Abstract
Skeletal muscle differentiation is controlled by interactions between myocyte enhancer factor-2 (MEF2) and myogenic basic helix—loop–helix transcription factors. Association of MEF2 with histone deacetylases (HDAC) -4 and -5 results in repression of MEF2 target genes and inhibition of myogenesis. Calcium/calmodulin-dependent protein kinase (CaMK) signaling promotes myogenesis by disrupting MEF2–HDAC complexes and stimulating HDAC nuclear export. To further define the mechanisms that confer CaMK responsiveness to HDAC4 and -5, we performed yeast two-hybrid screens to identify HDAC-interacting factors. These screens revealed interactions between HDAC4 and members of the 14-3-3 family of proteins, which function as signal-dependent intracellular chaperones. HDAC4 binds constitutively to 14-3-3 in yeast and mammalian cells, whereas HDAC5 binding to 14-3-3 is largely dependent on CaMK signaling. CaMK phosphorylates serines -259 and -498 in HDAC5, which subsequently serve as docking sites for 14-3-3. Our studies suggest that 14-3-3 binding to HDAC5 is required for CaMK-dependent disruption of MEF2–HDAC complexes and nuclear export of HDAC5, and implicate 14-3-3 as a signal-dependent regulator of muscle cell differentiation.
Members of the myocyte enhancer factor-2 (MEF2) family of transcription factors play critical roles in skeletal muscle differentiation and act as end points for diverse intracellular signaling pathways that control myogenesis and muscle hypertrophy (1). The four MEF2 proteins, MEF2A, -B, -C, and -D, share homology in an amino-terminal MCM1 agamous deficiens serum response factor (MADS) domain that mediates DNA-binding, dimerization, and cofactor interactions. MEF2 binding sites are present in the regulatory regions of a variety of muscle and growth factor-inducible genes.
The decision of a myoblast to differentiate depends on the association of MEF2 with positive or negative partners. Association of MEF2 with members of the MyoD family of skeletal muscle-specific basic helix–loop–helix proteins establishes a transcriptional code that activates muscle gene expression and myoblast fusion (2). In contrast, association of MEF2 with histone deacetylases (HDAC)-4 and -5 results in repression of myogenesis (3).
Histone acetylation/deacetylation represents a central mechanism for the control of gene expression (4). Histone acetyltransferases (HATs) catalyze the acetylation of core histones of nucleosomes, resulting in chromatin relaxation and transcriptional activation. The activity of HATs is antagonized by HDACs, which deacetylate histones and transcription factors, causing transcriptional repression. HDACs can be categorized into two classes, I and II, on the basis of size, sequence homology, and formation of distinct complexes. Class I HDACs (-1, -2, and -3) are expressed ubiquitously, whereas Class II HDACs (-4, -5, -6, and -7) are most abundant in heart, brain, and skeletal muscle (5–7), the same tissues that express MEF2 at highest levels (1). Class II HDACs contain a unique amino-terminal extension that mediates association with MEF2 factors (8–10).
Various signaling systems have been implicated in the control of MEF2 activity. Mitogen-activated protein kinases stimulate MEF2 transcriptional activity through phosphorylation of the MEF2 transactivation domain (11, 12). The calcium/calmodulin-dependent phosphatase calcineurin activates MEF2 by a posttranslational mechanism that may require association of MEF2 with the nuclear factor of activated T cells transcription factor (13–16). Recently, we reported that calcium/calmodulin-dependent protein kinase (CaMK) signaling stimulates MEF2 activity by disrupting MEF2–HDAC complexes, with resulting export of HDAC5 from the nucleus to the cytoplasm (10, 17). Nuclear export depended on CaMK-mediated phosphorylation of HDAC5 at two sites in its amino-terminal extension (17).
To further define the mechanisms that regulate the activity and subcellular localization of Class II HDACs, we performed yeast two-hybrid screens by using the amino-terminal regions of HDAC4 and -5 as bait. We found that HDACs interact with members of the 14-3-3 family of intracellular chaperones, which have been implicated in signal-dependent regulation of protein localization (18). In unstimulated yeast and mammalian cells, 14-3-3 efficiently associates with HDAC4, but not HDAC5. CaMK signaling promotes binding of 14-3-3 to HDAC5, and this binding appears to be required for CaMK-dependent disruption of MEF2–HDAC complexes, CaMK-mediated nuclear export of HDAC, and stimulation of myogenesis by CaMK.
Materials and Methods
Yeast Two-Hybrid Screens.
Mouse E10.5 and E17 embryo and adult heart cDNA libraries encoding GAL4-transactivation domain fusion proteins were screened with GAL4-DNA binding domain–HDAC baits in the yeast two-hybrid system (10). Positive clones were subjected to specificity tests and sequenced. After initial rescue of multiple clones encoding 14-3-3, a PCR-based strategy was used for high throughput screening for 14-3-3 family members among the HDAC-interacting clones.
