Abstract
Allopolyploid species form through the fusion of two differentiated genomes and, in the earliest stages of their evolution, essentially all genes in the nucleus are duplicated. Because unique mutations occur in each ancestor prior to allopolyploidization, duplicate genes in these species potentially are not interchangeable, and this could influence their genetic fates. This study explores evolution and expression of a simple duplicated complex—a heterodimer between RAG1 and RAG2 proteins in clawed frogs (Xenopus). Results demonstrate that copies of RAG1 degenerated in different polyploid species in a phylogenetically biased fashion, predominately in only one lineage of closely related paralogs. Surprisingly, as a result of an early deletion of one RAG2 paralog, it appears that in many species RAG1/RAG2 heterodimers are composed of proteins that were encoded by unlinked paralogs. If the tetraploid ancestor of extant species of Xenopus arose through allopolyploidization and if recombination between paralogs was rare, then the genes that encode functional RAG1 and RAG2 proteins in many polyploid species were each ultimately inherited from different diploid progenitors. These observations are consistent with the notion that ancestry can influence the fate of duplicate genes millions of years after duplication, and they uncover a dimension of natural selection in allopolyploid genomes that is distinct from other genetic phenomena associated with polyploidization or segmental duplication.
IN allopolyploid species, the complete genome of two species are fused, genes are duplicated, and the level of genetic redundancy that results is determined by divergence of protein coding and regulatory regions of genes in each ancestor and by epigenetic phenomena after allopolyploidization. In these genomes, natural selection and stochastic processes govern the genetic fate of each paralog—fates that include redundancy, subfunctionalization, neofunctionalization, and gene silencing (Ohno 1970; Hughes and Hughes 1993; Force et al. 1999; Lynch and Force 2000; Lynch et al. 2001; Gu 2003). After allopolyploidization, interactions between the subgenomes derived from each ancestral species included exchange of chromosome segments (Moscone et al. 1996; Skalická et al. 2005), concerted evolution (Wendel et al. 1995; Volkov et al. 1999), recombination (Zwierzykowski et al. 1998), and epistasis (Jiang et al. 2000). Restructuring can then alter the stoichiometry of protein interactions by modifying regulatory elements or changing gene copy number. Presumably these events trigger or permit compensatory responses including changed regulation, degeneration, or deletion of superfluous upstream or downstream paralogs. Modulation of the transcriptome can occur on an extraordinarily fine scale: a silenced paralog from one ancestor can be tightly linked to loci in which only the paralog from the other ancestor is silenced, or linked to loci in which paralogs from both ancestors are expressed (Lee and Chen 2001). Genomic changes such as subfunctionalization, gene deletion, and gene silencing can occur within generations after allopolyploidization, and these changes can be nonstochastic and repeatable, and biased by ancestry (Volkov et al. 1999; Ozkan et al. 2001; Shaked et al. 2001; Adams et al. 2003, 2004; Wang et al. 2004; Adams and Wendel 2005). Allopolyploidization can also lead to completely novel expression patterns that were not present in either parental species, perhaps as a result of dosage-dependent gene regulation (Wang et al. 2004). Expression can be influenced by the direction of the hybrid cross (Soltis et al. 2004), and coadapted proteins that were inherited from one ancestor may function more efficiently with one another than with proteins that were derived from a different ancestor (Comai 2000). Epigenetic phenomena, such as nucleolar dominance, gene silencing, alteration of cytosine methylation patterns, and activation of mobile elements, can also foment genetic change (Liu and Wendel 2002, 2003). However, variation exists among allopolyploid species in the extent of genomic rearrangement—genomic modulation is less common, for example, in the early stages of evolution in synthetic cotton allopolyploids (Liu et al. 2001) than in other allopolyploids such as wheat and Arabidopsis (Madlung et al. 2002; Levy and Feldman 2004; Lukens et al. 2004).
African clawed frogs (genera Xenopus and Silurana) offer a promising model with which to examine the impact of ancestry on gene fate in an allopolyploid genome because multiple independent instances of allopolyploidization occurred (Evans et al. 2004, 2005), and the long term effects of this type of genome duplication can be explored with replication. All extant species of clawed frogs in the genus Xenopus (i.e., those species with multiples of 2x = 18 chromosomes) share a common tetraploid ancestor (Evans et al. 2004, 2005). A definitive test of allopolyploid vs. autopolyploid origin of this ancestor is not currently possible because no extant Xenopus diploids (2x = 18) are known. However, this ancestor is suspected to have been an allotetraploid because (a) other polyploid clawed frogs are definitively allopolyploids (Evans et al. 2005), (b) Xenopus genomes are diploidized and duplicated pairs of chromosomes have visible differences in secondary constrictions (Tymowska 1991), (c) allopolyploid individuals can be created in the laboratory by crossing extant species (Kobel 1996), and (d) multiple unlinked loci indicate that the phylogenetic signal of many paralogs is not blended by recombination (Evans et al. 2005; Chain and Evans 2006; F. J. J. Chain, D. Ilieva and B. J. Evans, unpublished results). Silurana and Xenopus diversified from one another ∼53–64 million years ago (MYA) and tetraploidization in Xenopus occurred ∼21–41 MYA (Evans et al. 2004; Chain and Evans 2006).
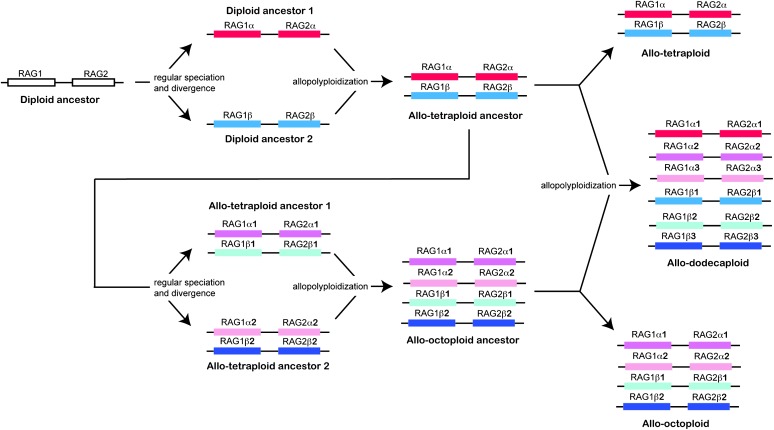
RAG1 and RAG2 proteins form a heterodimer that is crucial for the process of somatic rearrangement of DNA known as V(D)J recombination, making possible the extraordinary molecular variation of B-cell and T-cell antigen receptors that is needed to combat pathogen attack. The core region of RAG1, which, when paired with RAG2 is sufficient to carry out V(D)J recombination, spans human residues 386–1011 out of a total of 1040 amino acids in the protein (Sadofsky et al. 1993). The core region of RAG1 was derived from the Transib transposon superfamily, whereas the RAG2 and the N-terminal domain of RAG1 probably was derived from other sources (Kapitonov and Jurka 2005). These genes are tightly linked in jawed vertebrates; a recent build (version 4.1) of the complete genome sequence of the diploid clawed frog Silurana tropicalis indicates that there is only one copy of RAG1 and one of RAG2, and that each is convergently transcribed and tightly linked by an ∼6.5-kb intergenic region. This is consistent with findings in Xenopus laevis that suggest that this tetraploid was derived from the fusion of two diploid genomes and that each of these diploid genomes carried only one copy of RAG1 (Greenhalgh et al. 1993; Evans et al. 2005). Thus, in the absence of paralog deletion and degeneration, a null expectation is that the number of RAG1 and RAG2 paralogs corresponds with the ploidy level of a species: diploids should have one copy of each gene, tetraploids should have two, octoploids should have four, and dodecaploids should have six (Figure 1). Each of these paralogs would have two alleles.
