Abstract
In many plant species, exposure to a prolonged period of cold during the winter promotes flowering in the spring, a process termed vernalization. In Arabidopsis thaliana, the vernalization requirement of winter-annual ecotypes is caused by the MADS-box gene FLOWERING LOCUS C (FLC), which is a repressor of flowering. During the vernalization process, FLC is downregulated by alteration of its chromatin structure, thereby permitting flowering to occur. In wheat, a vernalization requirement is imposed by a different repressor of flowering, suggesting that some components of the regulatory network controlling the vernalization response differ between monocots and dicots. The extent to which the molecular mechanisms underlying vernalization have been conserved during the diversification of the angiosperms is not well understood. Using phylogenetic analysis, we identified homologs of FLC in species representing the three major eudicot lineages. FLC homologs have not previously been documented outside the plant family Brassicaceae. We show that the sugar beet FLC homolog BvFL1 functions as a repressor of flowering in transgenic Arabidopsis and is downregulated in response to cold in sugar beet. Cold-induced downregulation of an FLC-like floral repressor may be a central feature of the vernalization response in at least half of eudicot species.
IN winter-annual ecotypes of Arabidopsis thaliana, expression of the MADS-box transcription factor FLOWERING LOCUS C (FLC) during the first growing season creates a facultative vernalization requirement (Michaels and Amasino 1999; Sheldon et al. 1999). During prolonged cold treatments, FLC mRNA expression levels decrease, permitting the upregulation of genes that promote flowering (Lee et al. 2000; Samach et al. 2000). FLC is maintained at a repressed level following cold treatment by an epigenetic mechanism that involves histone modifications in FLC chromatin (Gendall et al. 2001; Bastow et al. 2004; Sung and Amasino 2004).
FLC belongs to a clade of six paralogs known from the Arabidopsis genome (Becker and Theissen 2003). All paralogs (FLC and MAF1–MAF5) function as repressors of flowering. FLC and MAF1–MAF4 are downregulated to varying degrees in response to cold whereas MAF5 is upregulated (Ratcliffe et al. 2001, 2003). Given that FLC-like genes share an upstream regulatory complex (He et al. 2004; Oh et al. 2004; Kim et al. 2005) and that at least two genes (FLC and MAF1) influence the same downstream regulator of flowering, SOC1 (Hepworth et al. 2002; Ratcliffe et al. 2003), the control of flowering time via FLC-like genes may be administered in a multilocus, dosage-dependent manner.
Summer-annual Arabidopsis ecotypes, which flower rapidly and do not respond to vernalization, have evolved on multiple occasions via disruption of FLC regulatory sequences by transposons (Michaels et al. 2003) and via deletions and frameshift mutations in FRIGIDA (FRI) (Johanson et al. 2000), a positive regulator of FLC (Michaels and Amasino 1999; Sheldon et al. 1999). An observed cline in flowering behavior in native European populations of Arabidopsis, characterized by winter annuals in the north and summer annuals in the south, has been attributed to a complex epistatic interaction between functional copies of FLC and FRI (Caicedo et al. 2004). Ironically, the maintenance of this latitudinal cline may be due to natural selection for increased water use efficiency, a pleiotropic consequence of the effect of FRI and FLC on flowering time (McKay et al. 2003), rather than mean winter temperature (Stinchcombe et al. 2004).
FLC-like genes have not been identified in taxa other than Arabidopsis, Brassica, and Raphanus, three genera within the Brassicaceae, in spite of targeted efforts to do so in numerous taxa (Schläppi and Patel 2001; Tadege et al. 2003; Hecht et al. 2005). Moreover, in wheat, vernalization causes downregulation of a class of flowering repressors distinct from FLC, the B-box flowering repressors VRN2 (ZCCT1) and ZCCT2 (Yan et al. 2003, 2004). The phenotypic distinction between “winter” varieties of wheat (which require vernalization to flower) and “spring” varieties (with no intrinsic vernalization requirement) may be a consequence of expression of a nonfunctional form of VRN2 in “spring” wheat cultivars (Yan et al. 2004). Thus the vernalization response in wheat is controlled, at least in part, by a mechanism that is analogous, not homologous, to the mechanism operating in Brassicaceae. While many species outside Brassicaceae or Poaceae (including the focus of this study, the sugar beet, Beta vulgaris) exhibit a vernalization response, it has not been established whether the response involves an FLC-mediated mechanism (i.e., downregulation of an FLC-like repressor of flowering), a VRN2-mediated mechanism, or some other, independently evolved mechanism.
The sugar beet and its wild progenitor sea beet (Beta vulgaris ssp. maritima) are facultative perennials that, under natural growing conditions, exhibit either an annual or a biennial flowering behavior. As in Arabidopsis, a significant association between life form and latitude has been observed among wild populations of B. vulgaris ssp. maritima. Along the Atlantic and Mediterranean coasts of France, biennial individuals are found with greater frequency in the north, while annuals predominate in the south (Van Dijk et al. 1997).
The difference in flowering phenology between annual and biennial sugar beets is determined by the genotype at the B locus, known as the “bolting gene” (Munerati 1931; Abegg 1936; Owen 1954; Abe et al. 1997). The bolting gene imposes a strict vernalization requirement in homozygous recessive individuals (bb) such that, under natural conditions, they perform as biennials. Biennial beets (bb) held under artificial long-day conditions will remain vegetative unless exposed to a lengthy period of cold (Stout 1945). A prolonged cold treatment is all that is required to cause biennial beets to flower, although long-day lengths during and after vernalization facilitate the transition (Fife and Price 1953). In annuals (BB or Bb), long days are necessary and sufficient to cause flowering. Under artificial short-day conditions, annual beets will grow vegetatively for an indefinite period of time without flowering. Thus the pathway to flowering in beet requires specific exogenous cues, whereas Arabidopsis will ultimately flower under most environmental conditions due to promotion by autonomous pathway genes (Koornneef et al. 1998).
In this study, we use a phylogenetic approach to identify FLC homologs in species outside the Brassicaceae, including the sugar beet. We describe the function and regulation of a sugar beet FLC homolog during vernalization and compare the response to FLC. Finally, we predict the taxonomic distribution of FLC-like genes in the angiosperms and propose that their role in the vernalization response has largely been conserved.
MATERIALS AND METHODS
Identification of FLC homologs by phylogenetic analysis:
A consensus MADS-box amino acid sequence was used to query expressed sequence tag (EST) databases from B. vulgaris, Populus spp., and Aquilegia formosa × pubescens. In total, five B. vulgaris and eight Populus spp. ESTs and 19 putative unique gene sequences from A. formosa × pubescens were analyzed. Twenty-two EST sequences representing Oryza sativa MIKC-type MADS-box unigenes were retrieved from the NCBI database. A Solanum lycopersicum EST sequence was identified as a possible FLC-like gene and was incorporated into the data set. Sixteen additional putative FLC-like genes that were distinct from those already integrated into the data set were identified using the Phytome database (Hartmann et al. 2006; gene family 51, subfamily 8).
Inferred amino acid sequences of all candidate nucleotide sequences were aligned to the “MIKCc” data set of Becker and Theissen (2003) using DIALIGN-t (Subramanian et al. 2005) and ClustalX version 1.83 (Thompson et al. 1997). The evolution of lineage-specific motifs within the MADS-box gene family could result in overestimation of branch support when using a global alignment such as that produced by ClustalX. Therefore, the local alignment procedure in DIALIGN-T was used for comparison. Regions of ambiguous alignment were identified by eye and eliminated. As recommended by the findings of Pařenicová et al. (2003), two Mδ-type MADS-box sequences (AGL30 and AGL94) were included as outgroups to root trees. The final DIALIGN-T and ClustalX alignments contained 142 and 138 amino acid characters, respectively. Both alignments included the complete MADS-box and the majority of the K-box. A total of 157 sequences were aligned, including a comprehensive sampling of MIKCc-type MADS-box genes from the Arabidopsis genome, as well as numerous characterized genes from other plant taxa, such that all described MADS-box gene lineages were represented. Aligned data sets are available as supplemental material at http://www.genetics.org/supplemental/.
