Abstract
We have determined by reverse Southern analysis and direct sequence comparisons that most of the dumpy gene has evolved in the dipteran and other insect orders by purifying selection acting on amino acid replacements. One region, however, is evolving rapidly due to unequal crossing over and/or gene conversion. This region, called “PIGSFEAST,” or PF, encodes in D. melanogaster 30–47 repeats of 102 amino acids rich in serines, threonines, and prolines. We show that the processes of concerted evolution have been operating on all species of Drosophila examined to date, but that an adjacent region has expanded in Anopheles gambiae, Aedes aegypti, and Tribolium castaneum, while the PF repeats are reduced in size and number. In addition, processes of concerted evolution have radically altered the codon usage patterns in D. melanogaster, D. pseudoobscura, and D. virilis compared with the rest of the dumpy gene. We show also that the dumpy gene is expressed on the inner surface of the micropyle of the mature oocyte and propose that, as in the abalone system, concerted evolution may be involved in adaptive changes affecting Dumpy's possible role in sperm–egg recognition.
IN recent years, sequence comparisons of orthologous genes and the use of programs designed to identify amino acid sites undergoing positive selection have identified a subset of rapidly evolving genes. In this subset are gene products that are involved in fertilization (reviewed by Swanson and Vacquier 2002). One example is the thick extracellular coat that surrounds all mammalian eggs, known as the zona pellucida (ZP), which is a specialized extracellular matrix that acts as a barrier for cross-species fertilization and is modified postfertilization to prevent multiple sperm from entering the same egg (Wassarman et al. 2001; Wassarman 2002). The zona pellucida is composed of ZP-domain-containing proteins, which form the sperm receptor. Surprisingly, despite being involved in such a fundamental process, these sperm receptor proteins are undergoing rapid and presumably adaptive evolution (Swanson and Vacquier 2002). This has led to suggestions that rapid coevolution of proteins involved in sperm and egg interactions may be important for reproductive isolation during speciation. The most striking example of this is the coevolution of another ZP domain protein, the abalone sperm receptor along with its ligand lysin. This sperm receptor has 22 tandem vitelline envelope receptors for lysin (VERL) repeats (150 aa each) N-terminal to its ZP domain (Galindo et al. 2002), and it is these repeats that bind to the sperm lysin. A mutation may occur in any one of these 22 consecutive VERL repeats. This change may be tolerated because of the redundancy among the repeats such that the mutations do not have significant effects on fitness. However, a mutation can spread through the array by gene conversion or unequal crossing over. This process, known as concerted evolution, acts to standardize the repeats within an organism. This creates a selective pressure on the sperm lysin to adapt to a constantly varying sperm-binding protein, and this coevolution between the sperm and egg may contribute to speciation among the abalone group (Swanson and Vacquier 1998).
We have previously identified in Drosophila melanogaster a ZP-domain-encoding gene of exceptionally large size known as dumpy, which is predicted to encode a 2.5-MDa protein (Wilkin et al. 2000). The dumpy gene encodes a membrane-inserted protein with a short cytoplasmic tail and an enormous extracellular region predicted to extend ∼1 μm in length. The latter consists of a membrane proximal ZP domain preceded N terminally by 308 epidermal growth factor (EGF) repeats, interspersed with 185 copies of a novel four-cysteine module that we call dumpy (DPY), consisting of 21 amino acids. Like other ZP domain proteins, Dumpy has a conserved Furin cleavage site between its ZP domain and transmembrane sequence, potentially allowing for the release of the extracellular domain from the rest of the protein. Dumpy has been shown to organize the chitinous layer of the cuticle. In its absence, there are defects in the innermost layer of the apical extracellular matrix (ECM), which detaches from the underlying epidermis (Bokel et al. 2005). Another ZP domain protein, Piopio, which is most similar to Endoglin (Jazwinska et al. 2003), has been shown to cooperate in this task with both Dumpy and Papillote, the latter containing a partial ZP domain (Bokel et al. 2005). Dumpy and Piopio also work together to organize the ECM in the developing trachea to allow proper reorganization of the cells into unicellular wide tubes (Jazwinska et al. 2003).
When the amino acid sequence of Dumpy from D. melanogaster was deduced from partial cDNA and genomic sequences, it was found to contain two additional repeated regions, one encoded by ∼4 kb, called the proline-rich (PR) region. The PR repeats are approximately two-thirds of the way from the amino terminus of the protein. The second repeated region is nearer the amino terminal end of the protein and consists of >30 repeats of 102 amino acids encoded by ∼14 kb of intron-less genomic DNA. These we call PIGSFEAST (PF) repeats since the single letter code for the 102 amino acids contains these two “words.” The PF sequence is very rich in serines and threonines and is likely to possess very little globular structure. We have proposed that the PR and PF repeats provide flexibility and/or elasticity to the protein in its role in cell–cell and cell–cuticle adhesion (Wilkin et al. 2000).
In this article, we show that the PF region of the Dumpy protein is evolving rapidly in different insects especially when compared with other domains in the protein. We determine that this region, like that of the VERL repeats of the abalone sperm receptor protein, is evolving by concerted evolution in several species from the genus Drosophila. We also show that a neighboring region is undergoing the same process in the homologous gene from Anopheles gambiae, which last shared a common ancestor with D. melanogaster ∼240 MYA (Yeates and Weigmann 1999; Weigmann et al. 2003). We also demonstrate that unequal crossing over has been and still is the driving force behind both sequence homogenization and Dumpy repeat expansion/contraction in these insects and that the repeats appear to be evolving under weak purifying selection. Furthermore, we show that Dumpy is strongly expressed in the developing micropyle, a tube structure through which sperm enters to fertilize the egg. Dumpy is therefore in a key location to act as a sperm receptor, making the parallels between Dumpy and the abalone sperm receptor system quite remarkable.
MATERIALS AND METHODS
Recovery of sequence data:
The D. melanogaster sequences used in the BLAST searches were obtained from the genomic sequence of dumpy in GenBank (CG33196, release 4.2.1, September 2005). Genomic DNA sequences from D. erecta (scaffold_4929, freeze 1), D. pseudoobscura (4_group 3, release 2.0, October 2005), D. virilis (scaffold_12963, freeze 1 assembly), D. ananassae (scaffold_12943 and scaffold_3099, freeze 1 assembly), D. persimilis (CH479187.1, gi:80982435), D. willistoni (scaffold_180703, freeze 1 assembly), D. mojavensis (scaffold_6500, 6148 and 6127 freeze 1 assembly), D. grimshawi (scaffold_15126, freeze 1 assembly), An. gambiae (AAAB0108980, gi:19612245), Aedes aegypti (supercontig_1.757), and Tribolium castaneum (AAJJ01000037.1, gi:73486610) were scanned with a BLAST search using parts of the D. melanogaster dumpy gene sequence surrounding the PF repeats as described below. The sequences from these species' dumpy genes were aligned using the Sequencher version 4.2 program or DNASTAR's Lasergene version 6 package, and the PF-like repeats were identified in the intervening region between the BLAST alignments. As shown below, the PF sequences from outside the melanogaster subgroup, except for D. pseudoobscura and D. persimilis, have evolved so rapidly that they cannot be aligned with those from D. melanogaster. The PF repeats in other melanogaster subgroup species can be aligned inter se, however.
PCR analyses of PF repeats from melanogaster subgroup species:
Isofemale lines or stocks of D. simulans, D. yakuba, and D. mauritiana were kindly provided by Charles Aquadro. Cultures of D. orena (stock no. 14021-0245.0), D. sechellia (14021-0248.1), D. takahashii (14022-0311.4), D. ananassae (14024-0371.0), and D. kikkawai (14028-0561.0) were obtained from the Drosophila species stock center at the University of Arizona, Tucson, Arizona.
Genomic DNA was prepared from 20–25 flies from each species using the Puregene kit from Gentra (Research Triangle Park, NC) systems. PCR was carried out using standard protocols and the Pfu turbo polymerase from Stratagene (La Jolla, CA). Degenerate primers or the primers specific for the D. melanogaster PF repeat, PIGS 1–4 (Table 1), were used initially to recover amplicons, which were then cloned and sequenced using the Invitrogen (San Diego) TOPO TA cloning kit and Cornell University's DNA sequencing facility. Species-specific primers were also designed and used to obtain additional PIGSFEAST repeats from the melanogaster subgroup species analyzed below. The degenerate and species-specific primers are in Table 1, in which the odd- and even-numbered primers were paired and used in the PCRs.
