Abstract
Association mapping focused on 36 genes involved in branch development was used to identify candidate genes for variation in shoot branching in Arabidopsis thaliana. The associations between four branching traits and moderate-frequency haplogroups at the studied genes were tested in a panel of 96 accessions from a restricted geographic range in Central Europe. Using a mixed-model association-mapping method, we identified three loci—MORE AXILLARY GROWTH 2 (MAX2), MORE AXILLARY GROWTH 3 (MAX3), and SUPERSHOOT 1 (SPS1)—that were significantly associated with branching variation. On the basis of a more extensive examination of the MAX2 and MAX3 genomic regions, we find that linkage disequilibrium in these regions decays within ∼10 kb and trait associations localize to the candidate genes in these regions. When the significant associations are compared to relevant quantitative trait loci (QTL) from previous Ler × Col and Cvi × Ler recombinant inbred line (RIL) mapping studies, no additive QTL overlapping these candidate genes are observed, although epistatic QTL for branching, including one that spans the SPS1, are found. These results suggest that epistasis is prevalent in determining branching variation in A. thaliana and may need to be considered in linkage disequilibrium mapping studies of genetically diverse accessions.
EVOLUTIONARY change in shoot architecture has played a central role in the morphological diversification of plant species, although relatively little is known about its molecular basis (Sussex and Kerk 2001). Only a small number of genes have been implicated in shoot architectural evolution (Bradley et al. 1997; Purugganan and Suddith 1998; Purugganan et al. 2000; Gallavotti et al. 2004; Yoon and Baum 2004; Vollbrecht et al. 2005), with the most comprehensively understood example coming from the maize teosinte branched 1 (tb1) gene (Doebley et al. 1997). In studies of morphological evolution under domestication, it has been demonstrated that tb1, a TCP class transcription factor, was a target of selection for reduced tillering during the evolutionary origin of maize from its wild ancestor teosinte (Wang et al. 1999; Clark et al. 2004, 2006). A fuller understanding of the evolutionary basis of plant shoot variation can be achieved only through the continued identification of the molecular mechanisms responsible for the vast diversity in plant architectures.
One broad component of shoot architecture is branching pattern. Developmental regulation of branching occurs at several levels, including (i) node patterning, (ii) meristem determination, and (iii) axillary meristem elongation (McSteen and Leyser 2005). Several genes have been shown to affect node patterning in the model plant species Arabidopsis thaliana, including LATERAL SUPPRESSOR (LAS) (Greb et al. 2003), SHOOT MERISTEMLESS (STM) (Long et al. 1996), REVOLUTA (REV) (Talbert et al. 1995), and the REGULATORS OF AXILLARY MERISTEMS (RAX) genes (Keller et al. 2006; Muller et al. 2006). The determination of inflorescence meristem identity is largely controlled by the floral identity genes TERMINAL FLOWER1 (TFL1) (Bradley et al. 1997) and LEAFY (LFY) (Weigel et al. 1992). Branch elongation is regulated by numerous phytohormones, such as auxin, cytokinin, and abscicic acid (Ward and Leyser 2004). Analyses of certain auxin signaling genes, such as AUXIN RESISTANT1 (AXR1) (Lincoln et al. 1990; Leyser et al. 1993; Stirnberg et al. 1999), as well of genes that appear to regulate auxin transport, such as the MORE AXILLARY GROWTH (MAX) genes (Stirnberg et al. 2002; Sorefan et al. 2003; Booker et al. 2005; Bennett et al. 2006), have shown that this hormone plays a crucial role in coordinating branch outgrowth with the plant developmental program.
In A. thaliana, it has been shown that significant variation in branch number and other quantitative aspects of shoot architecture exist (Ungerer et al. 2002). Quantitative trait locus (QTL) mapping has historically been used as the main approach to mapping genes responsible for variation in ecologically and evolutionary significant traits (Lynch and Walsh 1998) and QTL mapping experiments have identified numerous loci that may be responsible for branching variation (e.g., Ungerer et al. 2002, 2003). Recently, however, association or linkage disequilibrium (LD) mapping has emerged as a serious alternative to identifying genes underlying quantitative phenotypes and has been used with greater frequency in mapping traits in A. thaliana (Mitchell-Olds and Schmitt 2006), maize (Yu et al. 2006), Drosophila (Mackay 2004), and humans (Cardon and Abecasis 2003). Association studies in A. thaliana have covered a broad spectrum of genomic scales, ranging from candidate gene analysis (Hagenblad and Nordborg 2002; Caicedo et al. 2004; Hagenblad et al. 2004; Olsen et al. 2004) to genomewide scans (Aranzana et al. 2005; Zhao et al. 2007).
Since rarely, if ever, is it possible to pinpoint a priori which genes in a genetic pathway or network are likely to possess functional polymorphism(s), large-scale analyses of all genes in a trait's genetic network are a necessary next step in the candidate gene approach. The candidate gene approach is likely to be particularly useful in A. thaliana, since the relatively rapid decay of LD in its genome (<25 kb) suggests that trait associations will often span a few loci and possibly delimit functionally significant polymorphism(s) to appropriate candidate loci (Mitchell-Olds and Schmitt 2006). This mapping resolution will facilitate a more expedient characterization of the genetic basis of natural phenotypic variation in this species, although the promise of association mapping is dependent on our ability to differentiate significant results that are biologically informative from spurious associations. This continues to be a substantial challenge, as high false-positive rates for association mapping have been found in A. thaliana (Aranzana et al. 2005; Zhao et al. 2007).