Transfections.
10T1/2 and Cos cells were maintained in DMEM containing 10% (vol/vol) FBS. Transfections were performed by using the lipid-based reagent Fugene 6 (Roche Molecular Biochemicals). Epitope-tagged derivatives of 14-3-3ɛ, HDAC4, and HDAC5 containing amino-terminal FLAG or Myc tags, and MEF2C with a carboxy-terminal Myc tag were generated by using the pcDNA3.1 expression vector (Invitrogen). Mutagenesis was performed with the QuikChange kit (Stratagene). Expression plasmids for FLAG-tagged HDAC1 (pBJ) and HDAC3 (pcDNA6) have been described (5). The cDNA for CaMKI encodes a truncated protein that is constitutively active (19).
Protein–Protein Interaction and Immunofluorescence Experiments.
HDAC–14-3-3 interactions were examined by using glutathione S-transferase (GST)-pull down (20), coimmunoprecipitation (10), and indirect immunofluorescence assays (17), as previously described.
Results
Association of HDAC4 with 14-3-3 in Yeast.
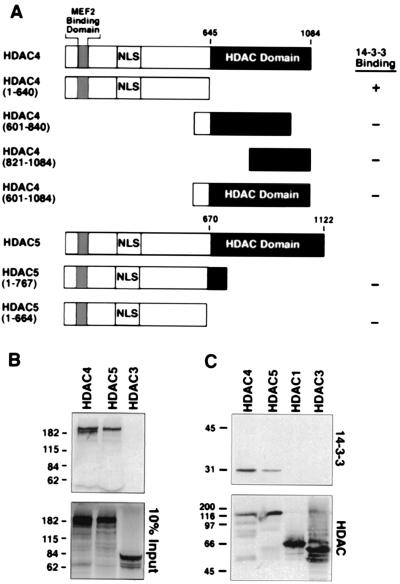
To begin to address the mechanism by which CaMK regulates HDAC function and subcellular localization, we performed yeast two-hybrid screens by using HDAC4 and -5 baits. The “prey” most frequently identified in these screens was 14-3-3, which specifically interacted with the first 640 amino acids of HDAC4 fused in-frame to the GAL4 DNA binding domain (Fig. 1A). From 462 positive clones, 391 were isoforms of 14-3-3 (Table 1). The 14-3-3–GAL4-activation domain fusion proteins interacted specifically with the HDAC4(1–640) bait and not with baits derived from the catalytic region of HDAC4 (Fig. 1A). Surprisingly, 14-3-3 failed to interact with either of two amino-terminal HDAC5 baits, which are ≈60% homologous to the corresponding HDAC4 bait (Fig. 1A). Because 14-3-3 typically binds phosphoproteins, these results suggest that HDAC4, but not HDAC5, is subject to phosphorylation by one or more yeast kinases in vivo. Other HDAC-interacting proteins identified in these screens will be described elsewhere (20).
Figure 1.
Association of HDAC4 and -5 with 14-3-3 in vivo and in vitro. (A) The indicated HDAC truncation mutants were fused to the GAL4-DNA binding domain and tested for interaction with a 14-3-3–GAL4-activation domain fusion protein in the yeast two-hybrid system. (B) GST–14-3-3ɛ fusion proteins were conjugated to glutathione-agarose beads and incubated with [35S]methionine-labeled HDACs. 14-3-3-associated proteins were resolved by SDS-PAGE, followed by autoradiography (Top). (Bottom) Ten percent of the input [35S]methionine-labeled HDAC was applied directly to the gel. (C) Cos cells were transfected with expression vectors encoding the indicated FLAG-tagged HDAC (1 μg), and FLAG-tagged proteins were immunoprecipitated from cell lysates with anti-FLAG antibody. Coimmunoprecipitating endogenous 14-3-3 proteins were detected by immunoblotting with polyclonal anti-14-3-3 antibodies (Top). The membrane was reprobed with anti-FLAG antibody to reveal total immunoprecipitated HDAC protein (Bottom).
Table 1.
14-3-3 clones recovered from yeast two-hybrid screens with HDAC4 bait
| 14-3-3 isoforms | Number of clones |
|---|---|
| ɛ | 117 |
| η | 101 |
| τ | 80 |
| σ | 76 |
| ξ | 17 |
Association of HDAC4 and -5 with 14-3-3 in Vitro.
To further characterize the interaction between HDACs and 14-3-3, binding assays were performed by using full-length HDACs translated in vitro and bacterially expressed GST-14-3-3 protein (20). HDAC4 and -5 interacted with GST-14-3-3 (Fig. 1B Top), but not with GST alone (data not shown). However, the Class I HDACs -1 and -3, which lack the amino-terminal extension required for MEF2 binding (8–10), failed to interact with 14-3-3 under these conditions (Fig. 1B, and data not shown). Of note, bacterially expressed HDACs failed to interact with 14-3-3 translated in vitro (data not shown), suggesting that these HDACs must be phosphorylated by a kinase in the reticulocyte lysate to associate with 14-3-3.
