Abstract
Nitric oxide (NO) bioactivity is mainly conveyed through reactions with iron and thiols, furnishing iron nitrosyls and S-nitrosothiols with wide-ranging stabilities and reactivities. Triiodide chemiluminescence methodology has been popularized as uniquely capable of quantifying these species together with NO byproducts, such as nitrite and nitrosamines. Studies with triiodide, however, have challenged basic ideas of NO biochemistry. The assay, which involves addition of multiple reagents whose chemistry is not fully understood, thus requires extensive validation: Few protein standards have in fact been characterized; NO mass balance in biological mixtures has not been verified; and recovery of species that span the range of NO-group reactivities has not been assessed. Here we report on the performance of the triiodide assay vs. photolysis chemiluminescence in side-by-side assays of multiple nitrosylated standards of varied reactivities and in assays of endogenous Fe- and S-nitrosylated hemoglobin. Although the photolysis method consistently gives quantitative recoveries, the yields by triiodide are variable and generally low (approaching zero with some standards and endogenous samples). Moreover, in triiodide, added chemical reagents, changes in sample pH, and altered ionic composition result in decreased recoveries and misidentification of NO species. We further show that triiodide, rather than directly and exclusively producing NO, also produces the highly potent nitrosating agent, nitrosyliodide. Overall, we find that the triiodide assay is strongly influenced by sample composition and reactivity and does not reliably identify, quantify, or differentiate NO species in complex biological mixtures.
Keywords: red blood cell vasodilation, S-nitrosohemoglobin, S-nitrosylation
The biological effects of nitric oxide (NO) are mediated in large part through binding to transition metals and cysteine thiols at active or allosteric sites within regulatory proteins (1), which elicits changes in protein activity, protein–protein interactions, and protein location (1). Within tissues, many dozens of S-nitrosylated proteins have been identified, and signatures of NO bound to nonheme and heme iron have been detected (1, 2). Additionally, NO can be transported in endocrine or paracrine fashion by reacting with heme iron and cysteine thiols in proteins [hemoglobin (Hb) and albumin] and peptides (glutathione and cysteinlyglycine) to form NO adducts with longer biological lifetimes (3–5); release of NO bioactivity from stable adducts is effected by allosteric and redox-based mechanisms that alter FeNO or S-nitrosothiol (SNO) reactivity (5, 6). An updated discussion of the factors influencing reactivity of S-nitrosohemoglobin, S-nitrosoalbumin, and low-molecular-weight SNO in the context of vasoregulation (5–15) can be found in supporting information (SI) Text.
The dynamic distribution of protein and low-molecular-weight NO compounds that subserve NO transport and signaling instantiate the variation in both FeNO and SNO reactivities (4, 6, 13, 16–24). Numerous factors are implicated: RSNO geometric isomers (cis vs. trans), Fe coordination number in FeNO complexes, oxidation states of SNO (SNO vs. SNO·H or RSN·HO) and FeNO [Fe(II)NO vs. Fe(III)NO], and stabilization of alternative resonance structures of RSNO (RSNO vs. RSδ+NOδ−) and FeNO (FeNO vs. FeNOδ+ or FeNOδ−) (5, 6, 13, 16–22, 25). Accordingly, bond dissociation energies of RSNO are reported to vary from ≈22 to 32 kcal·mol−1 (6, 26), and the dissociation constants of FeNO can vary by a factor of >106 (13, 23, 24), translating to intrinsic FeNO/SNO lifetimes ranging from seconds to years. Environmental factors that have been reported to influence SNO stability and reactivity, directly or through elicited conformational changes in proteins, include pH (low and high) (5, 6, 20, 26), metal ions (Ca, Mg, Cu, and Fe) (6, 14, 20, 27, 28), nucleophiles (ascorbate, thiolate, and amine) (6, 13), local hydrophobicity (denaturants) (29), oxidants and reductants (6, 19), proteolytic enzymes (30), alkylators (31), O2 tension (5, 32), and various intramolecular interactions (H-bonding, S-, N-, O- coordination, and aromatic residue interactions) (6, 16, 20, 22, 33–36). Many of these factors also affect FeNO stability (17, 23, 24). Tetrameric SNO-Hb stabilities are special cases; the constellation of heme oxidation and ligation states (valency hybrid), thiol functionalization, and other allosteric modulators appears to be very important (5, 37). This distribution of NO species reactivities is a fundamental feature of the biological situation and must be recognized in the development of assays.
A standard approach to assay NO species in biological systems involves liberation of molecular NO from the medium, followed by its detection via chemiluminescence accompanying its gas phase reaction with ozone. Assays can be divided into two classes: those that employ UV light to liberate NO photolytically and those that employ chemical reactions. To distinguish FeNOs from SNOs in photolysis-based assays, inorganic or organic mercury is added (in minimal excess over thiol at pH 7.4 to avoid protein precipitation or denaturation) (38, 39). Hg displaces NO from thiols in the form of nitrite, which goes largely undetected. Proteins are then desalted in physiological buffers to remove low-molecular-weight reactants. SNO levels are taken as the loss of signal caused by Hg. All SNOs are reactive to Hg, whereas FeNO is unreactive (38). Photolysis yields are largely independent of the chemical reactivities of FeNOs and SNOs.
In chemical assays, identification depends on differential reactivity of NO-compounds toward a series of chemical reagents. Triiodide methodology is popular because of the claim that it is uniquely capable of not only measuring SNO and FeNO but also nitrite and nitrosamines (40). In this assay, biological samples are placed in acid plus triiodide to liberate NO. Other reagents [potassium ferricyanide (FeCN), potassium cyanide, N-ethylmaleimide (NEM), sulfanilamide (SAA), and mercurous chloride] are incorporated to selectively eliminate or block formation of one or another NO species (41–45). For example, pretreatment of biological samples with FeCN (3–200 mM followed by desalting) is reported to selectively remove NO from hemes, and SAA/HCl is reported to selectively eliminate nitrite; NEM and cyanide supposedly stabilize SNOs by blocking reactive thiols and hemes, respectively. Triiodide chemiluminescence operates on the premise that the chemistry of triiodide, acetic acid, and added reagents are general across NO species of varying reactivities.
