Abstract
We present a strategy for the analysis of the yeast phosphoproteome that uses endo-Lys C as the proteolytic enzyme, immobilized metal affinity chromatography for phosphopeptide enrichment, a 90-min nanoflow-HPLC/electrospray-ionization MS/MS experiment for phosphopeptide fractionation and detection, gas phase ion/ion chemistry, electron transfer dissociation for peptide fragmentation, and the Open Mass Spectrometry Search Algorithm for phosphoprotein identification and assignment of phosphorylation sites. From a 30-μg (≈600 pmol) sample of total yeast protein, we identify 1,252 phosphorylation sites on 629 proteins. Identified phosphoproteins have expression levels that range from <50 to 1,200,000 copies per cell and are encoded by genes involved in a wide variety of cellular processes. We identify a consensus site that likely represents a motif for one or more uncharacterized kinases and show that yeast kinases, themselves, contain a disproportionately large number of phosphorylation sites. Detection of a pHis containing peptide from the yeast protein, Cdc10, suggests an unexpected role for histidine phosphorylation in septin biology. From diverse functional genomics data, we show that phosphoproteins have a higher number of interactions than an average protein and interact with each other more than with a random protein. They are also likely to be conserved across large evolutionary distances.
Keywords: yeast phosphoproteome, network analysis
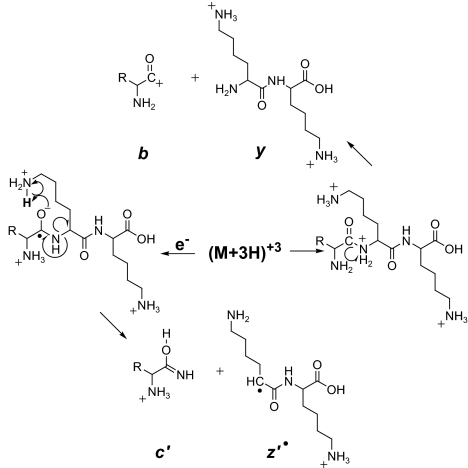
In an earlier study of the yeast phosphoproteome (1), we digested proteins from a whole cell lysate with trypsin, used immobilized metal-affinity chromatography (IMAC) to enrich the sample for phosphopeptides, and analyzed the resulting mixture by nano-flow HPLC interfaced to electrospray ionization tandem mass spectrometry (MS/MS). Low-energy collision-activated dissociation (CAD) was used to fragment the peptide backbone and to produce ions of types b and y (Fig. 1) required for successful sequence analysis and identification of phosphorylation sites. We detected >1,000 phosphopeptides but defined only 383 sites of phosphorylation, largely because the CAD process often promoted elimination of phosphoric acid from Ser and Thr residues without breaking the amide bonds along the peptide backbone. The resulting MS/MS spectra were essentially devoid of sequence information.
Fig. 1.
Peptide fragmentation. Scheme for generating type b and y ions by CAD and type c and z· ions by ETD.
To circumvent this problem, we have modified the LTQ mass spectrometer for ion/ion chemistry and now fragment both peptides (2) and intact proteins (3) by electron transfer dissociation (ETD). In this process, fluoranthene radical-anions are generated in a chemical ionization source and used as reagents to transfer an electron to a multiply charged peptide generated by electrospray ionization. This reaction is highly exothermic, reduces the peptide charge by one, and triggers fragmentation of the peptide backbone to produce a homologous series of complementary fragment ions of type c and z· (Fig. 1). Subtraction of m/z values for fragments within a given ion series that differ by a single amino acid affords the mass and thus the identity of the extra residue in the larger of the two fragments. The complete amino acid sequence of a peptide is deduced by extending this process to all homologous pairs of fragments within a particular ion series. ETD, unlike CAD, works exceptionally well on large multiply charged peptides and is indifferent to most posttranslational modifications, including phosphorylation.
Here we present a strategy for the analysis of the yeast phosphoproteome that uses endo-Lys C as the proteolytic enzyme, IMAC for phosphopeptide enrichment, ETD for peptide fragmentation, and the Open Mass Spectrometry Search Algorithm for phosphopeptide identification and assignment of phosphorylation sites. With this approach, we identify 1,252 phosphorylation sites on 629 proteins in a single experiment with 30 μg (≈600 pmols) of protein from a yeast whole cell lysate. We find that the identified phosphoproteins are encoded by a sample of genes that is representative of a wide variety of cellular processes. Expression levels for the identified phosphoproteins range from <50 to >1,200,000 copies per cell. We note that most protein-kinase recognition motifs predicted by SCANSITE are significantly enriched in our phosphorylation data and present evidence for a motif recognized by one or more unidentified yeast kinases. We analyze the identified phosphoproteins in the context of interaction networks and find that they have a significantly higher number of interactions than expected and interact with other phosphoproteins more than expected. We note that the observed phosphoproteins, but not individual phosphosites, are likely to be conserved across very large evolutionary distances.