TABLE 1.
Primer sequences (5′–3′)
| Degenerate primers | PFEASTP | CCNTTYGARCGNWSNACNCC |
| DOTTEST | GTNSWYTCNGTNGTYTGRTC | |
| Species-specific primers | ||
| D. melanogaster | PIGS-1 | TCTCCATCCGAAACTCCTGA |
| PIGS-2 | TCTGAAGGTAATGTTGTGGGTGTC | |
| PIGS-3 | TTGTTCTGTCACTTGCCCACC | |
| PIGS-4 | CCGAGCGAAGTCAGAAC | |
| D. sechellia | SECHSP-1 | TTGAACCGATTGGAACATTT |
| SECHSP-2 | ACGTGGGCAAGTGACAAGA | |
| D. yakuba | YAKUBASP-1 | TACGCACAACAAACGACTGAATCT |
| YAKUBASP-2 | GAAGGTAATGTTGTGGGTGTCTCC | |
| YAKUBASP-3 | ACCACCCCAAATGTTCTGAT | |
| YAKUBASP-4 | GCCCTTAGTGGTCTGGTCGTC | |
| D. orena | ORENASP-1 | CGTTGTGGGCGTCTCAG |
| ORENASP-2 | GTGGCGGAACAAACTACC | |
| ORENASP-3 | GTACAACCCGTGGACAAGTGG | |
| ORENASP-4 | TGGGTAGATGCCTCAAATGGT |
With the degenerate primers or with PIGS 1–4 on genomic DNA from other Drosophila species (i.e., not from D. melanogaster), following a 1-min denaturation at 94°, the annealing temperature was generally 48° for 30 sec with an extension for 1–2 min at 72° for 30 cycles. With species-specific primers, the annealing temperature varied from 50° to 55°, depending upon the particular species' genomic DNA.
Gel analysis of PF repeat numbers in different strains of D. melanogaster:
Genomic DNA was extracted from single flies or groups of 25 flies and restricted with HindIII or HphI (600 ng were digested for multiple fly preparations). As shown below, the PF repeats do not contain either restriction site, and the flanking restriction sites are conserved across the strains that we analyzed. Gel electrophoresis was carried out in 0.35% gels using Seakem gold agarose. Prior to denaturation and blotting, the gels were rinsed in 0.06 m HCl for 15 min. The filters were probed with a radiolabeled 1.3-kb EcoRI fragment (subclone 9B) from an insert from the PF region obtained in our chromosome walk through the dumpy gene (see Wilkin et al. 2000). This fragment contains only PF repeats. The washed filters were exposed to X-ray film for 1–3 days (multiple-fly extracts) or for 1 week (single-fly extracts).
Reverse Southern analyses:
Genomic DNA for reverse Southern analyses was prepared from frozen insects, viz. D. pseudoobscura, D. mettleri, the housefly Musca domestica, the honeybee Apis mellifera, and the Colorado potato beetle Leptinotarsa decemlineata, generously provided by James Fogleman, Jeff Scott, or Rick Roush. A total of 2.5 μg of DNA was radiolabeled with P32 by random primer extension at 37° for 6 hr and hybridized overnight to nylon filters blotted from gels containing the following plasmid subclones from the dumpy walk (Wilkin et al. 2000) and restricted with the indicated enzymes: 13F(XhoI and EcoRI); 5D (SacI and HindIII); 9A, 9B, and 9C (EcoRI); 26A (HindIII and EcoRI); 14A (EcoRI); and 56D (SalI and EcoRI). A total of 1 μg of DNA from each digestion or double digestion was loaded on each gel. The filters were exposed to X-ray film for 5, 22, or 33 days, depending upon the species DNA used as the probe.
Analytical methods:
We used the MEGA3 software program (Kumar et al. 2004) for the analyses of the PF sequences and the calculation of dN and dS values. The neighbor-joining (NJ) tree building method in that program was also used to infer insect species phylogenies or PF repeat clusters using the sequences obtained both in silico and experimentally. We also applied the McDonald–Kreitman test for selective neutrality (McDonald and Kreitman 1991) to the PF repeats from D. melanogaster and D. simulans using the DnaSP program, version 4.10.4.
In situ hybridization:
A digoxigenin-labeled RNA probe derived from near the C-terminal end of dumpy (see Figure 1a) was synthesized according to the manufacturer's instructions (Roche). Ovaries were dissected and the sheath removed in PBS + 0.1% Tween 20 (PBS-Tw) and fixed in 4% formaldehyde in PBS for 30 min at room temperature. These were rinsed in PBS-Tw and then transferred to ice-cold 100% methanol and stored at −20° for at least 24 hr. The in situ hybridization procedure was carried out on whole mounts as described in Cornell et al. (1999) using a hybridization temperature of 70° and detection with an alkaline-phosphatase-labeled antidigoxigenin antibody (Roche).
Figure 1.—
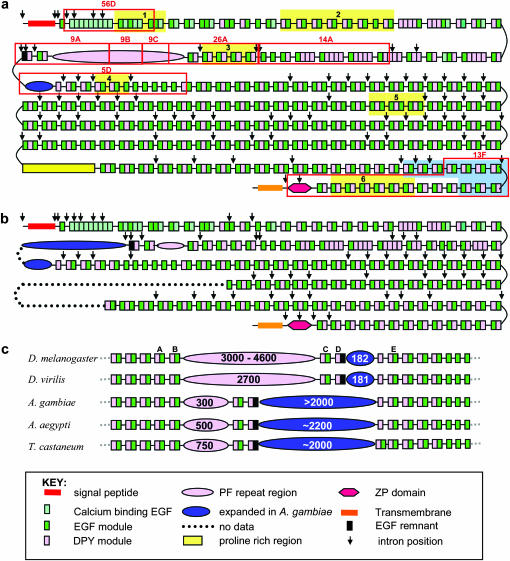
Comparison of the complete modular structure of D. melanogaster with that of An. gambiae, as well as a comparison across insects of the modular structure in the vicinity of the PF region. (a) The modular structure of D. melanogaster Dumpy is adapted from Wilkin et al. (2000). The majority of Dumpy is extracellular and composed of EGF domains (green rectangles) and a novel 21-amino-acid, four-cysteine module DPY (pink rectangles). Also marked as a black rectangle is a remnant EGF domain. Near its C terminus there is a single ZP domain depicted with a red hexagon. There are two main regions of low complexity sequence, one near the N terminus, shown in pink and referred to as the PF repeat region, and a second much nearer to the C terminus that is the proline-rich region. Shown as a blue ellipsoid is a charged proline-, serine-, threonine-rich spacer region. (b) The predicted modular structure of An. gambiae is basically identical to that of D. melanogaster in the region sequenced, except in the locality of the PF region, which has contracted, and in the large spacer region, which has expanded. (c) An enlargement of the vicinity of the PF region from D. melanogaster, D. virilis, An. gambiae, Ae. aegypti, and T. castaneum; numbers indicate the numbers of amino acids in the PIGSFEAST region. The modular structure of the DPY and EGF domains is conserved (except in T. castaneum, which has an extra EGF module following the region that is expanded in An. gambiae). In Drosophila, the PF region is expanded, and the adjacent spacer region is not, whereas in the mosquito and the red flour beetle, the reverse is true. In a, the regions demarcated by red boxes are the positions of subclones used for reverse Southern blots. The areas highlighted in yellow are utilized in Table 2 to assess the conservation of the dumpy gene across species. The region highlighted in blue marks the position of digoxygenin-labeled RNA probe used to detect dumpy in ovaries. In c, the positions of the DPY–EGF domains are marked to confirm the organization of the PF region in divergent species as discussed in the results.
