Abstract
Time-dependent selection causes the adaptive evolution of new phenotypes, and this dynamics can be traced in genomic data. We have analyzed polymorphisms and substitutions in Drosophila, using a more sensitive inference method for adaptations than the standard population-genetic tests. We find evidence that selection itself is strongly time-dependent, with changes occurring at nearly the rate of neutral evolution. At the same time, higher than previously estimated levels of selection make adaptive responses by a factor 10–100 faster than the pace of selection changes, ensuring that adaptations are an efficient mode of evolution under time-dependent selection. The rate of selection changes is faster in noncoding DNA, i.e., the inference of functional elements can less be based on sequence conservation than for proteins. Our results suggest that selection acts not only as a constraint but as a major driving force of genomic change.
Phenotypic adaptations build on genomic sequence substitutions driven by a positive fitness effect. The distribution of these fitness differences (selection coefficients) has been debated since the advent of neutral theory (1–6). Coding DNA evolves under considerable constraint, i.e., nonsynonymous substitutions take place at a lower rate than synonymous changes (7). This shows that selection on protein evolution is predominantly negative. However, there is also evidence that the nonsynonymous substitutions that do occur are in part driven by positive selection (8–10). The evolutionary role of noncoding DNA is less clear. A particularly intriguing idea is that phenotypic evolution is due in a large part to changes in gene regulation, whereas proteins evolve more slowly (11, 12). This hypothesis lacks quantitative evidence so far, but a number of recent studies have found fitness effects in noncoding DNA. Transcription factor binding sites in bacteria are under substantial selection for functionality (13), and putative regulatory regions in eukaryotes also show substantial selective constraints (14, 15). Evidence of positive selection has been reported for intergenic DNA in Drosophila (16, 17), albeit with near-neutral selection coefficients (17). Inference methods rely on various implicit assumptions, and they differ considerably in the inferred strength of selection and in its contribution to genomic change (6).
Our phenotypic concept of adaptation contains more than the mere presence of positive selection. Migration of a population followed by adaptation to a new habitat, the conquest of an ecological niche in coevolution, incipient sympatric speciation driven frequency-dependent selection: in all these examples, fitness itself is time-dependent, and the adaptive evolution of new functions is the response to this change.
Including the dynamics of selection into a quantitative picture of genome evolution is the purpose of this paper. To illustrate our rationale, let us first consider the case of static fitness, where intuition suggests that evolution reaches a balance between advantageous and deleterious substitutions. This point can be made more precise: the long-term dynamics of substitutions leads to an evolutionary equilibrium, where the probability of a (fixed, haplotype) sequence state a depends exponentially on its Malthusian fitness fa (scaled by the effective population size). This simple form of equilibrium, originally derived for a two-allele model (18), applies to arbitrary sequence spaces and genomic fitness landscapes (19). It is characterized by detailed balance: the likelihood of any substitution (i.e., the product of initial state probability and substitution rate) between sequence states with a scaled fitness difference f = fb − fa equals the likelihood of the corresponding backward process, which involves a fitness difference (−f). Hence, on average every deleterious substitution is offset by an advantageous mutation, the average fitness remains constant, and there are no adaptations. Now consider a simple switch in the genomic fitness function: at a single position of the sequence, the selection coefficients f of all point mutations change sign. If the magnitude of selection is substantial, the most likely genomic state before the switch is fixation of the fittest nucleotide. After the switch, this state becomes suboptimal, which gives rise to an adaptive substitution to the new fitness optimum. This point is generic: adaptation in a time-dependent fitness landscape involves a surplus of advantageous substitutions compared to equilibrium. Thus, we define adaptation as a pure nonequilibrium phenomenon, i.e., more restrictively than in much of the literature, where any substitution with a positive fitness effect is counted as an adaptation. However, our definition is in tune with the phenotypic view of adaptation sketched above and has a conceptual advantage to be made precise below: it provides an unambiguous statistical distinction of adaptive substitutions and near-neutral background changes. The adaptive surplus in substitutions can be traced in genomic data, which allows us to infer the dynamics of selection together with the genome's adaptive response.