Results and Discussion
Fig. 2A shows the base peak chromatogram for a 90-min analysis of yeast phosphopeptides obtained from a 30 μg (≈600 pmol) aliquot of total cell protein. Sample was digested with endo-Lys-C, enriched for phosphopeptides by IMAC, and analyzed by a combination of nanoflow-HPLC/electrospray ionization tandem MS, and ETD. From this experiment, we identify 1,252 phosphorylation sites on 629 proteins, ≈10% of the proteome. [The complete data set of identified yeast phosphopeptides is available in supporting information (SI) Table 2.] A subset of this data, phosphopeptides identified from yeast protein kinases, is shown in Table 1. Note that we find phosphorylation sites on a disproportionately high number (33 of 126, 26%) of these proteins, even though the expression levels for the yeast kinases and the other 593 phosphoproteins in SI Table 2 range from <50 to 1,200,000 copies per cell (4). It seems likely that many of the yeast kinases are, themselves, regulated by phosphorylation.
Fig. 2.
Phosphopeptide chromatogram and mass spectra. (A) Base peak, ion chromatogram for a 90 min separation of yeast phosphopeptides enriched from a 30-μg aliquot of yeast total cell protein. (B and C) ETD mass spectra recorded on (M + 3H)+3 ions from peptides eluting under peaks I and II, respectively, in A. Observed c and z· ions are indicated on the peptide sequence by ⌉ and ⌊, respectively.
Table 1.
Protein-kinase phosphopeptides from S. cerevisiae
| Protein* | Copies per cell† | Peptide sequence | E value‡ |
|---|---|---|---|
| YAL017W | 1,000 | EFAGRRSpSLPRTSASANHLMNRNK | 1.E-02 |
| KSpSSRISTGTLK | 1.E-305 | ||
| RGNQPVSTFLRpTPEK | 1.E-07 | ||
| YBR059C | 3,000 | QSSDPTISEQpSPRLNTQSLPQRQK | 1.E-03 |
| YDL101C | 3,500 | REHpSGDVTDpSpSFK | 1.E-03 |
| REHpSGDVTDpSSFK | 1.E-06 | ||
| REHpSGDVTDSpSFK | 1.E-07 | ||
| REHpSGDVTDSSFK | 1.E-306 | ||
| YDR122W | 300 | FGNIFRKLpSQRRK | 1.E-05 |
| FGNIFRKLpSQRRKK | 1.E-305 | ||
| IRNQQQRQpSHENIEK | 1.E-305 | ||
| pSVGHARREpSLK | 1.E-06 | ||
| pSVGHARRESLK | 1.E-04 | ||
| YDR283C | 300 | pSIQNVPRRRNFVK | 1.E-04 |
| YDR490C | ND | GNRpSLTEADHALLSK | 1.E-02 |
| YDR507C | 700 | KRQpSIpSSVSVSPpSK | 1.E-02 |
| KVSTpTPQRRRNRESLISVTpSpSRK | 1.E-06 | ||
| KVSTpTPQRRRNRESLISVTSpSRK | 1.E-03 | ||
| KVSTpTPQRRRNRESLISVTSSRK | 2.E-05 | ||
| QNpSQEGDQAHPK | 1.E-305 | ||
| SRHFpSEpSNK | 1.E-06 | ||
| YER129W | 800 | QRTHERpSRSLTVAELNEEK | 1.E-07 |
| YHL007C | 300 | pSKTSPIISTAHTPQQVAQSPK | 1.E-05 |
| SKTpSPIISTAHTPQQVAQpSPK | 1.E-04 | ||
| pSSTDIRRATPVSTPVISK | 1.E-03 | ||
| QTHAPTTPNRpTSPNRpSSISRNATLK | 1.E-02 | ||
| QTHAPTTPNRpTSPNRSpSISRNATLK | 1.E-05 | ||
| YHR082C | 1,300 | ELEQERRLpSMEQK | 1.E-03 |
| YRRPpSSSSYTGK | 1.E-04 | ||
| YHR205W | 4,000 | KKPLYTHRpSSSQLDQLNSCSSVTDPSK | 1.E-304 |
| YIL095W | 1,300 | SRQNTGDpSIRSAFGK | 1.E-03 |
| YJL095W | 100 | REAPKPPANTpSPQRTLSTSK | 1.E-07 |