Figure 1.—
Diploidization of allopolyploid genomes means that recombination between alleles of different paralogs of the same gene is rare. The evolutionary history of linked paralogs in diploidized allopolyploid genomes, therefore, can be inferred by combining information on synteny with information on the evolutionary relationships among paralogs of each gene. The RAG1 and RAG2 genes, for example, are tightly linked. The evolution of the expected number of paralogs of each of these genes in species with different ploidy levels (tetraploid, octoploid, and dodecaploid) if no deletion or degeneration occurred is depicted. An allotetraploid species inherits a linked set of RAG1 and RAG2 α paralogs from diploid ancestor 1 and a linked set of RAG1 and RAG2 β paralogs from diploid ancestor 2. An allo-octoploid species inherits two sets of linked RAG1 and RAG2 paralogs from two different allotetraploid ancestors. In these species the α1 and α2 paralogs of RAG1 and RAG2 are derived from diploid ancestor 1 and the β1 and β2 paralogs of RAG1 and RAG2 are derived from diploid ancestor 2. An allododecaploid inherits an allo-octoploid and an allotetraploid genome and has three α paralogs (α1, α2, α3) derived from diploid ancestor 1 and three β paralogs (β1, β2, β3) derived from diploid ancestor 2. Note that the observed number of functional RAG1 and RAG2 paralogs is less than this expectation in most (RAG1) or all (RAG2) species of Xenopus due to degeneration or deletion.
Contrary to this null expectation, a previous study reported that in different polyploid species of Xenopus, many RAG1 paralogs had become pseudogenes due to stop codons and frameshift insertion/deletions (Evans et al. 2005). These species of Xenopus were all ultimately derived from a tetraploid ancestor that probably formed through the amalgamation of two diploid genomes by allopolyploidization; this tetraploid ancestor therefore initially had two paralogs of RAG1 and two paralogs of RAG2. If recombination between paralogs was rare or absent after allotetraploidization, then all of the degenerate paralogs detected by Evans et al. (2005) were ultimately derived from only one of these diploid progenitors—diploid “β”—as opposed to being derived from both of the diploid ancestors (“α” and “β”).
This pattern of degeneration could be explained by (a) a shared ancestral degeneration of a coding or regulatory region of RAG1 paralog β that was not sequenced by Evans et al. (2005), (b) multiple independent and biased degenerations in different species, or (c) some combination of shared and independent degenerations (Evans et al. 2005). This study aims to further investigate these possibilities and track the evolutionary history of this heterodimer through multiple episodes of speciation and genome duplication. To this end, (a) new sequences were obtained from most or all upstream coding regions of RAG1 paralogs to test for shared coding-region degeneration, (b) expression analyses were performed on multiple species to test for ancestral silencing of paralogs, and (c) sequences were obtained from paralogs of the linked partner gene RAG2.
MATERIALS AND METHODS
DNA sequencing:
Sequence data were obtained from all or almost all RAG1 and RAG2 paralogs from all known species of clawed frog, averaging 2341 bp out of ∼3135 bp total per RAG1 paralog and 1123 bp out of 1575 bp total per RAG2 paralog (supplemental Figure 1 at http://www.genetics.org/supplemental/). The sequenced portion of each RAG1 paralog varied but generally covers most or all of the core region of RAG1 (supplemental Figure 2 at http://www.genetics.org/supplemental/). Some paralogs were not detected using a variety of primer combinations, and these may have been deleted (supplemental Figure 1). Sequencing of individual paralogs was accomplished through a combination of TA cloning (Invitrogen) and targeted amplification using paralog-specific primers (supplemental Figure 1). Allelic clones with the least number of autapomorphic mutations were selected for analysis. When both alleles of a paralog were directly sequenced with paralog-specific primers, polymorphic positions were analyzed using IUPAC degenerate nucleotide symbols. Some sequence data from expressed paralogs were obtained from nonvouchered individuals; these data were conatenated with sequences from the corresponding paralog from other individuals that usually were vouchered, as detailed in (Evans et al. 2004, 2005). Following Evans et al. (2005), paralogs of RAG1 and RAG2 that are closely related to the linked X. laevis paralogs identified by Greenhalgh et al. (1993) are referred to as β paralogs and the others are referred to as α paralogs. In Silurana, tetraploid paralogs closely related to the diploid S. tropicalis are designated α paralogs and the others are β paralogs.
Attempts to amplify and clone the α paralogs of RAG2 failed in all species of Xenopus using a variety of primer combinations (supplemental information 1 at http://www.genetics.org/supplemental/), even though both paralogs were detected in the Silurana tetraploids. To rigorously test whether RAG2 α paralogs were deleted in clawed frogs as opposed to just not being amplified, a systematic effort was made to coamplify both paralogs using seven pairwise combinations of three forward and three reverse primers (supplemental Figure 1 at http://www.genetics.org/supplemental/). S. tropicalis was used as a positive control to demonstrate that in addition to Xenopus RAG2 paralog β, these primers can successfully amplify RAG2 in a more distantly related lineage than Xenopus RAG2 paralog α.
Amplification of expressed RAG1 paralogs:
To determine which RAG1 paralogs are expressed in various species, cDNA was amplified across an intron in the 5′ untranslated region of the RAG1 transcript (Greenhalgh et al. 1993). Negative controls with no DNA and with genomic DNA were performed to ensure that only expressed and spliced paralogs were amplified; previously published and new primers were used (supplemental Figure 1 at http://www.genetics.org/supplemental/; Greenhalgh et al. 1993). RNA was extracted using the RNeasy mini kit (QIAGEN) and converted to cDNA using the Omniscript RT kit (QIAGEN). Individual expressed paralogs were amplified from cDNA generated from different tissues (brain, liver, spleen, testis, and/or bone marrow) from a variety of species (X. laevis, X. gilli, X. borealis, X. muelleri, X. amieti, X. andrei, X. new octoploid, and X. boumbaensis), and then cloned and sequenced. Additionally, cDNA from some tissues was directly sequenced and chromatograms were inspected for paralog-specific single nucleotide polymorphisms.
Phylogenetic analyses:
Phylogenetic analysis was performed on coding portions of RAG1 and RAG2 in three types of data configurations: (1) each locus was analyzed independently, (2) putatively linked paralogs were combined into single taxonomic units, and (3) for Xenopus only, a “synthetic” data set was constructed in which α and β paralogs of RAG1 and RAG2 that were derived from the same most recent tetraploid ancestor were combined into a single taxonomic unit. This third configuration exploits the redundant phylogenetic information available in co-inherited paralogs. For example, in the third data configuration, tetraploid α and β paralogs were combined, octoploid α1 and β1 paralogs were combined, but octoploid α1 and β2 paralogs were not combined. For the third analysis, S. tropicalis was used as an out-group to RAG1 α and RAG2 paralogs and no out-group sequence was used for the portion of the sequences that was composed of RAG1 β paralogs.
MrBayes version 3.1.2 was used for Bayesian phylogenetic analysis, and Bayes factors were used to select a model of evolution, as described by Nylander et al. (2004). Seven partitioned models were explored (supplemental Table 1 at http://www.genetics.org/supplemental/). These models were compared on the basis of the harmonic mean of the posterior probability of trees sampled after a conservative burn-in of 1 million generations from two independent MCMC runs, each of 2 million generations. A highly parameterized model of evolution was favored for phylogenetic analyses of RAG1 and RAG2 (supplemental Table 1), and this model was also used for the combined analyses, using 5 million generations and the same burn-in. Branch support was also evaluated with 2000 nonparametric bootstrapping replicates, each with a single replication of random taxon addition, a limit of 10 million rearrangements per replicate, and the maximum parsimony criterion using PAUP version 4.0b10 (Swofford 2002). Almost all of the well-supported relationships from the Bayesian analyses also have nonparametric bootstrap values of >80% (supplemental Figure 3 at http://www.genetics.org/supplemental/).