RESULTS
The dumpy gene is conserved in insect evolution:
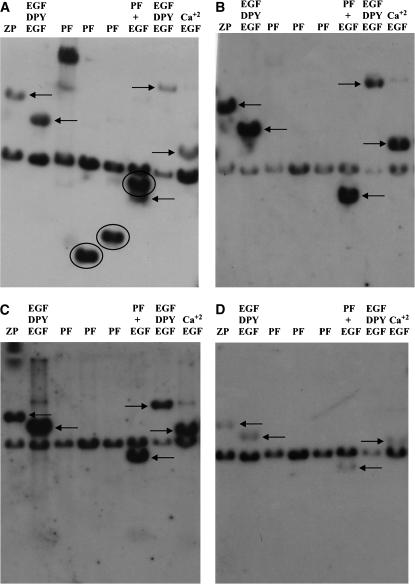
We probed digested plasmid subclones from the chromosomal walk through the gene (Wilkin et al. 2000) with radiolabeled genomic DNA from several different insect species at low stringency (50°–55°) (see Figure 1a for the locations of the subclones). The subclones were 13F, whose cloned fragment encodes the ZP domain at the C terminus of the protein as well as 17 EGF and DPY modules; 5D, which encodes a series of EGF motifs; 9A, 9B, and 9C from the PF array; 26A, which contains N-terminal PF repeats and adjacent EGF domains; 14A, which codes for triplet motifs of EGF and DPY motifs; and 56D, which encodes primarily calcium-binding EGF modules. The results of several of the reverse Southern analyses in Figure 2, A–D, show that the ZP domain and several EGF-containing motifs appear to be conserved in other Drosophila species, in the more distantly related dipteran M. domestica (data not shown), and in the coleopteran species L. decemlineata (the Colorado potato beetle). In contrast, the PF sequences, except in the sibling species D. simulans (Figure 2A), are apparently evolving so rapidly that they no longer hybridize with sequences encoding PF from D. melanogaster.
Figure 2.—
Reverse Southern analysis of dumpy gene evolution. Restriction enzyme fragments from the dumpy chromosomal walk (Wilkin et al. 2000) encoding different domains in the protein and subcloned in the plasmid vector, pBluescript (Stratagene), were subjected to electrophoresis and blotted to nylon filters as described in materials and methods. The subclones and their encoded domains are as follows: lane 1, subclone 13F encoding the zona pellucida domain; lane 2, subclone 5D encoding EGF–DPY–EGF modules; lanes 3–5, subclones 9A, 9B, and 9C encoding PF repeats; lane 6, subclone 26A encoding PF repeats and several adjacent EGF modules; lane 7, subclone 14A encoding EGF and DPY repeats; and lane 8, subclone 56D encoding predominately calcium-binding EGF domains. A filter was probed with radiolabeled genomic DNA from D. simulans (A), D. pseudoobscura (B), D. mettleri (C), and the Colorado potato beetle L. decemlineata (D). In each case, the 2.9-kb band containing the plasmid DNA hybridizes to contaminating bacterial DNA in the insect genomic DNA used as the probe. PF-containing sequences are detected only by D. simulans genomic DNA. Circled hybridizing bands contain PIGSFEAST repeats. Arrows point to hybridizing bands containing other parts of the dumpy gene indicated above each lane. Hybridizing bands, although very faint, are indicated in D by arrows. These specify, from left to right, the ZP domain (13F), EGF–DPY–EGF triplet motifs (5D and 26A), and calcium-binding EGF modules (56A).
Conserved homologs of the dumpy gene can be identified within the genomic sequences of all insects sequenced so far, although the full dumpy sequence is rarely available. This includes all the sequenced Drosophila species (D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi), the mosquitoes (both An. gambiae and Ae. aegypti), as well as the silkworm, honeybee, and red flour beetle (T. castaneum). We analyzed in detail the partial sequence of dumpy from An. gambiae, and Figure 1b shows that its domain structure and organization is very similar to that of D. melanogaster. Comparison of the spacing between the cysteines in the first EGF repeat within arrays of EGF–DPY–EGF units showed it to be almost identical between these two species and indicates that there has not been recent shuffling or rearrangements of these exons (supplemental Table 1 at http://www.genetics.org/supplemental/).
Amino acid sequence alignments across several different regions of the dumpy gene, indicated as A–E in Figure 1c (intron positions for D. melanogaster and An. gambiae are shown in Figure 1, a and b, respectively), display a high degree of identity among the different sequenced insects (Table 2). A and B are quite conserved; those in C are much less so. In between the short proline, threonine-rich spacer region, ∼182 amino acids in length (shown as D in Figure 1c), there are two exons that encode a DPY, and a remnant EGF domain that appears to be homologous. The region that is expanded in An. gambiae ends in a DPY motif in all species except the red flour beetle where there is also an EGF module. These DPY modules are not well preserved. In contrast, the DPY and EGF modules encoded by the following exon are highly conserved (E in Figure 1c). This indicates that the large, repetitive region in An. gambiae is not the equivalent of the D. melanogaster PF region, but is an expansion of a neighboring region C terminal to PIGSFEAST.
TABLE 2.
Percentage of amino acid identity in six regions of Dumpy between D. melanogaster and eight insect species
| Species | 1 | 2 | 3 | 4 | 5 | 6 | mean |
|---|---|---|---|---|---|---|---|
| D. simulans | 100 | 98a | 96 | NA | 100 | 99 | 99 |
| D. sechellia | 100 | 98 | NA | 98 | NA | 98 | 98 |
| D. yakuba | 99 | 98 | 98 | 98 | 99 | 98 | 98 |
| D. pseudoobscura | 91 | 90 | 91 | 91 | 91 | 88 | 90 |
| D. virilis | 90 | 83 | 91 | 88 | 89 | 84 | 87 |
| An. gambiae | 65 | 56 | 58 | 68 | 67 | 64 | 62 |
| Ae. aegypti | 68 | 58 | 62 | 74 | 70 | 66 | 65 |
| T. castaneum | 66 | 59 | 56 | 68 | 66 | 60 | 62 |
The percentage identity across six different regions of the D. melanogaster Dumpy protein with that of the translated genomic sequence from several different insect species. The regions chosen are shown in Figure 1a.
Sequence not available for certain regions of the locus, shown as NA.
In D. melanogaster, the PF region consists of 40 repeats and, as mentioned above, is flanked by DPY–EGF units at its N and C termini. Sequence analysis of other Drosophila species shows that the PF repeat region is normally >2000 amino acids long (D. erecta is 2100 aa, D. ananassae is >2500 aa, D. willistoni is 4086 aa, D. virilis is 2721 aa, and D. grimshawi is 2300 aa). In An. gambiae, however, the region expected to contain PF repeats, although clearly defined by the conservation of the adjacent domains (see Figure 1c), is considerably diverged. In addition, this region is considerably shorter than in D. melanogaster, being only 296 amino acids long. In contrast, the above-mentioned C-terminal spacer region, which is short in D. melanogaster, is much enlarged in An. gambiae into a highly repetitive structure of >2000 amino acids. We found a similar structure in Ae. aegypti and T. castaneum (Figure 1c). The unusual nature of this part of the protein prompted a closer investigation of the evolution of the PF-containing region.
The PF region within dumpy from D. melanogaster is evolving by unequal crossing over:
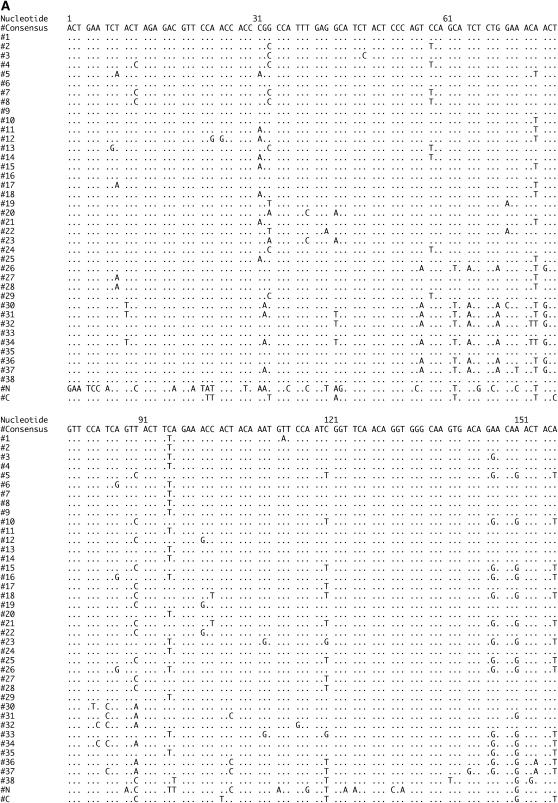
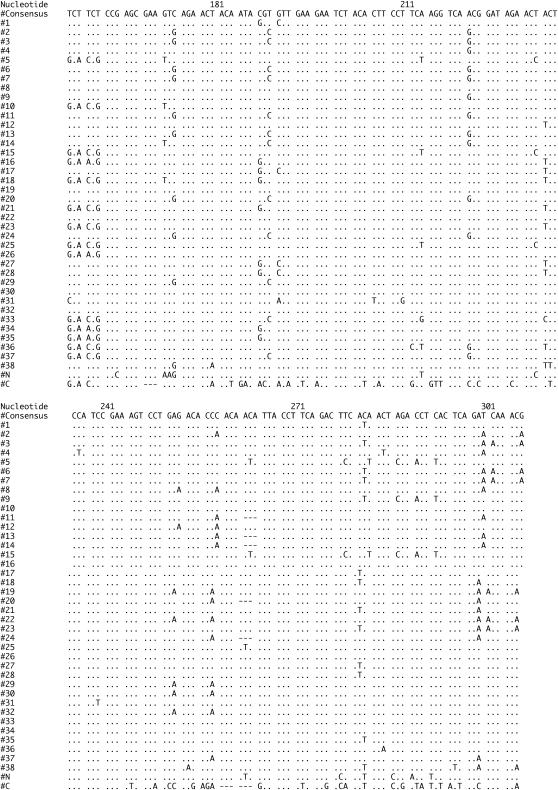
The sequence through the PIGSFEAST region in the D. melanogaster dumpy gene from release 3.1 consists of 40 almost identical repeats of either 303 or 306 nt. These repeats encode a proline-, serine-, and threonine-rich region that also contains several charged residues. Approximately 25% of the amino acid positions in each repeat is predicted to be suitable for O-linked glycosylation (Julenius et al. 2005). The consensus nucleotide and amino acid sequences of the 38 internal repeats are presented in Figure 3, A and B, along with variable sites where the repeats differ in sequence from the consensus. Also presented at the bottom of Figure 3, A and B, are the more highly diverged sequences from the N- and C-terminal repeats in the PF array.