Notably, the overall chemistry of triiodide that would specifically identify FeNOs has not been described (40–45); thus, the chemistry behind the putative effects of added reagents (oxidants, reductants, electrophiles, and nucleophiles) that are used to differentiate FeNO, SNO, and nitrite remains unclear. Moreover, few NO standards have actually been tested, and no basis has been provided for asserting that response of these standards captures the general behavior. Recovery of certain FeNO standards is reported to be as low as zero (40), and the one SNO-Hb standard that has been widely used (an R-structured Hb that contains ≈2 NO per tetramer) (40–46) is neither characteristic of general SNO-Hb reactivity nor of the reactivity of the micropopulation found in RBCs (a valency hybrid, estimated 1 NO per tetramer) (5, 47). Furthermore, the claimed effects of added reagents in triiodide assays, including FeCN and NEM (alone and in combination), are not supported, and they, along with the acidic and denaturing conditions of the triiodide assay, can alter the reactivities of SNO and FeNO as well as disrupt the partitioning of NO species within hydrophobic compartments and thus lead to their misidentification. A detailed discussion of these issues is provided in SI Text (4–15, 17, 20, 29, 31, 48).
By contrast with triiodide, validation of the photolysis assay has been performed not only through the analysis of a wide variety of standards but also through verification of NO mass balance with mixtures and complex-reactive systems and by direct measures of NO bioactivity. Specifically, we have used chemical reactions to interconvert FeNOs, nitrite, and SNOs, and we have balanced the changes in FeNO levels determined by photolysis, UV/visual light, and EPR spectroscopy against changes in SNO levels determined by colorimetric, chemiluminescence, and fluorometric assays (37, 48–52). In addition, we have used chemical reactions to interconvert SNO in endogenous Hb, as measured by photolysis, to nitrite as determined by two different chemical assays (12, 51). These latter assays involved removal of protein to avoid the potential errors introduced by side reactions with proteins. Early concerns that photolysis directly detects nitrite and nitrate (in the added presence of thiol) proved unfounded (47). Furthermore, we have shown that the amounts of SNO measured by photolysis directly predict vasodilatory activity of RBCs (47, 49, 50). The consistent mass balance obtained in these experiments and predictive value of the method in assessing vasoactivity validates the consistency of NO group recovery in photolysis analysis of complex samples.
Here, we directly compare the performance of photolysis vs. triiodide assays with an emphasis on complex species of biological significance. We find that photolysis consistently gives essentially quantitative recoveries of FeNOs and SNOs, whereas recovery in triiodide is highly variable and generally low (approaching zero for some species). We also detail significant effects of added chemical reagents, sample pH, and ionic composition that impact recoveries in the triiodide assay. In addition, we show that triiodide produces the highly potent nitrosating agent, nitrosyliodide (NOI). Overall, the results indicate that triiodide assays are strongly influenced by sample composition, rather than solely by the identity or quantity of NO species; the assay does not accurately identify, quantify, or differentiate NO species in complex biological mixtures. It is our hope that the results reported here will clarify the confusion and diminish the controversy that has hindered this field of inquiry.
Results
FeNO Detection.
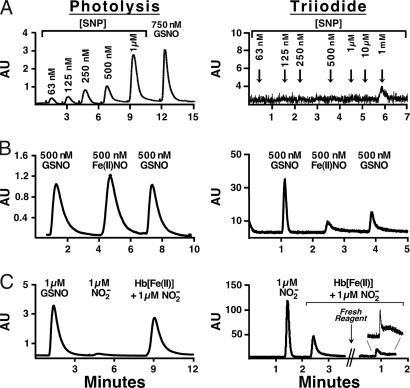
In previous work, we and others have called attention to the coupling of NO and heme redox chemistry in nitrosyl valency hybrids and the role of this coupling in steering chemical interactions of NO with Hb (5, 53, 54). It has been suggested that most heme iron-bound NO within the bloodstream could be a Fe(III)NO or Fe(II)NO+ complex (54). These important complexes (54), like the well studied Fe(III)NO complex sodium nitroprusside (SNP) (17, 20), have been reported to go undetected in triiodide assays (40). To our knowledge, the comparative performance of the photolysis assay for the detection of SNP has not been previously reported. We therefore examined a series of SNP standards with both photolysis and triiodide methods. As shown in Fig. 1A, photolysis liberates NO from SNP with excellent efficiency, as standardized against S-nitrosoglutathione (GSNO); SNP was detectable to low nanomolar levels. However, SNP was not detected by triiodide at concentrations up to 10 μM (Fig. 1A), levels that exceed the FeNO concentrations detected in vivo (≈1–5 μM) (35, 51, 54, 55). In the triiodide assay, 1 mM SNP yielded a signal equivalent to ≈10 nM GSNO standard, corresponding to a recovery of 0.001%.
Fig. 1.