Phylogenetic analysis was used to determine support for relationships between putative FLC-like sequences and known MADS-box gene lineages. Bootstrap consensus trees were constructed using neighbor-joining or maximum parsimony. One thousand replicates were used. Maximum parsimony searches used the TBR-M search strategy of DeBry and Olmstead (2000) in PAUP*4.0b10 (Swofford 1999). A mixed amino acid model was used (“aamodelpr=mixed”) for Bayesian Markov chain Monte Carlo analysis (Ronquist and Huelsenbeck 2003). Four chains were run for 1.5 × 107 generations. The analysis was repeated to ensure convergence onto a single stationary distribution of trees. A total of 1 × 107 generations were discarded as burn-in, after which 50,000 trees were sampled from each replicate run to determine the optimal consensus tree and posterior probabilities for relevant clades. Complete phylogenetic trees are available as supplemental material at http://www.genetics.org/supplemental/.
Isolation of full-length FLC-like cDNAs from B.vulgaris:
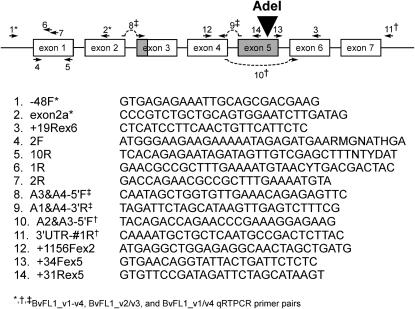
The inferred amino acid sequences of Arabidopsis FLC (AF537203), Arabidopsis MAF1 (AF342808), and a single B. vulgaris EST (BQ595637) identified by phylogenetic analysis to be an FLC homolog were used to design consensus-degenerate primers (Rose et al. 1998; Henikoff et al. 2000) to amplify a short region of BvFL1 exon 1, from which genomic DNA sequence (GenBank accession nos. DQ189214 and DQ189215) was obtained by genome walking using the Universal GenomeWalker kit (BD Biosciences, San Jose, CA). A complete genomic sequence (EF036526) was subsequently obtained by shotgun sequencing BAC clone SBA02L13 derived from cultivar USH20 (McGrath et al. 2004). Primer sequences and locations are shown in Figure 1.
Figure 1.—
Location and sequence of PCR primers targeting the sugar beet FLC homolog BvFL1. Primers that span intron/exon boundaries are indicated with a dashed line. The unique AdeI site in exon 5 is marked with a solid triangle. Putative alternatively spliced regions are shaded.
Total RNA was isolated from biennial FC606 (PI 590843) (Smith and Ruppel 1980) and annual SLC003 (PI 590811) (Owen 1950) germ plasm. Complete coding sequences of expressed B. vulgaris FLC-like genes were amplified using a 3′ RACE procedure (SMART RACE cDNA amplification kit, BD Biosciences) primed with −48F, which targeted the 5′-untranslated region (5′-UTR). PCR products representing full-length cDNAs were cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. Comparison of genomic and RACE product sequences allowed the prediction of intron/exon boundaries.
Alternative splicing PCR assay:
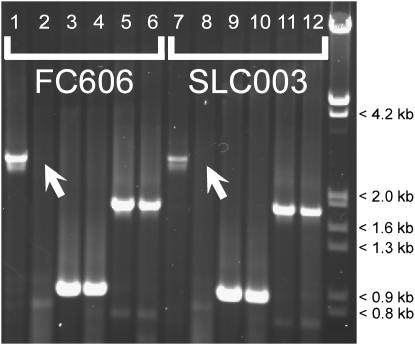
The mRNA variants BvFL1_v1 and BvFL1_v4 contained a 42-bp sequence corresponding to exon 5, whereas BvFL1_v2 and BvFL1_v3 did not (Figure 2a). To determine whether this distinction was attributable to transcription from multiple distinct genomic loci or alleles, or due to alternative splicing of a single transcribed locus, we performed the following assay: A single AdeI restriction site was identified in exon 5 (Figure 1). No other AdeI sites were present in the genomic BvFL1 sequence obtained by genome walking. Total genomic DNA from FC606 and SLC003 was digested with AdeI. Primers located in exon 4 (+1156Fex2) and the 3′-UTR (3′UTR-#1R) were used to amplify uncut (Figure 3, lanes 1 and 7) and AdeI-digested genomic DNA (lanes 2 and 8). Digestion with AdeI prevented amplification of the expected 2.9-kb product. Therefore, there must be an AdeI site between +1156Fex2 and 3′UTR-#1R in all amplifiable genomic sequences. To verify that the cut site was in exon 5, PCR was performed with primer pairs +1156Fex2/+31Rex5 and +34Fex5/3′UTR-#1R. +31Rex5 and +34Fex5 were positioned immediately adjacent to the AdeI site in exon 5. Appropriately sized products were amplified in both cut and uncut genomic DNA; thus the AdeI cut site must be located in exon 5, and not in the intervening regions between exon 5 primers and +1156Fex2 or 3′UTR-#1R, in all amplifiable genomic sequences. We therefore concluded that all amplifiable genomic sequences that contain exon 4 and the 3′-UTR also contain exon 5. This finding supports the hypothesis that variation for the presence of exon 5 seen in BvFL1 mRNA is attributable to alternative splicing since no genomic sequence that lacked the 42-bp exon 5 region could be identified.
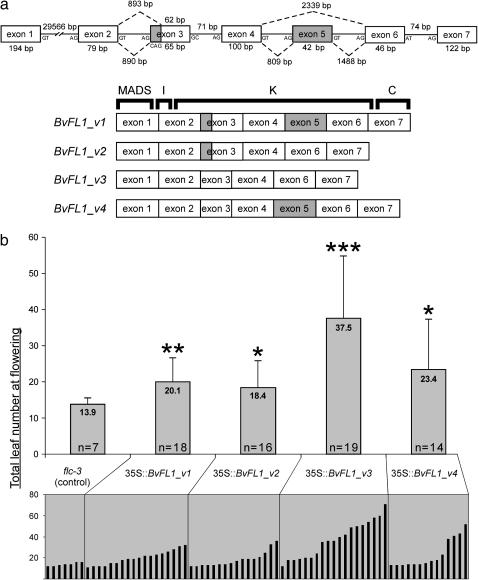
Figure 2.—
(a) Genomic structure of the sugar beet FLC homolog BvFL1. Dashed lines indicate the four putative alternative splicing possibilities required to generate mRNA variants BvFL1_v1–v4. Coding regions affected by splice site variation are shaded. Approximate boundaries of the MADS, I, K, and C domains are indicated for BvFL1_v1. (b) (Top) Alteration of flowering phenology caused by the introduction of BvFL1 transgenes into Arabidopsis. Expression of BvFL1_v1–v4 variants caused a statistically significant increase in time to flowering relative to the untransformed control (Welch's t-test, *α[0.05], **α[0.01], and ***α[0.001]). Error bars indicate 1 standard deviation from the mean. (Bottom) Distribution of total leaf number at flowering in individual T1 transformants shows that the only measurable effect of BvFL1 transgenes was to delay flowering relative to the average baseline value from flc-3, an FLC null mutant.
Figure 3.—
Agarose gel image showing results of alternative splicing PCR assay. Even-numbered lanes are AdeI-digested genomic DNA; odd-numbered lanes are uncut. Primers used: +1156Fex2/3′UTR-#1R (lanes 1, 2, 7, 8), +1156Fex2/+31Rex5 (lanes 3, 4, 9, 10), and +31Fex5/3′UTR-#1R (lanes 5, 6, 11, 12). Digestion of genomic DNA with AdeI prevented the amplification of a 2.9-kb fragment (open arrows), suggesting that variation for the presence of exon 5 observed in the mature mRNA is not caused by variation for the presence of exon 5 sequence in the encoding genomic DNA. Therefore, the four mature mRNA variants observed (BvFL1_v1–v4) are a likely consequence of alternative splicing.