Spurious associations typically occur when the demographic structure of a population is correlated with trait variation (Pritchard et al. 2000b), but this problem is more pronounced in A. thaliana due to the selfing nature of this species and the geographic isolation of subpopulations with distance (Zhao et al. 2007). Methods to account for demographic structure in association mapping panels have been developed by incorporating estimates of population ancestry (Pritchard et al. 2000b; Price et al. 2006) and kinship (Yu et al. 2006) into genotype–phenotype association tests. In general, these methods dramatically improve the power of association mapping to detect functional genetic variation, but do not totally resolve confounding demographic structure. Thus, results from association studies in this species must be cautiously evaluated and, ideally, replicated through other approaches, such as QTL mapping or transformation experiments.
We explore the genetics of branching variation in A. thaliana through association mapping focused on candidate genes involved in branch development. We initially sequenced ∼600-bp fragments of 36 genes involved in shoot architectural development from a set of 24 geographically diverse A. thaliana accessions. Common haplogroups (≥10%) of these genes were genotyped in 96 geographically restricted accessions for association mapping. Several genes show nominally significant associations with branching, and we localize two of these associations to the gene level through a more extensive analysis of linked genomic regions. None of the observed associations are detected as additive QTL within the Ler × Col or Cvi × Ler recombinant inbred lines (RILs), although reanalysis of the QTL mapping data shows numerous epistatic interactions underlying branching variation. These results provide an opportunity to compare candidate gene association studies with QTL mapping analyses and suggest the possibility that epistatic interactions should be considered in association-mapping investigations.
MATERIALS AND METHODS
Amplification and sequencing of the candidate genes and their genomic regions:
Candidate genes included in this study are listed in Table 1. Amplification primers were designed in Primer3 (http://fokker.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), using the Col-0 genome sequence. Accessions used for sequencing are noted in supplemental data at http://www.genetics.org/supplemental/. PCRs were conducted using standard reaction conditions and Perkin-Elmer (Norwalk, CT) thermalcyclers. Either Taq or Ex-Taq (TaKaRa, Otsu, Japan) polymerases were used for PCR amplification. PCR products were purified using QIAGEN (Hilden, Germany) PCR purification kits or QIAGEN gel extraction kits or ExoSAP (Invitrogen, Carlsbad, CA). Sequencing reactions were performed with BigDye terminators (ABI; Applied Biosystems, Foster City, CA). Sequencing was conducted at the North Carolina State University Genome Research Lab or at the New York University Genetic Analysis Center on ABI 3100, 3700, or 3730 capillary sequencers according to standard protocols.
TABLE 1.
Genes included in this study
| Gene | Abbreviation | Gene ID | Annotation |
|---|---|---|---|
| ABA INSENSITIVE 3 | ABI3 | At3g24650 | Transciption factor |
| ALTERED MERISTEM PROGRAM 1 | AMP1 | At3g54720 | Glutamate carboxypeptidase |
| AINTEGUMENTA | ANT | At4g37750 | Transcription factor |
| APETALA 1 | AP1 | At1g69120 | MADS transcription factor |
| AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE | ARGOS | At3g59900 | Auxin-inducible gene that controls lateral organ size |
| AUXIN RESISTANT 1 | AXR1 | At1g05180 | Ubiquitin-activating enzyme E1-related protein |
| AUXIN RESISTANT 2 | AXR2 | At3g23050 | Auxin-responsive protein/indoleacetic acid-induced protein 7 (IAA7) |
| AUXIN RESISTANT 3 | AXR3 | At1g04250 | IAA17 |
| AUXIN RESISTANT 6 | AXR6 | At4g02570 | Cullin family protein |
| BREVIPEDICELLUS | BP | At4g08150 | Homeobox protein knotted-1 like 1 |
| BUSHY AND DWARF 1 | BUD1 | At1g18350 | MAP kinase kinase7 |
| CAULIFLOWER | CAL | At1g26310 | MADS transcription factor |
| CYTOKININ-INDEPENDENT 1 | CKI1 | At2g47430 | Cytokinin-responsive histidine kinase |
| CYTOKININ RESPONSE 1 | CRE1 | At2g01830 | Histidine kinase AHK4 |
| EMBRYONIC FLOWER 1 | EMF1 | At5g11530 | Regulates reproductive development |
| ERECTA | ER | At2g26330 | Receptor protein kinase |
| ENHANCED RESPONSE TO ABA 1 | ERA1 | At5g40280 | β-subunit of farnesyl-trans-transferase, |
| LATERAL SUPPRESSOR | LAS | At1g55580 | GRAS transcription factor |
| LEAFY | LFY | At5g61850 | Transcription factor |
| MORE AXILLARY GROWTH 1 | MAX1 | At2g26170 | Cytochrome P450 (CYP711A) |
| MORE AXILLARY GROWTH 2 | MAX2 | At2g42620 | F-box protein |
| MORE AXILLARY GROWTH 3 | MAX3 | At2g44990 | Carotenoid cleavage dioxygenase 7 |
| MORE AXILLARY GROWTH 4 | MAX4 | At4g32810 | Carotenoid cleavage dioxygenase 8 |
| MONOPTEROS | MP | At1g19850 | IAA24, auxin response factor 5 (ARF5) |