Coimmunoprecipitation assays were also performed by using extracts from Cos cells overexpressing epitope-tagged derivatives of HDACs (10). HDAC4 efficiently interacted with endogenous 14-3-3 (Fig. 1C). Association of HDAC5 with 14-3-3 was also detected, but was consistently weaker and more variable than with HDAC4 (see Fig. 3 C and D). HDAC1 and -3 failed to interact with 14-3-3.
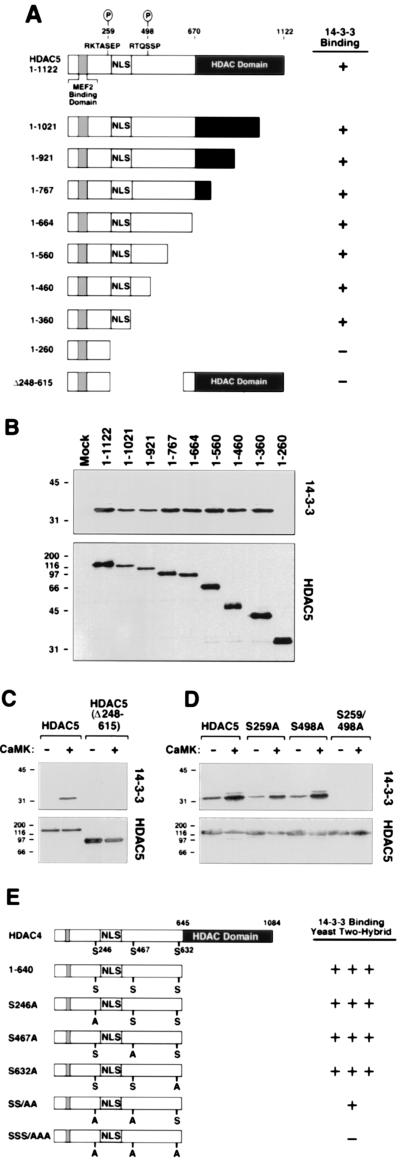
Figure 3.
Mapping of 14-3-3 binding sites on HDAC. (A) Summary of coimmunoprecipitation results. (B) Cos cells were cotransfected with expression vectors (1 μg each) for Myc-tagged 14-3-3ɛ, activated CaMKI, and the indicated FLAG-tagged HDAC protein. HDAC proteins were immunoprecipitated from cell lysates with a monoclonal anti-FLAG antibody, and coimmunoprecipitating 14-3-3 was detected by immunoblotting with anti-Myc antibody (Top). The membrane was reprobed with anti-FLAG antibody (Bottom). (C and D) Cos cells were transfected with expression vectors (1 μg each) for full-length HDAC5, a deletion mutant of HDAC5 lacking amino acids 248–615 (C) or the indicated alanine substitution mutants (D), all FLAG-tagged in the absence or presence of a construct for activated CaMKI (1μg). Association of endogenous 14-3-3 with FLAG-tagged HDACs was assessed as in Fig. 1C. (E) Association of 14-3-3 with the indicated point mutants of HDAC4 in the yeast two-hybrid system; +++, strong interaction; +, weak interaction; −, undetectable interaction.
CaMK Signaling Stimulates Binding of 14-3-3 to HDAC5.
Several observations suggest that HDAC4 and -5 are differentially regulated in vivo. Most notable are the distinct subcellular distributions of these proteins. In transfected fibroblasts, ectopic HDAC4 is cytoplasmic in 50–80% of the cells in which it is expressed (Fig. 2 A and B; ref. 9). In contrast, HDAC5 is exclusively nuclear under identical conditions, but is shuttled from the nucleus to the cytoplasm in response to CaMK signaling (Fig. 2 A and C; ref. 17). One possible explanation for these differences is that HDAC4 and -5 are differentially phosphorylated in vivo, with HDAC4 being constitutively phosphorylated and HDAC5 being phosphorylated in a signal-dependent manner. If 14-3-3 binding plays a role in cytoplasmic accumulation of HDACs, then it would be predicted that HDAC4 would bind constitutively to 14-3-3, whereas binding to HDAC5 would require a signal.
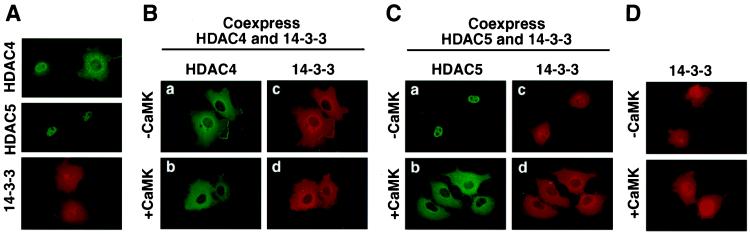
Figure 2.