Transformation of Arabidopsis with BvFL1:
The coding sequences of BvFL1_v1–v4 were cloned into pMDC32 via Gateway cloning (Curtis and Grossniklaus 2003). These constructs were introduced (Clough and Bent 1998) into the Arabidopsis FLC null mutant flc-3 and expressed under the constitutive 35S promoter. Time to flowering was measured in T1 plants as the total leaf number, including both rosette and cauline leaves, at flowering. Individual T1 transformants typically represent independent T-DNA insertional events (Desfeux et al. 2000); thus statistical analysis of the effect of BvFL1 expression on Arabidopsis flowering time was appropriate (i.e., the assumption that repeated measurements were independent was met). The number of T1 plants measured for each treatment group is shown in Figure 2b. Statistical differences between treatment groups were tested using Welch's t-test (Uitenbroek 1997) to correct for unequal variances.
Plant growth conditions and experimental design:
Biennial FC606 seeds were planted in growth chambers at 24° with constant illumination by high-pressure sodium and metal halide lamps. Vernalized plants were grown for 30 days in the growth chamber (to the 6- to 10-leaf stage), after which immature leaves were sampled for RNA (RNeasy plant mini kit, QIAGEN, Valencia, CA). Plants were then placed into a photothermal induction chamber and grown at 5° under continuous light for 90 days. Leaves were sampled for RNA after 45 days (at the 15- to 19-leaf stage) and again after 90 days (at the 20- to 24-leaf stage). A 90-day vernalization treatment is sufficient to induce flowering in FC606. Following the vernalization treatment, plants were directly returned to the 24° growth chamber, with no gradual temperature transition. Seventeen days later, a final leaf RNA sample was obtained (at the 27- to 41-leaf stage). Unvernalized plants were grown continuously in the 24° growth chamber and sampled for RNA at comparable developmental stages. Treatment of annual SLC003 plants was identical except for the timing of RNA sampling, which occurred at earlier leaf stages for the unvernalized treatment group to avoid sampling leaves from bolting individuals.
For shoot apex RNA preparations, unvernalized plants were grown as above, with apical excisions occurring at the two- to four-leaf stage. Vernalized plants were germinated in damp blotters at 24° for 3 days under constant illumination and then transplanted into planting mix when radicles were <2 cm and cotyledons had not yet emerged. Plants were placed into the 5° photothermal induction chamber immediately after transplantation and held for 77 days prior to excision of the shoot apical meristem region, when two to four leaves were present in the basal rosette.
Reverse transcriptase PCR:
The Quantitect SYBR Green reverse transcriptase–PCR (RT–PCR) kit and One-Step RT–PCR kit (QIAGEN) were used to perform RT–PCR using 100 ng of total RNA as template in 25-μl reactions. Primers −48F and +19Rex6, which flanked the length variable regions of BvFL1, were used. Qualitative assessment of BvFL1 transcript levels was performed by examining RT–PCR products on native polyacrylamide gels.
Quantitative real-time PCR:
Primers used for quantitative real-time PCR are shown in Figure 1. All primer pairs were tested to ensure that they specifically amplified the desired variants using clones containing BvFL1_v1–v4. With the exception of the pair targeted to BvFL1_v1/v4, all primer pairs were determined not to amplify contaminating genomic DNA using “no reverse transcriptase” controls. For BvFL1_v1/v4 assays, DNase-treated total RNA was used (Turbo DNA Free, Ambion, Austin, TX). Cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) was used as the reference gene (primer gapCex5/6F, GCTGCTGCTCACTTGAAGGGTGG and primer gapCex8R, CTTCCACCTCTCCAGTCCTT). Standard curves were created using three replicates each of five dilutions spanning a three order of magnitude (or greater) dilution series. R2 values for standard curves were >0.97 for all products. Empirically determined amplification efficiency values were similar among loci (BvFL1_v1–v4, E = 2.01; BvFL1_v1/v4, E = 2.07; BvFL1_v2/v3, E = 2.06; GapC, E = 2.01).
Quantitative RT–PCR (qRT–PCR) was performed using the Quantitect SYBR Green RT–PCR kit (QIAGEN) on a Cepheid SmartCycler. A total of 50 ng total RNA was used in each 25-μl qRT–PCR reaction. Cycling profiles for all loci used a 15-sec denaturation at 94° and a 30-sec extension at 72°. Annealing times were 30 sec, and temperatures were 60° for BvFL1_v1–v4 and BvFL1_v2/v3, 62° for BvFL1_v1/v4, and 55° for GAPC. Fluorescence levels were measured for 6 sec at 80° for BvFL1_v1/v4 and BvFL1_v2/v3, at 77° for BvFL1_v1–v4, and at 78° for GAPC. Two reactions were performed for each primer pair and a mean Ct value was computed (Ct threshold of 5). Reactions were then repeated for a total of four replicates/sample.
The magnitude (“fold change”) and statistical significance of differences in expression level between treatment groups were calculated using REST-XL (Pfaffl et al. 2002) and are reported in Table 1.
TABLE 1.
Statistical analysis of qRT–PCR data
| Linea | Tissue | n (control, sample)b | Average fold changec | Direction | Significanced | Target | Treatment |
|---|---|---|---|---|---|---|---|
| FC606 | Leaf | 6, 5 | 1.113 | Down | P ≥ 0.7236 | BvFL1_v2/v3 | 45 days vernalization |
| FC606 | Leaf | 6, 6 | 2.909 | Down | P ≤ 0.0377 | BvFL1_v2/v3 | 90 days vernalization |
| FC606 | Leaf | 6, 5 | 2.646 | Up | P ≤ 0.0064 | BvFL1_v2/v3 | 17 days postvernalization |
| FC606 | Shoot apex | 2, 2 | 1.222 | Down | P = 0.3323 | BvFL1_v2/v3 | 77 days vernalization |
| SLC003 | Leaf | 4, 4 | 2.948 | Down | P ≤ 0.0199 | BvFL1_v2/v3 | 90 days vernalization |
| SLC003 vs. FC606 | Shoot apex | 2, 2 | 1.532 | SLC003 > FC606 | P = 0.5027 | BvFL1_v2/v3 | No vernalization |
| FC606 | Leaf | 4, 4 | 1.732 | Down | P ≥ 0.1826 | BvFL1_v1/v4 | 90 days vernalization |
| FC606 | Shoot apex | 2, 2 | 1.023 | Down | P = 0.8347 | BvFL1_v1/v4 | 77 days vernalization |
| SLC003 | Leaf | 4, 4 | 1.961 | Down | P ≤ 0.0025 | BvFL1_v1/v4 | 90 days vernalization |
| SLC003 vs. FC606 | Shoot apex | 2, 2 | 1.089 | SLC003 > FC606 | P = 0.8347 | BvFL1_v1/v4 | No vernalization |
| FC606 | Leaf | 4, 4 | 1.101 | Up | P ≥ 0.7356 | BvFL1_v1–v4 | 90 days vernalization |
| FC606 | Leaf | 6, 4 | 1.297 | Up | P ≥ 0.4685 | BvFL1_v1–v4 | No vernalization |
| SLC003 vs. FC606 | Leaf | 4, 6 | 1.478 | SLC003 > FC606 | P ≥ 0.4443 | BvFL1_v1–v4 | Two- to seven-leaf stage |
FC606, biennial; SLC003, annual.
SLC003 used as control for comparisons between lines.
Average magnitude of change, calculated from the two independent qRT–PCR setups.
For statistically significant results (P < 0.05), the larger of the P-values calculated from replicated qRT–PCR setups is reported. For nonsignificant results, the smallest P-value is shown.