Comparison of the sensitivity and specificity of the photolysis and triiodide assays for paired samples of FeNO compounds. (A) SNP, a model Fe(III)NO compound, at the indicated concentrations, with GSNO shown as a standard. AU, arbitrary units. Note that, in this and subsequent figures, the magnitudes of signals generated by photolysis and by triiodide, expressed as arbitrary units, cannot be compared directly (the two methods exhibit equivalent sensitivity for NO). (B) A sequence of injections of a GSNO standard (500 nM), a Fe(II)NO Hb solution {1 mM Hb[Fe(II)] containing 500 nM Hb[Fe(II)NO]}, and a repetition of the GSNO standard after Hb. The repeat injection of GSNO in the triiodide gave a distorted, diminished signal. (C) Hb Fe(III)NO/Fe(II)NO+ equivalent (SNO precursor) (37). Photolysis accurately measures the transient formation of a Fe(III)NO/Fe(II)NO+ equivalent generated from 1 μM nitrite/1 mM deoxyHb (×5–10 s), with scant response from nitrite alone (1 μM GSNO shown for comparison). In contrast, nitrite produces a prominent signal in the triiodide assay, whereas its signal in the presence of 250 μM deoxyHb is markedly attenuated. Furthermore, the signal generated by such samples can be variable and difficult to quantify (the line shape of a second sample, which is magnified for clarity, hampers reliable integration).
In blood, heme is present in great excess over NO (10,000:1). It has been reported that Fe(II)NO signals are quenched in triiodide assays if physiological heme/NO ratios are present (56). Fe(II)NO Hb {500 nM in NO (derived from PROLI NONOate) in the presence of 1 mM deoxyHb[Fe(II)]} is detected with the same sensitivity as 500 nM GSNO by photolysis (≈100% recovery); additional injections of Hb have no effect on this quantitative recovery (Fig. 1B). In contrast, the peak height of the same Fe(II)NO Hb in the triiodide assay (paired sample) was attenuated nearly 4-fold relative to GSNO, and the signal duration was prolonged by 5-fold. Subsequent injections of GSNO showed progressive signal attenuation and prolongation of peaks. Overall, the total peak area produced by GSNO was often up to many times greater than that produced by Fe(II)NO Hb and many times greater than GSNO injected after Hb had been introduced, reflecting both variations in the experimental condition, NO/Hb concentrations, and integration procedures (see Methods). The mean area of the Fe(II)NO vs. original GSNO standard (see Methods) was attenuated by 55%.
Redox Hybrid Detection in Vitro and in Situ.
Nitrosylated redox hybrids of Hb (5), generated by the incubation of nitrite (1 μM) with deoxyHb (1 mM heme) for 10 s (37), are detected by photolysis with yield of ≈100% compared with a GSNO standard, whereas 1 μM nitrite itself is, as expected, hardly detected (1–2% yield, pH 7.4) (Fig. 1C). The NO-Hb species formed under these reaction conditions have been recently identified as Fe(III)NO/Fe(II)NO+ equivalents that convert to SNO after oxygenation at 50% yield (37). When assayed by the triiodide method, these species are underestimated by >40% (integrated signal), whereas nitrite is readily detected (Fig. 1C). Moreover, this NO-liganded micropopulation is not eliminated by addition of SAA/HCl (3 min; data not shown) provided solutions are kept strictly anaerobic. Triiodide assays of protein-NO adducts often show an extended line shape. Broadening and concomitant attenuation of peak height hampers reliable integration, especially for weak signals (Fig. 1C; see also Triiodide Cautionary Notes in SI Text).
Mixed nitrosyl/met Hb species derived from incubation with NO/nitrite are suggestive of the complexities of the sample composition encountered in actual biological samples. Given the disparity in NO recovery by the photolysis and triiodide assays for these species and for SNP, we undertook a further examination of their relative performance in samples derived from RBCs. Biological samples are known to show a large population variance in NO/Hb levels of ≈15–150 nM NO/100 μM Hb (49, 51). To highlight the contrast between photolysis and triiodide, we report results from RBC samples exhibiting high values. Venous blood was drawn and either immediately oxygenated or maintained under strictly anaerobic conditions. RBC lysates were purified by centrifugation and Hb desalted over G25 columns (50, 51). The photolysis assay gave total signals corresponding to ≈100–150 nM NO/100 μM Hb in both deoxygenated and oxygenated RBCs, with FeNO predominating in deoxygenated blood (Fig. 2 A) and SNO predominating in oxygenated samples (Fig. 2B). In contrast, recoveries in the triiodide assay (from the same sample) were again very modest, approaching zero (≈5–10 nM NO/100 μM deoxygenated Hb or oxygenated Hb) (Fig. 2). Interestingly, the small triiodide signal from the deoxygenated sample was not quenched by SAA (Fig. 2A), but the signal from the oxygenated sample was (Fig. 2B); thus, the former would be identified as a FeNO, whereas the latter would be misidentified as nitrite. Note that these data from RBCs illustrate not only problems in the recovery of FeNO by triiodide analysis but also in the recovery of Hb SNO.
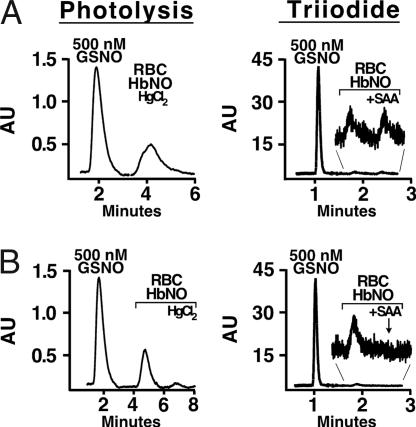
Fig. 2.
Comparison of photolysis and triiodide assays for endogenous nitrosyl Hb and S-nitroso Hb. (A) Deoxygenated venous blood FeNO Hb. Injections of 100 μM deoxyHb (RBC lysate after clarification and desalting) with or without Hg pretreatment gave a strong signal by photolysis vs. a greatly diminished signal in triiodide. A GSNO standard is shown for comparison. (Inset) Triiodide signal magnified. (B) Oxygenated blood SNO-Hb. Injections of 100 μM oxyHb (RBC lysate after clarification and desalting) yielded a strong signal by photolysis that is largely eliminated by Hg, whereas the signal is barely detected in triiodide. (Inset) The very small triiodide signal is eliminated by SAA.