To test hypotheses of fewer gene silencing events in RAG1 than was suggested by phylogenetic analysis, parametric bootstrap tests (Huelsenbeck et al. 1996; Goldman et al. 2000) were performed as in Evans et al. (2005). This procedure tests the fit of the data to alternative phylogenetic hypotheses that are different from the consensus tree that was obtained from the Bayesian analysis, and that would be consistent with fewer instances of independent gene degeneration (supplemental Figure 4 at http://www.genetics.org/supplemental/). To maximize the phylogenetic signal of these tests, simulations were performed according to the synthetic data configuration using Seq-Gen version 1.3.2 (Rambaut and Grassly 1997). A Perl script was written to modify the simulations to match the observed data in terms of the quantity and positions of missing data for each taxon.
A caveat to the interpretation that autapomorphic degenerations occurred independently is that recurrent substitutions could have erased an ancestral degeneration in one or both descendant paralogs. To explore this possibility, marginal ancestral reconstruction of ancestral character states was performed with a general time-reversible nucleotide model and a gamma-distributed rate heterogeneity parameter using the baseml program of PAML version 3.14 (Yang 1997). Reconstructed sequences were then translated into protein using MacClade version 4.08 (Maddison and Maddison 2000) and inspected for stop codons.
Testing for phylogenetic bias in RAG1 degeneration:
To explore whether there is significant bias in gene degeneration of RAG1 with respect to ancestry of each paralog, two approaches were taken. The first approach used a maximum likelihood framework to compare rates of degeneration (δ), using Discrete version 4.0 (Pagel 1994). This framework tested whether rates of degeneration in RAG1 and RAG2 were significantly different, and also whether rates of degeneration in the RAG1 α and β lineages were significantly different. The rate of resuscitation of degenerate paralogs was set to a negligible value, missing paralogs were coded as degenerate, and the most recent ancestor of expressed paralogs with autapomorphic degenerations was set as nondegenerate. Likelihood ratio tests were used to compare models with two degeneration parameters to models with only one, and these tests were performed with and without a gamma-distributed approximation for rate heterogeneity (γ). To be conservative, modified topologies were used in which the independent degenerations (numbered 1–12 in Figure 2) were each forced to be a clade. Branch lengths were estimated under the GTR+I+Γ model and imposing a molecular clock using PAUP* (Swofford 2002).
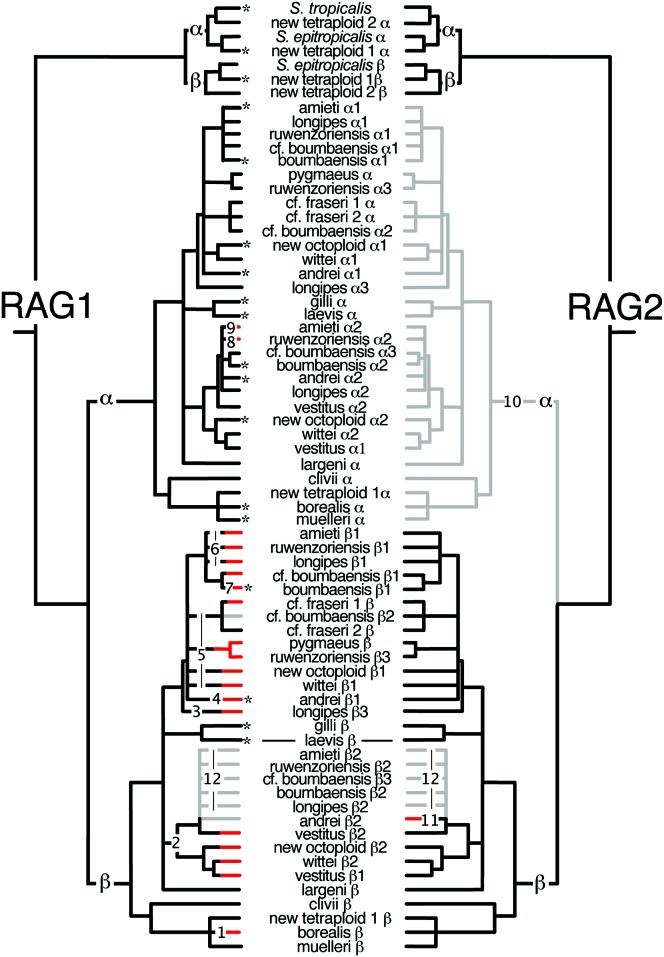
Figure 2.—
Combined phylogenetic analysis of RAG1 and RAG2 paralogs. Nodes with <95% posterior probability or that were inconsistent between the α and β lineages are collapsed, asterisks indicate RAG1 paralogs for which expression was confirmed, and linked X. laevis RAG1 and RAG2 paralogs (Greenhalgh et al. 1993) are connected by a line. Degenerate paralogs are in red and missing paralogs are in gray. Numbers 1–9 indicate the minimum independent degenerations of RAG1 mentioned in the text, and 10, 11, and 12 refer to degeneration of RAG2 lineage α, degeneration X. andrei RAG2 paralog β2, and a suspected ancestral deletion spanning paralogs of RAG1 and RAG2, respectively.
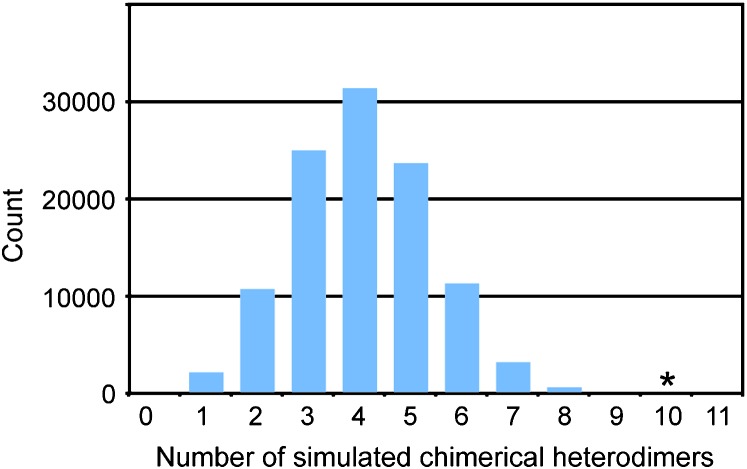
A second approach to test for bias in RAG1 degeneration used simulations to estimate the probability that the observed number of “chimerical” heterodimers—those composed of RAG1 and RAG2 proteins from different paralogous lineages—occurred by chance if there were no bias to RAG1 degeneration. It was assumed that the number of observed degenerate paralogs in each species follows a binomial distribution with a species-specific probability density for the mean of this distribution that was calculated from the observed data. It was also assumed that at least one RAG1 paralog must remain functional in each species, that no simulated degeneration could occur in species with no observed degeneration, that each nondegenerate paralog is expressed at the same intensity, time, and place, and that RAG1/RAG2 heterodimers form from paralogs from the same ancestral lineage (α or β) whenever possible. For eight extant or ancestral species with observed independent RAG1 degeneration (see results), 100,000 simulations drew k degeneration events from n − 1 paralogs, where n is the total number of nondegenerate paralogs inferred to be present when that species originated. Degeneration was modeled as a delayed transformation phenomenon wherein paralogs degenerated after allopolyploidization, and it was also performed in a phylogenetically independent manner such that ancestral degenerations were inherited and not “re-simulated” in descendant species. Chimerical heterodimers were quantified for each simulation on the basis of observed and suspected degenerations and deletions of RAG2 (i.e., degenerations 10–12 in Figure 2).