| YJL128C | 2,200 | RSApSVGSNQSEQDK | 1.E-09 |
| RTSpSTSSHYNNINADLHARVK | 1.E-02 | ||
| YJL141C | ND | FRRApSLNpSK | 1.E-03 |
| FRRApSLNSK | 1.E-09 | ||
| YJL165C | ND | RARpSLpSESIK | 1.E-04 |
| RARpSLSESIK | 1.E-305 | ||
| GDEKLpSRHpTSLK | 1.E-04 | ||
| YJR059W | 1,400 | STpSTNDFSENSLDAPHDQEVIHTSNPFLK | 1.E-08 |
| RPTSPSISGSGSGGNSPSSSAGARQRpSASLHRRK | 1.E-05 | ||
| RPpTSPSISGSGSGGNSPSSSAGARQRSASLHRRK | 1.E-04 | ||
| STSpTNDFSENSLDAPHDQEVIHTSNPFLK | 1.E-08 | ||
| YKL101W | <50 | LRLpSPENPSNTHMQK | 1.E-305 |
| RRPpSEEpSVNPK | 1.E-305 | ||
| RRPpSEESVNPK | 1.E-06 | ||
| SPpSRYSLSRRAIHASPpSTK | 1.E-305 | ||
| SPpSRYSLSRRAIHASPSTK | 1.E-05 | ||
| YKL126W | 3,000 | EEQQNNQATAGEHDASITRSpSLDRK | 1.E-03 |
| HSGFFHSSKKEEQQNNQATAGEHDASITRSpSLDRK | 1.E-304 | ||
| KEEQQNNQATAGEHDASITRpSSLDRK | 1.E-07 | ||
| YKL139W | 100 | RPLFGKRpSPNPQSLARPPPPK | 1.E-03 |
| YKL166C | 1,600 | MYVDPMNNNEIRKLpSITAK | 1.E-305 |
| YKL168C | 1,000 | RQQPVTRRVRpSFSESFK | 1.E-305 |
| YLR096W | ND | LTIPEQAHTSPpTSRK | 1.E-08 |
| QHSLPpSPKNESEILERQK | 1.E-07 | ||
| YLR248W | 1,800 | DVSQITSpSPKK | 1.E-305 |
| YMR001C | 1,500 | LEEYHQNRPFLPHSLpSPGGTK | 1.E-06 |
| YMR216C | 2,400 | RLQRHVSRSpSDITANDSSDEK | 1.E-04 |
| YMR291W | 10,000 | TLpSRQGpSSTpSVKK | 1.E-06 |
| KNpSpTFVLDPKPPK | 1.E-04 | ||
| TLpSRQGpSSTSVKK | 1.E-08 | ||
| DTPNFSFHPTIRRVSpSTASMHTLRSPSK | 1.E-06 | ||
| YNL020C | 1,500 | TRpSLGSYSTRGNIK | 1.E-09 |
| YOL016C | 7,500 | TLHDDREpSK | 1.E-305 |
| YOL100W | <50 | ATPQRSTpSNRNVGDLLLEK | 1.E-03 |
| HNpSFpSESINpSAK | 1.E-02 | ||
| VIERRTpSSSGRAIPK | 1.E-05 | ||
| YOL128C | ND | TYFCpSRFYRAPELLLNSK | 1.E-305 |
| YOR267C | 300 | KLpSMSQLRSKK | 1.E-03 |
| QTHpSMAELK | 1.E-09 | ||
| YPL150W | <50 | ISSQRAYSHSIAGpSPRK | 1.E-06 |
*Standard gene names from the Saccharomyces genome database, www.yeastgenome.org.
†Protein expression levels obtained from genome-wide protein affinity purification experiments (4), ND means no detected expression.
‡Expectation scores assigned by Open Mass Spectrometry Search Algorithm. An E value <0.05 is statistically significant. With current search parameters, spectra assigned scores higher than 1.E-04 should be subjected to manual interpretation.
Fig. 2 B and C shows the ETD spectra recorded on peptides eluting under peaks I and II, respectively, in Fig. 2A. Ion currents carried by (M + 3H)+3 ions for these two peptides differ by more than a factor of 1,000 (3E7 and 5E4), yet the resulting ETD spectra still contain a near full complement of c and z· type ions that facilitate sequence assignment and location of the single site of phosphorylation in each peptide. Note that fragments of type c and z· that contain the N and C termini of Pro, respectively, remain attached to the peptide backbone through other atoms in the proline ring and are thus not observed in ETD mass spectra (2).