RESULTS AND DISCUSSION
FLC homologs are present in the three major eudicot lineages:
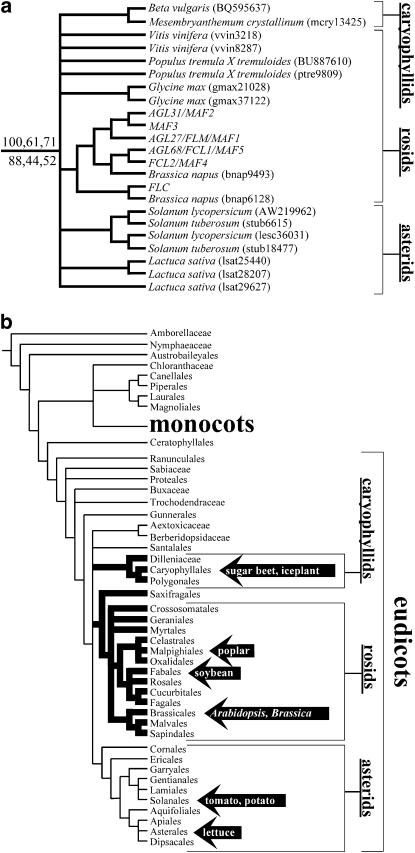
To determine whether genes belonging to the FLC-like subfamily are present outside Brassicaceae, we identified the MIKC-type MADS-box genes (including putative FLC-like sequences) present in EST databases of taxa representing major angiosperm lineages. These included monocots (O. sativa, rice; Triticum aestivum, wheat), Ranunculales (A. formosa × pubescens, columbine), caryophyllids (B. vulgaris ssp. vulgaris, sugar beet; Mesembryanthemum crystallinum, iceplant), rosids (Vitis vinifera, grape; Populus tremula × tremuloides, poplar; Glycine max, soybean), and asterids (S. lycopersicum, tomato; Solanum tuberosum, potato; Lactuca sativa, lettuce). Phylogenetic analysis of inferred amino acid sequences was used to evaluate relationships between EST sequences/unigenes and all MIKC-type MADS-box genes present in Arabidopsis. We identified ESTs from tomato (AW219962), beet (BQ595637), and poplar (BU887610) that were more closely related to Arabidopsis FLC-like genes than to any other MIKC-type MADS-box lineage known from Arabidopsis (Figure 4a and phylogenetic trees supplied as supplemental material at http://www.genetics.org/supplemental/). Likewise, unipeptides from iceplant, grape, soybean, potato, and lettuce, identified in the Phytome database (Hartmann et al. 2006) as putative relatives of FLC, were found here to belong to the clade containing Arabidopsis FLC-like genes. These relationships were found in the consensus tree regardless of the analytical method or alignment algorithm used. While bootstrap support values for the clade containing FLC-like genes were low (44–71%) by some standards, it should be recognized that bootstrap support is a relative measure of character support that is relevant only within a single data set. As shown in Table 2, the support values found here for the FLC-like clade were not significantly different from the values that we recovered for the A-, B-, C-, and E-class MADS-box gene lineages, which are generally accepted as monophyletic groups (Doyle 1994; Purugganan et al. 1995; Theissen et al. 1996; Becker and Theissen 2003). We therefore conclude that a previously undescribed lineage of FLC homologs exists. Given the accepted phylogenetic relationships among the taxa included, the lineage of FLC homologs must have originated prior to the diversification of the three major core eudicot clades (caryophyllids, rosids, and asterids) (Figure 4b). It should therefore be possible to identify FLC homologs in all taxa belonging to the caryophyllids, rosids, and asterids, barring secondary loss of the loci in particular lineages during the course of evolution.
Figure 4.—
(a) Strict consensus subtree showing the FLC-like gene subfamily. Support values (Bayesian posterior probability, maximum-parsimony bootstrap percentage, neighbor-joining bootstrap percentage) are indicated for the node that distinguishes the FLC-like subfamily from all other MADS-box gene lineages. Upper values were obtained from a global amino acid sequence alignment (ClustalX); lower values were from a local alignment (DIALIGN-T). GenBank accession numbers or Phytome unipeptide IDs are in parentheses. The presence of sequences from the caryophyllid, rosid, and asterid lineages suggests a core eudicot distribution of FLC homologs. (b) Phylogenetic relationships of angiosperms according to the Angiosperm Phylogeny Group (2003) and Judd and Olmstead (2004). Based on evidence from sugar beet and Arabidopsis, the predicted taxonomic distribution of vernalization-responsive FLC-like repressors of flowering is shown with thick branches. Arrows indicate taxa where FLC homologs were found.
TABLE 2.
Comparison of clade support values
| Clade/subfamilya | Becker and Theissen (2003) | Parsimony, ClustalX | Parsimony, DIALIGN-T | Neighbor-joining, ClustalX | Neighbor-joining, DIALIGN-T | Bayesian, ClustalXb | Bayesian, DIALIGN-T |
|---|---|---|---|---|---|---|---|
| flc | 100 | 61 | 44 | 71 | 52 | 100 | 88 |
| squa (A class) | 100 | 32 | 27 | 30 | 38 | 50 | 100 |
| def (B class) | 99 | 87 | 82 | 82 | 74 | 100 | 98 |
| glo (B class) | 54 | 46 | 32 | 61 | 38 | 79 | 36 |
| ag (C class) | 100 | 92 | 96 | 99 | 100 | 99 | 100 |
| agl2 (E class) | 94 | 31 | 74 | 94 | 80 | 73 | 98 |
| mean (A, B, C, E) | 89.4 | 57.6 | 62.2 | 73.2 | 66.0 | 80.2 | 86.3 |
| sd (A, B, C, E) | 19.9 | 29.8 | 30.9 | 28.2 | 27.4 | 20.7 | 28.4 |
| pc | 0.594 | 0.909 | 0.556 | 0.938 | 0.609 | 0.339 | 0.952 |
Subfamily names are based on Becker and Theissen (2003).
Bayesian posterior probabilities multiplied by 100 for uniformity.
Based on a one-sample z-test to determine whether the support value for the FLC-like clade was significantly different from the mean of the A-, B-, C-, and E-class clades, which are accepted as monophyletic.
Although the number of sequences present in EST/unigene databases of rice, wheat, and columbine was large (66,238 unipeptides, 88,853 unipeptides, and 84,960 ESTs, respectively), phylogenetic analysis failed to provide any strong indication that FLC homologs were present in these species. Weak support for the inclusion of three monocot sequences [rice unigene S16411843 (AK066160), rice FDRMADS5 (AAD38368), and wheat F90-A (CAE53900; Ciaffi et al. 2005)] at the base of the clade containing FLC-like sequences was found. However, this result was limited to the DIALIGN-T alignments; thus an alignment artifact cannot be ruled out. The two monocot sequences (osat72611 and taes20622) identified by Phytome to be close relatives of FLC appeared, in our analyses, to be more closely related to APETALA 1 and SQUAMOSA. While the absence of a sequence from an EST database cannot be held as conclusive evidence that a gene is not present in the genome, the results of our analyses provide an initial suggestion that the FLC-like gene lineage may have originated at the base of the core eudicot clade. Accordingly, recognizable FLC-like genes may be restricted to core eudicot taxa.
It is premature to describe the newly identified members of the FLC-like gene lineage as orthologs of FLC because (1) our identification strategy could not result in complete sampling of all MIKC-type MADS-box genes from each taxon and (2) the frequent occurrence of whole-genome duplication events (and subsequent lineage-specific gene extinction events) (Blanc and Wolfe 2004) and the complex birth–death pattern of diversification within the MADS-box gene family (Nam et al. 2004) during the evolution of the angiosperms frustrates a strict application of the concept of orthology. Hereafter we refer to the clade containing FLC homologs as the FLC-like gene subfamily.