| PINOID | PID | At2g34650 | Serine/threonine kinase |
| PINFORMED 1 | PIN1 | At1g73590 | Auxin efflux carrier protein |
| PINHEAD | PNH | At5g43810 | Translation initiation factor |
| REGULATOR OF AXILLARY MERISTEMS 1 | RAX1 | At5g23000 | Myb transcription factor 37 |
| REGULATOR OF AXILLARY MERISTEMS 2 | RAX2 | At2g36890 | Myb transcription factor 38 |
| REGULATOR OF AXILLARY MERISTEMS 3 | RAX3 | At3g49690 | Myb transcription factor 84 |
| REVOLUTA | REV | At5g60690 | Homeodomain-leucine zipper protein |
| SEUSS | SEU | At1g43850 | Transcriptional coregulator of AGAMOUS |
| SUPERSHOOT 1 | SPS1 | At1g16410 | Cytochrome P450 (CYP79F1) |
| SHOOT MERISTEMLESS | STM | At1g62360 | Knotted-like homeodomain protein |
| TERMINAL FLOWER 1 | TFL1 | At5g03840 | Controls inflorescence meristem identity |
| TRANSPORT INHIBITOR RESPONSE 1 | TIR1 | At3g62980 | F-box protein |
Annotations are from the Arabidopsis Information Resource (http://www.arabidopsis.org).
Sequence manipulation and analysis:
Sequence data were compiled into contigs using Phred/Phrap (CodonCode, Dedham, MA) and initially aligned in Biolign v2.0.9 (Tom Hall, Ibis Therapeutics, Carlsbad, CA). All polymorphisms were confirmed using the sequence trace files. Final sequence alignment was performed in Bioedit v7.0.5.2 (Tom Hall, Ibis Therapeutics).
Levels of polymorphism were determined on the basis of the average number of pairwise differences between alleles (π) (Tajima 1983) or on the basis of the number of segregating sites (S) in the sequence sample (θw) (Watterson 1975) in DnaSP v4.0 (Rozas et al. 2003). Nucleotide diversity values for nonsynonymous and synonymous sites (πn and πs, respectively) in coding regions, as well as Tajima's D (Tajima 1989), were also computed in DnaSP v4. The number of haplotypes (h) is also reported for each gene. Maximum-parsimony gene genealogies were created in MEGA v3.1 (Kumar et al. 2004).
LD in the MAX2 and MAX3 genomic regions was calculated as r2 (Hill 1974). Polymorphisms with minor allele frequencies ≤10% were excluded from analysis. The median LD decay plot was created by grouping r2-values into bins of 5 kb on the basis of the distances between markers. The median r2-value was taken from each bin and plotted against bin midpoint. LD was plotted in TASSEL v1.9.4 (Ed Buckler lab, Cornell University, Ithaca, NY). Significance for LD was determined through 10,000 permutations.
Genotyping:
Polymorphisms for genotyping were determined on the basis of haplogroups present in the gene genealogies. For both the candidate genes and the genomic region sequencing, one polymorphism was genotyped from any branch that separated ≥10% of the alleles from all others, using cleaved amplified polymorphic sequences (CAPS) or degenerate CAPS (dCAPS) markers. CAPS markers were chosen by comparison of the sequences of different alleles in NEBCutter v2.0 (http://tools.neb.com/NEBcutter2/index.php). dCAPS Finder v2.0 (http://helix.wustl.edu/dcaps/dcaps.html) was used to choose dCAPS markers. All primers for marker amplification were designed in Primer3. All restriction enzymes for digestion were purchased from New England Biolabs (Ipswich, MA). For BP and SEU, the alleles for these genes were sequenced from all 96 individuals, rather than genotyped with CAPS or dCAPS markers. Genotyped accessions and haplogroup assignments are provided in supplemental data at http://www.genetics.org/supplemental/.
Phenotype data for LD mapping:
Data are from a controlled growth chamber experiment conducted at North Carolina State University (Olsen et al. 2004). A lateral branch was defined as any elongated branch along the primary inflorescence. A basal branch was characterized as any branch emanating from the rosette. In large part, these basal branches extend from apical rosette nodes. Total branches were the sum of lateral branches and basal branches. Lateral branch nodes were considered any point along the primary inflorescence where a lateral branch could form and were counted as the number of cauline leaves. Least-squares (LS) means for trait values were used for the analyses in this article. Trait values were standardized prior to running the mixed model to improve convergence.
Association tests:
To decrease the possibility of spurious associations due to hidden population structure in our sample, we used a recently reported mixed-model method that incorporates population ancestry estimates from the program STRUCTURE v2 (Pritchard et al. 2000a) and pairwise kinship estimates from the program SPAGeDi v1.2 (Hardy and Vekemans 2002). STRUCTURE runs with a prior of four ancestral populations (K = 4), resulting in the highest likelihood value of all K-values (supplemental data at http://www.genetics.org/supplemental/). The ancestry estimates are based on previously described SNP data (Schmid et al. 2006). Mixed-model association tests were conducted in SAS v9.1.2 (SAS, Cary, NC) with a previously described program (Yu et al. 2006).