Recombination between alleles of different paralogs:
Recombination between alleles of different paralogs could blend their phylogenetic signal, and recombination between the intergenic regions of paralogous chromosomes could alter the synteny of paralogs (Greenhalgh et al. 1993). To test for evidence of recombination in Xenopus and Silurana sequences, multiple tests were used because their performance varies with the level of divergence, the extent of recombination, and among site-rate heterogeneity (Posada and Crandall 2001; Posada 2002). These tests included the recombination detection program, geneconv, chimera, bootscan, and siscan, as implemented by the Recombination Detection Program (Martin and Rybicki 2000). Details of these methods can be found elsewhere (Maynard Smith 1992; Salminen et al. 1995; Padidam et al. 1999; Gibbs et al. 2000; Martin and Rybicki 2000; Posada and Crandall 2001). A variety of parameter settings were explored for each method as in Evans et al. (2005).
RESULTS
New data lead to a reassessment of the evolutionary history of X. boumbaensis:
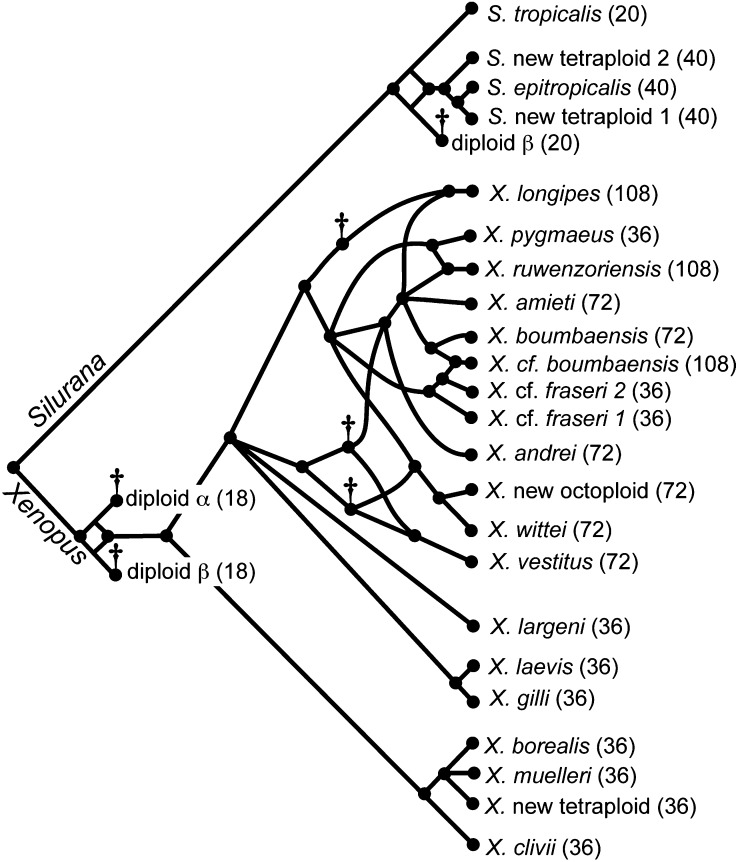
Evolutionary relationships inferred from RAG1 (this study; Evans et al. 2005), mitochondrial DNA (Evans et al. 2004), and RAG2 (Figure 2, supplemental Figure 3 at http://www.genetics.org/supplemental/) can be synthesized into a reticulate phylogeny (Figure 3). New data identified an exception to expected relationships among paralogs in an individual herein referred to as X. cf. boumbaensis. Additional sequencing from X. cf. boumbaensis identified a third RAG1 α paralog and two RAG2 paralogs whose relationships are suggestive of dodecaploidy (Figure 2). This suggests that this individual from Younde, Cameroon, was previously incorrectly classified as X. boumbaensis (Evans et al. 2004, 2005), which is an octoploid species (Loumont 1983; Tymowska 1991). It appears that X. cf. boumbaensis is a dodecaploid derived from allopolyploidization between an octoploid ancestor of X. boumbaensis and a tetraploid ancestor of X. cf. fraseri 2 (Figure 3). Data from an X. boumbaensis individual from the type locality of Moloundou, Cameroon, which were not included in Evans et al. (2005), are consistent with octoploidy (Figure 2 and 3) and lead to a re-evaluation of the evolutionary history of this species. Moreover, it appears that X. boumbaensis, X. ameiti, and X. andrei share a common octoploid ancestor, as opposed to each originating independently as was previously proposed (Evans et al. 2005). Thus these genealogies support three rather than five independent allopolyploid origins of most extant octoploids: (1) X. vestitus, (2) X. wittei and X. new octoploid, and (3) X. amieti and X. andrei and X. boumbaensis, but three rather than two independent origins of dodecaploids: (1) X. ruwenzoriensis, (2) X. longipes, and (3) X. cf. boumbaensis (Figure 3). This new information also changes the number of ancestral species that are predicted but for whom an extant descendant with the same ploidy level is unknown, from three diploids, three tetraploids, and one octoploid (Evans et al. 2005) to three diploids and three tetraploids (Figure 3). Mitochondrial DNA sequences from X. cf. boumbaensis are almost identical to a X. boumbaensis sample from the type locality (Evans et al. 2004), suggesting recent dodecaploidization of this individual.
Figure 3.—
Evolutionary relationships merge when a species evolves by allopolyploidization, and a reticulate phylogeny can be constructed from well-supported nodes in the genealogies of RAG1 and RAG2 in Figure 1. The ploidy level follows each species name including three diploids indicated with daggers whose existence is predicted in the past but for whom an extant descendant with the same ploidy level is not known. Additionally, three tetraploid ancestors (each with 4x = 36), also indicated by daggers, are predicted but not known by an extant representative.
Deletion of RAG2 and RAG1 paralogs:
Both paralogs of RAG2 were amplified, cloned, and sequenced from genomic DNA in Silurana tetraploids but only one divergent lineage of RAG2 was amplified in Xenopus. Systematic attempts to amplify additional RAG2 paralogs in Xenopus with seven combinations of other primers (supplemental Figure 1 at http://www.genetics.org/supplemental/) were unsuccessful, even though these primers successfully amplified a more divergent RAG2 ortholog in the diploid S. tropicalis. Southern blotting of X. laevis genomic DNA also detected only one RAG2 paralog but two RAG1 paralogs (Greenhalgh et al. 1993). This suggests that one RAG2 paralog was deleted in an early tetraploid ancestor (deletion 10 in Figure 2), as opposed to the alternatives that gene conversion occurred or that another paralog is present but undetected. All Xenopus species are therefore suspected to have half of the number of RAG2 paralogs than RAG1 paralogs.
Another suspected deletion of RAG2 occurred in an ancestral paralog from which paralog β2 of X. amieti, X. ruwenzoriensis, X. boumbaensis, and X. longipes and RAG2 paralog β3 of X. cf. boumbaensis would have descended (deletion 12 in Figure 2). These predicted paralogs were not detected in amplifications from genomic DNA, even after multiple attempts with different primer combinations (supplemental Figure 1 at http://www.genetics.org/supplemental/). Linked RAG1 paralogs from these species also were not detected (deletion 12 in Figure 2), suggesting that the deleted region spans both of them. Apart from these putative deletions, the only other observed gene degeneration in RAG2 was in X. andrei RAG2 paralog β2, which experienced a frameshift deletion.
Independent degeneration of RAG1 paralogs:
To test whether the unique degenerations in the 3′ region of RAG1 (Evans et al. 2005) could have occurred after a shared ancestral degeneration in the 5′ coding region, RAG1 sequences were obtained from most of the coding region of most or all RAG1 paralogs of all known species of clawed frog (supplemental Figure 2 at http://www.genetics.org/supplemental/). Including previously identified degenerate paralogs (Evans et al. 2005), a total of 17 degenerate RAG1 β paralogs and 2 degenerate RAG1 α paralogs were detected in various Xenopus species (Figure 2, supplemental Figure 2). The only shared stop codons or frameshift mutations that were identified were (1) one frameshift and one stop codon shared by X. pygmaeus paralog β and X. ruwenzoriensis paralog β3, (2) a stop codon shared by X. vestitus paralog β1 and X. longipes paralog β3, and (3) a stop codon shared by the X. ruwenzoriensis paralog β1 and X. vestitus paralog β2. Maximum likelihood reconstructions indicate that the first example is due to shared ancestry, but that the other two evolved independently.