High divergence within the FLC-like subfamily may explain why previous attempts to identify FLC homologs outside Brassicaceae have failed. Methods for homolog identification that rely on overall similarity (e.g., BLAST and Southern hybridization) are most effective within the context of a gene family when sequences diverge in a clock-like manner. Unlike the majority of MADS-box genes, Arabidopsis FLC-like genes have evolved under positive Darwinian selection (Martínez-Castilla and Alvarez-Buylla 2003), which can cause rapid divergence among orthologs and substantial deviation from uniform sequence divergence between gene lineages. In such cases, the application of models of sequence divergence that accommodate non-clock-like evolution (e.g., most phylogenetic analysis procedures) will be essential to distinguish putative orthologs from genes belonging to more distantly related paralogous lineages.
Cloning and characterization of the sugar beet FLC homolog BvFL1:
Several examples of gene co-option, or duplication followed by neo- or subfunctionalization, have been documented within the MADS-box gene family (Irish and Litt 2005). Membership of a MADS-box gene within a particular subfamily cannot necessarily be used to predict its function. Because all newly identified members of the FLC-like subfamily are uncharacterized [with the exception of the Brassica napus FLC ortholog (Tadege et al. 2001)], it is not known whether they perform a role similar to Arabidopsis FLC during vernalization. We have therefore examined some regulatory and functional attributes of the FLC homolog identified in sugar beet for purposes of comparison.
The complete coding sequence for the sugar beet FLC homolog corresponding to EST BQ595637 was determined. Four RNA variants were identified in immature leaf and shoot apex cDNA libraries from biennial sugar beet germ plasm FC606. The variants differed by two K-domain indels arranged in all four possible pairwise combinations. The inferred reading frame was identical between variants such that all variants shared the same translation stop codon. There were no nucleotide substitution differences among variants within the region sequenced, which included a portion of the 5′-UTR (24 bp), the complete coding region (603–648 bp), and the 3′-UTR (345 bp).
Comparison of the cDNA sequences with a genomic sequence obtained by genome walking (DQ189215) showed that the 3-bp indel located in exon 3 could be explained by two competing 3′ splice sites in intron 2. The 42-bp indel could be explained by the inclusion or omission of an exon 5 cassette (Figure 2a). A PCR assay that took advantage of a unique AdeI restriction site in exon 5 showed that all amplifiable genomic sequences that include exon 4 and the 3′-UTR also include exon 5. Therefore, the absence of exon 5 sequence in two mRNA variants cannot be explained by the absence of exon 5 sequence in the genomic locus that encodes them (Figure 3).
Additionally, we screened 28 sugar beet cultivars and 360 individuals from 30 wild European B. vulgaris ssp. maritima populations for genotypic variation at two microsatellite loci: one located within intron 1 and a second located ∼25 kb upstream of the BvFL1 start codon (data not shown). We found no evidence that multiple copies of this genomic region are routinely present in wild beet or sugar beet cultivars. Two or fewer alleles amplified in 99.5 and 98.4% of wild individuals for the intron 1 and upstream locus, respectively; two or fewer alleles amplified in all cultivars.
Finally, we screened a BAC library with a total of seven PCR primer pairs distributed across ∼60 kb of the sugar beet genome within and around the locus. The library had 6.1× genome coverage, mathematically sufficient to recover any sequence with >99% probability (McGrath et al. 2004). Three primer pairs targeted transcribed sequence and thus were known to be an exact match for all possible loci. PCR-based screens of the matrix-pooled library identified four BACs, three of which were subsequently determined via DNA sequencing to contain overlapping inserts derived from the same genomic region. The remaining BAC contained a putative pseudogene that lacked sequence-encoding exons 2–7 and exhibited some DNA substitution differences in regions flanking the stretch of sequence homologous to exon 1. Thus we found no evidence for more than one expressed locus in the BAC library.
Taken together, these lines of evidence suggest that the four mature mRNA variants are a result of differential processing of precursor transcripts coded by a single genomic locus. The unlikely alternative hypothesis is that the mRNA variants are encoded by two or more loci that are the result of a duplication event (or events) that occurred so recently that no substitutions have occurred in the transcribed regions and that insufficient change has accumulated at the intron 1 microsatellite locus to permit the identification of multiple loci. Alternative splice variants have been observed for all FLC-like genes in Arabidopsis (Scortecci et al. 2001; Ratcliffe et al. 2003; Caicedo et al. 2004) so the hypothesis is not without precedent. While the possibility that the mRNA variants represent differentially spliced duplicate loci cannot be categorically ruled out, we refer here to the four variants collectively as a single locus, BvFL1 (B. vulgaris, FLC-LIKE 1), and individually as putative splice variants BvFL1_v1–BvFL1_v4 (DQ189210–DQ189213).
BvFL1 is a repressor of flowering in transgenic Arabidopsis:
To test the degree of functional conservation between BvFL1 and FLC in a heterologous context, the ability of BvFL1_v1–v4 to repress flowering in Arabidopsis was evaluated. BvFL1_v1–v4 constructs driven by the constitutive 35S promoter were inserted into the genome of the Arabidopsis FLC null mutant flc-3 (Michaels and Amasino 1999). All constructs resulted in a statistically significant delay in the time to flowering (as measured by total leaf number) relative to untransformed flc-3 (Figure 2b). Therefore, BvFL1 functions as a repressor of flowering in transgenic Arabidopsis. The strongest repressor was BvFL1_v3, which caused a significantly greater (P < 0.05) delay in time to flowering than constructs containing the other three putative splice variants. The ability of BvFL1 to complement FLC suggests a likely function as a repressor of flowering in sugar beet. Accordingly, we hypothesize that repression of flowering may be a conserved function of genes belonging to the FLC-like gene subfamily. This hypothesis remains to be explicitly tested in sugar beet.
BvFL1 is downregulated by cold in sugar beet:
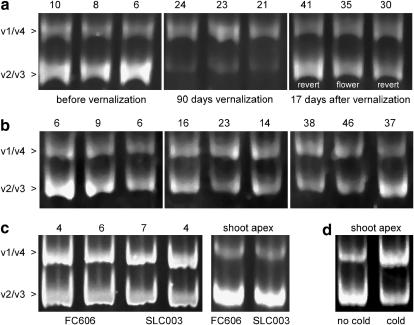
We examined whether BvFL1 was downregulated in response to a cold treatment in beet as most FLC-like genes are in Arabidopsis (i.e., FLC, MAF1–MAF4). Examination of BvFL1 RT–PCR products in FC606 individuals before and after a 90-day vernalization treatment suggested a substantial decrease in the level of BvFL1_v2 and BvFL1_v3 (hereafter BvFL1_v2/v3), the two transcripts that did not contain exon 5, relative to BvFL1_v1/v4 (Figure 5a). RT–PCR products measured at similar developmental stages in individuals that had not been exposed to a vernalization treatment showed no such response (Figure 5b). In contrast to BvFL1_v2/v3, qualitative changes in BvFL1_v1/v4 levels were not marked during cold treatment.
Figure 5.—
Expression of BvFL1 in vernalized (a) and unvernalized (b) biennial sugar beet FC606 as measured by RT–PCR. The two resolved bands each contain a pair of comigrating splice variants as labeled on the left. Qualitative expression levels are shown for three individuals, arranged in the same order as in a and b, for both treatments. In a, the center individual flowered following vernalization whereas the others reverted to vegetative growth. Images have been normalized such that the BvFL1_v1/v4 band is of roughly equal intensity between panels. To compare similar developmental stages between vernalized and unvernalized treatment groups, leaf number, counted upward from the first true leaf, was used. The leaf number from which RNA was taken is shown above each lane. Splice variants BvFL1_v2/v3 were downregulated relative to BvFL1_v1/v4 in immature leaves during cold treatment. BvFL1_v2/v3 was reset to prevernalization levels following cold treatment regardless of whether the plant ultimately flowered or reverted to vegetative growth. (c) Comparison of BvFL1 expression in immature leaves and shoot apexes of biennial FC606 and annual SLC003. The phenotypic distinction between annual and biennial sugar beets cannot be explained by differences in expression level of BvFL1 in immature leaves or the shoot apex. (d) The difference in BvFL1_v1/v4 and BvFL1_v2/v3 expression level between vernalized and unvernalized FC606 shoot apexes was not significant. The response to vernalization of BvFL1 was primarily confined to immature leaves.