Mapping epistatic QTL:
We analyzed previously published data for branching from the Ler × Col or Cvi × Ler RILs (Ungerer et al. 2002, 2003) in the program EPISTACY (Holland 1998). A significance threshold of P = 0.001 was used. Marker combinations exhibiting epistatic interactions were evaluated for linkage to determine the span of the detected interactions. Epistatic QTL intervals were determined on the basis of the physical positions of all linked markers that appear to represent the same epistatic interaction. In some cases, only a single marker pair was found to represent the epistatic interaction, in which case the physical spans of the epistatic QTL could not be determined.
RESULTS AND DISCUSSION
Levels of variation and haplogroup structure of candidate branching genes:
The 36 candidate genes used in this study were chosen to represent loci that control various developmental and physiological processes involved in shoot branching. One ∼600-bp fragment composed primarily of exons was sequenced from each candidate gene in a panel of 24 accessions from across the geographic range of the species. Nucleotide diversity levels at the 36 genes were lower than previously reported genomewide values for functional genes with π = 0.002 and θw = 0.003 (Nordborg et al. 2005; Schmid et al. 2005). PINOID (PID), which has no polymorphisms, has the lowest nucleotide diversity of all genes, while PINHEAD (PNH) possesses the highest diversity (π and θw = 0.014) (Table 2). In coding regions, nucleotide diversity values for nonsynonymous polymorphisms (π = 0.001 and θw = 0.002) were nearly 3.5-fold lower than for synonymous polymorphisms (π = 0.004 and θw = 0.005), and numerous genes were not polymorphic at their nonsynonymous sites.
TABLE 2.
Genetic variation at the sequenced genes
| Gene | Classa | n | Length | S | h | π | θw | πn | πs | Tajima's D |
|---|---|---|---|---|---|---|---|---|---|---|
| ABI3 | S | 23 | 766 | 7 | 7 | 0.0010 | 0.0025 | 0.0007 | 0.0015 | −1.883 |
| AMP1 | P | 24 | 765 | 7 | 9 | 0.0020 | 0.0025 | 0.0014 | 0.0043 | −0.606 |
| ANT | P | 21 | 805 | 4 | 4 | 0.0007 | 0.0014 | 0.0004 | 0.0019 | −1.458 |
| AP1 | D | 25 | 518 | 12 | 8 | 0.0029 | 0.0062 | 0.0000 | 0.0000 | −1.83 |
| ARGOS | P | 17 | 551 | 4 | 5 | 0.0022 | 0.0022 | 0.0004 | 0.0000 | 0.135 |
| AXR1 | S | 25 | 842 | 7 | 8 | 0.0013 | 0.0022 | 0.0021 | 0.0000 | −1.335 |
| AXR2 | S | 23 | 834 | 24 | 11 | 0.0082 | 0.0080 | 0.0034 | 0.0186 | 0.134 |
| AXR3 | S | 23 | 717 | 4 | 5 | 0.0010 | 0.0015 | 0.0003 | 0.0049 | −0.931 |
| AXR6 | S | 24 | 822 | 5 | 6 | 0.0010 | 0.0016 | 0.0005 | 0.0000 | −1.188 |
| BP | P | 22 | 413 | 10 | 6 | 0.0052 | 0.0068 | 0.0010 | 0.0115 | −0.806 |
| BUD1 | S | 23 | 487 | 4 | 7 | 0.0031 | 0.0022 | 0.0027 | 0.0046 | −1.13 |
| CAL | D | 25 | 520 | 7 | 8 | 0.0024 | 0.0036 | 0.0005 | 0.0020 | −1.035 |
| CKI1 | S | 24 | 794 | 2 | 3 | 0.0002 | 0.0007 | 0.0000 | 0.0009 | −1.515 |
| CRE1 | S | 25 | 788 | 1 | 2 | 0.0001 | 0.0003 | 0.0001 | 0.0000 | −1.158 |
| EMF1 | P | 17 | 487 | 2 | 3 | 0.0005 | 0.0012 | 0.0000 | 0.0022 | −1.504 |
| ER | S | 24 | 520 | 7 | 6 | 0.0019 | 0.0036 | 0.0003 | 0.0085 | −1.464 |
| ERA1 | S | 25 | 798 | 10 | 9 | 0.0016 | 0.0033 | 0.0013 | 0.0042 | −1.682 |
| LAS | P | 25 | 806 | 2 | 3 | 0.0002 | 0.0007 | 0.0000 | 0.0008 | −1.514 |
| LFY | D | 25 | 544 | 3 | 4 | 0.0006 | 0.0015 | 0.0000 | 0.0028 | −1.504 |
| MAX1 | S | 25 | 687 | 6 | 6 | 0.0019 | 0.0023 | 0.0008 | 0.0086 | −0.519 |
| MAX2 | S | 25 | 760 | 16 | 11 | 0.0062 | 0.0006 | 0.0055 | 0.0084 | 0.38 |
| MAX3 | S | 25 | 660 | 4 | 5 | 0.0015 | 0.0016 | 0.0009 | 0.0035 | −0.151 |
| MAX4 | S | 25 | 690 | 6 | 5 | 0.0019 | 0.0024 | 0.0036 | 0.0000 | −0.632 |
| MP | P | 25 | 808 | 4 | 5 | 0.0008 | 0.0013 | 0.0003 | 0.0029 | −1.019 |
| PID | S | 25 | 796 | 0 | 1 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | NA |
| PIN1 | S | 21 | 812 | 5 | 5 | 0.0007 | 0.0017 | 0.0000 | 0.0000 | −1.795 |
| PNH | P | 22 | 831 | 29 | 12 | 0.0093 | 0.0101 | 0.0022 | 0.0303 | −0.247 |