Expression of at least one RAG1 paralog was confirmed in heart, brain, liver, testes, spleen, and bone marrow (Table 1). Xenopus laevis and X. gilli each express both of their RAG1 paralogs and neither is degenerate. Likewise, no degeneration was observed at the DNA level in either RAG1 paralog of some other tetraploids including X. clivii, X. largeni, and X. muelleri (supplemental Figure 2 at http://www.genetics.org/supplemental/).
TABLE 1.
Expression of RAG1 paralogs in different Xenopus species
| Species (ploidy) | Tissue | Expected paralogs | Observed expressed paralogs | Cloned (C) or directly sequenced (DS) |
|---|---|---|---|---|
| S. new tetraploid 1 (4x = 40) | Brain | α, β | α, β | C |
| Testes | α, β | α, β | DS | |
| Liver | α, β | α, β | DS | |
| X. laevis (4x = 36) | Bone marrow | α, β | α, β | C, DS |
| Heart | α, β | α | DS | |
| Liver | α, β | α, β | DS | |
| Spleen | α, β | α, β | DS | |
| Liver | α, β | α, β | DS | |
| X. gilli (4x = 36) | Bone marrow | α, β | α, β | C |
| X. borealis (4x = 36) | Brain | α, β | α | DS |
| Liver | α, β | α | DS | |
| Testes | α, β | α | DS | |
| X. muelleri (4x = 36) | Brain | α, β | α | DS |
| Testes | α, β | α | DS | |
| Bone marrow | α, β | α | DS | |
| X. amieti (8x = 72) | Liver | α1, α2, β1, β2 | α1, α2a | C |
| Brain | α1, α2, β1, β2 | α1 | DS | |
| X. andrei (8x = 72) | Bone marrow | α1, α2, β1, β2 | α1, α2, β1a | C, DS |
| Spleen | α1, α2, β1, β2 | α1 | DS | |
| Brain | α1, α2, β1, β2 | α1 | DS | |
| X. new octoploid (8x = 72) | Bone marrow | α1, α2, β1, β2 | α1, α2 | C |
| Brain | α1, α2, β1, β2 | α1, α2 (low) | DS | |
| Heart | α1, α2, β1, β2 | α1 (low), α2 | DS | |
| Testes | α1, α2, β1, β2 | α1, α2 | C | |
| X. boumbaensis (8x = 72) | Bone marrow | α1, α2, β1, β2 | α1, α2, β1a | C |
| Brain | α1, α2, β1, β2 | α1, β1a | DS | |
| Heart | α1, α2, β1, β2 | β1a | DS |
Expression was confirmed by amplifying cDNA across an intron and either cloning (C) or directly sequencing (DS) the PCR product.
Expressed paralogs that are degenerate.
Degeneration of many paralogs appears to have occurred independently because (a) the stop codons and frameshift mutations are not shared with other paralogs and because (b) these degenerate paralogs are either still expressed or closely related to other expressed paralogs. X. andrei RAG1 paralog β1, X. boumbaensis RAG1 paralog β1, and X. amieti RAG1 paralog α2, for example, are expressed even though each one is degenerate (degenerations 4, 7, and 9, respectively, in Figures 2 and 4). Overall, under the assumptions of no resuscitation of silenced genes, phylogenetic analyses support a minimum of seven independent episodes of degeneration of the Xenopus RAG1 β paralogs and two independent episodes in the Xenopus RAG1 α paralogs (Figures 2 and 4).
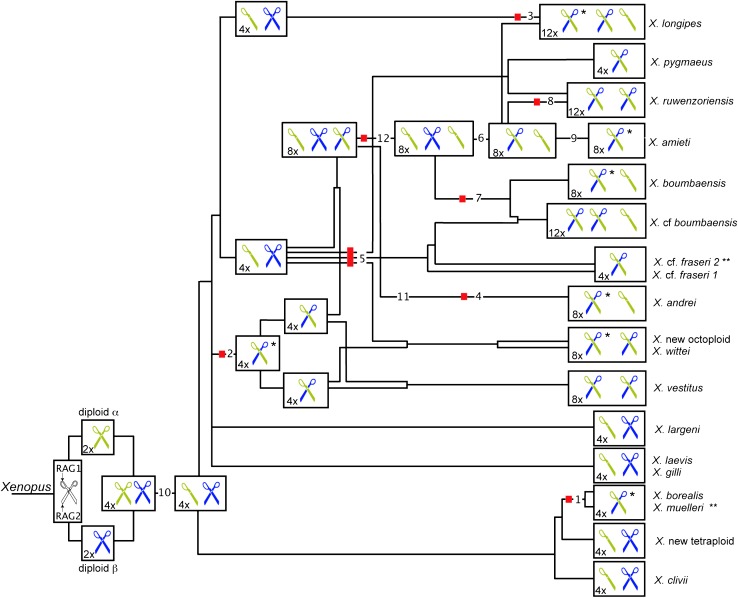
Figure 4.—
Evolution of RAG1 and RAG2 in Xenopus. The RAG1 and RAG2 heterodimer is depicted as a scissor and the evolutionary trajectory of this heterodimer is followed through the reticulate phylogeny that is depicted in Figure 3, including degeneration and deletions of paralogs encoding each protein that are depicted in Figure 2. Proteins encoded by paralogs inherited from diploid ancestor α are yellow-green and those encoded by paralogs inherited from diploid ancestor β are blue. Numbered deletion and degeneration events from Figure 2 are labeled and the ploidy of each ancestor or extant species (2x, 4x, 8x, or 12x) is indicated inside each box. Species and ancestors for which simulations were performed are indicated by a red box. As a result of biased degeneration of RAG1, heterodimers of many extant species must form between proteins that were ultimately inherited from different diploid species (i.e., the scissor is composed of a yellow-green RAG1 protein and a blue RAG2 protein). Single asterisks indicate a minimum of seven independently evolved chimerical heterodimers. Double asterisks indicate caveats that X. cf. fraseri had no observed degeneration of RAG1 paralog β, but that only a small fragment of this paralog was sequenced (supplemental Figure 2 at http://www.genetics.org/supplemental/), and that expression of paralog β was not detected in X. muelleri but no degeneration was observed in the coding region.
The minimum seven instances of gene silencing RAG1 β paralogs, listed by numbers corresponding with Figures 2 and 4, include (1) a heterozygous stop codon in X. borealis RAG1 paralog β—expression of this paralog also was not detected in multiple tissues (Table 1), (2) RAG1 paralog β of a tetraploid ancestor of both of the tetraploid ancestors of X. vestitus, (3) RAG1 paralog β3 of X. longipes or its tetraploid ancestor, (4) X. andrei RAG1 paralog β1, (5) X. wittei RAG1 paralog β1, (6) X. amieti RAG1 paralog β1, and (7) X. boumbaensis RAG1 paralog β1; the two examples of degeneration in the RAG1 α paralogs are (8) X. ruwenzoriensis paralog α2 and (9) X. amieti paralog α2 (Figure 2). Both instances of degeneration of RAG1 paralog α occurred very recently and potentially after interactions between RAG1 paralog α and RAG2 paralog β proteins were already established by pseudogenization of other RAG1 paralogs.