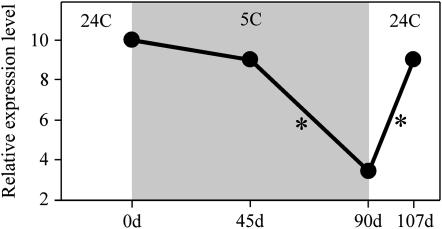
Quantitative RT–PCR confirmed these observations. A statistically significant threefold decrease in BvFL1_v2/v3 levels (measured simultaneously) relative to the GAPC reference gene occurred during the 90-day vernalization treatment, whereas no change in BvFL1_v1/v4 levels was detected (results of all qRT–PCR analyses are shown in Table 1). However, BvFL1_v2/v3 expression levels were not significantly different from prevernalization levels after only 45 days of vernalization, in agreement with our observations that a 45-day vernalization treatment is insufficient to induce flowering in most FC606 individuals. Figure 6 shows the relative changes in BvFL1_v2/v3 expression levels that occurred during the course of the experiment.
Figure 6.—
Relative change in BvFL1_v2/v3 expression levels during and after vernalization treatment of sugar beet germ plasm FC606 as measured by qRT–PCR. The x-axis indicates time in days. At 0 days, 30-day-old individuals grown at 24° were sampled for RNA and placed into a 5° photothermal induction chamber. RNA samples were taken after 45 and 90 days at 5°. After 90 days, plants were returned to 24° growing conditions. The y-axis indicates mRNA expression level, in relative units, with the starting level arbitrarily set to 10. BvFL1_v2/v3 levels decreased progressively during cold treatment and then returned to prevernalization levels following vernalization. Asterisks indicate intervals during which statistically significant changes in expression level occurred.
These results are consistent with the hypothesis that, in biennial sugar beet, the block on flowering imposed by the strong repressor BvFL1_v3 is progressively relieved during vernalization by selective downregulation of the BvFL1_v2/v3 mRNA variants. The vernalization response in biennial beet may therefore involve temperature-sensitive differential production of BvFL1 variants such that relatively weak repressors containing exon 5 (BvFL1_v1/v4) are favored during cold exposure and strong repressors lacking exon 5 (BvFL1_v3) predominate during warm growing conditions. This hypothesis requires further examination.
In Arabidopsis, FLC expression levels are modulated at the transcriptional level through the interaction of upstream regulators with promoter and intron 1 sequences (Sheldon et al. 2002; Bastow et al. 2004; Sung and Amasino 2004). While a change in the prevalence of alternative FLC transcripts has been observed during vernalization (Caicedo et al. 2004), no functional consequence has yet been ascribed to splice variation among Arabidopsis FLC-like genes. A possible distinction between beet and Arabidopsis in the regulatory mechanism underlying vernalization-induced promotion of flowering is intriguing because it implies that the same raw materials (e.g., genes belonging to the FLC-like subfamily) may come under the control of different regulatory processes during the course of evolution, yet elicit the same general phenotypic response.
The primary response of BvFL1 to cold occurs in immature leaves, not in the shoot apex:
Previous experiments showed that nonvernalized, vegetative shoot apexes of biennial sugar beet could be induced to make the transition to flowering by grafting them onto vernalized biennial stock or annual stock (Stout 1945; Curtis et al. 1964). These studies demonstrate that the signal to flower is transmissible and that a vegetative shoot apical meristem of biennial sugar beet need not be exposed to cold to be reprogrammed into an inflorescence meristem. The response of BvFL1 to vernalization at the shoot apical meristem was evaluated using RNA pools derived from vernalized and nonvernalized FC606 individuals. While generally indicative of temperature-sensitive differential RNA processing, the qualitative difference in the BvFL1 transcript profile between shoot apex treatments was not as marked as it was in leaves (Figure 5d). The difference in expression level, as measured by qRT–PCR, of BvFL1_v1/v4 and BvFL1_v2/v3 between meristem RNA pools was not statistically significant (P = 0.8347 and P = 0.3323, respectively). Thus, in FC606, the response of BvFL1 to cold was prominent in immature leaves but was not measurable in the shoot apical meristem. This result is consistent with studies showing that immature developing leaves are a site of cold perception in beet (Crosthwaite and Jenkins 1993) and with experiments demonstrating that changes in FLC expression levels in leaves are an essential component of the Arabidopsis vernalization response (Searle et al. 2006).
Cold-induced downregulation of BvFL1 is reversed by warm temperatures:
In Arabidopsis the effect of vernalization on FLC expression level is largely irreversible because repressed levels are mitotically stabilized by histone methylation (Bastow et al. 2004; Sung and Amasino 2004). Maintained repression of FLC leads to the so-called “memory of winter” in Arabidopsis wherein previously vernalized plants remain competent to flower long after the initial cold exposure (Michaels and Amasino 2000). A stable vernalized state is common to many species, most notably Hyoscyamus niger, where it has been studied extensively (Lang 1965). Vernalized sugar beets, however, are prone to inflorescence reversion—the return of the shoot apical meristem from a state permissive of flowering to a noninduced state of vegetative growth (Battey and Lyndon 1990). Reversion is dependent on the environmental conditions immediately following vernalization. It is promoted by short days and rapid increase in temperature and suppressed by long days and gradual warming (Chroboczek 1934; Owen 1940). In sugar beet, inflorescence reversion is indicated by the resumption of a rosette pattern of leaf development at the shoot apex following bolting or by the abortion of the elongated bolting stalk prior to the development of flowers and a return to vegetative growth by axillary meristems.
We examined whether repression of BvFL1_v2/v3 was stably maintained in sugar beet following cold treatment. The BvFL1 transcript profile examined 17 days after a 90-day vernalization treatment was qualitatively similar to the profile measured prior to the cold treatment (Figure 5a), suggesting that BvFL1_v2/v3 returns to prevernalization levels after vernalization. Quantitative RT–PCR confirmed a 2.6-fold increase in BvFL1_v2/v3 expression levels during the 17-day postvernalization period, thereby returning BvFL1_v2/v3 to a level approximately equal to that observed before vernalization (Figure 6). Moreover, BvFL1_v2/v3 was upregulated following vernalization regardless of whether the plant ultimately flowered or reverted to vegetative growth (Figure 5a). Thus, in contrast to FLC in Arabidopsis, a vernalization treatment does not result in the maintained repression of BvFL1_v2/v3 in sugar beet: BvFL1 is quickly reset to the unvernalized state following the return to warm growing conditions. This response is consistent with the propensity for inflorescence reversion observed in sugar beet.
In nature, a resetting BvFL1 response may prevent biennial B. vulgaris individuals from flowering following transient cold periods, akin to the proposed function of Arabidopsis MAF2 (Ratcliffe et al. 2003). Additionally, a vernalization mechanism that resets BvFL1 expression to a state that represses flowering could promote the resumption of vegetative growth following a single round of flowering in the spring, thereby suggesting one possible mechanism for the facultative perenniality observed in B. vulgaris.
