Abstract
Mendelian loci that control the expression levels of transcripts are called expression quantitative trait loci (eQTL). When mapping eQTL, we often deal with thousands of expression traits simultaneously, which complicates the statistical model and data analysis. Two simple approaches may be taken in eQTL analysis: (1) individual transcript analysis in which a single expression trait is mapped at a time and the entire eQTL mapping involves separate analysis of thousands of traits and (2) individual marker analysis where differentially expressed transcripts are detected on the basis of their association with the segregation pattern of an individual marker and the entire analysis requires scanning markers of the entire genome. Neither approach is optimal because data are not analyzed jointly. We develop a Bayesian clustering method that analyzes all expressed transcripts and markers jointly in a single model. A transcript may be simultaneously associated with multiple markers. Additionally, a marker may simultaneously alter the expression of multiple transcripts. This is a model-based method that combines a Gaussian mixture of expression data with segregation of multiple linked marker loci. Parameter estimation for each variable is obtained via the posterior mean drawn from a Markov chain Monte Carlo sample. The method allows a regular quantitative trait to be included as an expression trait and subject to the same clustering assignment. If an expression trait links to a locus where a quantitative trait also links, the expressed transcript is considered to be associated with the quantitative trait. The method is applied to a microarray experiment with 60 F2 mice measured for 25 different obesity-related quantitative traits. In the experiment, ∼40,000 transcripts and 145 codominant markers are investigated for their associations. A program written in SAS/IML is available from the authors on request.
THE recently developed microarray technology allows us to measure the expression of many genes or transcripts in a single chip. The transcript abundance can be treated as a classical quantitative trait and thus mapping can be done on the transcript. Mendelian loci in the genome that control the expression levels of transcripts are called expression quantitative trait loci (eQTL). In eQTL analysis, an expression trait is mapped to genomic locations represented by cis- or trans-loci. The cis-eQTL represent sequence variants that encode transcriptional differences (Hubner et al. 2005). The trans-eQTL, however, represent remote genes that regulate the expression of the gene being transcribed (Yvert et al. 2003). The purpose of a linkage study is to identify the cis- and trans-eQTL for each transcript. Results from the eQTL analysis may provide more detailed information about the biological processes of the gene network than the classical quantitative trait analysis. Regular quantitative traits are often gross clinical measurements and may be far remote from the biological processes giving rise to the clinical traits (Schadt et al. 2003).
In eQTL analysis, we often deal with thousands of expression traits simultaneously. Methods developed for multiple-quantitative trait QTL mapping may not apply here because of the high dimensionality of the model. Two simple approaches may be taken in eQTL analysis: (1) individual transcript analysis in which a single expression trait is mapped at a time and the entire eQTL mapping involves separate analysis of thousands of traits and (2) individual marker analysis where differentially expressed transcripts are detected on the basis of their association with the segregation pattern of an individual marker and the entire analysis requires scanning markers of the entire genome. The first approach requires only known single-trait QTL mapping procedures, e.g., interval mapping (Lander and Botstein 1989), composite-interval mapping (Jansen 1993; Zeng 1994), or multiple-QTL mapping (Kao et al. 1999). A common practice for handling thousands of transcript traits is to select a small number of target transcripts on the basis of some criterion for preselection and map QTL only for these prescreened transcripts (Lan et al. 2003). The second approach requires only a method for differential expression analysis, e.g., the regularized t-test (SAM) (Tusher et al. 2001), the hierarchical mixture model of Newton et al. (2004), or the model-based cluster analysis (Pan 2002). In differential expression analysis, one requires samples from at least two conditions, the control and the treatment. When applied to expression–marker association studies, the conditions become the marker genotypes, e.g., individuals carrying one genotype are arbitrarily designated as the control and those carrying the other genotypes as the treatments. When applied to an F2 design where three groups (genotypes) are present, differential expressions are presented in two forms. One is for the “additive” effect where control and treatment represent the two homozygotes. The other is for the “dominance” effect where control and treatment represent the homozygote (both types) and the heterozygote.
It appears that eQTL mapping has been treated as either a QTL mapping problem for multiple traits or a microarray differential expression problem for multiple treatment comparisons. Other than the method described in Kendziorski et al. (2006), no unique method has been particularly designed for eQTL analysis. Neither the first nor the second aforementioned simple approach is optimal because data are not analyzed jointly. The mixture over marker (MOM) approach developed by Kendziorski et al. (2006) is the first attempt to analyze transcripts and markers jointly. The method is called MOM because the expression level of a transcript is described by a mixture model over markers. A transcript is either associated with a marker or not associated with any markers at all. Given that the transcript is associated with a marker, it is associated with one and only one of the markers. We believe that the assumption of a transcript associated with at most one marker is too stringent and needs to be relaxed. The MOM approach is able to detect either the cis-locus or one of the trans-loci, but not both. This seriously limits the application of the MOM method.
In this study, we propose a novel statistical method that combines the two simple approaches into a single step of analysis so that parameters are inferred using multiple transcripts and markers simultaneously. This joint approach captures the maximum information from the microarray experiment. Like a regular quantitative trait, a transcript can be mapped to many different locations, including cis- and trans-loci. In multiple-QTL mapping, we face a variable selection problem. To avoid variable selection, we have adopted the Bayesian shrinkage analysis, in which marker loci of the entire genome are evaluated simultaneously (Wang et al. 2005). Markers with small effects are forced to shrink their effects to zero and markers with large effects are subject to no shrinkage. The shrinkage estimation is made possible through Bayesian hierarchical modeling (Gelman 2005). In this study, eQTL parameters are estimated under the framework of shrinkage analysis.