Nine additional instances of degeneration of RAG1 β paralogs are possible but their independence depends on whether ancestral gene silencing preceded autapomorphic degeneration of the coding regions of various paralogs (Figure 2). Expression of X. muelleri RAG1 paralog β, for example, was not detected in multiple tissues (Table 1) and expression of this paralog may have been silenced ancestrally prior to the speciation of X. muelleri and X. borealis and coding-region degeneration in X. borealis (degeneration 1 in Figures 2 and 4). Likewise, a lack of detected expression of X. new octoploid RAG1 paralog β2 could be explained by silenced expression of paralog β in one of the tetraploid ancestors of X. vestitus, X. wittei, and X. new octoploid. If this were the case, then degeneration of RAG1 paralog β1 and β2 in X. vestitus, paralog β2 in X. wittei, and paralog β2 in X. new octoploid should be considered a single event (degeneration 2 in Figures 2 and 4), even though no shared degeneration of the coding region of these paralogs was observed (supplemental information 2 at http://www.genetics.org/supplemental/).
Parametric bootstrap tests strongly reject hypotheses of fewer independent degenerations in the β lineage of RAG1 (P < 0.001). Of note is that the full sequence of X. boumbaensis RAG1 paralog β1 was not obtained (supplemental Figure 2 at http://www.genetics.org/supplemental/) so the possibility of shared degeneration with another closely related paralog cannot be completely dismissed. Also of interest is the observation that in X. new octoploid, the intensity of paralog-specific polymorphisms on sequence chromatograms suggests that expression of RAG1 paralog α1 was higher than paralog α2 in the brain, whereas the opposite was observed in amplifications of RAG1 from heart cDNA from this species (Table 1), a pattern of expression that is suggestive of subfunctionalization (Force et al. 1999).
Significant bias to RAG1 degeneration:
Comparison of alternative parameterizations of a model of stochastic degeneration indicates that the rate of degeneration of the RAG1 β lineage is significantly higher than the RAG1 α lineage with a gamma rate heterogeneity model (P = 0.0274, δRAG1α = 6.5519, δRAG1β = 50.0343, γRAG1α = 0.0082, γRAG1β = 0.0042, d.f. = 2), or without one (P = 0.0054, δRAG1α = 4.8927, δRAG1β = 34.4187, d.f. = 1). Additionally, the overall rate of degeneration of RAG1 is significantly higher than the rate of degeneration of RAG2 when modeled with a gamma rate heterogeneity model (P = 0.0431, δRAG1 = 31.2407, δRAG2 = 3.08511, γRAG1 = 0.0082, γRAG2 = 0.0096, d.f. = 2), or without one (P = 0.0323, δRAG1 = 7.8978, δRAG2 = 1.8141, d.f. = 1).
Because some species may have inherited RAG1 β paralogs that were already degenerate, even if subsequent degeneration of RAG1 were unbiased, chimerical heterodimers would still be expected to occur. However, under the assumptions discussed above and given the observed pattern of deletion in RAG2, simulations indicate that the observed number of chimerical heterodimers that resulted from RAG1 degeneration is significantly in excess of expectations if RAG1 degeneration were unbiased (P < 0.0001, Figure 5).
Figure 5.—
Simulations of the expected distribution of chimerical heterodimers (those formed between proteins encoded by from paralogs derived from different diploid ancestors) given the observed degeneration and deletion of RAG2 and phylogenetically independent and unbiased deletion of RAG1. Simulations were performed on the eight species or ancestors as indicated in Figure 4. An asterisk indicates the 10 observed chimerical heterodimers in the species for which the simulations were performed, including 7 that evolved independently, but not counting the one of the chimerical heterodimers in X. wittei that may have been inherited from one of the tetraploid ancestors that was simulated. The observed number of chimerical heterodimers depart significantly from expectations under no bias (P < 0.0001).
Recombination between RAG1 paralogs and between RAG2 paralogs is rare:
Using congruence of multiple tests as a criterion for credibility (Posada and Crandall 2001; Posada 2002), there is not convincing evidence of recombination between paralogs of RAG1, including when new data are included, or between paralogs of RAG2 (this study; Evans et al. 2005). No two tests recovered significant evidence of recombination for the same region of any paralog of RAG1 or RAG2 and most tests did not recover any signal of recombination at all. A few putative recombination events that had a parent sequence within the same species were individually identified by various tests but these were deemed not credible on the basis of a visual inspection of the data and a lack of corroboration by other tests (Posada and Crandall 2001; Posada 2002).
Linkage of RAG1 paralog β and RAG2 paralog β has been demonstrated (Greenhalgh et al. 1993) and a similar linkage structure is suggested by a putative deletion that spanned RAG1 and RAG2 paralogs of an ancestor of X. amieti, X. ruwenzoriensis, X. boumbaensis, X. cf. boumbaensis, and X. longipes (deletion 12 in Figure 2). The possibility that recombination occurred between the intergenic region that separates linked alleles of RAG1 and RAG2 is difficult to conclusively rule out because a diploid Xenopus (2x = 18), which could confirm the ancestral synteny of RAG1 and RAG2 paralogs, is not available. However, phylogenetic congruence among RAG1, RAG2, and mtDNA, and the internal consistency between the α and β genealogies of RAG1 support the contention that recombination between these paralogs is rare (Figure 2; Evans et al. 2004, 2005). Recombination among paralogous alleles is expected to be rare if diploidization of these allopolyploid genomes occurred soon after their formation, which is typical of allopolyploid cotton, for example (Cronn et al. 1999).
DISCUSSION
Evolution of a heterodimer that was duplicated by allopolyploidization:
The crucial process of V(D)J recombination requires the action of a heterodimer that is made up of the RAG1 and RAG2 proteins, which are encoded by tightly linked genes. The genes whose protein products form this heterodimer were duplicated, probably by allo- rather than autopolyploidization, to generate a tetraploid ancestor of all extant species in the genus Xenopus. Because allotetraploids inherit a complete genome from two different diploid species, this tetraploid ancestor initially had two linked sets of RAG1 and RAG2 paralogs. Immediately after allopolyploidization, the RAG1/RAG2 heterodimers probably had essentially identical functions, but their dosage could have varied as a result of unique mutations that occurred in each of the diploid ancestors. At this point the paralogous expression domains were also probably similar, making possible interactions between proteins encoded by paralogs that were inherited from different diploid ancestors. These linked sets of paralogs were then duplicated again when multiple independent episodes of allopolyploidization generated new octoploid and dodecaploid species (Figure 3; Evans et al. 2005).
Before the tetraploid Xenopus ancestor diversified to give rise to the extant species, it appears that one RAG2 paralog—paralog α—was deleted, leaving only the other RAG2 paralog—paralog β—but still two functional paralogs of RAG1, α and β (Figures 2 and 4). Later, after speciation of this tetraploid ancestor without change in genome size and also after further episodes of allopolyploidization, paralogs of RAG1 in different species independently degenerated, making their copy number similar or equal to RAG2. Surprisingly, until very recently degeneration of RAG1 paralogs occurred exclusively in closely related members of paralog lineage β. As a result, many Xenopus species have functional paralogs of RAG1 that are exclusively α paralogs, and their protein products must heterodimerize with proteins encoded by RAG2 β paralogs (Figures 2 and 4). Moreover, in many species it appears that the functional copy of RAG1 is linked to a region where a paralog of RAG2 was deleted, and the functional copy of RAG2 is linked to a paralog of RAG1 that either degenerated or was deleted. While the tetraploid species X. laevis and X. gilli still express functional linked β paralogs of RAG1 and RAG2 and also an α paralog of RAG1, in most other Xenopus species the only functional paralogs of RAG1 and RAG2 are unlinked, suggesting that each one was derived from a different diploid ancestor. In contrast, allotetraploid clawed frogs that evolved independently in the genus Silurana (4x = 40) retain both paralogs of RAG1 and of RAG2, all appear functional at the DNA level in the portions of these genes that were sequenced, and both RAG1 paralogs are expressed in at least one Silurana tetraploid (S. new tetraploid 1).