BvFL1 is not the “bolting gene”:
Vernalization-responsive winter-annual Arabidopsis ecotypes have higher FLC levels than their rapid-flowering, summer annual counterparts (Michaels et al. 2003). We examined whether biennial beets exhibit higher BvFL1 levels than annual beets, which possess no intrinsic vernalization requirement for flowering. The expression level of BvFL1_v1–v4 (all mRNA variants, measured simultaneously) in immature leaves of biennial FC606 was not significantly different from annual germ plasm SLC003 (P = 0.444) when grown under inductive day lengths. Similarly, the relative expression level of BvFL1_v1/v4 and BvFL1_v2/v3 in shoot apexes and leaves was indistinguishable between FC606 and SLC003 (Figure 5c). Therefore, the difference in vernalization requirement between biennial FC606 and annual SLC003 cannot be attributed to differences in the abundance of BvFL1 transcripts in immature leaves or the shoot apex. Furthermore, BvFL1 was downregulated in response to vernalization in SLC003 in a manner similar to biennial FC606 (Table 1). Thus, at least in terms of BvFL1 levels, the annual SLC003 retains an intact vernalization response. The lack of clear differences in the quantity or the regulation of BvFL1 between SLC003 and FC606 suggests that BvFL1 is not the bolting gene. Moreover, we have mapped BvFL1 to chromosome 6 of sugar beet. The bolting gene is located on chromosome 2 (Butterfass 1968; Boudry et al. 1994); thus BvFL1 cannot be the bolting gene.
In contrast to Arabidopsis, the presence of a vernalization requirement in sugar beet does not appear to be attributable to a difference in expression level of the FLC homolog BvFL1. Thus the primary mechanism (upregulation of FLC by FRI) hypothesized to cause the adaptive delay in flowering in fall-germinating winter-annual Arabidopsis ecotypes (Johanson et al. 2000; Michaels and Amasino 2000) may not be responsible for the evolution of the vernalization requirement (which plays a similar ecological role) in wild B. vulgaris. This should not be surprising. First, the FRI/FLC mechanism is only one component of a complex regulatory network that controls flowering time in Arabidopsis. A number of loci other than FRI and FLC play independent roles in the Arabidopsis vernalization response (Michaels and Amasino 2001; Ratcliffe et al. 2003; Werner et al. 2005a,b). During evolution, any number of mechanisms, both within and outside the vernalization pathway, could be recruited to produce a vernalization requirement. Second, by mutagenizing an annual line (BB) with ethyl methylsulfonate, Hohmann et al. (2005) produced beets that performed as biennials: they had lost the ability to flower under long days alone, but retained an obligate vernalization requirement, flowering only after cold treatment. Hohmann et al.'s (2005) results demonstrate that a vernalization requirement may be imposed in beet via loss-of-function mutations with no obvious impact on the vernalization response. Accordingly, we suggest that the bolting gene may lie outside the vernalization pathway. The recessive allele b could, for example, represent a loss-of-function allele in the photoperiod pathway. The obligate vernalization requirement of biennial beets could have evolved because of a failure to recognize a long-day flowering cue such that vernalization became the only effective pathway to flowering.
Summary and prospects:
Using phylogenetic analysis, we have identified homologs of Arabidopsis FLC in the three major eudicot lineages. These data represent the first report of FLC homologs outside the Brassicaceae and demonstrate that the FLC-like gene subfamily is old, having originated in the common ancestor of the core eudicots or earlier. We have shown that BvFL1, an FLC homolog in sugar beet, is a repressor of flowering in transgenic Arabidopsis and is downregulated during vernalization in sugar beet. We therefore propose that a key component of the Arabidopsis vernalization mechanism—downregulation of an FLC-like floral repressor—may be conserved in distantly related B. vulgaris. This attribute of the vernalization mechanism likewise may be conserved in the broad diversity of taxa descended from the common ancestor of Arabidopsis and B. vulgaris.
Higher-level phylogenetic relationships among the taxa shown to contain FLC-like genes have been well-studied (Angiosperm Phylogeny Group 2003; Judd and Olmstead 2004; Soltis and Soltis 2004). On the basis of these relationships, it is reasonable to predict that vernalization-responsive, FLC-like genes should be found, at a minimum, in the caryophyllid and rosid clades (Figure 4b). Together these two clades comprise approximately half of eudicot species diversity (one-third of angiosperm diversity). Included in the rosids and caryophyllids are numerous fruit, forage, vegetable, and seed crops where, as in sugar beet, flowering time is a trait of critical economic importance. Conservation of the FLC-mediated vernalization response creates the opportunity to control flowering time in a broad variety of crop species using molecular approaches or through conventional breeding practices that utilize the natural variation at FLC-like loci present in existing genetic resources. Furthermore, flowering time is a trait of clear ecological and evolutionary significance. The evolution of genetically based differences in flowering time is an important mechanism of speciation (Coyne and Orr 2004). Divergence in flowering phenology, for example, has been implicated as the most probable cause of reproductive isolation in the first documented cases of sympatric speciation in plants (Silvertown et al. 2005; Savolainen et al. 2006). By understanding the molecular mechanisms that determine flowering time in diverse taxa, crucial insight into the genetic basis of plant speciation may also be attained.
Acknowledgments
We thank M. McClintock for assistance with plant culture and helpful insights on sugar beet vernalization, A. Becker for sharing a comprehensive data set of MADS-box genes for phylogenetic analyses, M. McGrath and S. Shaw for assistance with genetic mapping and BAC library screening, and M. Simmons and R. Larson for helpful comments on the manuscript. Partial funding for BAC sequencing was provided by the Beet Sugar Development Foundation. Research in R.M.A.'s lab was supported by the College of Agricultural and Life Sciences of the University of Wisconsin and by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the National Science Foundation. R.J.S. was supported by the National Institutes of Health Predoctoral Training Program in Genetics 5 T32 GM07133.
References
- Abe, J., G.-P. Guan and Y. Shimamoto, 1997. A gene complex for annual habit in sugar beet (Beta vulgaris L.). Euphytica 94: 129–135. [Google Scholar]
- Abegg, F. A., 1936. A genetic factor for the annual habit in beets and linkage relationship. J. Agric. Res. 53: 493–511. [Google Scholar]
- Angiosperm Phylogeny Group, 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 141: 399–436. [Google Scholar]
- Bastow, R., J. S. Mylne, C. Lister, Z. Lippman, R. A. Martienssen et al., 2004. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Battey, N. H., and R. F. Lyndon, 1990. Reversion of flowering. Bot. Rev. 56: 162–189. [Google Scholar]
- Becker, A., and G. Theissen, 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry, P., R. Wieber, P. Saumitou-Laprade, K. Pillen, H. Van Dijk et al., 1994. Identification of RFLP markers closely linked to the bolting gene B and their significance for the study of the annual habit in beets (Beta vulgaris L.). Theor. Appl. Genet. 88: 852–858. [DOI] [PubMed] [Google Scholar]
- Butterfass, T., 1968. The assignment of the R locus of sugar beet (Hypocotyl color) to chromosome II. Theor. Appl. Genet. 38: 348–350 (in German). [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., J. R. Stinchcombe, K. M. Olsen, J. Schmitt and M. D. Purugganan, 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chroboczek, E., 1934. A study of some ecological factors influencing seed-stalk development in beets (Beta vulgaris L.). Cornell University Agricultural Experimental Station Memoir 154: 1–84. [Google Scholar]
- Ciaffi, M., A. R. Paolacci, E. D'Aloisio, O. A. Tanzarella and E. Porceddu, 2005. Identification and characterization of gene sequences expressed in wheat spikelets at the heading stage. Gene 346: 221–230. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Crosthwaite, S. K., and G. I. Jenkins, 1993. The role of leaves in the perception of vernalizing temperatures in sugar beet. J. Exp. Biol. 44: 801–806. [Google Scholar]
- Curtis, G. J., K. G. Hornsey and G. K. G. Campbell, 1964. Graft-transmissible induction of elongation and flowering in scions of sugar-beet bred for resistance to bolting. Nature 202: 1238. [Google Scholar]
- Curtis, M. D., and U. Grossniklaus, 2003. A Gateway cloning vector set for high-throughput functional analysis of genes in Planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debry, R. W., and R. G. Olmstead, 2000. A simulation study of reduced tree-search effort in bootstrap resampling analysis. Syst. Biol. 49: 171–179. [DOI] [PubMed] [Google Scholar]
- Desfeux, C., S. J. Clough and A. F. Bent, 2000. Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 123: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J. J., 1994. Evolution of a plant homeotic multigene family: toward connecting molecular systematics and molecular developmental genetics. Syst. Biol. 43: 307–328. [Google Scholar]
- Fife, J. M., and C. Price, 1953. Bolting and flowering of sugar beets in continuous darkness. Plant Physiol. 28: 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall, A. R., Y. Y. Levy, A. Wilson and C. Dean, 2001. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535. [DOI] [PubMed] [Google Scholar]
- Hartmann, S., D. Lu, J. Phillips and T. J. Vision, 2006. Phytome: a platform for plant comparative genomics. Nucleic Acids Res. 34: D724–D730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., M. R. Doyle and R. M. Amasino, 2004. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V., F. Foucher, C. Ferrándiz, R. Macknight, C. Navarro et al., 2005. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137: 1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, J. G., S. Pietrokovski, C. M. McCallum and S. Henikoff, 2000. Blocks-based methods for detecting protein homology. Electrophoresis 21: 1700–1706. [DOI] [PubMed] [Google Scholar]
- Hepworth, S. R., F. Valverde, D. Ravenscroft, A. Mouradov and G. Coupland, 2002. Antagonistic regulation of flowering time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21: 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann, U., G. Jacobs and C. Jung, 2005. An EMS mutagenesis protocol for sugar beet and isolation of non-bolting mutants. Plant Breed 124: 317–321. [Google Scholar]
- Irish, V. F., and A. Litt, 2005. Flower development and evolution: gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15: 454–460. [DOI] [PubMed] [Google Scholar]
- Johanson, U., J. West, C. Lister, S. Michaels, R. Amasino et al., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Judd, W. S., and R. G. Olmstead, 2004. A survey of tricolpate (eudicot) phylogenetic relationships. Am. J. Bot. 91: 1627–1644. [DOI] [PubMed] [Google Scholar]
- Kim, S. Y., Y. He, Y. Jacob, Y.-S. Noh, S. Michaels et al., 2005. Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., C. Alonso-Blanco, A. J. M. Peeters and W. Soppe, 1998. Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 345–370. [DOI] [PubMed] [Google Scholar]
- Lang, A., 1965. Physiology of flower formation, pp. 1371–1536 in Encyclopedia of Plant Physiology, edited by W. Ruhland. Springer-Verlag, Berlin.