The quantitative analysis of this joint evolution process is based on a statistical model of time-dependent selection: point mutations at individual positions have fitness effects f(t) = ±σ, which randomly and independently change sign at a rate γ. Thus, selection is characterized by two parameters, the strength σ and the fluctuation rate γ. The latter governs the relative weight of positive and negative selection: higher values of γ generate a larger function of sites under positive selection, as shown by the above example. In this picture, populations evolve under two stochastic forces: genetic drift and selection switches. The classical work on fluctuating selection by Wright, Kimura, Ohta, Gillespie, and others (2, 20–26) was aimed at describing ecological changes such as seasonality or frequency-dependent selection. For these micro-evolutionary fluctuations, the correlation time of selection does not exceed the characteristic time scale of genetic drift given by the effective population size, 1/γ ≤ N (2). Here we focus on macro-evolutionary fluctuations with much longer correlation times of the order of the mutation time scale, 1/γ ∼ 1/μ ≫ N, as discussed by Gillespie (26) and Cutler (27) as an explanation for the overdispersion of the molecular clock. We obtain analytical solutions for the joint statistics of polymorphisms and substitutions, which depends on the strength and fluctuation rate of selection. Based on these solutions, we infer time-dependent selection and the resulting adaptation by a systematic Bayesian procedure. From a computational point of view, our method is a model-based scoring system for hybrid intra- and interspecies sequence alignments. As we show by explicit benchmarking, it is more sensitive than the classical population-genetic tests for adaptation (8, 28), which address partial aspects of the polymorphism and substitution frequencies. Hence, time dependence is not just an additional facet of selection but a crucial part of its quantitative inference.
Applying this method to sequence data from Drosophila, we find evidence for genome-wide selection with substantial amplitudes (2Nσ0 > 1) and fluctuation rates of the order of the neutral mutation rate (γ ∼ μ). As will be discussed below, it is both characteristics together that establish adaptations as a major driving force of genomic evolution.
Theory of Fluctuating Selection
Mutation-Selection-Drift Model.
We first consider a single-locus model with two alleles a and b and denote by x the population frequency of allele b, various generalizations will be discussed later. In the diffusion approximation, the time-dependent haplotype frequency distribution p(x, t) obeys the Fokker–Planck equation
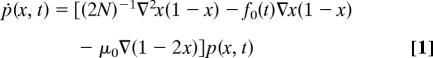 |
describing reproductive fluctuations (genetic drift) in a population of effective size N, selection with a Malthusian fitness difference 2N f0(t) ≡ fb(t) − fa(t) between the two alleles, and mutations with a rate μ0 per individual per generation (assumed to be equal for forward and backward changes). We consider a simple model of time-dependent selection,
with constant magnitude σ0 and fluctuating direction η(t) = ±1, which follows a Poisson process with rate γ0. This process defines a statistical ensemble given by the average and the covariance
of the variables η(t). Measuring time in units of the diffusion scale 2N introduces the rescaled evolutionary parameters f(t) = 2N f0(t), σ = 2σ0N, μ = 2μ0N, and γ = 2γ0N.
Stationary Evolution Under Constant Selection.
First recall well known results for time-independent selection, f(t) = σ (29). The Kimura equation (Eq. 1) has the form of a continuity equation, ṗ = −∇Jp with the rescaled probability current J = −∇x(1 − x) + σx(1 − x) + μ(1 − 2x). Two normalized and linearly independent stationary solutions pa(x) (a = ±1), which are eigenfunctions of J, can be defined by the boundary conditions that p+(x) remain finite at x = 1 and p−(x) remain finite at x = 0. Asymptotically for small μ, these solutions take the form
up to terms of order μ2, with normalization factors Za given in terms of hypergeometric functions [see supporting information (SI) Appendix]. Their current eigenvalues define the Kimura–Ohta substitution rates for these processes, i.e., Jpa(x) = auapa(x) with
A generic normalized stationary solution p(x) of Eq. 1 can be written as a linear combination
with 0 ≤ λa ≤ 1 and λ+ + λ− = 1. Evolutionary equilibrium defines the unique stationary solution peq(x) with a vanishing substitution current, Jpeq(x) = 0, i.e., there is detailed balance between forward and backward substitutions. The equilibrium distribution, given exactly for all values of μ by peq(x) = [x(1 − x)]−1+μeσx/Zeq with a normalization factor Zeq (30), has the form (Eq. 6) with
Quasistationary Evolution.
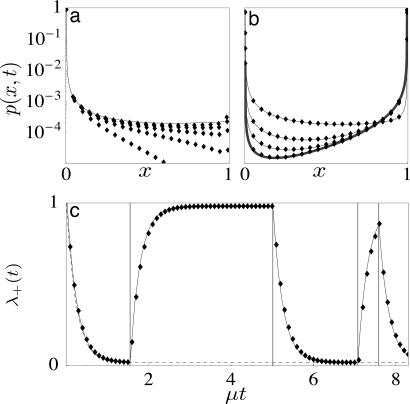
For time-dependent solutions of Eq. 1, no generic closed form exists even in the case of constant selection. For μ ≪ 1, however, an important simplification arises due to a separation of dynamical regimes, which is illustrated in Fig. 1. From an arbitrary initial distribution p(x, t = 0), the frequency distributions describing forward and backward processes reach their stationary shapes p+(x) and p−(x) within an initial time regime of order 2N generations (i.e., of order 1 in our rescaled time units); see Fig. 1a. For larger values of t, the distribution takes the quasistationary form (Eq. 6) with time-dependent coefficients λa(t) (a = ±1) up to correction terms of order exp(−t); see Fig. 1b. The long-term dynamics of substitutions is given by the rate equations
and governs the approach to equilibrium,
Fig. 1.