The rate of degeneration is significantly higher in RAG1 β paralogs than in RAG1 α paralogs and significantly higher in RAG1 than in RAG2. Simulations also indicate that, given the observed pattern of degeneration of RAG2, the probability of unbiased degeneration of RAG1 producing by chance such a high number of chimerical heterodimers derived from unlinked paralogs of RAG1 and RAG2 is very low. It is surprising that degeneration of RAG1 paralogs occurred in this manner because an allopolyploid origin of the tetraploid ancestor of Xenopus would suggest that unlinked paralogs share a shorter coevolutionary history than do the linked ones.
Unique characteristics of each subgenome could account for the biased degeneration of RAG1 paralog β. Epigenetic phenomena after allopolyploidization could contribute to this bias, for example, if paralog expression were differently affected by asymmetric mobility of transposable elements in each subgenome. Alternatively, genetic explanations for nonrandom degeneration of RAG1 paralogs include (1) “intraparalog” phenomena, such as differences in dosage of each RAG1 paralog, or (2) “intermolecular” phenomena involving selection on interactions between the protein products of specific RAG1 paralogs and other molecules. This second genetic explanation includes scenarios involving a disadvantage (negative selection) or an advantage (positive selection) to interactions between proteins derived from specific paralogs of RAG1 and other molecules, which may or may not include proteins encoded by specific paralogs of RAG2.
Dosage as an explanation for nonrandom gene silencing of RAG1:
One explanation for biased degeneration of RAG1 is that, after allotetraploidization in Xenopus, expression of the RAG1 paralog that was derived from diploid ancestor α was greater than the one that was derived from diploid ancestor β. If this difference in dosage meant that the RAG1 paralog α could operate alone in a polyploid genome whereas RAG1 paralog β could not, then the former would be both necessary and sufficient. Under this scenario, the function of each RAG1 paralog would be identical and interchangeable, and the difference driving degeneration of RAG1 lineage β would be one of quantity, not of quality, of proteins encoded by RAG1 paralogs. However, because copy number of RAG2 was reduced by deletion in an ancestor before any RAG1 copies degenerated, dosage constraints on RAG1 after allopolyploidization would have to have been imposed by other cofactors of RAG1.
Haplo-insufficient phenotypes result from reduced expression or activity of a heterozygous locus, and these phenotypes could stem from multiple mechanisms including altered enzymatic stoichiometry (Veitia 2002). The importance of stoichiometric requirements is suggested in yeast in that proteins that form complexes are rarely members of large gene families and underexpression or overexpression of these genes can be deleterious (Papp et al. 2003). In the same way, ancestral differences in gene dosage could influence allopolyploid phenotypes and ultimately affect the genetic fate of paralogs in an allopolyploid genome. For example, laboratory generated allopolyploids with two Z chromosomes from X. gilli and one W chromosome from X. laevis were mostly male whereas individuals with the same W chromosome but one Z chromosome from X. gilli and one Z chromosome from X. muelleri (or X. laevis) were only female (Kobel 1996). Dosage could also be an important factor in the evolution of dominance. That wild-type alleles are generally dominant over new mutations could be a byproduct of selection for surplus capability that is needed to operate under heterogeneous conditions (Wright 1934; Charlesworth 1979; Kacser and Burns 1981; Orr 1991; Forsdyke 1994).
Selection on interactions between specific paralogs of RAG1 and other molecules:
If the tetraploid ancestor of Xenopus evolved through allopolyploidization, negative selection on molecular interactions could arise from Dobzhansky–Muller incompatibilities (Dobzhansky 1937; Muller 1942). However, Dobzhansky–Muller incompatibilities between paralogs of RAG1 and RAG2 could explain the degeneration of specific RAG1 paralogs only if the paralogs that are currently functional were derived from the same diploid ancestor. But this does not appear to be the case because comparison to linked paralogs in X. laevis (Greenhalgh et al. 1993) suggests that in most species the only functional RAG1 and RAG2 paralogs are not in synteny. Thus, similar to the dosage hypothesis, if Dobzhansky–Muller incompatibilities drove degeneration of RAG1 paralog β, they would probably involve an interaction other than that with the protein product of the RAG2 gene. This could include interactions with other protein cofactors of V(D)J recombination or with the recombination signal sequences (RSSs) or the spacer regions between RSSs that flank variable, diversity, and joining segments (Sakano et al. 1979; Ramsden et al. 1994).
If the tetraploid ancestor of Xenopus was allopolyploid, positive selection could account for this pattern of gene silencing in at least two ways. First, coadaptation of paralogs of RAG1 and RAG2 could have occurred in one of the diploid ancestors and then these paralogs could have been unlinked by recombination after allopolyploidization. However, the coadaptation hypothesis is disfavored for the same reason that Dobzhansky–Muller incompatibilities between RAG1 and RAG2 are an unlikely explanation: recombination between alleles of different paralogs appears rare and functional RAG1 and RAG2 paralogs in many species therefore are probably derived from different diploid ancestors. A second possibility is that there is a performance advantage to protein–protein interactions between RAG1 and RAG2 paralogs that are derived from different diploid ancestors. This scenario posits an advantage, or heterosis, to a combination of two evolutionarily naive proteins over a heterodimer whose constituents have a longer period of coevolutionary history. Heterosis is also suggested, for example, in Xenopus polyploids that are resistant to parasites that infect both of their parental taxa (Jackson and Tinsley 2003).
Explanations for heterosis include overdominance, dominance, or epistasis. The overdominance hypothesis posits that heterozygous loci are more fit than homozygous loci (whether dominant or recessive) (Shull 1908; Hull 1945), and this could apply to allopolyploids that coexpress alleles from each parental species. In fact, allopolyploidization provides a way to maintain heterosis associated with heterozygosity because alleles from each parental species are forced to cosegregate in disomic allopolyploids. In the current case, however, the overdominance explanation for heterosis does not apply because, while many allopolyploids have a RAG1/RAG2 heterodimer composed of proteins derived from genes with different ancestry, each paralog that encodes the proteins in these heterodimers is homozygous with respect to their diploid ancestry. The dominance explanation for heterosis posits that if dominant alleles are more fit, hybrids (or allopolyploids) would benefit from the dominant alleles from each parent (Davenport 1908). Dominance can result from differences in dosage, which was discussed earlier, and could also be a consequence of epistasis, so these explanations for heterosis are not mutually exclusive. Epistasis could lead to heterosis in an allopolyploid if there were a performance advantage to interactions between proteins derived from different ancestors.
Taken together, these observations suggest that allopolyploid transcriptomes are sculpted by natural selection on each subgenome. Mutational or regulatory differences that accumulated in each ancestor may be advantageous or deleterious, and their paralogs can be preserved or discarded after genome fusion on the basis of their performance and their interactions with molecules from the other subgenome. In some cases, pseudogenization is strongly biased with respect to ancestry over millions of years, and chimerical interactions between proteins from different ancestors may be favored over interactions between proteins that share a longer period of coevolution.
Acknowledgments
I thank Brian Golding for helpful discussion about this project and for access to computational resources in his laboratory and to two anonymous reviewers for providing comments on this manuscript. This research was supported by the Canadian Foundation for Innovation, the National Science and Engineering Research Council, the Ontario Research and Development Challenge Fund, and McMaster University.