- Lee, H., S.-S. Suh, E. Park, E. Cho, J. H. Ahn et al., 2000. The AGAMOUS-LIKE MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Castilla, L. P., and E. R. Alvarez-Buylla, 2003. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 100: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, J. M., R. S. Shaw, B. G. de los Reyes and J. J. Weiland, 2004. Construction of a sugar beet BAC library from a hybrid with diverse traits. Plant Mol. Biol. Rep. 22: 23–28. [Google Scholar]
- McKay, J. K., J. H. Richards and T. Mitchell-Olds, 2003. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12: 1137–1151. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1999. Flowering locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 2000. Memories of winter: vernalization and the competence to flower. Plant Cell Environ. 23: 1145–1153. [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., Y. He, K. C. Scortecci and R. M. Amasino, 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munerati, O., 1931. L'eredità della tendenza all annualità nella comune barbabietola coltivata. Zeitschrift fur Züchtung. Reihe A. Pflanzenzüchtung. 17: 84–89. [Google Scholar]
- Nam, J., J. Kim, S. Lee, G. An, H. Ma et al., 2004. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101: 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S., H. Zhang, P. Ludwig and S. van Nocker, 2004. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, F. V., 1940. Photothermal induction of flowering in sugar beets. J. Agric. Res. 61: 101–124. [Google Scholar]
- Owen, F. V., 1950. The sugar beet breeder's problem of establishing male-sterile populations for hybridization purposes. Proc. Am. Soc. Sugar Beet Technologists 6: 191–194. [Google Scholar]
- Owen, F. V., 1954. The significance of single gene reactions in sugar beets. Proc. Am. Soc. Sugar Beet Technologists 8: 392–398. [Google Scholar]
- Pařenicová, L., S. de Folter, M. Kieffer, D. S. Horner, C. Favalli et al., 2003. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. W., G. W. Horgan and L. Dempfle, 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D., S. D. Rounsley, R. J. Schmidt and M. F. Yanofsky, 1995. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O. J., G. C. Nadzan, T. L. Reuber and J. L. Richmann, 2001. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126: 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O. J., R. W. Kumimoto, B. J. Wong and J. L. Riechmann, 2003. Analysis of Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F., and J. P. Huelsenbeck, 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum et al., 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 16: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., H. Onouchi, S. E. Gold, G. S. Ditta, Z. Schwarz-Sommer et al., 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Savolainen, V., M.-C. Anstett, C. Lexer, I. Hutton, J. J. Clarkson et al., 2006. Sympatric speciation in palms on an oceanic island. Nature 441: 210–213. [DOI] [PubMed] [Google Scholar]
- Schläppi, M., and M. Patel, 2001. Biennialism and vernalization-promoted flowering in Hyoscyamus niger: a comparison with Arabidopsis. Flowering Newsl. 31: 25–32. [Google Scholar]
- Scortecci, K. M., S. D. Michaels and R. M. Amasino, 2001. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26: 229–236. [DOI] [PubMed] [Google Scholar]
- Searle, I., Y. He, F. Turck, C. Vincent, F. Fornara et al., 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C. C., J. E. Burn, P. P. Perez, J. Metzger, J. A. Edwards et al., 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C. C., A. B. Conn, E. S. Dennis and W. J. Peacock, 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvertown, J., C. Servaes, P. Biss and D. Macleod, 2005. Reinforcement of reproductive isolation between adjacent populations in the Park Grass Experiment. Heredity 95: 198–205. [DOI] [PubMed] [Google Scholar]
- Smith, G. A., and E. G. Ruppel, 1980. Registration of FC 606 and FC 606 CMS sugarbeet germplasm. Crop Sci. 19: 300. [Google Scholar]
- Soltis, P. S., and D. E. Soltis, 2004. The origin and diversification of angiosperms. Am. J. Bot. 91: 1614–1626. [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J. R., C. Weinig, M. Ungerer, K. M. Olsen, C. Mays et al., 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. USA 101: 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout, M., 1945. Translocation of the reproductive stimulus in sugar beets. Bot. Gaz. 107: 86–95. [Google Scholar]
- Subramanian, A. R., J. Weyer-Menkhoff, M. Kaufmann and B. Morgenstern, 2005. DIALIGN-T: an improved algorithm for segment-based multiple sequence alignment. BMC Bioinformatics 6: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and R. M. Amasino, 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 1999. PAUP* Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0b10. Sinauer Associates, Sunderland, MA.
- Tadege, M., C. C. Sheldon, C. A. Helliwell, P. Stoutjesdikh, E. S. Dennis et al., 2001. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28: 545–553. [DOI] [PubMed] [Google Scholar]
- Tadege, M., C. C. Sheldon, C. A. Helliwell, N. M. Upadhyaya, E. S. Dennis et al., 2003. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1: 361–369. [DOI] [PubMed] [Google Scholar]
- Theissen, G., J. T. Kim and H. Saedler, 1996. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43: 484–516. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitenbroek, D. G., 1997. Binomial. SISA (http://home.clara.net/sisa/binomial.htm).
- Van Dijk, H., P. Boudry, H. McCombie and P. Vernet, 1997. Flowering time in wild beet (Beta vulgaris ssp. maritima) along a latitudinal cline. Acta Oecol. 18: 47–60. [Google Scholar]
- Werner, J. D., J. O. Borevitz, N. H. Uhlenhaut, J. R. Ecker, J. Chory et al., 2005. a FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J. D., J. O. Borevitz, N. Warthmann, G. T. Trainer, J. R. Ecker et al., 2005. b Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102: 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, A. Blechl, G. Tranquilli, W. Ramakrishna et al., 2004. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]