| RAX1 | P | 25 | 568 | 9 | 8 | 0.0022 | 0.0042 | 0.0014 | 0.0050 | −1.584 |
| RAX2 | P | 25 | 437 | 5 | 4 | 0.0028 | 0.0030 | 0.0027 | 0.0032 | −0.269 |
| RAX3 | P | 25 | 556 | 7 | 8 | 0.0018 | 0.0033 | 0.0008 | 0.0057 | −1.469 |
| REV | P | 21 | 720 | 9 | 9 | 0.0025 | 0.0035 | 0.0021 | 0.0073 | −1.003 |
| SEU | P | 24 | 535 | 9 | 9 | 0.0040 | 0.0050 | 0.0043 | 0.0030 | −0.622 |
| SPS1 | S | 24 | 700 | 11 | 14 | 0.0028 | 0.0042 | 0.0032 | 0.0005 | −1.125 |
| STM | P | 25 | 644 | 11 | 6 | 0.0024 | 0.0047 | 0.0000 | 0.0030 | −1.665 |
| TFL1 | D | 24 | 566 | 2 | 3 | 0.0004 | 0.0010 | 0.0000 | 0.0000 | −1.202 |
| TIR1 | S | 25 | 814 | 2 | 3 | 0.0004 | 0.0007 | 0.0000 | 0.0000 | −0.941 |
Functional groupings used for population genetic analyses: D, determination of meristem identity; P, shoot patterning; S, phytohormone signaling.
The average Tajima's D-value observed in this sequence data set is −1.023, the negative value arising from the prevalence of low-frequency mutations in this data set. The mean number of polymorphic sites observed per gene is 7.6, while the mean number of haplotypes per gene is 6.5, indicating that a large proportion of observed mutations are found in unique haplotypes. Most sequenced genes exhibit haplotype structure characterized by numerous low-frequency haplogroups that are differentiated from each other by a small number of mutations (Figure 1). Three genes—AUXIN RESISTANT 2 (AXR2), MORE AXILLARY GROWTH 2 (MAX2), and PINHEAD (PNH)—possess a relatively high number of moderate-frequency mutations in comparison to other loci. Only 27 of the 36 genes (75%) possess haplogroups that are present at a frequency ≥10%.
Figure 1.—
Maximum-parsimony gene genealogies of the 36 candidate genes, corresponding to (A) ABI3, (B) AMP1, (C) ANT, (D) AP1, (E) ARGOS, (F) AXR1, (G) AXR2, (H) AXR3, (I) AXR6, (J) BP, (K) BUD1, (L) CAL, (M) CKI1, (N) CRE1, (O) EMF1, (P) ER, (Q) ERA1, (R) LAS, (S) LFY, (T) MAX1, (U) MAX2, (V) MAX3, (W) MAX4, (X) MP, (Y) PID, (Z) PIN1, (AA) PNH, (AB) RAX1, (AC) RAX2, (AD) RAX3, (AE) REV, (AF) SEU, (AG) SPS1, (AH) STM, (AI) TFL1, and (AJ) TIR1. The shaded circles represent branches along which a SNP was genotyped. All genealogies are presented at the same scale.
We attempted to determine if genes involved in different developmental processes underlying shoot branching possess different levels of sequence variation. We categorized the 36 genes into three general groups on the basis of their roles in branch development: (i) node patterning, (ii) meristem identity determining, and (iii) phytohormone signaling genes. Signaling genes have the highest levels of nucleotide diversity (π = 0.004), with the other two classes possessing similar levels of polymorphism (π = 0.001 for both classes). There are also lower nonsynonymous diversity levels for the meristem identity and patterning genes (πn ≈ 0 for both classes) than for the signaling genes (πn = 0.002). Meristem identity genes also have a higher proportion of loci with no nonsynonymous site variation (75%) than either node patterning (21%) or hormone signaling loci (28%).
Characteristics of the mapping population:
Population structure in A. thaliana is extensive, with substantial differences in allele frequencies occurring across its geographic range (Nordborg et al. 2005; Schmid et al. 2005). This poses a serious challenge for association mapping since many traits in this species exhibit clines that are correlated with population structure (Aranzana et al. 2005). To minimize the confounding effect of population structure on association mapping, we use 96 accessions from a geographically restricted range in Central Europe between latitudes 45°N and 55°N and longitudes 4°E and 18°E. Other studies have shown that population structure within this region is less severe than the global population structure of the species (Nordborg et al. 2005; Schmid et al. 2005; Korves et al. 2007).