Endo-Lys-C cleaves proteins on the C-terminal side of Lys, but not Arg, and thus produces many fragments that are larger than those generated with trypsin. Of the phosphopeptides identified in the present study, 36% contain >20 residues. Fig. 3A shows the ETD mass spectrum recorded on the (M + 6H)+6 ion at m/z 683.3 for a 35-residue phosphopeptide (MW 4,093) derived from the 76.5-kDa, Ser/Thr kinase, YPK1 (JKL126W). This peptide contains eight Ser/Thr residues. Phosphorylation of these residues increases their mass by 80 Da. In the spectrum shown in Fig. 3A, the fragment that contains the last four amino acids of the peptide, z4, appears at the predicted m/z value of 543. The z5 ion contains the same four amino acids plus Ser31. Its m/z value is predicted to occur at m/z 630, but is observed at m/z 710. All larger singly and doubly charged z· ions undergo shifts of 80 and 40 Da, respectively. We conclude that Ser31 is the only site of phosphorylation in this peptide. Note that although the mass range of the LTQ instrument is currently only 2,000 Da, the presence of singly and doubly charged c and z· ions in the ETD spectrum usually makes it possible to sequence peptides that are 40–50 residues in length.
Fig. 3.
Phosphopeptide mass spectra. ETD mass spectra recorded on (M + 6H)+6 ions at m/z 683.3 for a 35 residue phosphopeptide of MW 4,093 (A), and (M + 3H)+3 ions from a pHis containing peptide at the C terminus of the septin protein, Cdc10 (B). Observed c and z· ions are indicated on the peptide sequence by ⌉ and ⌊, respectively. Observed doubly charged c and z· ions are indicated by an additional label, circle and asterisk, respectively.
Fig. 3B shows the ETD spectrum recorded on the (M + 3H)+3 ion at m/z 690.5 for an 18-residue phosphopeptide (MW 2,067) derived from Cdc10 (YCR002C), one of five septin proteins that form a filamentous ring structure and play a conserved role in both cytokinesis and morphogenesis. This peptide contains five Ser residues. Fragments containing the first nine residues from the N terminus, c9, and the last eight residues from the C terminus, z8, both occur at the predicted m/z values, 948 and 904, respectively. In contrast, fragments c10 and z10, which contain His314, are both observed at m/z values that are 80 Da higher than expected. We conclude that the Cdc10 peptide is phosphorylated on His rather than Ser.
Histidine kinases are common in bacteria and are part of a two-component system where a membrane-bound, sensor/kinase autophosphorylates a His residue and then transfers phosphate to an Asp residue on a cytoplasmic response regulator protein. The response regulators, in turn, function as molecular switches and control numerous cellular processes (5). In yeast, there is only one known histidine kinase (Sln 1), a membrane-bound sensor/kinase that regulates the response to both osmotic (6) and oxidative stress (7) through phosphorelay pathways involving the response regulator proteins, Ssk1 and Skn7, respectively. Sln1 undergoes autophosphorylation on His576, transfers the phosphate to Asp1144 in its own receiver domain, and then releases it to His64 on the small histidine phosphotransfer (HPt) protein, Ypd1. Ypd1 delivers the phosphate to specific Asp residues on the response regulator proteins, Ssk1 and Skn7, which then interact with proteins in the MAP kinase pathway. The finding that Cdc10 is phosphorylated on His suggests that it is a likely to be regulated by Sln1. This finding points to unexpected role for histidine phosphorylation in septin biology.
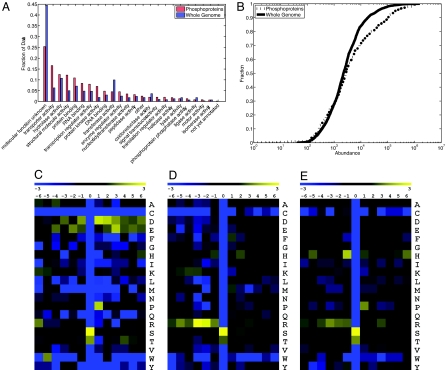
The 629 phosphoproteins presented in SI Table 2 were identified from Saccharomyces cerevisiae grown on rich medium containing glucose. These proteins represent a broad sample of yeast proteins covering most cellular processes (Fig. 4A and SI Table 3). The abundances of the phosphoproteins identified also span the entire range of known protein abundances (Fig. 4B). These abundances are estimated from genome-wide protein affinity purification experiments derived from cells grown under similar conditions to those used in the present study (4). Together, these data suggest that the identified phosphopeptides are encoded by a representative sample of genes corresponding to a wide variety of cellular processes and are observed in proportion to their expression within the yeast proteome.
Fig. 4.