Colocalization of 14-3-3 and HDACs. (A) Cos cells were transfected with expression vectors for FLAG-tagged derivatives of HDAC4 or -5, or a Myc-tagged version of 14-3-3ɛ (1 μg each). The subcellular distribution of ectopic proteins was determined by indirect immunofluorescence by using primary antibodies against the epitope tags and fluorescein (HDAC)- or rhodamine (14-3-3)-conjugated secondary antibodies. (B and C) Cos cells were cotransfected with expression plasmids (0.5 μg each) for FLAG-tagged HDAC4 (B) or HDAC5 (C) and Myc-tagged 14-3-3 in the absence (a, c) or presence (b, d) of an expression vector for constitutively active CaMKI (0.5 μg). Cells were costained with an anti-FLAG monoclonal antibody and polyclonal anti-Myc antibody, followed by fluorescein (HDAC)- or rhodamine (14-3-3)-conjugated secondary antibodies. (D) Subcellular distribution of ectopic14-3-3ɛ in the absence or presence of activated CaMKI.
To address this issue, indirect immunofluorescence experiments were performed (17). HDAC4 was predominantly cytoplasmic in transfected Cos cells coexpressing 14-3-3. Ectopic 14-3-3, which localized to both the nucleus and the cytoplasm in the absence of HDAC4 (Fig. 2A), was exclusively cytoplasmic in the presence of HDAC4 (Fig. 2B, a). In cells coexpressing HDAC5 and 14-3-3, the localization of these proteins was identical to that seen when they were expressed individually, with HDAC5 in the nucleus and 14-3-3 in both the nucleus and the cytoplasm (Fig. 2C, a and c). In contrast, in the presence of activated CaMK, both HDAC5 and 14-3-3 were colocalized in the cytoplasm (Fig. 2C, b and d). Together, these results suggest that 14-3-3 and HDAC4 associate constitutively, whereas CaMK signaling stimulates association between HDAC5 and 14-3-3 and nuclear export of both proteins. The subcellular distribution of 14-3-3 was unaltered by CaMK in the absence of HDAC5 (Fig. 2D), suggesting that HDAC5, not 14-3-3, was the target for CaMK signaling.
Identification of 14-3-3 Binding Sites on HDAC5.
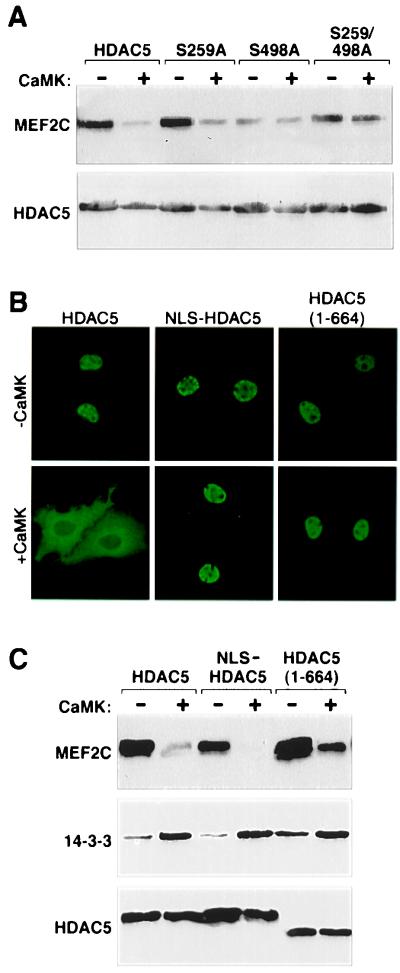
To map the 14-3-3 binding site(s) on HDAC5, a panel of HDAC5 deletion mutants was used in coimmunoprecipitation assays. Initial experiments were performed in the presence of activated CaMK and thus did not distinguish between basal and signal-dependent association of 14-3-3 with HDAC5. Deletion of carboxy-terminal sequences up to amino acid 360 had no noticeable effect on binding of 14-3-3 to HDAC5 (Fig. 3 A and B). However, removal of additional residues up to amino acid 260 led to a complete loss of 14-3-3 binding. In agreement with these findings, an HDAC5 mutant lacking amino acids 248–615 was incapable of binding endogenous 14-3-3 in the absence or presence of CaMK signaling (Fig. 3C).