Figure 3.—
PF repeat sequences from D. melanogaster. Variations from the consensus listed at the top in nucleotides (A) and amino acids (B) for each repeat in the array. The more diverged N- and C-terminal sequences are shown at the bottom whereas the internal 38 repeats are shown in their order within the array in the first column.
A possible origin of such a tandem repeat structure is the process of unequal crossing over at meiosis or in premeiotic germ cells. Two characteristic features have been shown to be diagnostic of this process: McAllister and Werren (1999) clearly showed that the termini of repeat arrays diverge in sequence as unequal crossing over makes internal repeats more similar, and Durfy and Willard (1989) experimentally showed that adjacent repeats within the primate α-satellite are more similar in sequence because of the exchange events within the array. We have calculated the nonsynonymous and synonymous differences per each kind of site, dN and dS, between all the internal repeats and between each pair of adjacent repeats within the array from D. melanogaster. Also, dN and dS values were calculated for each of the terminal repeats vs. the internal repeats. These values are shown in Table 3. Several significant features are apparent in these comparisons. First, the N- and C-terminal repeats are divergent as evidenced by the three- to five-fold higher dN and dS values obtained when the end repeats are compared with the internal repeats. Second, the average dN and dS values between all the internal repeats are higher than those obtained when adjacent repeats are compared. These two observations strongly indicate that unequal crossing over has been responsible for the evolution of the PF array in D. melanogaster. This is in contrast to the evolution of other repeat regions within dumpy, which appear to have arisen by duplication of gene segments. For example, we have previously demonstrated the existence of a super-repeat structure consisting of six tandem EGF–DPY–EGF units, which has apparently been duplicated at least 12 times (Wilkin et al. 2000).
TABLE 3.
dN and dS values for PIGSFEAST repeats from D. melanogaster
| Repeat | Average dN |
|---|---|
| N-terminal repeat to all internal repeats | 0.131 |
| C-terminal repeat to all internal repeats | 0.171 |
| Adjacent repeats | 0.038 |
| Internal repeats | 0.043 |
| Repeat | Average dS |
| N-terminal repeat to all internal repeats | 0.247 |
| C-terminal repeat to all internal repeats | 0.311 |
| Adjacent repeats | 0.095 |
| Internal repeats | 0.102 |
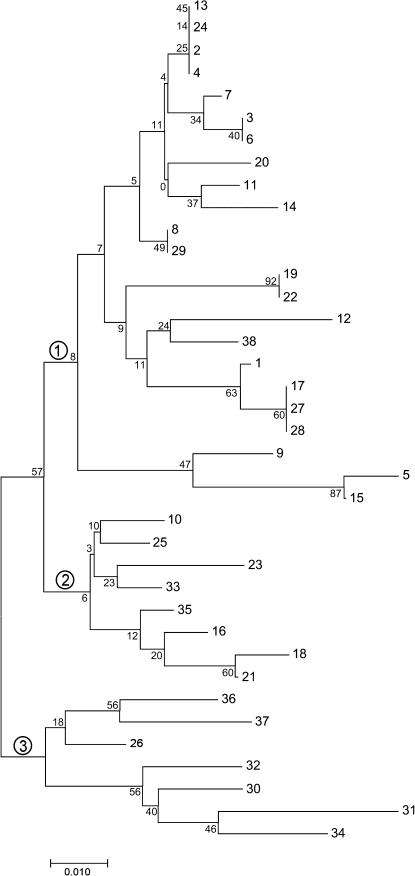
We constructed an NJ tree of the 38 internal PF repeats to trace the evolution of the PF array within the D. melanogaster lineage. The tree is shown in Figure 4. This approach was used by Desseyn et al. (2000) to cluster 73 related repeats in a tandem array in the human mucin gene, MUC5B. There are three major clusters of PF repeats, with cluster 1 consisting primarily of repeats from the N-terminal region of the array (hence with lower numbers). Cluster 2 contains mostly internal repeats, and cluster 3 includes repeats near the C terminus. This would be expected if the mispairing events preceding the unequal crossovers were in general only slightly out of register, i.e., by one or only a few repeats. The bootstrap values shown in Figure 4 indicate strong support (57%) for a closer relationship between the repeats in clusters 1 and 2 vs. those in cluster 3. When C- and N-terminal repeats or D. simulans repeats are added as outgroup sequences, similarly strong support (55%) is seen for cluster 3 (data not shown). There are several anomalies in the cluster patterns, however. For example, repeats 8 and 29 are nearly identical and repeat 38 is in the “N terminal” cluster. This could be due to a very “out of register” mispairing followed by a crossover, or perhaps to a recent gene conversion event.
Figure 4.—
Neighbor-joining tree of the nucleotide sequences of the PIGSFEAST repeats from D. melanogaster. The numbers refer to the position of each repeat in the array, with number 1 being the most N-terminal internal repeat and 38 being the most C-terminal internal repeat. The divergent N- and C-terminal repeats are not included in this analysis. The circled numbers refer to the three major clusters of repeats discussed in the results.
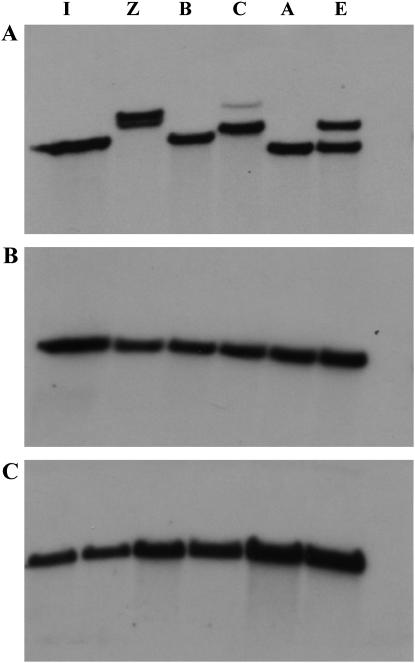
Another indication that unequal crossing over is operating on the PF repeats is their variable numbers in different geographic strains of D. melanogaster. We restricted genomic DNA from eight different strains with either HindIII or HphI, which cut outside and/or just inside the array; HindIII cuts 848 and 1037 nt away from the N- and C-terminal ends, respectively, and HphI, 9 and −86 nt from the two ends. The fragments were separated on low-percentage agarose gels, blotted and probed with a cloned PF repeat. Figure 5 shows the patterns of the hybridizing band or bands in six different strains. Size standards (not shown) indicate that Australia has the smallest number of repeats, ∼30, and Zimbabwe the most, 46–47. Note in Figure 5A that three strains, Zimbabwe, California, and Ecuador, appear to be polymorphic for length variants of the PF array. B and C in Figure 5 show that the size differences between PF-hybridizing fragments are not due to polymorphisms in the flanking restriction sites. The same filter was stripped and reprobed first with a subclone containing the HindIII fragment adjacent to the N-terminal side of the PF array (B) and then with subclone 5D, which contains the HindIII fragment on the C-terminal side of the array (C).