Allele frequency evolution under constant and fluctuating selection. The distribution p(x, t) of a two-allele Fisher–Wright model involving mutations, selection, and genetic drift with parameters μ = 0.025, σ = 4, and a discretization value N = 400 (shown as dots) is obtained by numerical propagation using the exact continuous time Master equation. (a) Initial regime (t < 1) for constant selection: From an initially monomorphic allele-a population, the solution p(x, t)(shown here for t = 0.2, 0.4, 0.6, 0.8, 1.0, bottom to top) builds up the spectrum of “forward” polymorphisms towards the stationary shape p+(x) (solid line) as given by Eq. 4. (b) Quasistationary regime (t > 1) for constant selection: The solution p(x, t) (shown here for μt = 0.05, 0.45, 0.80) follows the quasistationary form (thin lines) in excellent approximation and approaches the equilibrium distribution peq(x) (thick line) for large values of μt. (c) Quasistationary evolution of the coefficient function λ+(t) for constant selection (dashed line) and fluctuating selection with κ = 0.5 (solid line), which follows Eq. 9 in each interval of constant selection for a given realization of switching events (shown as vertical lines).
Evolution Under Fluctuating Selection.
In the full model given by Eqs. 1–3, allele frequencies evolve under two stochastic forces: genetic drift and selection fluctuations. The joint statistics of both processes is very complicated in general (2). The problem becomes again tractable in the quasistationary approximation, which can be applied if μ ≪ 1 and γ ≪ 1, i.e., if selection fluctuations are macro-evolutionary. This is demonstrated in Fig. 1c for the coefficient function λ+(t), which follows Eq. 9 in each interval of constant selection for a given fluctuation history η(t). Generalizing Eq. 6, we define the joint probability Λaϵ(t) (a, ϵ = ±1) of allele a and the direction of selection η(t) = ϵ. These probabilities follow the quasistationary evolution equations
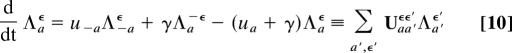 |
defining the 4 × 4 rate matrix U. Given initial probabilities at an earlier time t0, we obtain
with the transition matrix T(t − t0) = exp[(t − t0)U ]. In the following, we focus on key properties of this process relevant for cross-species sequence comparisons.
Nonequilibrium Stationarity.
The asymptotic limit of Eq. 11 defines a nonequilibrium stationary state, which describes the long-term average over selection fluctuations and genetic drift for a family of genomic loci evolving independently. The approach to stationarity is governed by the rate u+ + u− as in Eq. 9. In the data discussed below, it is much faster than for neutral evolution due to substantial levels of selection, with u+ + u− ≃ σμ. The statistics of the stationary state depends on two scaled selection parameters, the strength σ and the fluctuation rate in units of the neutral mutation rate, κ = γ/μ. The stationary allele probabilities counted in their contemporary direction of selection are given by the asymptotic state probabilities limt → ∞ Λaϵ = λ̄a·ϵ with
and should be compared with the equilibrium probabilities (Eq. 7) for constant selection. They determine the efficiency of the genomic response to time-dependent selection as measured by the degree of adaptation
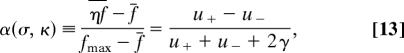 |
where ηf is the average fitness of the genomic nucleotide, f̄ is the average fitness of a random nucleotide, and fmax = fϵ·a is the fitness of the preferred nucleotide. Sequence turnover takes place at an average substitution rate
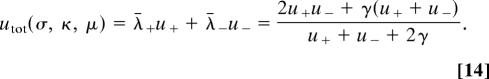 |
The rate of adaptations is given as the surplus of advantageous substitutions over deleterious ones,
and these produce an average fitness flux, i.e., a gain in fitness
per unit of time. We can thus decompose the sequence turnover into near-neutral substitutions due to genetic drift and adaptive substitutions generated by fluctuating selection. The fraction of adaptive substitutions at selection amplitude σ > 0 is
Cross-Species Correlations.