CaMK stimulates nuclear export of HDAC5 by phosphorylating Ser-259 and Ser-498 (17). Inspection of the sequences flanking these residues revealed the presence of motifs (RXXSXP) similar to those targeted by 14-3-3 in other proteins (Fig. 3A; ref. 18). To determine whether these sites are required for 14-3-3 binding to HDAC5, mutants of HDAC5 containing alanine in place of serine at position 259 and/or 498 were tested in coimmunoprecipitation assays. Replacement of either Ser-259 or Ser-498 with alanine (mutants S259A and S498A) had no discernible effect on CaMK-inducible binding of endogenous 14-3-3 to HDAC5 (Fig. 3D). However, simultaneous disruption of both Ser-259 and Ser-498 (mutant S259/498A) led to a complete loss of 14-3-3 binding. These results strongly suggest that both Ser-259 and Ser-498 of HDAC5 serve as sites for CaMK-inducible binding to 14-3-3.
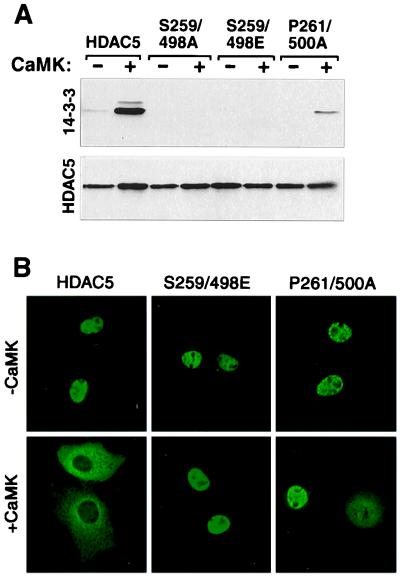
The S259/498A mutant of HDAC5 is completely resistant to CaMK-mediated nuclear export, suggesting a requisite role for 14-3-3 in this process (17). However, it remained possible that phosphorylation of Ser-259 or Ser-498, not 14-3-3 binding, was sufficient to promote HDAC nuclear export. In an attempt to distinguish between these possibilities, two types of HDAC mutants were generated: (i) Ser-259 and/or Ser-498 were substituted with glutamate or (ii) Pro-261 and/or Pro-500 were replaced with alanine (see Fig. 3A). Prior studies have established that glutamate can act as a phosphoserine mimetic by virtue of its negative charge (21), and proline at position +2 relative to phosphoserine is a critical determinant of 14-3-3 binding to various proteins (18). Single glutamate or alanine substitution mutants retained the capacity to bind 14-3-3 in a CaMK-dependent manner (data not shown). However, binding of 14-3-3 to the double-glutamate mutant (S259/498E) was completely lost, indicating that phosphate groups are required for 14-3-3 binding and binding to the double-alanine mutant (P261/500A) was significantly diminished (Fig. 4A). In agreement with a role for 14-3-3 in CaMK-mediated nuclear export of HDAC5, both double mutants remained nuclear in the presence of activated CaMK, however, some cytoplasmic staining of the P261/500A mutant was observed, consistent with its modest 14-3-3 binding (Fig. 4 A and B). Taken together, these results suggest that CaMK-dependent binding of 14-3-3 to HDAC5 regulates nuclear export of this transcriptional repressor.
Figure 4.
CaMK-mediated nuclear export of HDAC5 correlates with 14-3-3 binding. (A) Cos cells were transfected with expression vectors for FLAG-tagged derivatives of the indicated HDAC5 protein in the absence or presence of a construct for activated CaMKI (1 μg). To compensate for CaMK-mediated increases in expression from the CMV-driven expression plasmids, cells receiving CaMKI were transfected with 0.5 μg of HDAC plasmid, compared with 1 μg in those lacking CaMKI. Association of FLAG-tagged HDAC with endogenous 14-3-3 was assessed as in Fig. 1C. (B) The subcellular distribution of wild-type HDAC5 and the indicated HDAC5 mutants in the absence and presence of activated CaMKI in transfected Cos cells was assessed by indirect immunofluorescence employing anti-FLAG antibody and a fluoroscein-conjugated secondary antibody.
Three Serines in HDAC4 Regulate Binding to 14-3-3 in Yeast.
Recently, 14-3-3 was shown to bind HDAC4 in mammalian cells, and this binding depended on phosphorylation of HDAC4 at three serine residues: Ser-246, Ser-467, and Ser-632 (22, 23). Ser-246 and Ser-467, respectively, are analogous to Ser-259 and Ser-498 in HDAC5. To determine whether these sites mediate the binding of 14-3-3 to HDAC4 in yeast, HDAC4 mutants containing alanine in place of Ser-246, Ser-467, and Ser-632 were generated and fused to the yeast GAL4-DNA binding domain (Fig. 3E). Disruption of any single serine had no effect on binding of HDAC4 to 14-3-3. However, disruption of both Ser-246 and Ser- 467 led to a significant reduction in binding, and simultaneous mutation of all three serines led to a complete loss of 14-3-3 binding. These results suggest that 14-3-3 binding to HDAC4 in yeast is mediated by phosphorylation of these serines, consistent with the results obtained in mammalian cells.
14-3-3 Binding to HDAC5 Prevents Formation of MEF2–HDAC Complexes.