Figure 5.—
PIGSFEAST repeat number variation in strains of D. melanogaster. Genomic DNA from flies from Israel (I), Zimbabwe (Z), Beijing (B), California (C), Australia (A), and Ecuador (E) were digested with HindIII, subjected to electrophoresis in 0.35% agarose gels and blotted, and the filter was probed with subclone 9C (A); 3A, which is a HindIII fragment from subclone 26A (B); and 5D (C) from the chromosome walk through the dumpy gene (Wilkin et al. 2000). Subclone 9C contains only PIGSFEAST repeats whereas 3A and 5D contain the N- and C-terminal flanking HindIII fragments on either side of the PIGSFEAST array.
Concerted evolution of Dumpy PIGSFEAST repeats in other Drosophila species:
To determine which species have detectable PF repeats and estimate their rates of evolutionary change, we used the degenerate primers, or PIGS 1–4 shown in Table 1, to amplify contiguous repeats from the species' arrays. We obtained PCR products from D. simulans, D. sechellia, D. mauritiana, D. yakuba, and D. orena, but not D. ananassae, D. takahasii, and D. kikkawai despite repeated attempts at varying conditions. The latter are species that are in the melanogaster species group, but not the subgroup of the same name. Genomic Southerns containing DNA from these three species, probed with cloned PF repeats from D. melanogaster, showed that only D. takahashii contains hybridizing PF-like sequences (data not shown).
We relied on divergence within the PF arrays from the species of the melanogaster subgroup to prevent the binding of the primers to each adjacent PF repeat. Hence, we could amplify and clone multimers of the repeats, as shown in Figure 6 for D. simulans and D. yakuba. Here the products were blotted to a filter and probed with a cloned PF repeat from D. melanogaster. PCR products with six tandem repeats can be readily detected in D. simulans. In addition, the size of the D. simulans monomer appears to be smaller than that of D. melanogaster, which was verified by subsequently obtained sequence data. The most intense band in the D. yakuba lane is 2.5 repeats caused by one of the primers apparently annealing in the middle of a monomer. This approach allowed us to clone and sequence adjoining repeats to assess diversity within each species array. For example, using the degenerate primers, we obtained 16 full repeats and 22 half repeats from D. simulans, 6 full repeats and 32 half repeats from D. mauritiana, and 3 full and 34 half repeats from D. yakuba. The total numbers of repeats cloned and analyzed for each species are listed in Table 4. We could also then design the species-specific primers shown in Table 1 and use them to amplify larger numbers of monomers and multimers. The sequences from these repeats are also included in the diversity estimates presented below. In addition, we were able to analyze the genomic sequences for other sequenced Drosophila species, including D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi. The repeats from within a single species can be aligned, and the amount of intraspecific diversity in their arrays can be determined. Furthermore, the consensus amino acid sequences of the PF repeats from the melanogaster group, with the exception of D. ananassae, and the obscura group can be aligned (the alignments are shown in Figure 7). The PF-like sequences from the more distantly related species D. willistoni, D. mojavensis, D. virilis, and D. grimshawi cannot readily be aligned with those from the melanogaster subgroup. The consensus sequences for these species are also shown in Figure 7.
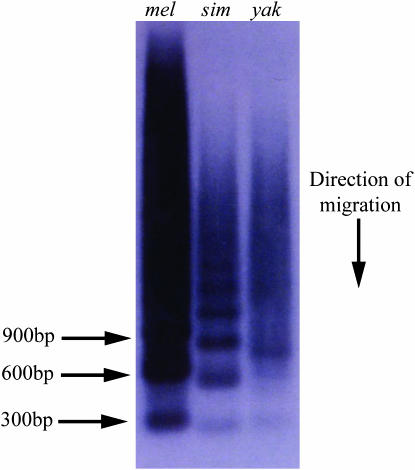
Figure 6.—
PIGSFEAST-containing PCR products generated from D. melanogaster (mel), D. simulans (sim), and D. yakuba (yak) genomic DNA with primers PIGS 1 and PIGS 2 (see Table 1). The products were separated by gel electrophoresis and blotted, and the filter was probed with subclone 9C from the dumpy chromosome walk. This subclone encodes only PF repeats. The ladder of hybridizing bands contains monomers (∼300 bp) and multimers of increasing numbers of repeats. Note the smaller size of the repeat in D. simulans and the prevalence of a multimer with 2.5 repeats in D. yakuba.
TABLE 4.
PF repeats analyzed from Drosophila and An. gambiae species
| Species | Full repeats analyzed | Partial repeats analyzed |
|---|---|---|
| D. melanogastera | 38 | |
| D. simulans | 16 | 22 |
| D. mauritiana | 6 | 32 |
| D. sechellia | 3 | 19 |
| D. yakuba | 3 | 34 |
| D. orena | 4 | 19 |
| D. erectaa | 18 | |
| D. ananassaea | 49 | |
| D. pseudoobscuraa | 17 | |
| D. persimilisa | 22 | |
| D. willistonia | 51 | |
| D. mojavensisa | 21 | |
| D. virilisa | 27 | |
| D. grimshawia | 16 | |
| An. gambiaea | 39 |
Obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/).
Figure 7.—
Consensus amino acid sequences. In D. melanogaster, the PF repeat appears to have arisen as a duplication of a more primitive repeat. The C-terminal half of this sequence is aligned beneath the N-terminal half of the D. melanogaster repeat and is labeled “c term.” There is a 14-amino-acid deletion in the PF repeat that has become fixed in D. simulans, D. sechellia, and D. mauritiana, and a 9-amino-acid deletion is fixed in the D. yakuba array. There are additional polymorphic additions and deletions not shown in the arrays from D. yakuba, D erecta, D. willistoni, D. mojavensis, and D. orena. Although the An. gambiae repeat is from a region adjacent to the Dumpy PF region, note that their amino acid compositions are similar.
The phylogenetic relationships between the species in the melanogaster subgroup are very well understood (Ashburner et al. 2005). We have examined their phylogeny with regard to several deletions that arose in certain lineages in the melanogaster subgroup and spread throughout the PF array. Thus, the 14-codon deletion arose and spread in a common ancestor of the mauritiana–simulans–sechellia clade. A deletion of 9 amino acids is fixed in all the PF repeats in D. yakuba, whereas a 6-codon deletion is polymorphic in the D. orena array. Note that the presence of all these codons in D. melanogaster must be the ancestral condition and identifies these indels as deletions. The latter conclusion is further confirmed by the presence of a D. melanogaster-sized PF repeat in D. erecta. Interestingly, the 101/102 amino acid PF repeat of D. melanogaster may have arisen from two tandem 51-amino-acid repeats, which can be aligned (Figure 7). This is also consistent with the short repeats of D. pseudoobscura and D. persimilis aligning with half of a D. melanogaster PF repeat.
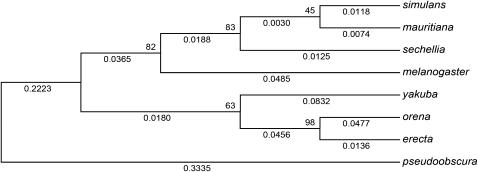
When the PF array consensus sequences from the different species are used to predict their phylogenetic relationships, the tree shown in Figure 8 is recovered. For this tree, the NJ algorithm was used, but qualitatively similar trees are obtained using the other algorithms in the MEGA3 program. PF nucleotide sequences, in general, provide a reasonably accurate view of evolution of the melanogaster subgroup species.
Figure 8.—
Phylogenetic analysis of PIGSFEAST repeats. Neighbor-joining tree of the melanogaster subgroup species generated from the dnucleotide values as described in the results. Bootstrap values are shown at each subsequent node.