SNO Detection.
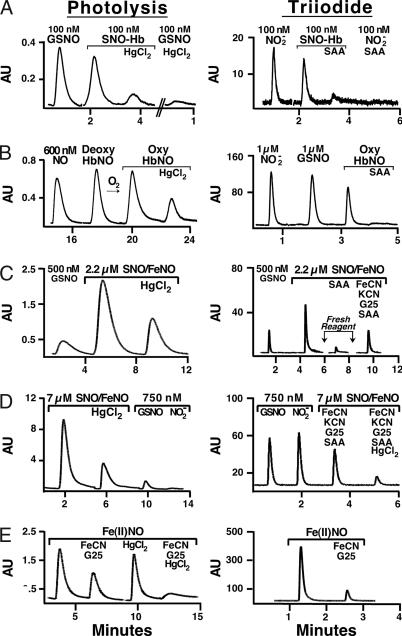
The reactivity of S-nitrosylated Hb depends on the redox valency state, the ligation state of the hemes, and the number of SNO/Hb (5, 12, 37, 47–49). SNO-Hb standards previously used in triiodide assays (40–46) were synthesized with a large excess of low-molecular-weight SNO, which produces SNO-Hbs with ≈2 SNOs per tetramer; heme valency and ligation states of the standards were not characterized, and the samples were not assayed at physiological ratios of SNO/Hb. As an initial validation, we synthesized stock solutions of SNO oxyHb (50–100 μM SNO/100 μM Hb as assayed by photolysis; ≈15% Met) and verified under these conditions that amounts of SNO when measured by photolysis, triiodide, and Saville assays (39) were in good agreement (values by triiodide and photolysis were within 5%, P was not significant). As previously described, stock solutions were relatively resistant to SAA in the triiodide assay (46). However, upon dilution of the stock directly into Hb (100 μM) (1:10 to 1:1,000) the SNO became progressively sensitive to SAA; at a ratio of 1:1,000 (100 nM SNO-Hb/100 μM Hb), simulating RBCs (see Fig. 2 and related text), most of the signal (n = 6; 85 ± 1%) was eliminated by SAA in triiodide, ostensibly identifying it as nitrite (Fig. 3A). In contrast, the SNO yield (and sensitivity to Hg) was unaffected by dilution in the photolysis assay (Fig. 3A). Moreover, pretreatment of the diluted sample with Hg followed by G25 eliminated the signal in triiodide (n = 3; data not shown), confirming its identity as SNO.
Fig. 3.
Comparison of the sensitivity and specificity of the photolysis and triiodide assays for SNO-Hb compounds. (A) SNO-Hb (100 nM SNO/100 μM Hb) synthesized by brief exposure to excess S-nitrosocysteine is largely eliminated by Hg in the photolysis assay. A 100 nM GSNO standard is similarly quenched by Hg. The same SNO-Hb sample is quenched by SAA in triiodide, ostensibly identifying it as nitrite. Nitrite is shown as a standard. (B) An SNO/FeNO valency hybrid synthesized from native HbA (0.4 mM heme) by using NO solution (method 1) contains 0.65 μM NO (≈0.4 μM SNO), as measured by photolysis. Note that native HbA contains ≈50 nM NO. Shown in sequence are the following: amount of NO added (0.6 μM), amount of NO bound to deoxyHb after NO addition (Deoxy HbNO), and amount NO bound to Hb after oxygenation with or without Hg (Oxy HbNO). The same oxyHbNO sample gives 100% yield by triiodide (≈0.65 μM NO); however, the signal is eliminated with SAA, to which both FeNO and SNO are reportedly impervious. (C) An SNO/FeNO valency hybrid derived from NO solution by using method 2 contains 2.2 μM NO (1.0 μM SNO, 1.2 μM FeNO, and 1 mM heme), as measured by photolysis. The sample is underestimated (1.6 μM NO) and misidentified by triiodide as nitrite (eliminated by SAA) and as FeNO (eliminated by FeCN/G25) and SNO (residual). (D) An SNO/FeNO valency hybrid measured by photolysis (5.0 μM SNO, 2 μM FeNO, and 1 mM heme). The sample was pretreated with FeCN and KCN to obtain an SNO value of 480 nM by triiodide (a yield of <10%). (E) Exposure of tetranitrosyl Hb (1 μM NO) to FeCN leads to the production of SNO-Hb (≈500 nM) as shown by photolysis, whereas the sample is identified as FeNO by triiodide.
We have reported that the reactivity of (SNO)2-OxyHb is different from SNO-Hbs synthesized with limiting NO and that the latter exhibit reactivities more representative of the native SNO in RBCs (5, 47). We analyzed SNO-Hb[FeNO] hybrids generated by rapid oxygenation of deoxygenated Hb (0.4–1 mM heme) immediately (≈5–10 s) after the addition of sub- to low micromolar aqueous NO (NO solution methods 1 and 2, respectively; amounts of NO added were precisely verified by both electrochemical and chemiluminescence methodologies; see SI Text) (47, 49, 52). The HbNOs were then desalted across G25 columns. SNO-Hb[FeNO] hybrids assayed by photolysis contained ≈0.6–2.2 μM NO, 40–66% of which was removed by Hg treatment (≈0.3–1 μM SNO-Hb) (Fig. 3 B and C). By comparison, paired samples measured with triiodide yielded 0.6–1.6 μM total NO (32–100% of photolysis). Moreover, as with SNO in RBCs (Fig. 2B) and with SNO measured at physiological ratios of SNO:Hb (Fig. 3A), the entire signal was eliminated by SAA, ostensibly identifying it as nitrite (Fig. 3 B and C). (Use of fresh triiodide with every measurement did not change the result.) Thus, not only are the HbNO signals frequently attenuated in the triiodide assay, but by following the methodology of Gladwin and coworkers (41–45), they are misidentified. These data may explain why Rassaf et al. (57) have claimed that SNO-Hb and FeNO do not exist in human RBCs and that NO signals in RBCs derive entirely from nitrite. In addition, 50% of the signal that is eliminated from these hybrids by SAA (in triiodide assays) is also eliminated by FeCN (followed by G25), a treatment that ostensibly identifies Hb[FeNO] (Fig. 3C) (41, 46). Thus, the same sample is identified in the triiodide assay as either nitrite or FeNO and SNO, depending on the reagents added.