Phosphoprotein and amino acid frequencies. (A) Comparison of GO Slim (33) term frequencies between the whole genome and the phosphoproteins. (B) Comparison of protein abundances between the entire genome and the phosphoproteins. The distribution of phosphoprotein abundances is comparable to that of the genomic background. (C–E) Log2 ratios of per-site amino acid frequencies relative to the genomic background. (C) Sites identified as acidophilic by SCANSITE. (D) Sites identified as basophilic by SCANSITE. (E) Sites not recognized by SCANSITE.
We find the identified phosphoproteins to be enriched in a small number of GO (8) processes, particularly fermentation, protein synthesis, and phosphorylation-related processes (SI Table 3). This is not surprising, given that the cells were grown in rich medium under conditions favoring rapid growth and fermentation. Interestingly, the genes are also enriched in a subset of cell-division specific processes; namely, budding, polarity, and cytokinesis. This finding was unexpected and suggests that there is a high degree of phosphorylation-dependent regulation of these events. Phosphorylated proteins were significantly more likely to themselves possess known phosphotransferase activity. This observation supports the fact that phosphorylation is a common regulation mechanism for kinases in yeast (9).
We further analyzed the kinase–substrate relationships in our data set by using SCANSITE to predict motifs within the phosphorylated proteins from which the peptides were derived (10). Nearly every protein kinase recognition motif predicted by the SCANSITE was significantly enriched in our phosphorylation data (SI Fig. 6). A summary of the SCANSITE kinase target-groups found in our data, using medium stringency criteria, appears in SI Table 4. Basophilic sites make up the largest group of motifs, and acidophilic and proline-directed motifs are also well represented. These results are in agreement with data from other large datasets (11, 12). In addition to these known sites, our assay identifies 381 phosphorylation sites not found by SCANSITE. The relative occurrence of amino acids flanking the sites of phosphorylation appears as a heat map in Fig. 4 C–E. The acidophilic (Fig. 4C) and basophilic (Fig. 4D) maps are as predicted from SCANSITE. The heat map for the 381 sites not found by SCANSITE (Fig. 4E) shows a moderate enrichment of proline and histidine at positions that are +1 and −1, respectively, to the site of phosphorylation. We suggest that this likely represents a new motif for one or more protein kinases in yeast.
The majority of the phosphorylation sites in our data are on serine (82.3%); the remainder are on threonine (17.5%) and tyrosine (0.027%). This finding confirms the extremely low extent of tyrosine phosphorylation in yeast (13). There are no true protein tyrosine kinases in yeast, but there are seven dual-specificity kinases predicted on the basis of sequence similarity, including the MAPKK proteins (Ste7, Mkk1, Mkk2, and Pbs2) and three kinases that regulate the cell cycle (Rad53, Mps1, and Swe1) (14). None of the tyrosine-phosphorylated proteins are obviously related by functional annotation. However, one of the proteins, Cnm67, is a structural component of the spindle pole body and is an excellent candidate for a substrate of Mps1, a protein kinase that regulates spindle pole body duplication (15, 16).
We took advantage of diverse functional genomic and proteomic data to analyze protein kinase targets in the context of interaction networks. We began with affinity precipitation and yeast two-hybrid data consisting of 13,325 interaction pairs and 4,697 proteins (17–20). We found that phosphoproteins have a significantly higher number of physical interactions than would be expected by chance and, in particular, interact with other phosphoproteins more than with a random protein (Fig. 5B and SI Table 5). One explanation for this is that signaling cascades are often organized on molecular scaffolds that promote the physical association of proteins participating in the signal-transduction pathway (21). Among genetic interaction data (4,775 interaction pairs, 1,469 proteins), we found that genes encoding phosphoproteins also exhibit a strong tendency toward a high degree of interaction and exhibit enriched interactions with other phosphoproteins in genetic interaction networks. This effect has been previously observed for essential proteins and has been implicated for conserved proteins (22). Interestingly, phosphoproteins are not statistically likely to be essential, but they are highly enriched for strongly conserved genes (Fig. 5C). We propose that nonessential phosphoproteins play a central role in biological processes making their conservation evolutionarily advantageous. and, in particular, interact with other phosphoproteins more than with a random protein (Fig. 5B and SI Table 5). One explanation for this is that signaling cascades are often organized on molecular scaffolds that promote the physical association of proteins participating in the signal-transduction pathway (21). Among genetic interaction data (4,775 interaction pairs, 1,469 proteins), we found that genes encoding phosphoproteins also exhibit a strong tendency toward a high degree of interaction and exhibit enriched interactions with other phosphoproteins in genetic interaction networks. This effect has been previously observed for essential proteins and has been implicated for conserved proteins (22). Interestingly, phosphoproteins are not statistically likely to be essential, but they are highly enriched for strongly conserved genes (Fig. 5C). We propose that nonessential phosphoproteins play a central role in biological processes making their conservation evolutionarily advantageous.
