A model of the insertion domain (residues 248–390) of Ec ProRS was generated similarly; however, in this case, residues 14–158 of the template YbaK protein were used in the alignment.
Supplementary Material
Acknowledgments
We thank Dr. Kathryn Splan and Ms. Kelly Hoadley for assistance with some of the experiments related to this work. This work was supported by National Institutes of Health Grant GM49928.
Abbreviations
- ProRS
prolyl-tRNA synthetase
- CP1
connective polypeptide 1
- AlaRS
alanyl-tRNA synthetase
- INS
insertion
- Sc
Saccharomyces cerevisiae
- Ec
Escherichia coli
- Hi
Haemophilus influenzae.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611110104/DC1.
References
- 1.Berg P. Annu Rev Biochem. 1961;30:293–324. [Google Scholar]
- 2.Fersht AR, Dingwall C. Biochemistry. 1979;18:2627–2631. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- 3.Lin L, Hale SP, Schimmel P. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 4.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 5.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt E, Schimmel P. Science. 1994;264:265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- 7.Silvian LF, Wang J, Steitz TA. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 8.Nomanbhoy TK, Hendrickson TL, Schimmel P. Mol Cell. 1999;4:519–528. doi: 10.1016/s1097-2765(00)80203-8. [DOI] [PubMed] [Google Scholar]
- 9.Hati S, Ziervogel B, SternJohn J, Wong F-C, Nagan MC, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K. J Biol Chem. 2006;281:27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 10.Starzyk RM, Webster TA, Schimmel P. Science. 1987;237:1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- 11.Chen JF, Guo NN, Li T, Wang ED, Wang YL. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 12.Mursinna RS, Lincecum TL, Jr, Martinis SA. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt E, Schimmel P. Biochemistry. 1995;34:11204–11210. doi: 10.1021/bi00035a028. [DOI] [PubMed] [Google Scholar]
- 14.Ribas de Pouplana l, Schimmel P. Cold Spring Harbor Symp Quant Biol. 2001;66:131–166. doi: 10.1101/sqb.2001.66.161. [DOI] [PubMed] [Google Scholar]
- 15.Beuning PJ, Musier-Forsyth K. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 17.Tsui WC, Fersht AR. Nucleic Acids Res. 1981;9:4627–4637. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beebe K, Ribas De Pouplana L, Schimmel P. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn CS, Ehresmann C, Moras D. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 20.Sankaranarayanan R, Dock-Bregeon AC, Rees B, Bovee M, Caillet J, Romby P, Francklyn CS, Moras D. Nat Struct Biol. 2000;7:461–465. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- 21.Roy H, Ling J, Irnov M, Ibba M. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beebe K, Merriman E, De Pouplana LR, Schimmel P. Proc Natl Acad Sci USA. 2004;101:5958–5963. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korencic D, Ahel I, Schelert J, Sacher M, Ruan B, Stathopoulos C, Blum P, Ibba M, Söll D. Proc Natl Acad Sci USA. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dwivedi S, Kruparani SP, Sankaranarayanan R. Nat Struct Mol Biol. 2005;12:556–557. doi: 10.1038/nsmb943. [DOI] [PubMed] [Google Scholar]
- 25.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. J Biol Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 26.Ahel I, Stathopoulos C, Ambrogelly A, Sauerwald A, Toogood H, Hartsch T, Söll D. J Biol Chem. 2002;277:34743–34748. doi: 10.1074/jbc.M206928200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Huang K, Li Z, Banerjei L, Fisher KE, Grishin NV, Eisenstein E, Herzberg O. Proteins. 2000;40:86–97. doi: 10.1002/(sici)1097-0134(20000701)40:1<86::aid-prot100>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 28.Ahel I, Korencic D, Ibba M, Söll D. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An S, Musier-Forsyth K. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Ruan B, Söll D. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 32.Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structure (London) 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Lue SW, Kelley SO. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 34.Beuning PJ, Musier-Forsyth K. J Biol Chem. 2001;276:30779–30785. doi: 10.1074/jbc.M104761200. [DOI] [PubMed] [Google Scholar]
- 35.Geslain R, Ribas de Pouplana L. Trends Genet. 2004;20:604–610. doi: 10.1016/j.tig.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 36.Schimmel P, Ribas De Pouplana L. Trends Biochem Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 37.Campanacci V, Dubois DY, Becker HD, Kern D, Spinelli S, Valencia C, Pagot F, Salomoni A, Grisel S, Vincentelli R, et al. J Mol Biol. 2004;337:273–283. doi: 10.1016/j.jmb.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 38.An S, Musier-Forsyth K. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 39.Stehlin C, Heacock DH, Jr, Liu H, Musier-Forsyth K. Biochemistry. 1997;36:2932–2938. doi: 10.1021/bi962295s. [DOI] [PubMed] [Google Scholar]
- 40.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 41.Parker R, Simmons T, Shuster EO, Siliciano PG, Guthrie C. Mol Cell Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiestl RH, Gietz RD. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 43.Heacock D, Forsyth CJ, Shiba K, Musier-Forsyth K. Bioorg Chem. 1996;24:273–289. [Google Scholar]
- 44.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 46.Hou YM, Westhof E, Giegé R. Proc Natl Acad Sci USA. 1993;90:6776–6780. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Peterson R, Kessler J, Musier-Forsyth K. Nucleic Acids Res. 1995;23:165–169. doi: 10.1093/nar/23.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Proc Natl Acad Sci USA. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DW, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.