The D. virilis and D. erecta arrays, like that of D. melanogaster, have diverged repeats at their ends. Indeed, two repeats at the N-terminal end are highly diverged in D. virilis in addition to a diverged single repeat at its C-terminal end. The genomic sequence from D. pseudoobscura contains only one end of the PF array and, here, also the final repeat is quite divergent. This suggests that unequal crossing over has also been involved in PF region evolution in these other Drosophila species. The dN and dS values calculated from within species repeat comparisons are shown in Table 5A. It is also possible to align the consensus sequences for the melanogaster subgroup species and that from D. pseudoobscura, allowing us to also calculate interspecific dN and dS values. The interspecific dN and dS values (Table 5B) are larger than the intraspecific values shown in Table 5A. This implies that processes of concerted evolution homogenize the repeats within the species from the melanogaster subgroup. Nenoi et al. (2000) “quantified” the effect of concerted evolution by estimating the ratio of the interspecific dS values to the mean of the two intraspecific dS values (i.e., dxy/[(dx + dy)/2], where x and y refer to the two species being compared). Since only weak selection appears to be operating on nonsynonymous sites (see below), we combined nonsynonymous and synonymous sites and calculated both intraspecific and interspecific differences as dnucleotide or simply d values as shown in Table 5C. Note the generally low d values for the intraspecific comparisons in Table 5C. This results in ratios of the interspecific d value to the mean of the intraspecific d values generally being >1, especially over longer phylogenetic distances, as seen in Table 5C. This indicates that the PF repeats have been evolving in a concerted fashion for at least 30 million years or since the divergence of D. pseudoobscura and the melanogaster subgroup species from a common ancestor. Indeed, it seems very likely, given the small dN and dS values for the within-species comparisons of the D. virilis repeats (Table 5A), that concerted evolution has been going on in the PF region of the dumpy gene since the origin of the genus.
TABLE 5.
dN and dS values for repeats within the PF regions analyzed and between concensus sequences from the melanogaster subgroup species, D. pseudoobscura, D. virilis, and An. gambiae
| A. Intraspecific comparisons | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| D. melanogaster | D. simulans | D. mauritiana | D. sechellia | D. yakuba | D. orena | D. erecta | D. pseudoobscura | D. virilis | An. gambiae | |
| dN | 0.043 | 0.029 | 0.037 | 0.042 | 0.093 | 0.052 | 0.148 | 0.128 | 0.099 | 0.021 |
| dS | 0.102 | 0.033 | 0.077 | 0.061 | 0.135 | 0.101 | 0.168 | 0.193 | 0.150 | 0.019 |
| dN/dS | 0.422 | 0.879 | 0.481 | 0.689 | 0.689 | 0.515 | 0.881 | 0.663 | 0.660 | 1.11 |
| B. Interspecific comparisons: dN values above the diagonal; dS values below the diagonal | ||||||||
|---|---|---|---|---|---|---|---|---|
| melanogaster | simulans | mauritiana | sechellia | yakuba | orena | erecta | pseudoobscura | |
| melanogaster | 0.063 | 0.063 | 0.069 | 0.146 | 0.144 | 0.128 | 0.371 | |
| simulans | 0.107 | 0.021 | 0.016 | 0.144 | 0.159 | 0.144 | 0.345 | |
| mauritiana | 0.107 | 0.013 | 0.016 | 0.120 | 0.148 | 0.134 | 0.342 | |
| sechellia | 0.094 | 0.053 | 0.040 | 0.126 | 0.154 | 0.139 | 0.354 | |
| yakuba | 0.253 | 0.187 | 0.172 | 0.187 | 0.133 | 0.112 | 0.370 | |
| orena | 0.247 | 0.173 | 0.159 | 0.173 | 0.234 | 0.045 | 0.375 | |
| erecta | 0.233 | 0.130 | 0.116 | 0.130 | 0.161 | 0.094 | 0.349 | |
| pseudoobscura | 0.563 | 0.619 | 0.627 | 0.598 | 0.623 | 0.612 | 0.596 | |
| C. Interspecific dnucleotide (d) values above the diagonal; intraspecific dnucleotide values underlined and on the diagonal | ||||||||
| melanogaster | simulans | mauritiana | sechellia | yakuba | orena | erecta | pseudoobscura | |
| melanogaster | 0.062 | 0.080 | 0.080 | 0.080 | 0.200 | 0.197 | 0.176 | 0.630 |
| simulans | 1.79 | 0.033 | 0.019 | 0.027 | 0.175 | 0.184 | 0.155 | 0.627 |
| mauritiana | 1.41 | 0.45 | 0.052 | 0.023 | 0.149 | 0.169 | 0.141 | 0.627 |
| sechellia | 1.43 | 0.65 | 0.45 | 0.050 | 0.159 | 0.179 | 0.151 | 0.627 |
| yakuba | 2.30 | 2.43 | 1.82 | 1.96 | 0.111 | 0.182 | 0.137 | 0.667 |
| orena | 3.03 | 3.57 | 2.78 | 3.03 | 2.02 | 0.069 | 0.061 | 0.667 |
| erecta | 1.49 | 1.50 | 1.25 | 1.35 | 0.96 | 0.50 | 0.174 | 0.613 |
| pseudoobscura | 5.55 | 6.25 | 5.88 | 5.88 | 4.83 | 5.88 | 3.63 | 0.164 |
dxy/[(dx + dy)/2] values are below the diagonal where x and y refer to two different species.
There are, however, several interesting exceptions in Table 5C, viz. the comparisons among the three closely related species D. simulans, D. mauritiana, and D. sechellia and the comparison between D. orena and D. erecta. Here the interspecific differences are smaller than the intraspecific d values. Apparently mutational variants accumulated in a common ancestor of the three species from the simulans clade and were fixed by the process of concerted evolution, but the subsequent and very recent speciation events have precluded the addition of any substantial lineage-specific mutations. The low interspecific dnucleotide value for D. orena and D. erecta is remarkable, given that these two species are thought to have diverged ∼7 MYA (Tamura et al. 2004).
We also analyzed the PF region in some other non-Drosophila insect species. In both the mosquitoes, An. gambiae and Ae. aegypti, there are no discernible repeats within their PF regions, although the sequences are still charged and enriched in proline, serine, and threonine, and in T. castaneum there are only remnants of repeats. All these species have short PF regions, and the area that is expanded is C terminal (see Figure 1c). In Anopheles, this region is composed of >45 repeats of 44 amino acids that are also apparently undergoing concerted evolution (see Figure 7).
Codon usage changes resulting from concerted evolution in the dumpy gene:
It is clear that concerted evolution has contributed substantially to altering the patterns of codon usage in the PF exon vs. the rest of the dumpy gene. The raw data and G-test results—when codon numbers in the PF exon are compared to those expected, given the percentages of codons used in the rest of the dumpy gene—are presented in supplemental Table 2 (http://www.genetics.org/supplemental/). The differences in the majority of cases are highly significant when there are a sufficient number of codons for a particular amino acid in the PF exon. Thus, 16 of 17, 14 of 16, and 15 of 17 codon usage patterns are significantly different at the 0.001 level in D. melanogaster, D. pseudoobscura, and D. virilis, respectively. Looking at the preferred codons (see supplemental Table 3 at http://www.genetics.org/supplemental/) within each species, different codons for the six-, four-, and three-codon families are preferred in 25 of 26 comparisons; e.g., GCC is the preferred alanine codon in non-PF exons in the D. melanogaster gene, but GCA is used 78% of the time in the PIGSFEAST exon. These data are summarized in Table 6A, and, even in the two-codon families, a different codon is preferred in the PF exon 50% of the time.
TABLE 6.
Intraspecific and interspecific comparisons of preferred codons in PF and in non-PF exons from D. melanogaster, D. pseudoobscura, and D. virilis
| A. Intraspecific comparisons | ||
|---|---|---|
| Same codons preferred | Different codons preferred | |
| Six-codon families (ARG, LEU, SER) | 0 | 9 |
| Four- or three-codon families (ALA, GLY, ILE, PRO THR, VAL) | 1 | 17 |
| Two-codon families (ASN, ASP, CYS, GLN, GLU, HIS, LYS, PHE, TYR) | 10 | 10 |
| Total | 11 | 36 |
| B. Interspecific comparisons | |||
|---|---|---|---|
| Same preferred codon in the compared species | Different preferred codon(s) in the compared species | ||
| Six-, four- and three-codon families | Non-PF exons | 4 | 5 |
| PF exonsa | 0 | 9 | |
| Two-codon families | Non-PF exons | 7 | 2 |
| PF exonsa | 5 | 3 | |
For the PF exon, only two species could be compared in the two-codon families for ASN, HIS, PHE, and TYR, and none could be compared for CYS.