Fig. 5.
Phosphoprotein interactions and conservation. (A) A subset of the KEGG sce04110 cell cycle pathway (34). Proteins phosphorylated in this study appear as bold nodes. Known physical interactions are represented by blue edges, and known genetic interactions are shown as red edges. (B) A comparison of phosphoprotein interactions to those of random genomic samples. Clique interactions represent genetic or physical interactions between phosphoproteins (or within random subsamples), and total interactions contain all known genetic or physical interactions between phosphoproteins/sampled proteins and the yeast genome. (C) A representation of the number of model organisms (A. gossypi, C. elegans, D. melanogaster, H. sapiens, and A. thaliana) across which yeast proteins are conserved with significant BLASTP hits. Phosphoproteins are much more likely than a random yeast protein to be conserved (leftmost bars), and conserved phosphoproteins are much more likely to be conserved in all five genomes examined (rightmost bars). Conservation in just one genome is largely explained by the data from the closest organism to S. cerevisiae, A. Gossypi (overlay in darker colors). Error bars represent ± 1 standard deviation.
One example of the propensity of phosphorylated proteins to be hubs and to interact with other phosphorylated proteins is shown in Fig. 5A. The network involves proteins required for three successive stages of the cell cycle and there are two interconnecting hubs that are phosphorylated protein kinases having multiple interactions. Dbf4 is the regulatory subunit of the Cdc7 kinase that regulates the initiation of DNA synthesis, and Cdc5 is the polo-like kinase that is an important mitotic regulator. The network is especially interesting given that the cell must coordinate DNA metabolism with mitosis. Dbf4 interacts with two general classes of proteins, one (light cyan) required for the initiation of DNA synthesis (Cdc7, Cdc45, Mcm2, Orc2, Orc3, Orc5, Orc6, and Swi5) and the other (dark cyan) required for the DNA damage checkpoint (Chk1, Ddc1, Mec3, Rad9, Rad17, Rad24, and Rad53). Similarly, Cdc5 also interacts with two general classes of proteins; one (dark blue) is required for chromosome structure and executing anaphase (Mcd1, Smc1, Smc3, and Swe1) and the other (light blue) for the exit from mitosis (Cdc15 and Mob1). One interesting possibility is that the phosphorylation status of the hub dictates which of the two classes of interactions occurs. Alternatively, the phosphorylation status may regulate interactions between the hubs.
Phosphorylation is expected to be evolutionarily conserved given the overall importance of phosphorylation in cell signaling and regulation. Indeed, we find that phosphorylated proteins are significantly more conserved as compared with other proteins in the proteome, even across large evolutionary distances (Fig. 5C). Fifty-five of these conserved phosphoproteins are themselves kinases that are extremely well conserved (>90% are present in all five organisms). However, even after the removal of these conserved kinases, the remaining phosphoproteins are still observed in all five organisms at a level ≈3.5 standard deviations above random. Given the presumed importance of phosphorylation sites in the functionality of these proteins, it is expected that the phosphorylation sites themselves would be more strongly conserved than the surrounding protein. We compared the conservation of phosphorylation sites within the sequenced fungal genomes (including separate tests against sensu stricto and sensu lato that span 10 and 300 million years of evolution, respectively) as well as with more distant model organisms (SI Fig. 7) (23). Surprisingly, phosphorylated serines and threonines were not found to be significantly more conserved than similar residues in the surrounding protein, regardless of evolutionary distance.
Summary
We have presented a strategy for the analysis of phosphoproteomes that uses endo-Lys C as the proteolytic enzyme, IMAC for phosphopeptide enrichment, ETD for peptide fragmentation, and the Open Mass Spectrometry Search Algorithm for phosphopeptide identification and assignment of phosphorylation sites. With this approach, we identified 1,252 phosphorylation sites on 629 proteins in a single experiment on 30 μg (≈600 pmol) of protein from a yeast whole cell lysate. Expression levels of identified phosphoproteins varied from <50 to >1,200,000 copies per cell. By implementing the ETD technology on LTQ-orbitrap and LTQ-FTMS instruments, it should be possible to sequence still larger phosphopeptides and, possibly, intact phosphoproteins on a chromatographic time scale and thus locate multiple phosphorylation sites that occur on the same protein molecule. Coupling the ETD technology with protein separation by isoelectric focusing (IEF) (24) and phosphopeptide fractionation by either IEF (25) immunity affinity pull downs (26–28), ion exchange chromatography (11), or both ion exchange and immobilized metal affinity chromatography (29), should facilitate in depth analyses of human cellular proteomes (30, 31) in the near future.