THEORY AND METHODS
Hierarchical linear model:
Let G be the number of transcripts measured from N subjects of a mapping population. Let M be the number of markers whose genotypes are measured for all the subjects. We now define the following variables. For the kth individual, let yik be the expression level of transcript i and xjk be the genotype indicator of marker j, for 
 and
and  The genotype indicator variable is defined as xjk = {−1, 1} for a backcross (BC) individual or xjk = {−1, 0, 1} for an F2 individual for
The genotype indicator variable is defined as xjk = {−1, 1} for a backcross (BC) individual or xjk = {−1, 0, 1} for an F2 individual for  Note that, for simplicity, only the additive effect is considered for an F2 population in the current study. The expression level yik can be expressed as
Note that, for simplicity, only the additive effect is considered for an F2 population in the current study. The expression level yik can be expressed as
 |
(1) |
where αi is the intercept for the ith transcript, γij is the effect of the jth marker on the ith transcript, and ɛik is the residual error with an assumed N(0, σ2) distribution. Model 1 can be rewritten in a matrix notation,
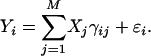 |
(2) |
In model 2,  is an N × 1 vector for the expression levels of transcript i, and
is an N × 1 vector for the expression levels of transcript i, and 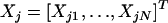 is an N × 1 vector for the genotype indicator variables of locus j, where both Yi and Xj are assumed to have been centered (deviations of the original measurements from the mean) and thus
is an N × 1 vector for the genotype indicator variables of locus j, where both Yi and Xj are assumed to have been centered (deviations of the original measurements from the mean) and thus  and
and  for
for  and
and  Because of this, there is no intercept in the current linear model. Let ɛi be an N × 1 vector of the residual errors, and ɛi ∼ N(0, Rσ2). Note that R is a known positive definite matrix. It is not an identity matrix because covariances among the residual errors have been introduced when the intercept is removed Qu and Xu (2006). Given all the γij variables, the probability density of Yi is
Because of this, there is no intercept in the current linear model. Let ɛi be an N × 1 vector of the residual errors, and ɛi ∼ N(0, Rσ2). Note that R is a known positive definite matrix. It is not an identity matrix because covariances among the residual errors have been introduced when the intercept is removed Qu and Xu (2006). Given all the γij variables, the probability density of Yi is
 |
(3) |
where we use a notation Normal(x; b, d) or N(x; b, d) to denote a normal density for vector x with mean b and variance matrix d. In subsequent sections, we adopt the same notation for other probability densities, i.e.,
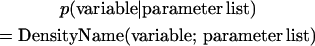 |
with different parameters in the list separated by commas. Because the model is hierarchical, each of the regression coefficients is assumed to be a random variable sampled from a distribution. In this study, we assign a mixture distribution to γij as originally suggested by George and Mcculloch (1993),
 |
(4) |
where δ = 10−4 (a small positive number) and  is an unknown variance assigned to the jth locus. Variable ηij = {0, 1} is used to indicate whether γij is sampled from a N(0, δ) or a N(0,
is an unknown variance assigned to the jth locus. Variable ηij = {0, 1} is used to indicate whether γij is sampled from a N(0, δ) or a N(0, 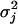 ) distribution. If it comes from the first normal distribution, γij is virtually fixed at zero. If it comes from the second normal distribution (the distribution with a large variance), γij has a nontrivial value and should be estimated from the data. Therefore, ηij = 1 means that locus j is an eQTL for transcript i. In some of the literature (e.g., Zhang et al. 2005), the mixture distribution is described as
) distribution. If it comes from the first normal distribution, γij is virtually fixed at zero. If it comes from the second normal distribution (the distribution with a large variance), γij has a nontrivial value and should be estimated from the data. Therefore, ηij = 1 means that locus j is an eQTL for transcript i. In some of the literature (e.g., Zhang et al. 2005), the mixture distribution is described as
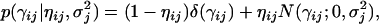 |
(5) |
where δ(γij) is the Dirac delta function (originally introduced by the British theoretical physicist Paul Dirac) that has the value of ∞ for γij = 0 and the value zero elsewhere. The Dirac delta function is actually a probability density function. Therefore, the integral of the delta function from −∞ to +∞ is 1. The above mixture distribution says that γij has a nonzero probability mass at the value zero and a quite flat normal distribution around zero provided that  is large. This distribution is called the spike and slab distribution (Mitchell and Beauchamp 1988). When δ = 0, the two forms of mixture distribution are identical. We further describe ηij by a Bernoulli distribution with probability ρj, denoted by Bernoulli(ηij; ρj). Because of the hierarchical nature of the model, we can further describe ρj by a Dirichlet distribution, denoted by Dirichlet(ρj; 1, 1). The parameter ρj will control the proportion of the transcripts that are associated with marker j. All transcripts that are assigned to the second component of the mixture distribution are associated with the jth marker. The variance of the genes in this cluster is assigned a scaled inverse chi-square distribution, denoted by
is large. This distribution is called the spike and slab distribution (Mitchell and Beauchamp 1988). When δ = 0, the two forms of mixture distribution are identical. We further describe ηij by a Bernoulli distribution with probability ρj, denoted by Bernoulli(ηij; ρj). Because of the hierarchical nature of the model, we can further describe ρj by a Dirichlet distribution, denoted by Dirichlet(ρj; 1, 1). The parameter ρj will control the proportion of the transcripts that are associated with marker j. All transcripts that are assigned to the second component of the mixture distribution are associated with the jth marker. The variance of the genes in this cluster is assigned a scaled inverse chi-square distribution, denoted by  where d0 = 5 and ω0 = 50 are used in this study. The residual variance is assigned a vague prior; i.e., Inv − χ2(σ2; 0, 0) = 1/σ2. This vague prior may cause an improper posterior (Hobert and Casella 1996). In practice, this vague prior has been used all the time and people rarely see any problems. We used a proper scaled inverse gamma distribution for one case of the simulation to test the difference between the vague prior and the proper prior and did not see any notable difference (data not shown).
where d0 = 5 and ω0 = 50 are used in this study. The residual variance is assigned a vague prior; i.e., Inv − χ2(σ2; 0, 0) = 1/σ2. This vague prior may cause an improper posterior (Hobert and Casella 1996). In practice, this vague prior has been used all the time and people rarely see any problems. We used a proper scaled inverse gamma distribution for one case of the simulation to test the difference between the vague prior and the proper prior and did not see any notable difference (data not shown).
Markov chain Monte Carlo:
Given the likelihood and the prior of parameters, we are ready to infer the posterior distribution of the parameters. Because the form of the posterior distribution is intractable, we use Markov chain Monte Carlo (MCMC) sampling to draw a posterior sample from which empirical posterior means of interested parameters can be found. The most important parameters in the eQTL mapping are ηij and ρj. The posterior mean of ηij represents the probability that transcript i is associated with marker j. The posterior mean of ρj reflects the proportion of transcripts that are associated with marker j. In addition to these parameters, other parameters may also be interesting. For example, the posterior mean of γij represents how strongly marker j affects the expression of transcript i, i.e., the size of the eQTL.