CaMK signaling prevents efficient formation of MEF2–HDAC5 complexes (10). Coimmunoprecipitation experiments were performed to address the potential role of 14-3-3 binding to Ser-259 and Ser-498 in this process. Consistent with our prior findings, association between wild-type HDAC5 and MEF2C was significantly reduced in the presence of activated CaMK (Fig. 5A). Likewise, the S259A mutant of HDAC5 failed to efficiently associate with MEF2C in the presence of CaMK (Fig. 5A). In contrast, release of MEF2 from HDAC5 was largely blocked by substitution of Ser-498 with alanine, suggesting a key role for this site in the disruption of MEF2–HDAC interactions.
Figure 5.
CaMK-dependent disruption of MEF2–HDAC complexes. (A and C) Cos cells were transfected with expression vectors for FLAG-tagged derivatives of the indicated HDAC protein and Myc-tagged MEF2C in the absence or presence of a plasmid for activated CaMKI (1 μg), as in Fig. 4. FLAG-tagged proteins were immunoprecipitated from cell lysates and associated MEF2C was measured by immunoblotting with anti-Myc antibody (Top). The membrane was reprobed with anti-FLAG antibody (Bottom). (B) Subcellular distribution of wild-type HDAC5 and the indicated HDAC5 mutants in the absence and presence of CaMKI in transfected Cos cells was assessed by indirect immunofluorescence employing anti-FLAG antibody and a fluoroscein-conjugated secondary antibody.
Disruption of HDAC–MEF2 Complexes in the Absence of HDAC Nuclear Export.
Because experiments addressing the consequences of CaMK signaling on MEF2–HDAC complexes were performed with transfected cells overexpressing active CaMK, and CaMK stimulates nuclear export of HDAC5, it remained possible that the observed loss of MEF2 binding to HDAC5 was because of the reduced nuclear concentration of HDAC5, rather than disruption of MEF2–HDAC complexes. To distinguish between these possibilities, we generated mutants of HDAC5 that bind 14-3-3 in a CaMK-dependent manner, but are resistant to CaMK-mediated nuclear export. A derivative of full-length HDAC5 containing the nuclear localization signal from the SV40 large T antigen fused to its amino terminus (NLS-HDAC5) was completely resistant to CaMK-mediated nuclear export (Fig. 5B). Likewise, a carboxy-terminal truncation mutant () remained largely nuclear in the presence of activated CaMK, because of the absence of a nuclear export signal (NES) (17). Both mutants efficiently formed complexes with MEF2 as assessed in coimmunoprecipitation experiments (Fig. 5C). However, in the presence of activated CaMK, binding to MEF2 was significantly diminished, whereas association with 14-3-3 was elevated (Fig. 5C Middle). These results demonstrate that CaMK signaling disrupts MEF2–HDAC complexes, even in the absence of HDAC nuclear export.
Control of Skeletal Myogenesis by HDAC Nuclear Export and 14-3-3 Binding.
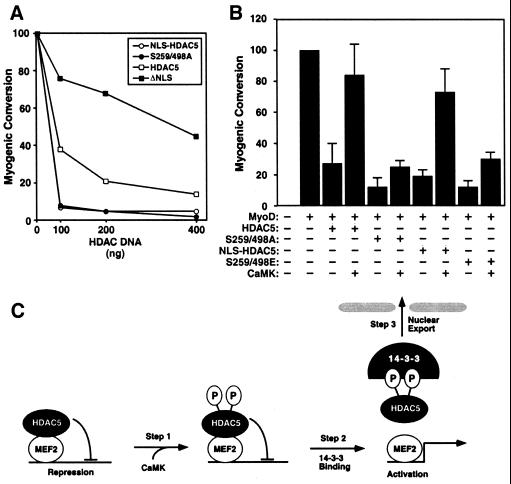
HDAC5 blocks muscle differentiation by associating with MEF2, and CaMK signaling overcomes this inhibition by disrupting MEF2–HDAC complexes, with resulting HDAC nuclear export (3, 10, 17). To address the relative contributions of HDAC phosphorylation, 14-3-3 binding, and nuclear export to stimulation of myogenesis by CaMK, we examined the consequences of overexpressing HDAC5 mutants deficient in each of these steps on MyoD-dependent conversion of fibroblasts to muscle. Transfection of 10T1/2 cells with MyoD expression plasmid resulted in expression of myosin heavy chain. Overexpression of wild-type HDAC5 interfered with this process in a dose-dependent manner (Fig. 6A). The constitutively nuclear HDAC5 mutants, NLS-HDAC5 and S259/498A, blocked muscle cell conversion significantly more efficiently that wild-type HDAC5. In contrast, the capacity of a constitutively cytoplasmic mutant of HDAC5 lacking an NLS (HDAC5ΔNLS) to inhibit myogenesis was severely impaired (Fig. 6A).
Figure 6.