The triiodide assay has also been used to differentiate SNO-Hb and Hb[FeNO] in vivo on the basis of reactivity toward FeCN/KCN (41–46). We tested the validity of this approach by analysis of another SNO-Hb redox valency hybrid containing Fe(III) (synthesized with NO solution by using method 1). Photolysis after Hg addition (pH 7.4) gave a SNO content of 5 μM (calibrated against a GSNO standard) (Fig. 3D). The same sample was assayed by triiodide (41–45): 0.2 M FeCN and 0.2 M KCN was used to eliminate FeNO; the sample was desalted on a G25 spin column to remove nitrite; Hg was used to displace NO from SNO, 0.5% SAA/1 M HCl was added to verify nitrite/SNO. Substantial sample precipitation leading to losses during desalting could not be avoided, as previously noted (56). The SNO-Hb yield by triiodide (i.e., NO that survives FeCN and SAA and is reactive to Hg) was 430 nM or 8.4% of SNO measured by photolysis (Fig. 3D) (56).
The FeCN treatment applied above was aimed at selectively oxidizing hemes (41–45). However, we have previously reported that FeCN converts βFe(II)NO into SNO (48). In Fig. 3E, we show the results of experiments in which we treated Hb[Fe(II)NO]4 with excess FeCN (50 mM). The product yield as determined by photolysis shows that as much as 1 SNO-Hb is produced for every two hemes that are oxidized (overall yield of SNO, 20–50%), confirming previous reports that used the Saville method (48). This side reaction makes the same analysis by triiodide difficult to interpret (Fig. 3E).
Synergistic Effects of Added Reagents.
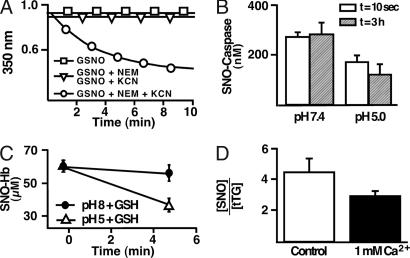
As suggested by the above experiments with FeCN (Fig. 3 C–E), assertions that the many reagents added in triiodide have selective effects on either FeNO and SNO have not been verified. Furthermore, the effects of these compounds in combination and when added to physiological systems have not been tested, potentially leading to unforeseen problems. For example, NEM and KCN are used in triiodide assays to stabilize SNO (41–45). We observed that GSNO incubated with either 5 mM NEM or 200 mM KCN remains stable for >10 min. However, addition of both NEM and KCN led to a rapid decay of GSNO (decrease in absorbance at 335 nm, corrected for initial NEM absorbance) (Fig. 4A). This decrease fit a double exponential decay, indicating that both GSNO and NEM are consumed (data not shown; see Eqs. 4–6 in SI Text). Similar concerns arise when adding NEM together with other nucleophiles (e.g., SAA).
Fig. 4.
Instability of protein SNOs under conditions of the triiodide assay. (A) Spectrophotometric analysis reveals GSNO degradation in the presence of KCN and NEM. GSNO (1 mM) is stable when incubated in PBS with 0.1 mM DTPA (□), PBS/DTPA and 5 mM NEM, or PBS/DTPA and 200 mM KCN (identical trend lines; ▽). However, the combination of NEM and KCN rapidly degrades GSNO (○). (B and C) SNO-caspase (B) and SNO-Hb (in the presence of 1 mM glutathione) (C) degrade with decreases in pH. (D) Tissue transglutaminase (tTG) SNO content decreases after a change in Ca2+ concentration.
SNO-Protein Stability.
It has been asserted that SNOs and FeNOs in general and SNO-Hbs in particular are stable in acid (46). However, it is well know that acid can destabilize metal nitrosyls (17, 58) and SNOs (Eqs. 4–6 in SI Text) (6, 20). High effective molarity of acidic residues (H+) that reside adjacent to SNO in proteins and/or conformational changes induced by H+ ions (e.g., an R-to-T state shift in Hb) may influence RSNO pKa and thus greatly alter the reactivity of SNO (5, 35). To illustrate the destabilizing effects of low pH, we present the results of changes in pH on SNO-procaspase-3 and SNO-Hb. SNO-caspase-3 (≈2–3 SNOs per protein) was subjected to a pH change from 7.4 to 5 (Fig. 4B). Lowering pH resulted in an immediate loss of ≈1 SNO; the remaining SNOs were stable at pH 5 (Fig. 4B, t = 3 h). Similarly, it is known that lowering pH promotes the T state in Hb, which increases SNO reactivity (9). SNO-Hb (synthesized with S-nitrosocysteine excess, pH 8) appeared stable at both pH 8 and pH 5; however, the addition of 1 mM glutathione led to rapid decay of SNO at pH 5, whereas it had little effect at pH 8 (Fig. 4C). Lowering pH below 5 will lead to protein precipitation and denaturation and to a general loss of cellular architecture. SNO-proteins in vivo would be exposed to the effects of proteolytic enzymes (30) and calcium, which is released from intracellular stores. We have previously shown that tissue transglutaminase is regulated by poly-S-nitrosylation and that Ca2+ determines the stoichiometry of S-nitrosylation (28). As shown in Fig. 4D, addition of 1 mM Ca2+ to poly-S-nitrosylated tissue transglutaminase decreases the SNO content from 4 to 3 mol of SNO per mol of protein.