The time-dependent transition probabilities (Eq. 11) determine genomic correlations between species due to their common ancestry, which depend on their divergence time t. The joint probability of states a, ϵ and a′, ϵ′ for two species is a sum over the states a″, ϵ″ of their last common ancestor,
assuming stationarity of the ancestor states and subsequent divergent evolution with independent selection fluctuations along the two branches of the phylogeny. Unlike the standard notion of derived alleles, we do not make any ad hoc assumption that the outgroup carries the ancestral or the preferred nucleotide, which would introduce a bias in the predicted frequencies. The cross-species correlations between allele frequencies at orthologous loci are then given by the joint distribution
To analyze genomic polymorphism data, we have to compute the expected allele frequency distributions in a finite sample of individuals randomly drawn from a population (31). For two species with divergence time t, the joint probability Q(k, k′|m, m′) of finding k alleles b (k = 0, 1, …, m) in a random sample of m individuals of the first species and k′ alleles b (k′ = 0, 1, …, m′) at the orthologous locus in a random sample of m′ individuals of the second species is
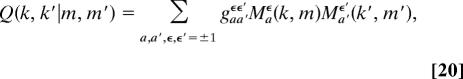 |
where Maϵ(k, m) are the binomial moments of the elementary stationary distributions (Eq. 4),
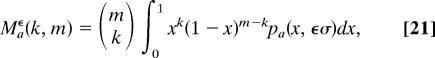 |
given in terms of hypergeometric functions (see SI Appendix). To leading order in t and in the limit of neutral evolution, the expressions (Eq. 20) reduce to the well known sampling formulae of ref. 31. Here we use the full time-dependence of (Eq. 18), which allows for multiple substitutions at the same site, since the short-time approximation can produce a spurious signal of selection.
Data Analysis
Sequence Data and Alignments.
We have aligned 271 Drosophila melanogaster sequence fragments, which are scattered across the X chromosome and are sampled from 12 individuals of a Zimbabwe population (17, 32, 33), to a single Drosophila simulans outgroup sequence. The aligned loci are binned into five broad genomic categories: 4-fold synonymous sites and nonsynonymous substitutions in protein coding DNA, intergenic regions, introns, and UTRs, similarly to the classification in ref. 17. In each category, we count k-fold single-nucleotide polymorphisms for k = 1, …, 11 (i.e., positions where k ingroup sequences differ from the nucleotide of the outgroup sequence at the orthologous position) as well as conserved positions (k = 0) and point substitutions within the sample (k = 12). Normalizing these counts per unit sequence length then defines the frequency distributions Q̂(k) (k = 0, …, 12), which are stable with respect to alignment changes and, in particular, do not overestimate the number of substitutions (for details and count tables, see SI Appendix). These genomic distributions can be compared to symmetrized pair probabilities Q(k) ≡ Q(k, 0|m, m′) + Q(m − k, m′|m, m′) with m = 12 and m′ = 1 as given in ref. 20.
Bayesian Inference of Evolutionary Parameters.
In a given genomic category, the distribution Q(k) has to be averaged over an a priori unknown distribution Ω(σ) of selection amplitudes,
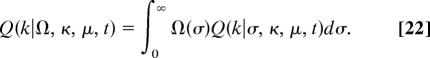 |
To infer this distribution, we use simple parameterizations which contain the average level of selection σave ≡ ∫ σ Ω(σ)dσ and the fraction of selected sites cs = ∫2∞ Ω(σ)dσ (with the threshold 2 chosen by convention) as independent parameters, see SI Appendix. The likelihood scoring function is then defined as
where we choose as reference distribution Q0 the best neutral model for 4-fold synonymous sites. (This choice does not influence our inferences, which are based on score differences.) A sequence category consisting of L independent loci with allele counts k1, …, kL has the total score
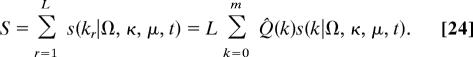 |
We then infer cs, σave, κ, and μ for each genomic category and the common parameter t by maximizing S summed over all categories (see ref. 34 for a similar approach). Confidence intervals follow from sampling of the Bayesian posterior probability.
Model Discrimination, P Values.