First, we choose an initial value for each variable. We then derive the distribution of one variable conditional on the data and values of all other variables. This distribution is called the conditional posterior distribution, denoted by p(θk | data, θ−k), where θk is the current variable of interest and θ−k is the list of the remaining variables (excluding θk). This distribution usually has a simple form from which a realized value for θk is sampled. Once each and every variable is updated, we complete one iteration or sweep. The sampling process continues until the Markov chain reaches its stationary distribution. Convergence diagnosis was conducted using the R package “coda” (Raftery and Lewis 1992). We discard a number of iterations from the beginning of the chain (burn in) and save one observation in every 10 sweeps of the remaining iterations to form a posterior sample until the posterior sample is sufficiently large to allow an accurate estimate of the posterior mean for each variable. We now describe the sampling algorithm for each variable:
- Variable ηij is simulated from Bernoulli(ηij; πij), where
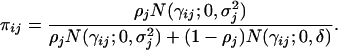
(6) - Variable γij is simulated from N(γij; μγ,
 ), where
), where
(7)
and
(8)
which is called the offset of Yi adjusted for the jth marker effect.
(9) - Sample
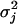 from
from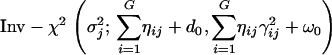
- Sample σ2 from

- Simulate ρj from
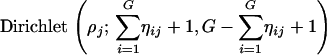
So far, every variable has been updated. Let  be the posterior mean of variable ηij, where Np is the posterior sample size. Transcript i is said to be associated with marker j if the posterior probability
be the posterior mean of variable ηij, where Np is the posterior sample size. Transcript i is said to be associated with marker j if the posterior probability  is greater than some prespecified threshold. It has been shown that the false discovery rate (FDR) can be controlled at α · 100% if the appropriate threshold is the smallest posterior probability such that the average posterior probability of all transcript–marker linkage exceeding the threshold is >1 − α (Efron 2004; Newton et al. 2004). In the current study, 0.8 is taken as the cutoff point. The 0.80 criterion is somewhat arbitrary and another value, say 0.90, may be used. In the application section, we show that any value between 0.6 and 1 is reasonable. This criterion does not affect the MCMC process other than the list of transcripts declared as being associated to marker j.
is greater than some prespecified threshold. It has been shown that the false discovery rate (FDR) can be controlled at α · 100% if the appropriate threshold is the smallest posterior probability such that the average posterior probability of all transcript–marker linkage exceeding the threshold is >1 − α (Efron 2004; Newton et al. 2004). In the current study, 0.8 is taken as the cutoff point. The 0.80 criterion is somewhat arbitrary and another value, say 0.90, may be used. In the application section, we show that any value between 0.6 and 1 is reasonable. This criterion does not affect the MCMC process other than the list of transcripts declared as being associated to marker j.
Missing markers:
The algorithm described above applies to the situation where the genotypes of markers are known for all individuals. In reality, the genotypes of some markers may be missing for some individuals. This means that some elements of Xj are also variables (data not observed) and subject to the same MCMC sampling as other variables. In this case, we adopt the usual sampling procedure in Bayesian mapping to simulate the missing genotypes (Rao and Xu 1998). First, all missing genotypes are sampled from their Mendelian prior distribution: 50% probability of taking either genotype for a BC individual or {25%, 50%, 25%} probability of taking one of the three genotypes for an F2 individual. Given the sampled missing genotypes, all marker genotypes are presumably known. We can now start the MCMC process by updating the missing values one at a time from its conditional posterior distribution given the two flanking marker genotypes and all other information available at the current stage of the MCMC process. The process of marker genotype imputation is the same as that described by Sen and Churchill (2001) and implemented in R/QTL (Broman et al. 2003). The conditional posterior distribution is briefly described below.
Consider that the genotype of marker j is missing for individual k. Let p(Y | X) be the probability density of the expression levels for all transcripts measured from individual k given the genotype at marker j. Of course, X can take up to two different values for a BC individual or three different values for an F2 individual. Let p(ML | X) and p(MR | X) be the probabilities for the left and the right flanking markers, respectively, conditional on X. Note that the order of the three markers is ML < X < MR. Let p(X) be the Mendelian prior probability for the genotype of the missing marker. The conditional posterior probability is defined as
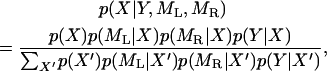 |
(10) |
where  means summation over all possible values of X′. This probability is used to simulate a realized value of X.
means summation over all possible values of X′. This probability is used to simulate a realized value of X.
Heritability for a transcript:
In the eQTL analysis, each transcript has been treated as a quantitative trait. Since the variance of a quantitative trait can be partitioned into a genetic variance component and an environmental variance component, the variance of an expression trait can be similarly partitioned. We now give the formula for calculating the variance components for each expression trait as a function of the eQTL effects and then further derive the average variance components for all expression traits in the entire microarray experiment. These variance components are further used to calculate the “heritability” of a transcript.
Let 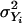 be the variance of transcript i over all subjects (a total of N subjects). Let
be the variance of transcript i over all subjects (a total of N subjects). Let  be the variance of variable Xj over all subjects for marker j. Let us further define
be the variance of variable Xj over all subjects for marker j. Let us further define  as the covariance between Xj and Xj′ across subjects, i.e., the covariance between markers j and j′ for j′ ≠ j. The variance of transcript i is expressed as
as the covariance between Xj and Xj′ across subjects, i.e., the covariance between markers j and j′ for j′ ≠ j. The variance of transcript i is expressed as
 |
(11) |
For a BC design,  and
and  whereas for an F2 design,
whereas for an F2 design,  and
and 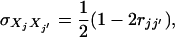 where rjj′ is the recombination fraction between markers j and j′. Let
where rjj′ is the recombination fraction between markers j and j′. Let
 |
(12) |
be the genetic component of  The heritability for transcript i is
The heritability for transcript i is
 |
(13) |
The average heritability of all transcripts may simply take the average 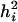 across all transcripts. However, there is a different definition for the heritability of all the transcripts. This type of overall heritability is defined as follows. First, we need to partition the expected variance of a transcript into variance components as shown below:
across all transcripts. However, there is a different definition for the heritability of all the transcripts. This type of overall heritability is defined as follows. First, we need to partition the expected variance of a transcript into variance components as shown below:
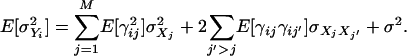 |
(14) |
The expectation is taken with respect to the eQTL effects. Let  be the overall variance of the transcripts. The mixture distribution of γij leads to
be the overall variance of the transcripts. The mixture distribution of γij leads to  where ρj is the proportion of transcripts that are linked to locus j. Because γij and γij′ are independent, we have E[γijγij′] = 0. Therefore, the above equation becomes
where ρj is the proportion of transcripts that are linked to locus j. Because γij and γij′ are independent, we have E[γijγij′] = 0. Therefore, the above equation becomes
 |
(15) |
Let 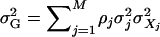 be the genetic variance component. The overall heritability is
be the genetic variance component. The overall heritability is
 |
(16) |
This overall heritability of transcripts is different from the average heritability across all transcripts.