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., R. Cronn, R. Percifeld and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100 4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., R. Percifeld and J. F. Wendel, 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain, F. J. J., and B. J. Evans, 2006. Molecular evolution of duplicate genes in Xenopus laevis is consistent with multiple mechanisms for their retained expression. PLoS Genet. 2 e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1979. Evidence against Fisher's theory of dominance. Nature 278 848–849. [Google Scholar]
- Comai, L., 2000. Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43 387–399. [DOI] [PubMed] [Google Scholar]
- Cronn, R. C., R. L. Small and J. F. Wendel, 1999. Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 96 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, C. B., 1908. Degeneration, albinism and inbreeding. Science 28 454–455. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Evans, B. J., D. B. Kelley, R. C. Tinsley, D. J. Melnick and D. C. Cannatella, 2004. A mitochondrial DNA phylogeny of clawed frogs: phylogeography on sub-Saharan Africa and implications for polyploid evolution. Mol. Phylogenet. Evol. 33 197–213. [DOI] [PubMed] [Google Scholar]
- Evans, B. J., D. B. Kelley, D. J. Melnick and D. C. Cannatella, 2005. Evolution of RAG-1 in polyploid clawed frogs. Mol. Biol. Evol. 22 1193–1207. [DOI] [PubMed] [Google Scholar]
- Force, A., M. Lynch, B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsdyke, D. R., 1994. The heat-shock response and the molecular basis of genetic dominance. J. Theor. Biol. 167 1–5. [DOI] [PubMed] [Google Scholar]
- Gibbs, M. J., J. S. Armstrong and A. J. Gibbs, 2000. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16 573–582. [DOI] [PubMed] [Google Scholar]
- Goldman, N., J. P. Anderson and A. G. Rodrigo, 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49 652–670. [DOI] [PubMed] [Google Scholar]
- Greenhalgh, P., C. E. M. Olesen and L. A. Steiner, 1993. Characterization and expression of Recombination Activating Genes (RAG-1 and RAG-2) in Xenopus laevis. J. Immunol. 151 3100–3110. [PubMed] [Google Scholar]
- Gu, X., 2003. Evolution of duplicate genes versus genetic robustness against null mutations. Trends Genet. 19 354–356. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., D. M. Hillis and R. Jones, 1996. Parametric bootstrapping in molecular phylogenetics: applications and performance, pp. 19–45 in Molecular Zoology: Advances, Strategies, and Protocols, edited by J. D. Ferraris and S. R. Palumbi. Wiley-Liss, New York.
- Hughes, M. K., and A. L. Hughes, 1993. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 10 1360–1369. [DOI] [PubMed] [Google Scholar]
- Hull, F. G., 1945. Recurrent selection and specific combining ability in corn. J. Am. Soc. Agronomy 37 134–145. [Google Scholar]
- Jackson, J. A., and R. C. Tinsley, 2003. Parasite infectivity to hybridising host species: A link between hybrid resistance and allopolyploid speciation? Int. J. Parasitol. 33 137–144. [DOI] [PubMed] [Google Scholar]
- Jiang, C.-X., P. W. Chee, X. Draye, P. L. Morrell, C. W. Smith et al., 2000. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution 54 798–814. [DOI] [PubMed] [Google Scholar]
- Kacser, H., and J. A. Burns, 1981. The molecular basis of dominance. Genetics 97 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V., and J. Jurka, 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3 e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel, H. R., 1996. Allopolyploid speciation, pp. 391–401 in The Biology of Xenopus, edited by R. C. Tinsley and H. R. Kobel. Clarendon Press, Oxford.
- Lee, H.-S., and Z. J. Chen, 2001. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A. A., and M. Feldman, 2004. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. 82 607–613. [Google Scholar]
- Liu, B., and J. F. Wendel, 2002. Non-mendelian phenomena in allopolyploid genome evolution. Curr. Genomics 3 489–505. [Google Scholar]
- Liu, B., and J. F. Wendel, 2003. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29 365–379. [DOI] [PubMed] [Google Scholar]
- Liu, B., C. L. Brubaker, G. Mergeai, R. C. Cronn and J. F. Wendel, 2001. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44 321–330. [PubMed] [Google Scholar]
- Loumont, C., 1983. Deux especes nouvelles de Xenopus du Cameroun (Amphibia, Pipidae). Rev. Suisse Zool. 90 169–177. [Google Scholar]
- Lukens, L. N., P. A. Quijada, J. Udall, J. C. Pires, M. E. Schranz et al., 2004. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 82 665–674. [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., M. O'Hely, B. Walsh and A. Force, 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, D. R., and W. P. Maddison, 2000. MacClade. Sinauer Associates, Sunderland, MA.
- Madlung, A., R. Masuelli, B. Watson, S. H. Reynolds, J. Davison et al., 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D., and E. Rybicki, 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16 562–563. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34 126–129. [DOI] [PubMed] [Google Scholar]
- Moscone, E. A., M. A. Matzke and A. J. M. Matzke, 1996. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105 231–236. [PubMed] [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Nylander, J. A. A., F. Ronquist, J. P. Huelsenbeck and J. L. Nieves-Aldrey, 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53 47–67. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin.
- Orr, H. A., 1991. A test of Fisher's theory of dominance. Proc. Natl. Acad. Sci. USA 88 11413–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan, H., A. A. Levy and M. Feldman, 2001. Allopolyploidy-induced rapid genomic evolution of the wheat (Aegilops-Triticum) group. Plant Cell 13 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam, M., S. Sawyer and C. M. Fauquet, 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265 218–225. [DOI] [PubMed] [Google Scholar]
- Pagel, M., 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. Ser. B 255 37–45. [Google Scholar]
- Papp, B., C. Pál and L. D. Hurst, 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424 194–197. [DOI] [PubMed] [Google Scholar]
- Posada, D., 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19 708–717. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A., and N. C. Grassly, 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13 235–238. [DOI] [PubMed] [Google Scholar]
- Ramsden, D. A., K. Baetz and G. E. Wu, 1994. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 22 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky, M. J., J. E. Hesse, J. F. McBlane and M. Gellert, 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano, H., K. Huppi, G. Heinrich and S. Tonegawa, 1979. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280 288–294. [DOI] [PubMed] [Google Scholar]
- Salminen, M. O., J. K. Carr, D. S. Burke and F. E. McCutchan, 1995. Identification of breakpoints in intergenotypic recombinants of HIV-1 by bootscanning. AIDS Res. Hum. Retroviruses 11 1423–1425. [DOI] [PubMed] [Google Scholar]
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull, G. H., 1908. The composition of a field of maize. Am. Breed. Assoc. 4 296–301. [Google Scholar]
- Skalická, K., K. Y. Lim, R. Matyasek, M. Matzke, A. R. Leitch et al., 2005. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 166 291–303. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., P. S. Soltis, J. C. Prires, A. Kovarik, J. A. Tate et al., 2004. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Lin. Soc. 82 485–501. [Google Scholar]
- Swofford, D. L., 2002. Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Tymowska, J., 1991. Polyploidy and cytogenetic variation in frogs of the genus Xenopus, pp. 259–297 in Amphibian Cytogenetics and Evolution, edited by D. S. Green and S. K. Sessions. Academic Press, San Diego.
- Veitia, R. A., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24 175–184. [DOI] [PubMed] [Google Scholar]
- Volkov, R. A., N. V. Borisjuk, I. I. Panchuk, D. Schweizer and V. Hemleben, 1999. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol. 16 311–320. [DOI] [PubMed] [Google Scholar]
- Wang, J., L. Tian, A. Madlung, H.-S. Lee, M. Chen et al., 2004. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 168 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., A. Schnabel and T. Seelanan, 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 92 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1934. Molecular and evolutionary theories of dominance. Am. Nat. 63 24–53. [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS 13 555–556. [DOI] [PubMed] [Google Scholar]
- Zwierzykowski, Z., R. Tayyar, M. Brunell and A. J. Lukaszewski, 1998. Genome recombination in intergeneric hybrids between tetraploid Festuca pratensis and Lolium multiflorum. J. Hered. 89 324–328. [Google Scholar]