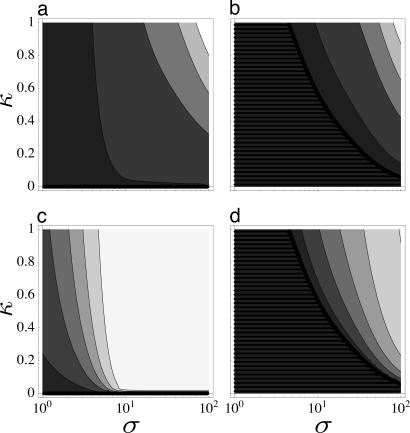
For a set of allele counts k1, …, kL drawn from a distribution Q(k|σ, γ, μ, t), any other model Q′ has a likelihood given by the corresponding difference of scores (Eq. 24), P ∼ exp[−β(S − S′)], with β = 1 for independent counts and a β < 1 for the actual datasets due to the partial linkage of loci (see SI Appendix). In particular, we can quantify the evidence for adaptive evolution by the P value of the best equilibrium model Q′, which is obtained by maximizing S′ with the constraint κ = 0. The resulting score difference per locus, Δs ≡ (S − S′)/L, is shown in Fig. 2a as a function of the parameters σ and κ of the input model Q. This can be compared with the analogous score difference ΔsMK of a McDonald–Kreitman (MK) test, which is based on the overall frequencies of polymorphisms Qp ≡ Σ k=1m−1 Q(k) and of substitutions Qs ≡ Q(m), and on their counterparts Q0p, Q0s in a neutral reference class, see Fig. 2b and SI Appendix for details. The increase in statistical power is significant: (i) The full score difference Δs is positive and, thus, produces evidence for adaptation for all parameters σ, κ > 0, whereas the MK test does not infer adaptations in the region where QsQ0p/QpQ0s ≤ 1 and hence ΔsMK = 0. (ii) Δs is always higher than ΔsMK, which implies that the same P value is reached with (sometimes considerably) less loci. (iii) Our method correctly reconstructs the fraction α̃ of adaptive substitutions as given by Eq. 17, whereas estimate α̃MK = 1 − QpQ0s/QsQ0p (6, 9) based on the MK test leads to a systematic underestimation; see Fig. 2 c and d. As shown in SI Appendix, there is a similar difference in statistical power compared to all inferences based only on the poly morphism spectrum, such as Tajimas D test and its variants (28).
Fig. 2.
Inference of adaptive evolution under fluctuating selection. (a) Statistical evidence for adaptation, as given by the likelihood score difference per locus, Δs, between the input model and the best equilibrium (κ = 0) model as a function of the input selection parameters σ and κ (input μ and t as in the Drosophila data sets below). Contours from bottom to top: 0 (thick line), 5 × 10−4, 5 × 10−3, 0.01, 0.015 (thin lines). (b) Analogous score difference ΔsMK/L based on the McDonald–Kreitman test. Contours as in a, no-inference region (ΔSMK = 0) marked by stripes. (c) Fraction of adaptive changes α̃(σ, κ) as given by Eq. 17, which is correctly reconstructed by the full maximum-likelihood procedure for a sufficiently large number of loci. Contours from bottom to top: 0 (thick line), 0.1, 0.3, 0.7, 0.9 (thin lines). (d) Estimated fraction α̃MK (9) using the McDonald–Kreitman test, which leads to a systematic underestimation. Contours as in c, no-inference region (α̃MK = 0) marked by stripes.
Demographic Effects.
The history of a population enters the allele frequency data through a time-dependent population size N(t) = Nν(t) (where N is today's effective population size), leading to deviations from equilibrium even for stationary selection. These demographic effects have been studied extensively for different Drosophila populations in the recent literature (32, 33, 35, 36). In particular, reduced population sizes for D. melanogaster in the past can lead to an increased number of substitutions, and recent variations in population size will also influence the polymorphism spectrum. As an alternative model to fluctuating selection, we consider the evolution under time-independent selection (κ = 0) and a bottleneck on the ingroup branch with strength νb, initial time ti, and duration tf − ti (0 ≤ ti ≤ tf ≤ t). The corresponding distributions Q(k|σ, μ, t, ti, tf, νb) have a selection-dependent increase in the substitution frequency. These distributions are obtained numerically, older bottlenecks (tf < t − 1) can also be treated analytically using the quasistationary approximation (Eq. 8) with scaled selection coefficients σ(t) = σν(t). The demography is shared between the different genomic categories and is hence treated with global parameters in the maximum-likelihood analysis (for further details, see SI Appendix).
Linkage Between Loci, Hitchhiking.
The sequence data originate from a number of different contiguous fragments. Loci on different fragments can be assumed independent, but loci on the same fragment are partially linked. These correlations can confound the inference of selection through hitchhiking effects, by which a locus under sufficiently strong positive selection influences the polymorphism spectrum at nearby loci, which are not under positive selection themselves (37). The characteristic length of reduced variance due to a hitchhiking event is ξ ∼ σ/ρ, where ρ is the recombination rate per 2N generations. The expected distance from a polymorphic site to the nearest adaptive change taking place during the lifetime of the polymorphism is ℓ ≈ 1/csγ. A scaling argument then suggests that the importance of hitchhiking is measured by the average ratio
which can be estimated from our inferred values of Φ and independent values of ρ. This is discussed in SI Appendix, where we show by a simulation of partially linked loci that hitchhiking may lead to an underestimation of some selection parameters but leaves the conclusions unaffected. In the same way, we verify numerically that other features not explicitly contained in our model, such as differences in neutral mutation rates and four-allele loci, do not confound our parameter inference.
Results and Discussion
Fluctuating Selection in Drosophila.