Control of skeletal myogenesis by HDAC nuclear export and 14-3-3 binding. (A) 10T1/2 fibroblasts were cotransfected with expression vectors for MyoD (0.5 μg) and the indicated HDAC5 proteins (100–400 ng). Cells were transferred to differentiation medium two days posttransfection and stained with anti-myosin antibodies after four additional days in culture. Myogenic conversion refers to the percentage of myosin positive cells in HDAC transfectants relative to cultures expressing MyoD alone (100 represents ≈500 myosin-positive cells per 35-mm dish). 10T1/2 fibroblasts were cotransfected with expression vectors for MyoD (0.5 μg) and the indicated HDAC5 protein (0.5 μg) in the absence or presence of a plasmid for activated CaMKI (0.5 μg). Values represent the mean +/− standard deviation from at least two experiments. (C) A model for signal-dependent regulation of myogenesis by CaMK and 14-3-3.
CaMK signaling efficiently rescued the block to myogenesis imposed by HDAC5 (Fig. 6B; ref. 3). In contrast, in the presence of the S259/498A mutant, which cannot be phosphorylated, bound by 14-3-3, or exported to the cytoplasm, CaMK-dependent rescue of myogenesis was blocked (Fig. 6B). Similar results were obtained with cells expressing the S259/498E mutant, which fails to bind 14-3-3 or undergo nuclear export. In contrast, the ability of cells to differentiate into muscle was restored by CaMK in the presence of NLS-HDAC5, which fails to undergo nuclear export, but binds 14-3-3 and is released from MEF2 in the presence of CaMK signaling. These results suggest that 14-3-3 binding to HDAC, with resulting disruption of HDAC–MEF2 complexes is a requisite step in the myogenic program.
Discussion
The results of this study demonstrate that 14-3-3 binding to HDAC4 and -5 has important consequences on activation of MEF2 and muscle differentiaion. This work also demonstrates that, whereas both HDAC4 and -5 associate with 14-3-3 in a phosphorylation-dependent manner, these transcriptional repressors are subject to differential regulation.
Differential Regulation of HDAC4 and -5.
Our discovery of the association of 14-3-3 with HDAC proteins came from the finding that multiple 14-3-3 isoforms interacted with the amino-terminal region of HDAC4 in yeast two-hybrid assays. Through systematic mutagenesis of consensus sequences for 14-3-3 binding in HDAC4, we demonstrated that Ser-246, Ser-467, and Ser-632 were responsible for 14-3-3 binding in yeast. All three sites were also shown recently to mediate binding of HDAC4 to 14-3-3 in mammalian cells (22, 23). Because the sites in HDAC4 responsible for interaction with 14-3-3 are partially conserved in HDAC5, it was concluded in those studies that a similar mechanism regulated HDAC5. However, our results demonstrate that HDAC5 fails to associate with 14-3-3 in yeast, and interacts with 14-3-3 in a largely CaMK-dependent manner in mammalian cells. These results are striking, given the high degree of identity between the relevant sites in HDAC4 and -5, and suggest that these HDACs are differentially regulated by distinct kinases.
Recently, we found that CaMK signaling rescues MEF2 from repression by HDAC4, and overcomes the block to myogenesis imposed by HDAC4. These results may seem paradoxical, given the results presented here, in which HDAC4 is constitutively bound to 14-3-3 and localized to the cytoplasm. However, it should be noted that MEF2 binding to HDAC4 is sufficient to drive this repressor into the nucleus (9), and nuclear HDAC4 is subject to CaMK-mediated nuclear export (unpublished results). Thus, in certain contexts, the phosphorylation status of HDAC4 is likely altered, allowing it to acquire sensitivity to CaMK signaling.
Regulation of HDAC Nuclear Export.
How does association with 14-3-3 result in nuclear exclusion of HDAC5? The simplest possibilities would be that (i)14-3-3 masks the NLS in HDAC5, which is flanked by 14-3-3 binding sites, (ii) 14-3-3 unmasks an NES in HDAC5, or (iii) it provides an NES in trans. In studies of HDAC4, Grozinger and Schreiber (22) concluded that 14-3-3 prevents nuclear import by masking the HDAC4 NLS, thereby preventing association with the nuclear import factor, importin α. Although it is likely that this represents a part of the mechanism, our results lead us to conclude that 14-3-3 also stimulates nuclear export of HDAC5. Two observations support this conclusion. First, a mutant of HDAC5 lacking its carboxy-terminal NES binds 14-3-3 in a CaMK-dependent manner, but remains nuclear in CaMK-expressing cells. Second, in cells overexpressing HDAC5, 14-3-3, and activated CaMK, exposure to leptomycin B, which blocks Crm-1-dependent nuclear export (24), leads to rapid relocalization of cytoplasmic HDAC5 to the nucleus (17). Together, these results suggest that 14-3-3 binding to HDAC5 is necessary but not sufficient to redistribute HDAC5 from the nucleus to the cytoplasm, and raise the possibility that 14-3-3 bound to the amino-terminus of HDAC5 crosstalks with carboxy-terminal sequences in HDAC5 to induce nuclear export. Of note, 14-3-3 proteins possess NESs that function in certain contexts (18). However, the existence of HDAC5 carboxy-terminal truncation mutants that bind 14-3-3 but remain nuclear in CaMK-expressing cells argues against a role for these sequences in HDAC nuclear export (Fig. 5B).