Chemistry of Triiodide.
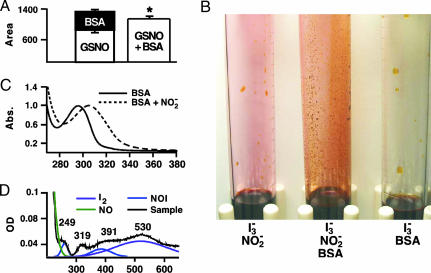
We noted that commercially purified BSA contains various amounts of nitrite as well as protein-bound NO and SNO. When nitrite (data not shown) or GSNO was added to BSA, the resulting signal in the triiodide assay was less than additive vs. signals from BSA and GSNO (or nitrite) alone (Fig. 5A). Furthermore, addition of nitrite or GSNO plus BSA to the reaction vessel led to precipitation of protein, and a purple gas was released from the solution (Fig. 5 A and B). Exclusion of triiodide eliminated both the precipitate and the gas. The precipitated protein, redissolved in 1 N NaOH, appeared yellow in color (UV absorbance peak shifted from 290 to 305 nm) (Fig. 5C). Taken together, the observations are consistent with production of a gaseous reactant that either nitrosates or oxidizes [or iodinates (59)] BSA. We considered the possibility that the mechanisms by which triiodide generates NO from nitrite or GSNO (60) would also allow formation of NOI:
The presence of NOI was indicated in UV/visual light spectroscopy of the gas phase above the triiodide/nitrite reaction (Fig. 5D). NOI is among the most labile, indiscriminate, and potent nitrosating agents, reacting with many substrates at close to diffusion-limited rates (20). Thus, although pure NOI readily generates stoichiometric NO in the absence of reactants, the products of NOI reactions in more complex systems will depend on the nature and concentrations of the reactants.
Fig. 5.
The triiodide assay produces potent nitrosating species and precipitates proteins: impact on NO yield. (A) GSNO and BSA were assayed by triiodide (I3−) individually (stacked bar) or as a mixture of GSNO/BSA (open bar); GSNO/BSA is consistently lower. (B) A purple gas is formed when nitrite is added to triiodide (left tube), and BSA forms a colored precipitate when added with nitrite/triiodide (middle tube), whereas BSA alone remains soluble in the reagent (right tube). (C) Spectrum of BSA precipitate redissolved in 1 N NaOH (dashed line) vs. BSA/triiodide without nitrite subjected to the same protocol (solid line). (D) NOI signature in the gas phase above the triiodide reagent after the addition of nitrite. I2 gives the broad absorption at 530 nm and contributes to the low wavelength edge (purple line), whereas the peaks at ≈250 and ≈390 are characteristic of NOI (blue line). The green line with a maximum at 230 nm corresponds to NO.
Discussion
It has been previously reported that levels of protein-bound NO in vivo, including Hb[FeNO], SNO-Hb, and SNO-albumin, are much lower when measured by triodide (4, 15, 40–46) than by other methods: photolysis chemiluminescence, a modified fluorescence assay, chemical and EPR-based approaches, mass spectrometry, electrochemistry, and the newly developed 3C assay of Doctor et al. (see refs. 4 and 5 for review of the subject as well as refs. 14, 39, 49, 51, 54, 55, 57, 61–63). Although preparative differences may partly account for these discrepancies and a recent modification of the triiodide assay may improve NO recovery (at the expense of the ability to discriminate between NO species) (56), concerns with triiodide remain (4, 5, 15, 56, 62), including denaturation, precipitation, and denitrosylation of proteins in acid; quenching of NO by reactants; and the failure of the few standards used to reflect the range of reactivities of SNO and FeNO species in vivo. In addition, the chemistry of the triiodide assay is not well understood, and it has been unclear how to adequately control for the many reagents used alone and in combination. To our knowledge, NO mass balance has not previously been demonstrated with triiodide in complex biological mixtures. Here, we have generated multiple nitrosylated standards over a range of reactivities and compared triiodide with photolysis chemiluminescence. We have found that yields by triiodide are generally low and, by contrast to photolysis, that the method is unable to accurately differentiate between SNOs, metal nitrosyls, and nitrite in either complex in vitro systems or endogenous samples. In addition, we observed the production of NOI, a potent and promiscuous nitrosating agent, whose fate, including its yield of NO, critically depends on sample composition. These results militate against the use of triiodide to assay nitrosylated species or nitrite in biological mixtures and suggest that previous results obtained with this methodology should be reassessed.
Materials and Methods
For details of experimental procedures used in these experiments see SI Text. The triiodide method, originally described by Samouilov and Zweier (60) was elaborated as detailed by Gladwin and colleagues (41–46). Photolysis chemiluminescence was as developed by Stamler and colleagues (38, 39). NO purified from PROLI NONOate (Cayman Chemical) and saturated NO (aqueous) stock solutions were prepared as described in ref. 52 (see also SI Text). Nitrosylated R-structured Hb and unsaturated Hb[Fe(II)NO] preparations were prepared under anaerobic conditions. S-nitrosylation of procaspase-3 and tissue transglutaminase and related treatments (pH changes, Ca2+) were as described in ref. 28 and in SI Text. [SNO]-oxyHb and SNO/FeNO valency hybrids were generated as described in SI Text. Samples analyzed with triiodide often exhibited significant prolongation of migration times and changes in peak morphology in comparison to GSNO or nitrite standards. Quantification of signals [area under the curve (AUC)] is described in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. Irwin Fridovich for critical review of the manuscript. This work was supported in part by National Heart, Lung, and Blood Institute Grants 5P01-HL424444 and 1P01-HL75443, National Institute of Environmental Health Sciences Grant 419-ES012496, National Science Foundation Grant MCB0981228, and by a Sandler Award.