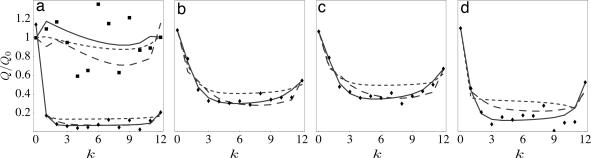
The model distributions obtained from Eq. 20 produce a good fit to the observed distributions Q̂(k) in all genomic categories (see Fig. 3).As expected, the frequency distribution of 4-fold synonymous sites in coding DNA is near-neutral (σave ∼ 1). In all other categories, we find substantial levels of selection affecting a substantial fraction of sites. The average selection strength over all sites in one category ranges from σave ∼ 14 in intronic regions to σave ∼ 100 for replacement substitutions in coding DNA (see Table 1). The fractions of selected sites are cs > 0.6, but these may be overestimated due to the possible presence of hitchhiking (37, 38) (see SI Appendix). The Bayesian posterior yields likelihood values P < 10−37 for neutral evolution, indicating that the presence of selection is statistically significant. Perhaps more surprisingly, the evolution is not only far from neutrality but also far from equilibrium. The inferred fluctuation rates of selection are of the same order of magnitude as the neutral mutation rates (0.1 < κ < 0.5). Hence, the fitness function at individual genomic sites is not static: it evolves at the same tempo as nucleotide changes, resembling more a seascape than a landscape. Because fluctuations at different sites are independent, these rates translate into a number of selection switches of order one per year in the Drosophila genome. The time-dependence of selection is statistically significant, with likelihood values P < 10−17 for any model with static selection (κ = 0) at equilibrium and P < 10−10 with only demographic deviations from equilibrium by a time-dependent effective population size, see Fig. 3 and SI Appendix.
Fig. 3.
Polymorphisms and substitutions in D. melanogaster. Genomic categories are shown. (a) Four-fold synonymous sites in coding DNA (squares), and nonsynonymous changes in coding DNA (diamonds). (b) Intergenic regions. (c) Introns. (d) UTRs. Data points indicate frequency distributions Q̂(k) (dots) of conserved positions (k = 0), k-fold polymorphisms (k = 1, …, 11), and substitutions (k = 12) per unit sequence length, relative to a reference distribution Q0(k) obtained as the best neutral fit for 4-fold synonymous sites. Maximum-likelihood model distributions Q(k) shown as lines to guide the eye:equilibrium model (κ = 0) (short-dashed lines) and demographic model (with a 100-fold reduced D. melanogaster population size between speciation and 0.65 of divergence time and time-independent selection, κ = 0) (long-dashed lines), producing a poor joint fit to all genomic categories (P < 10−17 and P < 10−10, respectively). Fluctuating-selection model, with parameters given in Table 1, produces a good joint fit to all genomic categories (solid lines).
Table 1.
Selection parameters and adaptive response
| 4-Fold synonymous | Nonsynonymous | Intergenic | Intronic | UTRs | |
|---|---|---|---|---|---|
| σave | 1.4 [0, 5] | 115 [52, >200] | 17 [9, 27] | 14 [6, 34] | 40 [14, 84] |
| κ | 0.0 [0.0, 2.8] | 0.12 [0.06, 0.27] | 0.27 [0.15, 0.46] | 0.44 [0.23, 0.83] | 0.38 [0.20, 0.93] |
| α | 0.0 [0.0, 0.78] | 1.0 [0.99, 1.0] | 0.97 [0.90, 0.99] | 0.94 [0.78, 0.98] | 0.98 [0.89, 0.99] |
| Φ | 0.0 [0.0, 0.13] | 0.37 [0.17, 0.82] | 0.15 [0.07, 0.22] | 0.18 [0.07, 0.30] | 0.50 [0.19, 0.88] |
σave, average magnitude of selection scaled by effective population size; κ, fluctuation rate of selection in units of the neutral mutation rate; α, degree of adaptation given by Eq. 13; Φ, fitness flux per site given by Eq. 16; brackets indicate 90% confidence interval, from sampling of the Bayesian posterior distribution.
Genetic Drift and Adaptations.
If genomic evolution is driven by fluctuations in reproduction and selection, can we disentangle the effects of these two stochastic forces? At a given selection amplitude σ, Eq. 17 provides an unambiguous decomposition of the observed sequence divergence into drift-generated background changes and adaptations. Background substitutions are mostly near-neutral, and they have selection coefficients f = +σ and f = −σ with equal frequencies, because advantageous changes merely compensate deleterious ones. For κ > 0, there is a surplus of substitutions with f = +σ, signaling the adaptive response to time-dependent selection. Averaging over the inferred distribution of selection amplitudes in a genomic category, we find substantial fractions of adaptive changes, α̃ave ≡ uad, ave/utot, ave > 0.6, again with a caveat due to possible hitchhiking. Yet, it appears that adaptations and background changes contribute to genomic evolution in comparable measure, a salomonic note in the neutralist–selectionist debate. Moreover, both classes are strongly intertwined: neutral changes can open new paths for subsequent adaptations.