A Three-Step Mechanism for 14-3-3-Dependent Control of Myogenesis.
The behavior of a series of HDAC mutants has enabled us to identify three distinct steps in the regulation of MEF2 activity by CaMK (Fig. 6C). In step 1, HDAC5 is phosphorylated at Ser-259 and Ser-498 in response to CaMK activation. In step 2, 14-3-3 associates with phosphoserines -259 and -498; binding to Ser-498 results in dissociation of HDAC from MEF2, possibly by inducing a conformational change in HDAC. In this regard, a recent study demonstrated that calmodulin is capable of associating with the MEF2 binding domain of HDAC4, resulting in disruption of MEF2–HDAC complexes (25). Whether Ser-498 of HDAC5 crosstalks with the MEF2 binding domain to allow calmodulin binding remains to be determined. In step 3, HDAC5 is exported to the cytoplasm, in a process that depends on 14-3-3 binding to either Ser-259 or Ser-498. Because CaMK is capable of activating MEF2 in the presence of HDAC mutants that are resistant to nuclear export, we conclude that nuclear export is not essential for activation of MEF2, but rather serves as a reinforcing mechanism allowing for sustained expression of MEF2 target genes.
Although these and other findings strongly suggest a role for CaMK in the transmission of signals that converge on Class II HDACs, the existence of distinct signaling mechanisms that control HDACs remains likely. Indeed, our results suggest that HDAC4 and -5, are subject to differential phosphorylation in vivo.
MEF2 is emerging as a key regulator of gene programs that control muscle differentiation and stress responses in cardiac muscle, neurons, and lymphocytes. As such, further elucidation of the signaling pathways that regulate this transcription factor may lead to specific pharmacological agents that target MEF2-responsive genes by means of HDAC.
Acknowledgments
We thank S. Schreiber and A. Means for expression constructs. We are grateful to A. Tizenor and S. Johnson for graphics, T. Gemelli for technical assistance, J. Page for editorial assistance, and W. Garrard and J. Spencer for critical reading of the manuscript. E.N.O. was supported by grants from National Institutes of Health, The Robert A. Welch Foundation, The D.W. Reynolds Foundation, and Myogen, Inc. T.A.M. is a Pfizer Fellow of the Life Sciences Research Foundation.
Abbreviations
- MEF2
myocyte enhancer factor-2
- MADS
MCM1 agamous deficiens serum response factor
- CaMK
calcium/calmodulin-dependent protein kinase
- HDAC
histone deacetylase
- GST
glutathione S-transferase
- NES
nuclear export signal
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260501497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260501497
References
- 1.Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 2.Molkentin J D, Black B L, Martin J F, Olson E N. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, McKinsey T A, Zhang C L, Olson E N. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 4.Kuo M H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 7.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 8.Sparrow D B, Miska E A, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun T J. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, McKinsey T A, Nicol R L, Olson E N. Proc Natl Acad Sci USA. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. . (First Published March 28, 2000; 10.1073/pnas.080064097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 12.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J D. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao Z, Wiedmann M J. J Biol Chem. 1999;274:31102–31107. doi: 10.1074/jbc.274.43.31102. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Naya F J, McKinsey T A, Mercer B, Shelton J M, Chin E R, Simard A R, Michel R N, Bassel-Duby R, Olson E N, Williams R S. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaeser F, Ho N, Prywes R, Chatila T A. J Biol Chem. 2000;275:197–209. doi: 10.1074/jbc.275.1.197. [DOI] [PubMed] [Google Scholar]
- 16.Youn H D, Chatila T A, Liu J O. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinsey T A, Zhang C L, Lu J, Olson E N. Nature (London) 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu H, Subramanian R R, Masters S C. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 19.Haribabu B, Hook S S, Selbert M A, Goldstein E G, Tomhave E D, Edelman A M, Snyderman R, Means A R. EMBO J. 1995;14:3679–3686. doi: 10.1002/j.1460-2075.1995.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, C. L., McKinsey, T. A., Lu, J. & Olson, E. N. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 21.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grozinger C M, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. . (First Published June 27, 2000; 10.1073/pnas.140199597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang A H, Kruhlak M J, Wu J, Bertos N R, Vezmar M, Posner B I, Bazett-Jones D P, Yang X J. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 25.Youn H D, Grozinger C M, Liu J O. J Biol Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]