APPLICATIONS
Simulation studies:
We carried out two simulation experiments to compare the performance of the proposed method (hereafter designated as BAYES) and the MOM approach (Kendziorski et al. 2006). In the first experiments, 10 markers were evenly placed on a 360-cM genome, i.e., 40 cM per interval. Genotypes of markers for each subject were simulated on the basis of the Haldane map function (Haldane 1919). Four eQTL were placed at markers 1, 3, 6, and 10; i.e., the segregation of these four markers affected the expression of some transcripts. We simulated 50 subjects and 1000 transcripts among which transcripts 605–610 were affected by eQTL at marker 1, transcripts 601–604 were affected by eQTL at marker 3, transcripts 961–1000 were affected by eQTL at marker 6, and transcripts 1–50 were affected by eQTL at marker 10. Note that a transcript was either associated with one eQTL or not associated with any of them. In eQTL analysis, a marker is claimed as an eQTL if it affects at least one transcript and a linkage is defined as a transcript–marker association. For example, in the first simulation experiment, there were a total of four eQTL and 100 linkages. The eQTL effects for the 100 linked transcripts (γij) were simulated from Normal(γij; 0, 32). The residual error was sampled from Normal(ɛij; 0, 0.12), such that the expected heritability for a transcript was 0.90. We chose this small residual variance because MOM did not work well when the residual variance was large. When the residual variance was large, MOM either declared all transcripts as differentially expressed or did not run by throwing out an error message. The experiment was replicated 20 times. We used two methods to analyze the 20 replicated data sets. For the BAYES method, the threshold may be chosen to bound the posterior expected FDR at α100% as described in theory and methods. The average threshold value of the 20 replicates was 0.55 when the expected FDR was set to 1%, indicating that any value between 0.6 and 1 can serve as a reasonable threshold value to achieve the controlled FDR. We used 0.8 as the threshold value throughout the entire experiments.
The length of the chain required for convergence was determined by the R package coda. In the current MCMC diagnosis, the quantile to be estimated was set to 0.1, the margin of error of the estimate was 0.05, the probability of obtaining an estimate in the desired interval was 0.95, and the precision for the estimate of time to convergence was 0.001. The diagnosis indicated that 1800 iterations were sufficient to achieve convergence. Another issue that sometimes arises is the high posterior correlations or the so-called “stickiness” of the MCMC algorithm. The commonly utilized approach to circumvent this problem is to use every kth simulation draw, for some value of k such as 10, 20, or 50 (see Raftery and Lewis 1992 and Gelman et al. 1995 for more details). In our study, we deleted 1000 iterations as a “burn-in” period and thereafter kept one observation in every 10 iterations until the posterior sample size reached 200. The overall length of the chain was 3000, much longer than the required length of 1800 reported from the coda diagnosis.
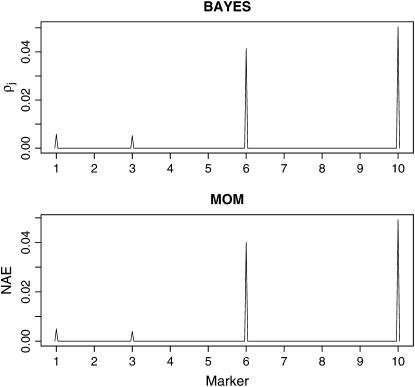
The MOM analysis actually consists of two steps: (1) identifying differentially expressed (DE) transcripts and (2) localizing eQTL for each differentially expressed transcript. In the first step, a transcript is identified as DE if the posterior probability of equivalent expression (EE) is smaller than some threshold, where thresholds are chosen to control the FDR at 5%. In the second step, the 90% highest posterior density (HPD) region was used to specify linkages between each transcript and markers. Both methods successfully detected four eQTL and 98 linkages on the basis of the analysis of 20 replicates. The two missed linkages were represented by associations between transcript 605 and marker 1 and between transcript 26 and marker 10, respectively. The true effects for the two linkages were 0.035 and 0.014, which are too small to be detectable with any reasonable analysis methods. Neither method detected any false linkages. The estimated proportions (ρj) of transcripts associated with each of the 10 markers using the proposed method (BAYES) are displayed visually in Figure 1 along with the results of MOM. Note that the plot for the MOM method was the normalized average evidence of linkage (see the bottom plot in Figure 1). The normalized average evidence (NAE) of linkage was defined as the average posterior probability over transcripts and normalized by the sum of the evidence over all markers (Kendziorski et al. 2006). The estimated proportions (ρj) in the BAYES method, which are equivalent to the NAE used in the MOM analysis, can be used to sift hot-spot regions where the transcripts are mapped. This simulation demonstrates that both methods are adequate if a transcript is affected by at most one eQTL. The estimated parameters and the effects of the 98 detected linkages used in the simulation experiments are provided in Table 1 and also illustrated in Figure 2. The estimated parameters and the effects of eQTL are very close to the true values used to generate the data. Note that the sample size for the linked transcripts was very small (≤50) for each eQTL. As a result, the estimated  's showed some deviations from the true value of 9.
's showed some deviations from the true value of 9.
Figure 1.—
Hot-spot regions along the chromosome for the first simulation experiment. NAE, normalized average evidence.
TABLE 1.
Posterior mean and posterior variance (in parentheses) of ρj and  from the Bayes analysis for the first simulation experiment
from the Bayes analysis for the first simulation experiment
| Parameter
|
|||
|---|---|---|---|
| Marker | ρj | 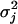 |
|
| 1 | True | 0.006 | 9 |
| Estimate | 0.006 (5.8E-6) | 2.74 (3.1E-4) | |
| 3 | True | 0.004 | 9 |
| Estimate | 0.005 (6.2E-6) | 11.7 (1.6E-3) | |
| 6 | True | 0.040 | 9 |
| Estimate | 0.041 (3.3E-5) | 6.83 (2.1E-4) | |
| 10 | True | 0.050 | 9 |
| Estimate | 0.050 (4.7E-5) | 7.85 (2.2E-4) | |
The true σ2 is 0.01. The posterior mean and variance of σ2 are 0.01 and 4.6E-9, respectively.