Evolvability by Adaptation.
For all genomic categories in D. melanogaster except 4-fold degenerate sites, we find average levels of selection σave about an order of magnitude higher than previously reported values (17, 31, 34, 39), due to the increased statistical sensitivity of our method discussed above. These substantial levels are crucial for adaptations to be an efficient mode of evolution under time-dependent selection: fitness changes occur at about the rate of neutral evolution, but adaptive responses are faster by a factor σ/κ. For example, a selection change in either D. melanogaster or D. simulans at the time of speciation ≈3 million years ago will have generated an adaptive differentiation between today's lineages in >60% of the affected sites with σ > 20 but in <10% of weakly selected sites (σ < 2). Evolvability by adaptation can be quantified by the degree of adaptation at stationarity, α, which is defined in (Eq. 13). Selected sites in all of the genomic categories in D. melanogaster except 4-fold synonymous sites have values α > 0.9 (see Table 1), due to their substantial average values of σ. This justifies our conclusion that the deviations from equilibrium in the observed distributions Q̂(k) are due to ongoing selection fluctuations in a nonequilibrium steady state, rather than just a poorly adapted genomic state approaching equilibrium in a static local fitness function. The time-dependence of selection implies a fitness cost due to temporary misadaptation (23). Thus, the high values of α imply that most of the selection switches readily trigger an adaptive response, resulting in a number of adaptive substitution of order one per year in the Drosophila genome. Importantly, evolvability by adaptation does not require evolutionary tuning of the neutral mutation rate to the fluctuation rate of selection, provided selection amplitudes are sufficiently high.
Fitness Flux Quantifies Adaptations.
The relative contribution of genomic categories to phenotypic adaptations can be estimated on the basis of their fitness flux, i.e., the expected fitness gain per unit time. This weighted measure is more appropriate than the mere number of changes, since substitutions differ by orders of magnitude in their fitness effects and, hence, in their putative phenotypic consequences. According to Eq. 16, the fitness flux Φ per unit sequence length is proportional to the selection parameters σ and κ for strong selection, but the contribution of weakly advantageous substitutions is suppressed: these either happen too slowly (if f < κ) or are too unstable against reversal (if f < 1). All genomic categories of the Drosophila except 4-fold synonymous sites are well above this threshold and have comparable values of Φ (see Table 1). Hence, the total flux in noncoding DNA, obtained by multiplying Φ with the number of sites in those categories, may indeed outweigh that of protein evolution, as suggested previously for the numbers of adaptive changes (17). Notably, the fitness flux in UTRs and intergenic DNA results from more frequent changes with a lower value of f each than for replacement changes in coding DNA, where selection is stronger but more static. This may point to the role of regulation in the adaptive differentiation between species.
Fitness Is Correlated Between Sites and in Time.
The time-dependence of selection can be put in perspective by comparison with known evolution models, which are related to the fluctuating-selection model. For κ = 0, individual sites evolve independently under time-independent selection. If a substitution at a given site has selection coefficient f, its backward substitution has selection coefficient (−f), i.e., the fitness effects of subsequent substitutions at the same site are correlated. On the other hand, the evolution of long genomic sequences is often described by the infinite-sites model, regarding any two subsequent substitutions as independent and neglecting correlations between their selection coefficients f. This approximation is justified to the extent that fitness correlations between subsequent substitutions at the same site have decayed, either because of external fluctuations or due to in-between substitutions at other sites that have a fitness effect on the site in question (epistasis). This is precisely what the fluctuation parameter κ measures in the generic case: the fitness changes at a given site driven by external causes or by the coupled evolution of the remainder of the genome. Fitness interactions changing the direction of selection for substitutions at a given site, so-called sign epistasis (41), occur in a number of observations and models of protein, RNA, and regulatory evolution (19, 40, 42–46). Thus, the higher values of κ in UTRs, intronic and intergenic DNA shown in Table 1 are in accordance with the expected ubiquity of sign epistasis in regulatory sequences (13, 19). Our analysis relates epistasis to temporal fitness correlations at individual genomic sites. It suggests that sign epistasis may be pervasive, indicating a genome-wide rugged fitness landscape.
Genomic evolution emerges as a complex stochastic process, shaped jointly be the driving force of time-dependent selection, fitness interactions between sites, and the ongoing background of near-neutral changes. Much more remains to be learned about the interplay of these evolutionary forces: in a large and strongly coupled system, one external signal can trigger an avalanche of subsequent compensatory responses, which build up an evolutionary innovation. This dynamics seems now within reach of genomic sequence analysis.