Previous association studies in our laboratory used a published AFLP data set (Sharbel et al. 2000) to correct for population structure (Caicedo et al. 2004; Olsen et al. 2004), but this data set indicated minimal population stratification. More recent analyses based on SNP data have shown that, in fact, substantial population stratification is present in A. thaliana (Nordborg et al. 2005; Schmid et al. 2005), requiring the reevaluation of our previous association results (Korves et al. 2007). In this study, we utilized 115 genomewide SNPs from Schmid et al. (2005), using the program STRUCTURE (Pritchard et al. 2000a), which confirms that there is population stratification in the accessions used in this study, with the most likely number of populations (K) being four. The percentage of membership to populations one through three is spatially correlated, with population one membership correlated to both latitude and longitude of origin of the accessions (F2,93 = 10.97, P < 0.001), whereas population two and three correlated to longitude (F1,94 = 7.48, P = 0.008) and latitude (F1,94 = 13.06, P < 0.001), respectively. Thus, despite sampling at a smaller geographic scale, there is still detectable genealogical and geographic structure in this accession panel.
We determined the distribution of shoot branching across these 96 accessions for four branching traits under long and short day controlled growth chamber conditions: (i) lateral branch number, (ii) basal branch number, (iii) total branch number, and (iv) lateral branch node number. Substantial quantitative variation was found in these shoot-branching traits (Figure 2), and broad sense heritabilities (H2) range from 0.09 for basal branches in short day to 0.41 for lateral branches in long day, with higher H2-values generally observed in long day than in short day.
Figure 2.—
Trait distributions for the 96 accessions used for association mapping. Aa-0 and Li-6, which have the highest and lowest number of lateral branches in long day, respectively, are shown as reference accessions across all environment–trait combinations. (A) Lateral branches in long day; (B) lateral branches in short day; (C) basal branches in long day; (D) basal branches in short day; (E) total branches in long day; (F) total branches in short day; (G) lateral branch nodes in long day; (H) lateral branch nodes in short day. LD, long day; SD, short day.
Branching traits exhibit geographic clines in our A. thaliana sample, with the majority of traits showing significant correlations with either the latitude or the longitude of origin of the accessions or with both. Long day lateral branches (ANOVA, F1,94 = 7.38, P = 0.008), short day lateral branch nodes (ANOVA, F1,94 = 6.94, P = 0.010), and short day basal branches (ANOVA, F1,94 = 6.95, P = 0.010) are all correlated with longitude, while long day lateral branch nodes are also associated with latitude (ANOVA, F1,94 = 5.56, P = 0.020).
Associations between candidate gene polymorphisms and branch architecture:
Of the 36 genes we initially analyzed, only 27 contained moderate-frequency (≥10%) haplogroups that were subsequently genotyped in the association-mapping panel of 96 accessions. We assessed the level of population structure in our mapping sample using these candidate gene haplogroups and found that unlike in the genomewide SNP set, the most likely number of putative ancestral populations (K) was found to be two (supplemental data at http://www.genetics.org/supplemental/). This suggests that our candidate genes exhibit lower levels of population stratification than genomewide markers in the set of 96 accessions.
Mixed-model association tests were conducted on the basis of haplogroup genotyping results, except for AINTEGUMENTA (ANT) for which no variation was found in our mapping sample. The observed nominally significant associations (P < 0.05) per environment–trait combination ranged from one to four (Table 3), with 7 genes (27%) exhibiting a nominally significant association in at least one environment–trait combination, while 5 genes (19%) are nominally significant in two or more environment–trait combinations. It has been shown that even with the use of the mixed-model approach there is still an excess of false-positive associations in A. thaliana (Aranzana et al. 2005; Zhao et al. 2007). In an effort to be conservative in our evaluation of results, we recomputed association probabilities by ranking the nominal P-values for the 26 genes. We accepted an association as significant only if (i) the mixed-model result was nominally significant (P < 0.05) and (ii) the association P-value was at the lower 5% tail of the observed P-value distribution of all the candidate genes. In essence, this latter criterion allows us to use all the candidate gene associations as an empirical distribution for P-values obtained from the mixed-model method (i.e., if ∼20 genes are used, then one may be considered actually significant).
TABLE 3.