In addition to producing different preferred codons for non-PF and PF exons within a species, the processes of concerted evolution also produce more extreme codon bias in the PF exons themselves from each of the three species. Summarizing the percentage of usages in supplemental Table 3 (http://www.genetics.org/supplemental/), we find that, in D. melanogaster, the average percentage of usage for preferred codons across all codon families is 48% for non-PF exons but 65% for the PF exons. The same values for D. pseudoobscura are 56 and 69% and for D. virilis, 47 and 65%, respectively.
The lineage-specific concerted evolutionary events occurring during the evolution of the three Drosophila species has resulted in more interspecific diversity in the preferred codons in the PF exons than in the non-PF exons. This is especially true in the six-, four-, and three-codon families as shown in Table 6B. Here the same preferred codon is used in non-PF exons about half the time when the three species are compared, whereas in no case is the same preferred codon used in all three species in the PF exon. The comparison is much less meaningful for two-codon families for obvious reasons.
The PF region is evolving in a near-neutral fashion with weak purifying selection:
The variation between internal repeats within the arrays from each species, both in terms of nonsynonymous differences per nonsynonymous site (dN) and in terms of synonymous differences per synonymous site (dS), is fairly small, ranging from 0.03 to 0.13 for dN and from 0.03 to 0.193 for dS. These values are presented in Table 5A. The ratios of dN to dS are <1 with the exception of the distinctive Anopheles repeat region. A dN-to-dS ratio of <1 generally implies the action of purifying selection, although positive selection can still be acting at individual sites (Massingham and Goldman 2005). Consistent with this finding, we note a number of sequence positions in the PF repeat amino acid sequence that are highly conserved or show conservative replacements throughout the melanogaster group (Figure 7), which seems to indicate a requirement to maintain a functional interaction and/or some structural integrity. Note, however, in Table 5A, that the dN-to-dS ratios are fairly close to 1, indicating that, if purifying selection is operating on the repeats within the species' arrays, it is relatively weak. We further subjected the data on PF repeats from D. melanogaster and D. simulans to the McDonald–Kreitman test (McDonald and Kreitman 1991) for neutrality, treating the repeats within each species as if they were from different geographic strains. Under neutrality, the ratio of replacement to synonymous substitutions fixed between species will not be significantly different from the same ratio of replacement and synonymous sites polymorphic within the two species. We found that, with zero fixed synonymous differences and 34 synonymous polymorphic sites along with three fixed nonsynonymous substitutions and 65 polymorphic nonsynonymous sites, the P-value (0.549) indicates that the PF repeats are evolving under very little or no selection. In this regard, we could not search for positively selected residues in the repeats using likelihood-based tests such as PAML (Nielsen and Yang 1998) or the SLR method (Massingham and Goldman 2005). These tests assume that the sites in the repeat are evolving independently, but the ongoing processes of concerted evolution make that assumption invalid.
Expression of Dumpy during micropyle formation:
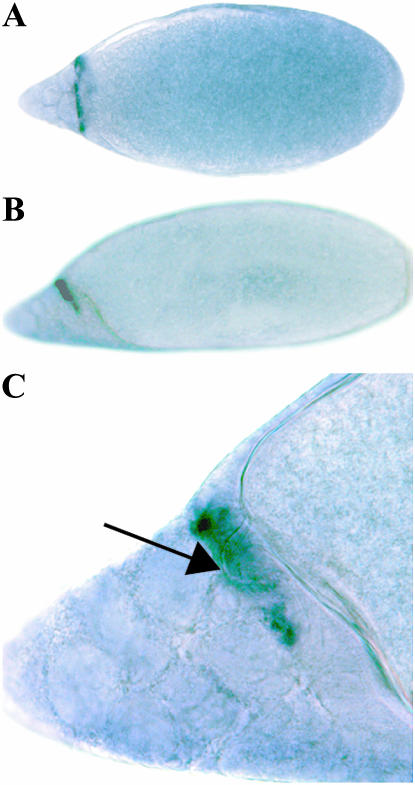
Given the precedent afforded by the abalone sperm receptor (Swanson and Vacquier 1998), we were interested in determining whether dumpy may have a role in insect egg formation. We therefore performed an in situ hybridization on the D. melanogaster ovary using a digoxygenin-labeled RNA probe toward the C-terminal end of Dumpy (see Figure 1a). This showed that dumpy is highly and specifically expressed in stage 12 oocytes in the follicle cells that surround the developing micropylar apparatus, a tube through which sperm enter the egg (Figure 9). This staining is somewhat reminiscent of the slightly later expression of Yellow G and Yellow G2, presumed cuticular proteins expressed in the same structure (Claycomb et al. 2004). Other Drosophila ZP genes have been found to be expressed in the follicle cells surrounding the egg chamber and may have roles in the formation of the egg envelope (Jazwinska and Affolter 2004), although none of these were shown to be expressed in the developing micropyle.
Figure 9.—
In situ hybridization of a digoxygenin-labeled dumpy RNA probe to stage 12 oocytes shows that dumpy is expressed in the developing micropylar apparatus. (A) A dorsal view of a stage 12 oocyte and (B) a lateral view, with dorsal up; in both cases, anterior to the left. (C) The micropylar region in B at ×4 higher magnification. The developing micropyle structure can seen through interference contrast (arrow).
DISCUSSION
We show that the dumpy gene has been conserved in insect evolution, preserving its domain content and organization. Dumpy is a protein that, in D. melanogaster, is expressed in regions of specialized cuticle epithelial interactions and probably plays an organizational and strengthening role at the interface between certain epithelial cells and the insect cuticle. For example, Dumpy is required for normal tracheal development and at locations where tendon epithelial cells link muscles to the overlying cuticle and at the interface between wing epithelial cells and the overlying apical matrix. Conservation of the Dumpy domain structure is likely to imply functional conservation in other insects. Functional genetic studies of dumpy have revealed it to be a complex genetic locus, suggesting that different regions might be functionally specialized (Grace 1980). Interestingly, our study has revealed that while most of the dumpy gene is evolving by a standard evolutionary mechanism by accumulated divergence from a common ancestor, a defined region comprising some 18% of its coding sequence, is undergoing rapid and concerted evolution. This concertedly evolving region is composed of a highly repetitive tandem modules region, called the PF repeats, and is recognizably present at approximately the same position in the gene in all the Drosophila species that we were able to examine. This region is undergoing far higher rates of evolution than other regions of the dumpy gene and this is occurring with near-neutral or weak purifying selection. Perhaps selection is needed only to maintain a low level of complexity in the PF repeat by preserving its richness in codons for the three most abundant amino acids—serine, threonine, and proline—as well as charged residues. Concerted evolution is indicated by the high ratios of interspecific dN and dS values to the mean of the same intraspecific values in almost all the pairs of species compared.
In addition, unequal crossing over is likely to be the driving force in the concerted evolution of the PF repeats, as indicated by the variable numbers of repeats in different strains of D. melanogaster, the increased variation in the terminal repeats in D. melanogaster, D. erecta, and D. virilis, and the greater sequence similarities between adjacent repeats in the arrays from D. melanogaster and D. virilis.
We also observe a region-specific codon usage bias within the PF repeats compared to the remainder of dumpy. One can envision how differential and biased codon usage is engendered by concerted evolution, especially if one assumes that a single repeat sweeps to fixation in an array by unequal crossing over, perhaps in conjunction with gene conversion events. Indeed, the fixation of an indel such as the 14-codon deletion in D. simulans and its related species implies that single repeats have indeed been fixed during the evolution of the PF array. If the fixation occurs over a short period of time, a codon for a single amino acid in the repeat, e.g., the single histidine codon in the PF repeat from D. melanogaster, will also become fixed and its percentage of usage in the array will be, initially at least, 100%. Codons for similarly rare amino acids will also show more extreme biased usage. Given enough time, codon usage patterns should converge to those found in the rest of the gene, assuming that there is at least some selection pressure acting on codon usage in Drosophila. This process would take a long time and presumably could be reversed by intervening episodes of concerted evolution. To our knowledge, this is the first documentation of the effects of concerted evolution on codon usage in a protein-coding gene.