Abbreviations
- FeCN
potassium ferricyanide
- GSNO
S-nitrosoglutathione
- NEM
N-ethylmaleimide
- NOI
nitrosyliodide
- SAA
sulfanilamide
- SNO
S-nitrosothiol
- SNP
sodium nitroprusside.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611191104/DC1.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Stamler JS, Lamas S, Fang FC. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 3.Foster MW, McMahon TJ, Stamler JS. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 4.Stamler JS. Circ Res. 2004;94:414–417. doi: 10.1161/01.RES.0000122071.55721.BC. [DOI] [PubMed] [Google Scholar]
- 5.Singel DJ, Stamler JS. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 6.Stamler JS, Toone EJ. Curr Opin Chem Biol. 2002;6:779–785. doi: 10.1016/s1367-5931(02)00383-6. [DOI] [PubMed] [Google Scholar]
- 7.Rafikova O, Sokolova E, Rafikov R, Nudler E. Circulation. 2004;110:3573–3580. doi: 10.1161/01.CIR.0000148782.37563.F8. [DOI] [PubMed] [Google Scholar]
- 8.Rafikova O, Rafikov R, Nudler E. Proc Natl Acad Sci USA. 2002;99:5913–5918. doi: 10.1073/pnas.092048999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon TJ, Exton Stone A, Bonaventura J, Singel DJ, Solomon Stamler J. J Biol Chem. 2000;275:16738–16745. doi: 10.1074/jbc.M000532200. [DOI] [PubMed] [Google Scholar]
- 10.Luchsinger BP, Rich EN, Yan Y, Williams EM, Stamler JS, Singel DJ. J Inorg Biochem. 2005;99:912–921. doi: 10.1016/j.jinorgbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Lipton AJ, Johnson MA, Macdonald T, Lieberman MW, Gozal D, Gaston B. Nature. 2001;413:171–174. doi: 10.1038/35093117. [DOI] [PubMed] [Google Scholar]
- 12.Jia L, Bonaventura C, Bonaventura J, Stamler JS. Nature. 1996;380:221–226. doi: 10.1038/380221a0. [DOI] [PubMed] [Google Scholar]
- 13.Holmes A, Williams DLH. J Chem Soc Perkin Trans. 2000;2:1630–1644. [Google Scholar]
- 14.Gandley RE, Tyurin VA, Huang W, Arroyo A, Daftary A, Harger G, Jiang J, Pitt B, Taylor RN, Hubel CA, Kagan VE. Hypertension. 2005;45:21–27. doi: 10.1161/01.HYP.0000150158.42620.3e. [DOI] [PubMed] [Google Scholar]
- 15.Foster MW, Pawloski JR, Singel DJ, Stamler JS. Hypertension. 2005;45:15–17. doi: 10.1161/01.HYP.0000150160.41992.71. [DOI] [PubMed] [Google Scholar]
- 16.Bartberger MD, Mannion JD, Powell SC, Stamler JS, Houk KN, Toone EJ. J Am Chem Soc. 2001;123:8868–8869. doi: 10.1021/ja0109390. [DOI] [PubMed] [Google Scholar]
- 17.Ford PC, Fernandez BO, Lim MD. Chem Rev. 2005;105:2439–2455. doi: 10.1021/cr0307289. [DOI] [PubMed] [Google Scholar]
- 18.Sharma VS, Isaacson RA, John ME, Waterman MR, Chevion M. Biochemistry. 1983;22:3897–3902. doi: 10.1021/bi00285a026. [DOI] [PubMed] [Google Scholar]
- 19.Oae S, Shinhama K. Org Prep Proc Int. 1983;15:165–198. [Google Scholar]
- 20.Williams DLH. Nitrosation. Cambridge, UK: Cambridge Univ Press; 1988. [Google Scholar]
- 21.Yonetani T, Tsuneshige A, Zhou Y, Chen X. J Biol Chem. 1998;273:20323–20333. doi: 10.1074/jbc.273.32.20323. [DOI] [PubMed] [Google Scholar]
- 22.Zhao YL, Houk KN. J Am Chem Soc. 2006;128:1422–1423. doi: 10.1021/ja057097f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma VS, Ranney HM. J Biol Chem. 1978;253:6467–6472. [PubMed] [Google Scholar]
- 24.Moore EG, Gibson QH. J Biol Chem. 1976;251:2788–2794. [PubMed] [Google Scholar]
- 25.Zhao YL, McCarren PR, Houk KN, Choi BY, Toone EJ. J Am Chem Soc. 2005;127:10917–10924. doi: 10.1021/ja050018f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro A, Williams DLH. J Chem Soc Perkin Trans. 1999;2:1989–1993. [Google Scholar]
- 27.Aracena P, Sanchez G, Donoso P, Hamilton SL, Hidalgo C. J Biol Chem. 2003;278:42927–42935. doi: 10.1074/jbc.M306969200. [DOI] [PubMed] [Google Scholar]
- 28.Lai TS, Hausladen A, Slaughter TF, Eu JP, Stamler JS, Greenberg CS. Biochemistry. 2001;40:4904–4910. doi: 10.1021/bi002321t. [DOI] [PubMed] [Google Scholar]
- 29.Nedospasov A, Rafikov R, Beda N, Nudler E. Proc Natl Acad Sci USA. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chvanov M, Gerasimenko OV, Petersen OH, Tepikin AV. EMBO J. 2006;25:3024–3032. doi: 10.1038/sj.emboj.7601207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 32.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 33.Nunez L, Vaquero M, Gomez R, Caballero R, Mateos-Caceres P, Macaya C, Iriepa I, Galvez E, Lopez-Farre A, Tamargo J, Delpon E. Cardiovasc Res. 2006;72:80–89. doi: 10.1016/j.cardiores.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 35.Stamler JS, Jia L, Eu JP, McMahon TJ, Demchenko IT, Bonaventura J, Gernert K, Piantadosi CA. Science. 1997;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- 36.Baciu C, Cho KB, Gauld JW. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2005;109:1334–1336. doi: 10.1021/jp0443759. [DOI] [PubMed] [Google Scholar]