Figure 2.—
Effects of 98 linked transcripts in the first simulation experiment. Solid and dashed bars represent the true and the estimated values of γij, respectively.
In the second simulation experiment, we used the same marker map and eQTL setting as in the first experiment and generated 20 replicates. In this case, however, we let the eQTL at marker 1 control transcripts 1–20 and transcripts 971–990 and let the eQTL at marker 3 control transcripts 17–20. The transcripts controlled by the eQTL at markers 6 and 10 remained the same as in the first experiment. The purpose of the second simulation experiment was to allow some transcripts to be controlled by more than one marker. For example, transcripts 1–16 were controlled by markers 1 and 10, transcripts 971–990 were controlled by markers 1 and 6, and transcripts 17–20 were controlled by markers 1, 3, and 10. Again, we sampled all the eQTL effects from Normal(γij; 0, 32) and the residual error from Normal (ɛij; 0, 0.12), such that the expected heritability for a transcript is 0.92. For the sake of comparison, we used the empirical type I and type II errors and the empirical statistical power to evaluate the performance of our method (BAYES) and the MOM method. The current simulation experiment contains 134 linkages. The BAYES analysis detected 133 linkages and no false positives were declared. Therefore, the empirical type I error was zero, the type II error was  and the empirical power was 1 − 0.007 = 0.993. When we examined the single missed linkage (transcript 2 associated with marker 10), we found that the true marker effect for this transcript was 0.055, which again might be too small to be detected by any reasonable methods. The true parameters used in the simulation and their estimated vales obtained from the proposed method are given in Table 2. The true and the estimated effects for the 133 detected linkages are illustrated in Figure 3. The estimates agree well with the true values. The hot-spot regions represented by the estimated proportions (ρj) of transcripts associated with the 10 markers are shown in Figure 4 (the top plot). All 4 markers controlling the expression of transcripts have been identified. A striking advantage of the new method over MOM is that an individual linkage picture can be obtained when we focus on a specific transcript. The estimated marker effects for 6 of the detected transcripts are plotted in Figure 5, showing a close agreement to the true values. In the MOM analysis, however, many true linkages have been missed (see Table 3) because each transcript was allowed to link to at most one locus. The MOM method worked well if a transcript is linked to only one marker, e.g., transcript 997 associated with marker 6 (see Figure 5). When a transcript is controlled by more than one marker, the linkage signal occurs only at the position where the greatest eQTL resides. For example, transcript 10 was controlled by markers 1 and 10. The linkage occurred only at marker 1 (true effect 4.462) with marker 10 (true effect 1.510) being completely missed. Transcript 12 was also controlled by markers 1 and 10 whose true effects were −0.997 and 4.489, respectively. MOM detected marker 10 for this transcript. If the effects of markers controlling the same transcript are similar, MOM generates a confusing result, as demonstrated by transcripts 971 and 18. In the bottom plot of Figure 4, markers 5 and 7 were falsely identified as eQTL by MOM. That marker 6 had a higher peak than marker 1 was also incorrect. The empirical type I error was
and the empirical power was 1 − 0.007 = 0.993. When we examined the single missed linkage (transcript 2 associated with marker 10), we found that the true marker effect for this transcript was 0.055, which again might be too small to be detected by any reasonable methods. The true parameters used in the simulation and their estimated vales obtained from the proposed method are given in Table 2. The true and the estimated effects for the 133 detected linkages are illustrated in Figure 3. The estimates agree well with the true values. The hot-spot regions represented by the estimated proportions (ρj) of transcripts associated with the 10 markers are shown in Figure 4 (the top plot). All 4 markers controlling the expression of transcripts have been identified. A striking advantage of the new method over MOM is that an individual linkage picture can be obtained when we focus on a specific transcript. The estimated marker effects for 6 of the detected transcripts are plotted in Figure 5, showing a close agreement to the true values. In the MOM analysis, however, many true linkages have been missed (see Table 3) because each transcript was allowed to link to at most one locus. The MOM method worked well if a transcript is linked to only one marker, e.g., transcript 997 associated with marker 6 (see Figure 5). When a transcript is controlled by more than one marker, the linkage signal occurs only at the position where the greatest eQTL resides. For example, transcript 10 was controlled by markers 1 and 10. The linkage occurred only at marker 1 (true effect 4.462) with marker 10 (true effect 1.510) being completely missed. Transcript 12 was also controlled by markers 1 and 10 whose true effects were −0.997 and 4.489, respectively. MOM detected marker 10 for this transcript. If the effects of markers controlling the same transcript are similar, MOM generates a confusing result, as demonstrated by transcripts 971 and 18. In the bottom plot of Figure 4, markers 5 and 7 were falsely identified as eQTL by MOM. That marker 6 had a higher peak than marker 1 was also incorrect. The empirical type I error was  and the power was 1 −
and the power was 1 − 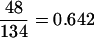 for the MOM analysis. The power would be even lower if more multiple linkages had been simulated.
for the MOM analysis. The power would be even lower if more multiple linkages had been simulated.
TABLE 2.
Posterior mean and posterior variance (in parentheses) of ρj and 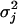 from the BAYES analysis for the second simulation experiment
from the BAYES analysis for the second simulation experiment
| Parameter
|
|||
|---|---|---|---|
| Marker | ρj |  |
|
| 1 | True | 0.040 | 9 |
| Estimate | 0.042 (2.9E-5) | 5.72 (3.3E-4) | |
| 3 | True | 0.004 | 9 |
| Estimate | 0.005 (6.1E-6) | 6.43 (1.1E-3) | |
| 6 | True | 0.040 | 9 |
| Estimate | 0.041 (3.7E-5) | 10.9 (2.1E-4) | |
| 10 | True | 0.050 | 9 |
| Estimate | 0.049 (4.5E-5) | 13.3 (2.9E-4) | |
The true σ2 is 0.01. The posterior mean and variance of σ2 are 0.01 and 3.8E-9, respectively.
Figure 3.—
Effects of 133 linked transcripts in the second simulation experiment. Solid and dashed bars represent the true and the estimated values of γij, respectively.
Figure 4.—
Hot-spot regions along the chromosome for the second simulation experiment. NAE, normalized average evidence.
Figure 5.—
Estimated marker effects γij, for six detected transcripts by BAYES (the second simulation experiment). The triangle in each plot indicates the position of eQTL discovered by MOM.