Materials and Methods
Sample Preparation.
Total protein, free of nucleic acids, was extracted from a lysate of Saccharomyces cerevisiae using the TRIzol reagent as described (1). The protein pellet was resuspended in 6 M guanidine HCl, dialyzed against 6 M guanidine HCl using Slyde-A-Lyzer, 10,000 MW cutoff, (Pierce, Rockford, IL) to remove small molecules, and stored at −80°C. A 30-μg (≈600 pmol) aliquot of this material was diluted 10-fold in 100 mM ammonium bicarbonate (pH 8.9), reduced with DTT, carboxyamidomethylated with iodoacetamide, and then digested with endo-Lys-C (Roche, Indianapolis, IN) (enzyme/substrate = 1/20) overnight at 37°C in a sand bath. To remove small molecules, the resulting solution of peptides was loaded onto a 360 μm o.d. × 200 μm i.d. fused-silica column packed with irregular reverse-phase C8 beads (10–30 μm, 120 A ODS-AQ; Waters, Milford, MA). After the beads were rinsed with 20 column volumes of 0.1% acetic acid, peptides were eluted with solvent consisting of acetonitrile (70%) (Mallinckrodt, Paris, KY) and 0.1% acetic acid (30%) (Sigma–Aldrich, St. Louis, MO). Solvent was removed on a SpeedVac (Savant Instruments, Farmingdale, NY) and residues were dissolved in 100 μl of freshly prepared methanolic HCl. This reagent was prepared by adding 40 μl of anhydrous thionyl chloride (Sigma–Aldrich) dropwise to 1 ml of anhydrous methanol (Alltech, Deerfield, IL). Esterification was allowed to proceed for 1 h at room temperature, solvent was removed on a SpeedVac, and the resulting peptide methyl esters were then redissolved in 60 μl of a solution containing equal parts of methanol, acetonitrile and 0.01% acetic acid.
Chromatography.
IMAC columns were constructed as described (1), with modification. Briefly, fused-silica columns (360 μm o.d. × 200 μm i.d.) (Polymicro Technologies, Phoenix, AZ) were packed with 8 cm of Poros MC (PerSeptive Biosystems, Framingham, MA) and then washed with (i) 50 mM EDTA (pH 9) for 5 min to remove metal ions, (ii) NANOPure (Barnstead, Dubuque, IA) water for 5 min to bring the pH back to neutral, (iii) 100 mM FeCl3 (Sigma–Aldrich) at a flow rate of 2 μl/min for 10 min to activate the column, and (iv) 0.01% acetic acid, 5–10 column volumes, to remove excess FeCl3. Peptides derived from the digestion of 30 μg (≈600 pmol) of yeast protein were loaded at a flow rate of 1 μl/min and the column was then washed with (i) 1:1:1, methanol: acetonitrile: 0.01% acetic acid (60 μl) to remove peptides bound nonspecifically and (ii) 0.01% acetic acid (20 μl) to remove organic solvent. A fused-silica precolumn (360 μm o.d. × 100 μm i.d.) packed with irregular C8 beads (Waters) was then connected to the IMAC column using a Teflon sleeve (0.012 in o.d. × 0.060 in i.d.; Zeus, Orangeburg, SC). Phosphopeptides were eluted from the IMAC column to the precolumn with 50 mM ascorbic acid (25 μl) (Sigma–Aldrich), and the precolumn was washed with 0.1% acetic acid (10 min) to remove ascorbic acid. A Teflon sleeve was used to connect to an analytical HPLC column (360 μm o.d. × 75 μm i.d.) packed with 5 cm of regular C8 beads (5 μm, Waters) and equipped with a laser-pulled, electrospray, emitter tip (2–4 μm in diameter). An Agilent 1100 Series binary HPLC system (Palo Alto, CA) was used to generate a gradient 0–7% B in 5 min, 7–45% B in 70 min, 45–100% B in 15 min, 100–0% B in 5 min (A = 0.1 M acetic acid; B = 70% acetonitrile in 0.1 M acetic acid) and to elute peptides to the mass spectrometer at a flow rate of 60 nl/min.
Instrument Modification and Operation.