In contrast, in insect species outside of Drosophila that we analyzed, we found that it is a second region just C terminal to the PF region that is enlarged and, in the case of An. gambiae, evolving concertedly. Meanwhile, the region in the location equivalent to that of the PF is much reduced in size such that the overall size of the two regions combined is similar. It is interesting to speculate as to whether this second region has also replaced the function of the PF domain region for an activity in which a highly repeated structure is a key feature. It is also noteworthy that there are a few exceptions, discussed in the results, where the PF or the flanking region have ceased to evolve in a concerted fashion, implying that different evolutionary pressures are at work.
The processes underlying the concerted evolution of DNA sequences, primarily unequal crossing over and gene conversion, have been investigated for a number of years (Dover 1982; Averbeck and Eickbush 2005). Initial studies involved rDNA repeats (Arnheim et al. 1970; Coen et al. 1982) and satellite DNA sequences (Willard 1991; Cardone et al. 2004). More recently, concerted evolution has been documented within protein-coding genes (e.g., Swanson and Vacquier 1998; Desseyn et al. 2000; Nenoi et al. 2000; Meeds et al. 2001; Johanneson et al. 2005). Interestingly, the products of such genes are often found on the surfaces of cells where they interact with other proteins, e.g., sperm lysins (Swanson and Vacquier 1998), mucins (Desseyn et al. 2000), spicule matrix proteins (Meeds et al. 2001), and fungal outer-cell-wall glycoproteins (Johannesson et al. 2005). Importantly, concerted evolution of such proteins can drive the coevolution of interacting partners. For example, in the abalone system, the lysin receptor on the egg surface or VERL is, like the Dumpy PF region, changing by processes of concerted evolution with only weak purifying selection. This rapid change is accompanied by the coevolution under positive selection of the sperm lysin itself, apparently resulting in the maintenance of species specificity in the sperm–egg recognition process (Swanson and Vacquier 1998). Since ZP domain proteins are frequently involved at the egg–sperm interface of many organisms (Mengerink and Vacquier 2001; Galindo et al. 2002; Sawada et al. 2002), we were interested in determining if there was additional similarity between Dumpy and the abalone egg receptor paradigm. Remarkably, we found that dumpy is expressed in the border cells, which are responsible for forming the micropyle of the oocyte, a tube-like structure that is the sperm entry site to the egg. Although we cannot say at present whether Dumpy contributes to the fertilization mechanism or whether the PF region is involved in sperm discrimination, the micropyle localization of Dumpy is such that it could interact with sperm proteins. Therefore, as in the abalone system, coevolution of Dumpy with sperm-specific proteins could play a role in the maintenance of species boundaries. It will be important now to determine whether there are interacting partners in the PF region of Dumpy that are rapidly coevolving to understand any impact that the rapid changes occurring within the dumpy gene may have on insect evolution.
Acknowledgments
We thank Carlos Bustamante for important advice on tests for selective neutrality on the PF repeats. We also thank former technical assistants Linda Hook D'Innocenzo, Jennifer Rosen, and Tashana Williams for their invaluable help in cloning and sequencing the PCR products from the species of the melanogaster subgroup.
References
- Arnheim, N., M. Krystal, R. Schnickel, G. Wilson, O. Ryder et al., 1970. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. USA 77: 7323–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., K. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Averbeck, K., and T. Eickbush, 2005. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics 171: 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokel, C., A. Prokop and N. H. Brown, 2005. Papillote and Piopio: Drosophila ZP-domain proteins required for cell adhesion to the apical extracellular matrix and microtubule organization. J. Cell Sci. 118(3): 633–642. [DOI] [PubMed] [Google Scholar]
- Cardone, M. F., L. Ballarate, M. Ventura, M. Rocchi, A. Marozzi et al., 2004. Evolution of beta satellite DNA sequences: evidence for duplication-mediated repeat amplification and spreading. Mol. Biol. Evol. 21: 1792–1799. [DOI] [PubMed] [Google Scholar]
- Claycomb, J. M., M. Benasutti, G. Bosco, D. D. Fenger and T. L. Orr-Weaver, 2004. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev. Cell 6(1): 145–155. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., J. Thoday and G. Dover, 1982. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature 295: 564–568. [DOI] [PubMed] [Google Scholar]
- Cornell, M., D. A. P. Evans, R. Mann, M. Fostier, M. Flasza et al., 1999. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desseyn, J. L., J. Paulert, N. Porchet and A. Laine, 2000. Evolution of the large secreted gel forming mucins. Mol. Biol. Evol. 17: 1175–1184. [DOI] [PubMed] [Google Scholar]
- Dover, G., 1982. Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. [DOI] [PubMed] [Google Scholar]
- Durfy, S., and H. Willard, 1989. Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics 5: 810–821. [DOI] [PubMed] [Google Scholar]
- Galindo, B. E., G. W. Moy, W. J. Swanson and V. D. Vacquier, 2002. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene 288(1–2): 111–127. [DOI] [PubMed] [Google Scholar]
- Grace, D., 1980. Genetic analysis of the dumpy complex locus in Drosophila melanogaster: complementation, fine structure and function. Genetics 94: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska, A., and M. Affolter, 2004. A family of genes encoding zona pellucida (ZP) domain proteins is expressed in various epithelial tissues during Drosophila embryogenesis. Gene Expr. Patterns 4(4): 413–421. [DOI] [PubMed] [Google Scholar]
- Jazwinska, A., C. Ribeiro and M. Affolter, 2003. Epithelial tube morphogenesis during Drosophila tracheal development requires Piopio, a luminal ZP protein. Nat. Cell Biol. 5(10): 895–901. [DOI] [PubMed] [Google Scholar]
- Johannesson, H., J. P. Townsend, C. Y. Hung, G. Cole and J. W. Taylor, 2005. Concerted evolution in the repeats of an immunomodulating cell surface protein, SOWgp, of the human pathogenic fungi, Coccidoides immitis and C. posadosii. Genetics 171: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius, K., A. Mølgaard, R. Gupta and S. Brunak, 2005. Prediction, conservation analysis and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15: 153–164. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Massingham, T., and N. Goldman, 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169: 1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, B., and J. Werren, 1999. Evolution of tandemly repeated sequences: What happens at the end of an array? J. Mol. Evol. 48: 469–481. [DOI] [PubMed] [Google Scholar]
- McDonald, J., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Meeds, T., E. Lockhard and B. Livingston, 2001. Special evolutionary properties of genes encoding a protein with a simple amino acid repeat. J. Mol. Evol. 53: 180–190. [DOI] [PubMed] [Google Scholar]
- Mengerink, K. J., and V. D. Vacquier, 2001. Glycobiology of sperm-egg interactions in deuterostomes. Glycobiology 11(4): 37R–43R. [DOI] [PubMed] [Google Scholar]
- Nenoi, M., S. Ichimura and K. Mita, 2000. Interspecific comparison in the frequency of concerted evolution at the polyubiquitin gene locus. J. Mol. Evol. 51: 161–165. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., and Z. Yang, 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, H., N. Sakai, Y. Abe, E. Tanaka, Y. Takahashi et al., 2002. Extracellular ubiquitination and proteasome-mediated degradation of the ascidian sperm receptor. Proc. Natl. Acad. Sci. USA 99(3): 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W., and V. Vacquier, 1998. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281: 710–712. [DOI] [PubMed] [Google Scholar]
- Swanson, W., and V. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kamura, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Wassarman, P. M., 2002. Sperm receptors and fertilization in mammals. Mt. Sinai J. Med. 69(3): 148–155. [PubMed] [Google Scholar]
- Wassarman, P. M., L. Jovine and E. S. Litscher, 2001. A profile of fertilization in mammals. Nat. Cell Biol. 3(2): E59–E64. [DOI] [PubMed] [Google Scholar]
- Weigmann, B., D. Yeates, J. Thorne and H. Kishino, 2003. Time flies: a new molecular time-scale for Brachyceran fly evolution without a clock. Syst. Biol. 52: 745–756. [PubMed] [Google Scholar]
- Wilkin, M., M. Becker, D. Mulvey, I. Phan, A. Chao et al., 2000. Drosophila dumpy is a gigantic extracellular protein required to maintain tension at epidermal-cuticle attachment sites. Curr. Biol. 10: 559–567. [DOI] [PubMed] [Google Scholar]
- Willard, H., 1991. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1: 509–514. [DOI] [PubMed] [Google Scholar]
- Yeates, D., and B. Weigmann, 1999. Congruence and controversy: toward a higher level phylogeny of Diptera. Annu. Rev. Entomol. 44: 397–428. [DOI] [PubMed] [Google Scholar]