Supplementary Material
Acknowledgments
We thank Joachim Hermisson, Diethard Tautz, and Thomas Wiehe for discussions and the anonymous referees for helpful comments. This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grant SFB 680 and STIPCO European Network Contract HPRN-CT-2002-00319.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This paper contains supporting information online at www.pnas.org/cgi/content/full/0607105104/DC1.
References
- 1.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge UK: Cambridge Univ Press; 1983. [Google Scholar]
- 2.Gillespie JH. The Causes of Molecular Evolution. Oxford: Oxford Univ Press; 1991. [Google Scholar]
- 3.Ohta T. Annu Rev Econ Syst. 1992;23:263–286. [Google Scholar]
- 4.Nei M. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr HA. Nat Rev Genet. 2005;6:120–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 6.Eyre-Walker A. Trends Ecol Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 7.Li WH. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 8.McDonald J, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 9.Smith NG, Eyre-Walker A. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 10.Fay JC, Wyckoff GJ, Wu CI. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 11.Monod J, Jacob F. Cold Spring Harbor Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 12.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 13.Mustonen V, Lässig M. Proc Natl Acad Sci USA. 2005;102:15936–15941. doi: 10.1073/pnas.0505537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin C-S, Chuang JH, Li H. Genome Res. 2005;15:205–213. doi: 10.1101/gr.3243305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keightley PD, Lercher MJ, Eyre-Walker A. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohn MH, Fang S, Wu CI. Mol Biol Evol. 2004;21:374–383. doi: 10.1093/molbev/msh026. [DOI] [PubMed] [Google Scholar]
- 17.Andolfatto P. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. Cold Spring Harbor Symp Quant Biol. 1955;20:33–53. doi: 10.1101/sqb.1955.020.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Berg J, Willmann S, Lässig M. BMC Evol Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright S. Evolution (Lawrence, Kans) 1948;2:279–294. doi: 10.1111/j.1558-5646.1948.tb02746.x. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. Genetics. 1954;39:280–295. doi: 10.1093/genetics/39.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta T. Genet Res. 1972;19:33–38. [PubMed] [Google Scholar]
- 23.Ohta T. J Mol Evol. 1972;1:305–314. [PubMed] [Google Scholar]
- 24.Takahata N, Ishii K, Matsuda H. Proc Natl Acad Sci USA. 1975;72:4541–4545. doi: 10.1073/pnas.72.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahata N, Kimura M. Proc Natl Acad Sci USA. 1979;76:5813–5817. doi: 10.1073/pnas.76.11.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillespie JH. Genetics. 1993;134:971–981. doi: 10.1093/genetics/134.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutler DJ. Genetics. 2000;154:1403–1417. doi: 10.1093/genetics/154.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewens WJ. Mathematical Population Genetics. New York: Springer; 2004. [Google Scholar]
- 30.Rouzine IM, Rodrigo A, Coffin JM. Micro Mol Biol Rev. 2001;65:151–185. doi: 10.1128/MMBR.65.1.151-185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawyer SA, Hartl DL. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glinka S, Ometto L, Mousset S, Stephan W, De Lorenzo D. Genetics. 2003;165:1269–1278. doi: 10.1093/genetics/165.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ometto L, Glinga S, De Lorenzo D, Stephan W. Mol Biol Evol. 2005;22:2119–2130. doi: 10.1093/molbev/msi207. [DOI] [PubMed] [Google Scholar]
- 34.Bustamante C, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- 35.Haddrill PR, Halligan D, Charlesworth B, Andolfatto P. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorton K, Andolfatto P. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fay JC, Wu C-I. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillespie JH. Evolution (Lawrence, Kans) 2001;55:2161–2169. doi: 10.1111/j.0014-3820.2001.tb00732.x. [DOI] [PubMed] [Google Scholar]
- 39.Piganeau G, Eyre-Walker A. Proc Natl Acad Sci USA. 2003;100:10335–10340. doi: 10.1073/pnas.1833064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bastolla U, Roman HF, Vendruscolo M. J Theor Biol. 1999;200:49–64. doi: 10.1006/jtbi.1999.0975. [DOI] [PubMed] [Google Scholar]
- 41.Weinreich DM, Watson RA, Chao L. Evolution (Lawrence, Kans) 2005;59:1165–1174. [PubMed] [Google Scholar]
- 42.Schuster P, Fontana W, Stadler PF, Hofacker IL. Proc R Soc London B. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y-H, D'Souza LM, Fox GE. J Mol Evol. 1997;45:278–284. doi: 10.1007/pl00006231. [DOI] [PubMed] [Google Scholar]
- 44.Kondrashov AS, Sunyaev S, Kondrashov FA. Proc Natl Acad Sci USA. 2002;98:548–552. [Google Scholar]
- 45.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 46.Remold SK, Lenski RE. Nat Genet. 2004;36:423–426. doi: 10.1038/ng1324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.