TABLE 3.
Number of linkages detected by BAYES and MOM for the second simulation experiment
| Marker
|
|||||
|---|---|---|---|---|---|
| Method | 1 | 3 | 6 | 10 | |
| True | 40 | 4 | 40 | 50 | |
| BAYES | Estimate | 40 | 4 | 40 | 49 |
| MOM | Estimate | 15 | 1 | 30 | 40 |
The residual variance used in the above two simulation experiments was too small, leading to a irrationally high expected heritability. In the following simulation experiments, we kept everything the same as that used for the second simulation experiment except residual variance. We varied the residual variance from 0.12 to 32, with σ2 = 0.52, 12, 1.52, 22, 2.52, 32. Again, for each of the six scenarios, we generated 20 replicated data sets. The corresponding expected heritabilities are 0.81, 0.56, 0.32, 0.17, 0.15, and 0.07, respectively. We used only the proposed method (BAYES) to analyze these data sets because MOM did not work when σ2 > 0.12. From Figure 6, we can see that the empirical power decreased dramatically as the residual variance increased. The empirical type I error increased accordingly, but not as much as the decrease in the empirical power due to the stringent control for the FDR.
Figure 6.—
The changes of empirical type I error (top) and empirical power (bottom) as the residual variance increases.
Mice data analysis:
We analyzed a mice data set published by Lan et al. (2006). The data are publicly available at gene expression omnibus (GEO) with accession no. GSE3330. The data consist of 40,738 transcripts whose expression levels were measured from 60 F2 (ob/ob) mice in an obesity-related research. The expression levels were normalized and background was corrected by the robust multiarray average method (Irizarry et al. 2003). Genotypes for 145 markers (distributed over 19 chromosomes) and phenotypes for 25 obesity-related traits were collected from the 60 mice. We noted that the expression levels of most transcripts are constant across the 60 individuals. Those transcripts may not provide any information on the eQTL analysis and thus should be eliminated prior to the analysis. We sorted all transcripts by their variances across individuals and deleted the transcripts with variances <0.12, leaving 1576 most varying transcripts for further analysis. Figure 7 shows the variations of 6 transcripts across 60 individuals. The 3 transcripts on the left had large variances and thus were kept in the data for further analysis. The remaining three transcripts (on the right) were deleted because their variances were small (<0.12).
Figure 7.—
Plots of expression for six selected transcripts with distinct variances across individuals.
For the BAYES method, we used the same length of the Markov chain as used in the simulation studies. The chain was diagnosed for convergence using coda.
Because there were 145 markers, the model for each transcript contained 145 effects. These 145 effects were estimated simultaneously. Of the 1576 transcripts included in the analysis, 843 of them were linked to at least one marker on the mouse genome. Of the 145 markers, 129 of them were claimed to control the expression of the transcripts to some extent. Figure 8 shows the proportion of transcripts associated with each of the 145 markers. Five markers with the highest proportions of linked transcripts are indicated by triangles. The five highest peaks of the profile (hot-spot regions) are different from the 5 strongest markers mapped by Kendziorski et al. (2006). Marker D4Mit237 was the largest eQTL on the mouse genome detected with MOM. However, we found that only 2 transcripts were linked to this locus. The marker with the highest proportion of linked transcripts identified by our method was D15Mit63, a locus proved to be associated with the trait of higher early life body weight (Miller et al. 2002). In addition, the hottest marker on chromosome 4 was actually D4Mit149. Another highly ranked eQTL declared by MOM was marker D2Mit241, which resides on chromosome 2. However, only 3 transcripts were claimed to be linked to this position with the BAYES method. The most influencing marker on chromosome 2 identified by the new method was D2Mit274. It is close to marker D2Mit9, which is an obesity-modifier locus recently discovered by Stoehr et al. (2004). We also found that marker D2Mit9, itself an eQTL identified by the BAYES method, affected the expression of 7 transcripts. Markers D5Mit1 and D8Mit249 are known to affect triglyceride level (Colinayo et al. 2003) and fat content (Naggert et al. 1995), both of which are obesity-related traits. These two markers were successfully identified by both the BAYES and the MOM methods.
Figure 8.—
Plots of the hot-spot regions for the mouse genome obtained from BAYES. The triangles indicate five markers with the highest proportions of linked transcripts.
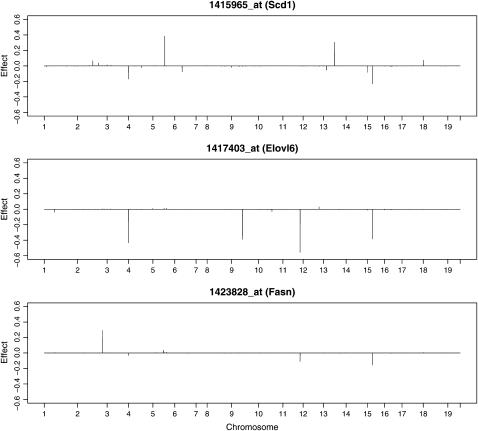
Researchers usually start with a particular gene that has a known function and then quickly identify other genes that are associated with the known gene. Further research is then performed to discover the biological functions for the unannotated genes. The new method developed in this study provides such a tool to detect these genes. For example, a recent study has shown that stearoyl-CoA desaturase-1 (Scd1) is an important gene for lipid metabolism and insulin sensitivity (Lan et al. 2006). The linkage profile (Figure 9, top) shows that the obesity-related gene Scd1 was associated with four markers represented by the largest four eQTL effects. Among the four markers, D15Mit63 was a very important locus that also controls the expression of 520 other transcripts. Note that marker D15Mit63 was the largest eQTL on the mouse genome identified with our method and it has been proved to be an obesity-related locus (Miller et al. 2002). We plotted the estimated effects for two of the transcripts that also link to this locus (see Figure 9). One gene was ELOVL family member 6 (Elovl6), a gene in charge of elongation of long-chain fatty acids. The other was a gene that encodes fatty acid synthase (Fasn). These genes are likely to be involved in metabolism related to obesity.
Figure 9.—
Effects of markers of the mouse genome for three selected transcripts estimated from the BAYES analysis.