Nominal P-values for mixed-model association tests
| Long day |
Short day |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Lateral branches | Basal branches | Total branches | Lateral branch nodes | Lateral branches | Basal branches | Total branches | Lateral branch nodes |
| AMP | 0.664 | 0.138 | 0.398 | 0.789 | 0.915 | 0.972 | 0.896 | 0.481 |
| AP1 | 0.793 | 0.227 | 0.255 | 0.226 | 0.200 | 0.370 | 0.146 | 0.705 |
| ARGOS | 0.225 | 0.8297 | 0.687 | 0.797 | 0.478 | 0.592 | 0.440 | 0.364 |
| AXR1 | 0.898 | 0.580 | 0.942 | 0.531 | 0.276 | 0.696 | 0.159 | 0.465 |
| AXR2 | 0.211 | 0.445 | 0.644 | 0.668 | 0.073 | 0.031 | 0.236 | 0.060 |
| AXR3 | 0.151 | 0.493 | 0.378 | 0.193 | 0.733 | 0.359 | 0.901 | 0.124 |
| AXR6 | 0.791 | 0.786 | 0.922 | 0.953 | 0.416 | 0.646 | 0.527 | 0.069 |
| BP | 0.500 | 0.776 | 0.586 | 0.739 | 0.736 | 0.129 | 0.889 | 0.717 |
| BUD1 | 0.646 | 0.057 | 0.101 | 0.156 | 0.454 | 0.595 | 0.384 | 0.995 |
| CAL | 0.318 | 0.566 | 0.616 | 0.213 | 0.862 | 0.861 | 0.899 | 0.775 |
| ER | 0.935 | 0.582 | 0.839 | 0.455 | 0.459 | 0.051 | 0.194 | 0.092 |
| ERA1 | 0.359 | 0.524 | 0.134 | 0.063 | 0.024 | 0.427 | 0.013 | 0.035 |
| MAX1 | 0.503 | 0.251 | 0.737 | 0.863 | 0.513 | 0.124 | 0.460 | 0.138 |
| MAX2 | 0.234 | 0.659 | 0.335 | 0.384 | <0.001* | 0.736 | <0.001* | 0.113 |
| MAX3 | 0.058 | 0.302 | 0.008* | 0.037* | 0.322 | 0.053 | 0.286 | 0.135 |
| MAX4 | 0.510 | 0.942 | 0.471 | 0.706 | 0.713 | 0.325 | 0.775 | 0.385 |
| MP | 0.490 | 0.879 | 0.375 | 0.212 | 0.267 | 0.327 | 0.358 | 0.820 |
| PNH | 0.964 | 0.299 | 0.947 | 0.966 | 0.767 | 0.518 | 0.632 | 0.788 |
| RAX1 | 0.133 | 0.186 | 0.898 | 0.667 | 0.299 | 0.235 | 0.192 | 0.487 |
| RAX2 | 0.989 | 0.514 | 0.504 | 0.648 | 0.473 | 0.101 | 0.241 | 0.966 |
| RAX3 | 0.089 | 0.453 | 0.357 | 0.219 | 0.050 | 0.117 | 0.183 | 0.104 |
| REV | 0.346 | 0.591 | 0.108 | 0.248 | 0.605 | 0.307 | 0.401 | 0.352 |
| SEU | 0.735 | 0.612 | 0.588 | 0.437 | 0.018 | 0.616 | 0.026 | 0.195 |
| SPS1 | 0.373 | 0.0158* | 0.109 | 0.645 | 0.207 | 0.003* | 0.244 | 0.014* |
| STM | 0.303 | 0.846 | 0.497 | 0.705 | 0.562 | 0.542 | 0.729 | 0.072 |
| TIR1 | 0.287 | 0.359 | 0.783 | 0.154 | 0.612 | 0.976 | 0.568 | 0.082 |
P < 0.05 based on ranking of nominal P-values within environment–trait combination.
On the basis of both our criteria, one single significant association was found for each environment–trait combination, except for long day lateral branches, which had no nominally significant associations. Three genes—MORE AXILLARY GROWTH 2 (MAX2), MORE AXILLARY GROWTH 3 (MAX3), and SUPERSHOOT1 (SPS1)—exhibited significant haplogroup–phenotype associations after the P-values were reassigned. The significant associations were: (i) MAX2 and lateral and total branches in short day, (ii) MAX3 and lateral branch nodes and total branches in long day, and (iii) SPS1 and basal branches in long day and short day and lateral branch nodes in short day. The SPS1 association with basal branching was the only gene–trait-significant association across environmental conditions.
Patterns of LD and genotype–phenotype associations across two linked genomic regions:
The patterns of polymorphism and linkage disequilibrium in the genome are the primary determinants of the resolution of association mapping (Gaut and Long 2003). Although the extent of LD in A. thaliana has been characterized above 25-kb scales, we were interested in documenting it below this level of resolution across our significant genes. We sequenced ∼600-bp fragments from three genes both upstream and downstream of MAX2 and MAX3; these two genes were chosen because they exhibited multiple trait associations and are linked on chromosome 2, permitting the analysis of trait association patterns at both the fine (∼35-kb) and coarse (∼800-kb) genomic scales. These two regions display a fivefold difference in polymorphism levels, with the MAX2 region having a mean π = 0.005, while the mean π of the MAX3 region is 0.001. The range of nucleotide diversity is also much higher in the MAX2 (πmin ≈ 0, πmax = 0.011) than in the MAX3 region (πmin ≈ 0, πmax = 0.002).