All experiments were performed on a Finnigan LTQ mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nano-flow HPLC microelectrospray ionization source and modified to perform ETD (2). The instrument was operated in the data-dependent mode and cycled through acquisition of a full MS spectrum plus ETD/MS/MS spectra on the 10 most abundant ions in the initial full MS spectrum every 5 s (mass window, 3 Da; dynamic exclusion, 45-s; repeat counts, 1). Instrument control software (ITCL) was modified to facilitate the following sequence of events after selection and storage of the precursor peptide cation; (i) injection of fluoranthene radical anions (≈2 ms) (3); (ii) isolation of fluoranthene radical anions (m/z 202) (10 ms); (iii) electron transfer (ion/ion reaction time of 65 ms); (iv) removal of excess fluoranthene radical anions, and (v) mass analysis of positively charged fragment ions.
Data Analysis.
MS/MS spectra were preprocessed with in-house software to determine the charge states of peptide precursor ions and to eliminate charged reduced, but nondissociated, ions as well as fragments formed from the parent ion by loss of small molecules rather than cleavage of the peptide backbone. The resulting spectra were then searched against the yeast protein database (www.yeastgenome.org) with Open Mass Spectrometry Search Algorithm software (version 1.0.5) (32) (http://pubchem.ncbi.nlm.nih.gov/omssa). Parameters for the search included the following: endo-Lys C specificity; static modifications of 14 Da on Asp, Glu, and the peptide C terminus (formation of methyl esters) and 57 Da on Cys (alkylation with iodoacetamide); differential modification of 80 Da on Ser, Thr, and Tyr (phosphorylation) and 16 Da on Met (oxidation). The window for the precursor ion mass was set as ± 1.5 Da and the fragment–ion mass tolerance was set as ± 0.5 Da. Peptide sequence assignments in SI Table 2 were validated by manual interpretation of the corresponding ETD mass spectra. Nonphosphorylated peptides were not detected under the above experimental conditions.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM37537 and National Science Foundation Grant MCB-0209793 (to D.F.H.).
Abbreviations
- IMAC
immobilized metal-affinity chromatography
- MS/MS
tandem MS
- CAD
collision-activated dissociation
- ETD
electron transfer dissociation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607084104/DC1.
References
- 1.Ficarro SB, McLeland ML, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 2.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 5.West AH, Stock AM. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 6.Posas F, Wurler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Cell. 1996;88:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. EMBO J. 1998;23:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi M, Elion EA. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 10.Obenauer JC, Cantly LC, Jaffe MB. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modesti A, Bini L, Carraresi L, Magherini F, Liberatori S, Pallini V, Manao G, Pinna LA, Raugei G, Ramponi G. Electrophoresis. 2001;22:576–585. doi: 10.1002/1522-2683(200102)22:3<576::AID-ELPS576>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Hunter T, Plowman GD. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 15.Lauze E, Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaerer F, Morgan G, Winey M, Philippsen P. Mol Biol Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bader GD, Donaldson I, Wolting C, Francis Ouellette BF, Pawson T, Hogue GWV. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitkreutz BJ, Stark C, Tyers M. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin A-C, Bosche M, Krause R, Grandl P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutiller K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya RP, Reményi A, Good MC, Bashor CJ, Falick AM, Lim WA. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 22.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 23.Balakrishnan R, Christie KR, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Nash R, et al. Nucleic Acids Res. 2005;33:D374–D377. doi: 10.1093/nar/gki023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moritz RL, Ji H, Schutz F, Connolly LM, Kapp EA, Speed TP, Simpson RJ. Anal Chem. 2004;76:4811–4824. doi: 10.1021/ac049717l. [DOI] [PubMed] [Google Scholar]
- 25.Malmström J, Lee H, Nesvizhskii AI, Shteynberg D, Mohanty S, Brunner E, Ye M, Weber G, Eckerskorn C, Aebersold R. J Proteome Res. 2006;5:2241–2249. doi: 10.1021/pr0600632. [DOI] [PubMed] [Google Scholar]
- 26.Salomon A.R., Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, et al. Proc Natl Acad Sci USA. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brockl A, Peters EC. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 28.Kim J-E, White FM. J Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 29.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NF, Mann M, Jensen ON. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Moser K, White FM. J Proteome Res. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- 31.Kim J-E., Tannenbaum SR, White FM. J Proteome Res. 2005;4:1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 32.Geer LY, Markey SP, Kowalak JA, Wagner L, Maynard DM, Yang X, Shi W, Bryant SH. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 33.Dwight SS, Balakrishnan R, Christie KR, Costanzo MC, Dolinski K, Engel SR, Feierbach B, Fisk DG, Hirschman J, Hong EL, et al. Brief Bioinform. 2004;5:9–22. doi: 10.1093/bib/5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Nat Biotechnol. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.