Two steps are usually taken to infer the functions of transcripts. The first step is to map QTL for the traits of interest. The second step is to perform eQTL analysis only for the markers detected in the QTL analysis. If a QTL regulating a trait of interest is also an eQTL for some transcripts, then the functions of the transcripts can be inferred. The proposed Bayesian analysis allows us to infer functions of transcripts jointly in a single step. In the mouse experiment (Lan et al. 2006), 25 obesity-related traits were measured. We simply treated the 25 traits as 25 additional transcripts and added them to the existing list of 1576 transcripts, making a list of 25 + 1576 = 1601 traits. These 25 traits were subjected to the same eQTL mapping. If a marker is identified as being associated with both a transcript and a regular quantitative trait, the transcript is claimed to be associated with the quantitative trait. However, quantitative traits are measured in different scales from the transcripts. Therefore, the effects of QTL have different scales from the effects of eQTL. This problem can be solved by rescaling the quantitative traits. In the mouse data analysis, we sorted the 1576 transcripts by the variance across the 60 mice in a descending order and calculated the average variance of the top 5% transcripts. We then used this average variance to rescale each of the 25 traits so that each of them had a variance equal to the average variance after the rescaling. There were 5% missing phenotype measurements in the mouse data. The missing phenotypes were imputed using the multiple-imputation method (Rubin 1987). The proposed Bayesian analysis shows that each one of the 25 traits was associated with at least one marker of the mouse genome and 12 traits were linked to marker D15Mit63. A total of 521 transcripts were also linked to marker D15Mit63, implying that these transcripts may alter the 12 obesity-related traits. A complete list of the associated transcripts, markers, and phenotypes is provided in the supplemental table at http://www.genetics.org/supplemental/.
DISCUSSION
MOM is so far the only statistical method specifically developed for analyzing expression data and marker data jointly. The advantage of the MOM approach is that its computational efficiency allows all expression traits to be accounted for in the analysis. However, the assumption of a single eQTL per expression trait is too strong. Although HPD can be used to identify multiple eQTL, it does not perform well in general. For example, if eQTL are adjacent to one another and their effects are of the same size, HPD is able to detect them all by placing posterior probabilities evenly on each eQTL (see the top plot in Figure 10). However, if neither of the two conditions is satisfied, the MOM method will generate incomplete or misleading results (see middle and bottom plots in Figure 10).
Figure 10.—
Linkage maps for three simulated transcripts obtained from BAYES. The triangles represent the positions of eQTL specified by MOM. The numbers next to the triangles are the posterior probabilities assigned to the eQTL specified by MOM.
The BAYES method proposed in this study is a multiple-eQTL model, in which a transcript is allowed to be linked to more than one locus. The results presented in this study showed that a multiple-eQTL model is more desirable than a single-eQTL model. However, an investigator needs to trim down the transcript space to a reasonable size. For the BAYES analysis, we suggested selecting transcripts on the basis of the variances of their expression levels; i.e., a cutoff value is subjectively chosen and transcripts with variances less than this value are excluded from analysis. Usually, 1000–2000 transcripts might be a good choice because the expression levels of most transcripts do not change across experimental subjects. The preliminary screening aimed at reducing the computational burden and had no effects on the result. We noted that results obtained from the analysis of 1500 selected transcripts were the same as those obtained from 5000 selected transcripts for mice data (data not shown). A similar prescreening scheme was used in a recent microarray data analysis (Ghazalpour et al. 2006). In reality, we may run the computationally quicker MOM approach first to estimate the number of differentially expressed transcripts and then use this information for transcript screening before the BAYES method is applied. In addition, the results obtained from MOM may be useful when setting priors for the BAYES analysis.
The hyperparameters for the scaled inverse chi-square priors in this study are chosen in a subjective way as done in Ishwaran and Rao (2005). According to Ishwaran and Rao (2003), these values do not need to be tuned for each data set and can be fixed. No substantial differences have been observed when several different sets of priors were tried in a simulation study (data not shown). The vague prior (1/σ2) we chose as the prior for the residual variance has not caused any problem, since the MCMC diagnosis indicated a satisfactory convergence of the posterior sample. However, as Hobert and Casella (1996) warned, the marginal posterior distribution for the variance may not exist. Therefore, the hyperparameters for the scaled inverse chi-square prior should be chosen so that it is proper, which will lead to a proper posterior.
In the simulation studies, we sampled the effects of linkages from Normal(γij; 0, 32), where 32 was chosen in a subjective manner. The performance of the proposed method does not depend on the choice of the variance for linkage effects, but rather on the ratio of this variance to the residual variance that affects the expected heritability for a transcript. As we demonstrated in the applications section, our method is robust given different residual variances. We also carried out one more simulation experiment, where the effects of linkages were sampled from a uniform distribution with a wide range (from −10 to 10). The performance of the BAYES method was still very satisfactory (data not shown).
We introduced a Bayes method for joint analysis of transcripts and markers. If a marker is associated with at least one transcript, this marker is considered as a candidate eQTL. However, it is rare that an eQTL sits exactly at a marker position. Therefore, the marker analysis will provide biased estimates for both the locations and the sizes of the eQTL unless the marker density is sufficiently high. The method can be extended so that eQTL can be mapped to arbitrary positions between markers. This is equivalent to the extension of individual marker analysis to interval mapping for quantitative trait loci. Note that QTL locations were often treated as fixed in QTL analyses (Hoeschele and VanRaden 1993a,b). In a very recent interval mapping technique, such as that in Wang et al. (2005), a QTL was allowed to take a position varying within a marker interval, leading to locating QTL and estimating their effects more precisely. This extension will add extra complexity to the existing method because eQTL positions become parameters also and are subject to similar Monte Carlo sampling in the Bayesian analysis. Two approaches can be taken for such an extension. One is the fixed-interval approach where one potential eQTL is assumed in each marker interval. The prior distribution of the location of the putative eQTL is uniform within the interval. The posterior distribution can be inferred and a realized location can be sampled using the Metropolis–Hastings method (Metropolis et al. 1953; Hastings 1970). If an interval has no eQTL, the posterior distribution will remain uniform and the eQTL effect will be shrunken to zero. If an interval does cover an eQTL, the posterior distribution of the eQTL location will be peaked at the true location and the eQTL effect will be estimated subject to no shrinkage (Wang et al. 2005). When the marker density is high and markers are unevenly distributed along the genome, the variable-interval approach may be adopted to sample the eQTL location (Wang et al. 2005), where the number of eQTL included in the model can be substantially less than the number of marker intervals. The eQTL genotypes are always missing, but realized genotypes can be sampled from the distribution given in Equation 10 of the Missing markers section.