- 37.Angelo M, Singel DJ, Stamler JS. Proc Natl Acad Sci USA. 2006;103:8366–8371. doi: 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon TJ, Stamler JS. Methods Enzymol. 1999;301:99–114. doi: 10.1016/s0076-6879(99)01073-3. [DOI] [PubMed] [Google Scholar]
- 39.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Proc Natl Acad Sci USA. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feelisch M, Rassaf T, Mnaimneh S, Singh N, Bryan NS, Jourd'Heuil D, Kelm M. FASEB J. 2002;16:1775–1785. doi: 10.1096/fj.02-0363com. [DOI] [PubMed] [Google Scholar]
- 41.Gladwin MT, Ognibene FP, Pannell LK, Nichols JS, Pease-Fye ME, Shelhamer JH, Schechter AN. Proc Natl Acad Sci USA. 2000;97:9943–9948. doi: 10.1073/pnas.180155397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gladwin MT, Wang X, Reiter CD, Yang BK, Vivas EX, Bonaventura C, Schechter AN. J Biol Chem. 2002;277:27818–27828. doi: 10.1074/jbc.M203236200. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Tanus-Santos JE, Reiter CD, Dejam A, Shiva S, Smith RD, Hogg N, Gladwin MT. Proc Natl Acad Sci USA. 2004;101:11477–11482. doi: 10.1073/pnas.0402201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X, Cho M, Spencer NY, Patel N, Huang Z, Shields H, King SB, Gladwin MT, Hogg N, Kim-Shapiro DB. Proc Natl Acad Sci USA. 2003;100:11303–11308. doi: 10.1073/pnas.2033883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang BK, Vivas EX, Reiter CD, Gladwin MT. Free Radic Res. 2003;37:1–10. doi: 10.1080/1071576021000033112. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Bryan NS, Macarthur PH, Rodriguez J, Gladwin MT, Feelisch M. J Biol Chem. 2006;281:26994–27002. doi: 10.1074/jbc.M603953200. [DOI] [PubMed] [Google Scholar]
- 47.McMahon TJ, Ahearn GS, Moya MP, Gow AJ, Huang YC, Luchsinger BP, Nudelman R, Yan Y, Krichman AD, Bashore TM, et al. Proc Natl Acad Sci USA. 2005;102:14801–14806. doi: 10.1073/pnas.0506957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luchsinger BP, Rich EN, Gow AJ, Williams EM, Stamler JS, Singel DJ. Proc Natl Acad Sci USA. 2003;100:461–466. doi: 10.1073/pnas.0233287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pawloski JR, Hess DT, Stamler JS. Proc Natl Acad Sci USA. 2005;102:2531–2536. doi: 10.1073/pnas.0409876102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawloski JR, Hess DT, Stamler JS. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 51.McMahon TJ, Moon RE, Luschinger BP, Carraway MS, Stone AE, Stolp BW, Gow AJ, Pawloski JR, Watke P, Singel DJ, Piantadosi CA, Stamler JS. Nat Med. 2002;8:711–717. doi: 10.1038/nm718. [DOI] [PubMed] [Google Scholar]
- 52.Gow AJ, Stamler JS. Nature. 1998;391:169–173. doi: 10.1038/34402. [DOI] [PubMed] [Google Scholar]
- 53.Herold S, Rock G. Arch Biochem Biophys. 2005;436:386–396. doi: 10.1016/j.abb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 54.Nagababu E, Ramasamy S, Abernethy DR, Rifkind JM. J Biol Chem. 2003;278:46349–46356. doi: 10.1074/jbc.M307572200. [DOI] [PubMed] [Google Scholar]
- 55.Kirima K, Tsuchiya K, Sei H, Hasegawa T, Shikishima M, Motobayashi Y, Morita K, Yoshizumi M, Tamaki T. Am J Physiol. 2003;285:H589–H596. doi: 10.1152/ajpheart.01010.2002. [DOI] [PubMed] [Google Scholar]
- 56.Rogers SC, Khalatbari A, Gapper PW, Frenneaux MP, James PE. J Biol Chem. 2005;280:26720–26728. doi: 10.1074/jbc.M501179200. [DOI] [PubMed] [Google Scholar]
- 57.Rassaf T, Bryan NS, Maloney RE, Specian V, Kelm M, Kalyanaraman B, Rodriguez J, Feelisch M. Nat Med. 2003;9:481–482. doi: 10.1038/nm0503-481. and author reply (2003) 9:482–483. [DOI] [PubMed] [Google Scholar]
- 58.Wasser IM, de Vries S, Moenne-Loccoz P, Schroder I, Karlin KD. Chem Rev. 2002;102:1201–1234. doi: 10.1021/cr0006627. [DOI] [PubMed] [Google Scholar]
- 59.Deng H. J Pept Sci. 2006 Nov 20; doi: 10.1002/psc.806. [DOI] [Google Scholar]
- 60.Samouilov A, Zweier JL. Anal Biochem. 1998;258:322–330. doi: 10.1006/abio.1998.2609. [DOI] [PubMed] [Google Scholar]
- 61.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, et al. Proc Natl Acad Sci USA. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doctor A, Gaston B, Kim-Shapiro DB. Blood. 2006;108:3225–3226. doi: 10.1182/blood-2006-05-026047. and author reply (2003) 108:3226–3227. [DOI] [PubMed] [Google Scholar]
- 63.Tsikas D. Circ Res. 2004;94:e106. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.