Mapping eQTL is an important subject in the field of statistical genomics. Current methods for eQTL mapping rely mostly on either a microarray analysis procedure or a QTL mapping statistic since no well thought out statistical method has emerged. The proposed joint analysis is one of the very few studies particularly targeting this subject. The field of eQTL mapping is very young and needs substantial effort from scientists across multidisciplinary fields to become a mature science. The proposed BAYES method may still be very crude, but it provides a starting point from which more comprehensive techniques can be developed.
Acknowledgments
This research was supported by National Institutes of Health grant R01-GM55321 and National Science Foundation grant DBI-0345205 to S.X.
References
- Broman, K. W., H. Wu, S. Sen and G. A. Churchill, 2003. R/qtl: Qtl mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Colinayo, V. V., J. H. Qiao, X. P. Wang, K. L. Krass, E. Schadt et al., 2003. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm. Genome 14: 464–471. [DOI] [PubMed] [Google Scholar]
- Efron, B., 2004. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J. Am. Stat. Assoc. 99: 96–104. [Google Scholar]
- Gelman, A., 2005. Analysis of variance - why it is more important than ever. Ann. Stat. 33: 1–31. [Google Scholar]
- Gelman, A., J. Carlin, H. Stern and D. Rubin, 1995. Bayesian Data Analysis. Chapman & Hall/CRC Press, New York.
- George, E. I., and R. E. Mcculloch, 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88: 881–889. [Google Scholar]
- Ghazalpour, A., S. Doss, B. Zhang, S. Wang, C. Plaisier et al., 2006. Integrating genetic and network analysis to characterize genes related to mouse weight. Plos Genet. 2: 1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Hastings, W. K., 1970. Monte-Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97–109. [Google Scholar]
- Hobert, J. P., and G. Casella, 1996. The effect of improper priors on Gibbs sampling in hierarchical linear mixed models. J. Am. Stat. Assoc. 91: 1461–1473. [Google Scholar]
- Hoeschele, I., and P. M. VanRaden, 1993. a Bayesian analysis of linkage between genetic markers and quantitative trait loci. I. Prior knowledge. Theor. Appl. Genet. 85: 953–960. [DOI] [PubMed] [Google Scholar]
- Hoeschele, I., and P. M. VanRaden, 1993. b Bayesian analysis of linkage between genetic markers and quantitative trait loci. II. Combining prior knowledge with experimental evidence. Theor. Appl. Genet. 85: 946–952. [DOI] [PubMed] [Google Scholar]
- Hubner, N., C. A. Wallace, H. Zimdahl, E. Petretto, H. Schulz et al., 2005. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 37: 243–253. [DOI] [PubMed] [Google Scholar]
- Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis et al., 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Ishwaran, H., and J. S. Rao, 2003. Detecting differentially expressed genes in microarrays using Bayesian model selection. J. Am. Stat. Assoc. 98: 438–455. [Google Scholar]
- Ishwaran, H., and J. S. Rao, 2005. Spike and slab gene selection for multigroup microarray data. J. Am. Stat. Assoc. 100: 764–780. [Google Scholar]
- Jansen, R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C. H., Z-B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendziorski, C. M., M. Chen, M. Yuan, H. Lan and A. D. Attie, 2006. Statistical methods for expression quantitative trait loci (eqtl) mapping. Biometrics 62: 19–27. [DOI] [PubMed] [Google Scholar]
- Lan, H., J. P. Stoehr, S. T. Nadler, K. L. Schueler, B. S. Yandell et al., 2003. Dimension reduction for mapping mRNA abundance as quantitative traits. Genetics 164: 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, H., M. Chen, J. B. Flowers, B. S. Yandell, D. S. Stapleton et al., 2006. Combined expression trait correlations and expression quantitative trait locus mapping. PloS Genet. 2: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis, N., A. W. Rosenbluth, M. N. Rosenbluth, A. H. Teller and E. Teller, 1953. Equation of state calculations by fast computing machines. J. Chem. Phys. 21: 1087–1092. [Google Scholar]
- Miller, R. A., J. M. Harper, A. Galecki and D. T. Burke, 2002. Big mice die young: early life body weight predicts longevity in genetically heterogeneous mice. Aging Cell 1: 22–29. [DOI] [PubMed] [Google Scholar]
- Mitchell, T. J., and J. J. Beauchamp, 1988. Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83: 1023–1036. [Google Scholar]
- Naggert, J. K., L. D. Fricker, O. Varlamov, P. M. Nishina, Y. Rouille et al., 1995. Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase-e mutation which reduces enzyme-activity. Nat. Genet. 10: 135–142. [DOI] [PubMed] [Google Scholar]
- Newton, M. A., A. Noueiry, D. Sarkar and P. Ahlquist, 2004. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics 5: 155–176. [DOI] [PubMed] [Google Scholar]
- Pan, W., 2002. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics 18: 546–554. [DOI] [PubMed] [Google Scholar]
- Qu, Y., and S. Xu, 2006. Quantitative trait associated microarray gene expression data analysis. Mol. Biol. Evol. 23: 1558–1573. [DOI] [PubMed] [Google Scholar]
- Raftery, A. E., and S. M. Lewis, 1992. One long run with diagnostics: implementation strategies for Markov chain Monte Carlo. Stat. Sci. 7: 493–497. [Google Scholar]
- Rao, S. Q., and S. Xu, 1998. Mapping quantitative trait loci for categorical traits in four-way crosses. Heredity 81: 214–224. [DOI] [PubMed] [Google Scholar]
- Rubin, D. B., 1987. Multiple Imputation for Nonresponse in Surveys. John Wiley & Sons, New York.
- Schadt, E. E., S. A. Monks, T. A. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehr, J. P., J. E. Byers, S. M. Clee, H. Lan, I. V. Boronenkov et al., 2004. Identification of major quantitative trait loci controlling body weight variation in ob/ob mice. Diabetes 53: 245–249. [DOI] [PubMed] [Google Scholar]
- Tusher, V., R. Tibshirani and C. Chu, 2001. Significance analysis of microarrays applied to ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Y. M. Zhang, X. M. Li, G. L. Masinde, S. Mohan et al., 2005. Bayesian shrinkage estimation of quantitative trait loci parameters. Genetics 170: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert, G., R. B. Brem, J. Whittle, J. M. Akey, E. Foss et al., 2003. Trans-acting regulatory variation in saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., M. T. Wells, C. D. Smart and W. E. Fry, 2005. Bayesian normalization and identification for differential gene expression data. J. Comput. Biol. 12: 391–406. [DOI] [PubMed] [Google Scholar]