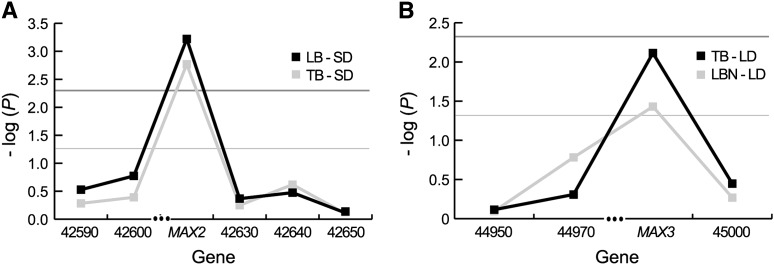
The patterns of LD are similar for both the MAX2 and the MAX3 genomic regions, with SNP correlations primarily observed between adjacent genes. In the MAX2 region, three haplotype blocks are distinguishable (Figure 3), with only slight LD (r2 ≈ 0.5) detectable between a pair of SNPs found in two blocks. As for the MAX3 region, all LD with r2 > 0.4 is found intragenically, except for LD that is detected between MAX3 and its downstream neighbor At2g45000 (Figure 3). There is no detectable disequilibrium between the MAX2 and MAX3 regions. These results suggest that phenotypic associations at MAX2 and MAX3 should not span >10 kb, encompassing at most two to three genes (Figure 4).
Figure 3.—
Linkage disequilibrium (LD) across the MAX2 and MAX3 regions. The MAX2 (A) and MAX3 (B) regions are shown according to scale. Fragments that were not polymorphic or were without common polymorphisms do not have lines connecting the LD plots to the physical maps. The At2g prefixes for all numbered genes are omitted in this and subsequent figures. Gene annotations are presented below the gene numbers. “Exp Prot,” expressed protein.
Figure 4.—
Decay of LD in MAX2 and MAX3 regions. Median r2 is plotted by the midpoint of each pairwise marker distance bin.
To test whether our haplogroup associations at MAX2 and MAX3 extend to linked genes, we employed the mixed-model association method on moderate-frequency haplogroups (≥10%) found across these two genomic regions. These results confirm that the trait associations span short genomic distances, localizing both the MAX2 and the MAX3 associations to these genes and not to the linked loci (Figure 5).
Figure 5.—
Trait associations across the MAX2 (A) and MAX3 (B) regions. P-values are plotted as −log(P). Genes with no polymorphisms at ≥10% frequency are not included. The horizontal line with light shading denotes P = 0.05, while the horizontal line with dark shading represents P = 0.005. LB, lateral branches; TB, total branches; LBN, lateral branch nodes; LD, long day; SD, short day.
Comparison of LD to QTL mapping:
To replicate our candidate gene association results, we attempted to determine if our gene–trait associations correspond to previously observed additive QTL in the Ler × Col and Cvi × Ler RIL populations. For lateral branches and lateral branch nodes combined, 5 QTL were observed in both short day and long day in the Ler × Col RILs, whereas 11 and 14 were observed in the Cvi × Ler RILs in short day and long day, respectively (Ungerer et al. 2002, 2003). The loci exhibiting trait associations with lateral branches or lateral branch nodes were sequenced in the parental lines of each set of RILs. The Ler × Col RILs have MAX2 haplotypes that are members of significantly different haplogroups in the association-mapping study (contrasts of the haplogroups containing the two alleles are significant for both lateral branches and total branches in short day, with P = 0.030 and P = 0.047, respectively), while the Cvi × Ler RILs possesses relevant haplotypes for SPS1 (contrast of the two relevant haplogroups for lateral branch nodes results in P = 0.042). However, for neither MAX2 nor SPS1 are overlapping additive QTL present in the appropriate RIL mapping populations.
The absence of additive QTL in the Ler × Col and Cvi × Ler mapping populations (Ungerer et al. 2002, 2003) encompassing either SPS1 or MAX2 was unexpected, given that we used a mixed-model approach that previous reports suggested had a relatively low false-positive rate (Yu and Buckler 2006; Zhao et al. 2007), a geographically restricted mapping panel to further minimize population stratification, and a conservative recomputation of P-values by comparison of nominal probabilities across all tested candidate genes. There are two possibilities to account for this discrepancy. First, despite our efforts to minimize false positives, we are unable to completely remove all residual population structure that can still give rise to spurious associations. That association mapping is fundamentally confounded by the demographic structure of a population has been well documented (Zhao et al. 2007), and although we have been methodologically conservative so as to mitigate population structure effects, cryptic population structure may continue to be present that leads to spurious associations.
The second possibility is that these genes indeed underlie natural variation in shoot branching, but in primarily epistatic interactions rather than as direct additive effects. To assess whether epistatic QTL involving the MAX2 and SPS1 genomic regions are present, we determined the number and locations of epistatic interactions involved in branching in the Ler × Col and Cvi × Ler RILs. Seven and nine branching epistatic interactions were found in the Ler × Col RILs in short day and long day, respectively, while six and seven were found in the Cvi × Ler RILs under the same growth conditions (Tables 4 and 5). Epistatic interactions were indeed found to overlap both genes in the appropriate RILs (Figure 6). Epistatic QTL that overlap MAX2 are observed in the Ler × Col mapping population, but are for a different branching trait–environment combination than was observed by the candidate gene association-mapping study. For SPS1, however, an overlapping epistatic QTL was detected in the Cvi × Ler lines that replicated the association-mapping results.
TABLE 4.
Epistatic QTL detected in the Ler × Col RILs
| Trait | IDa | Marker 1 | Position | Range | Marker 2 | Position | Range |
|---|---|---|---|---|---|---|---|
| Long day environment | |||||||
| Lateral branches | E1 | CATTS039 | I, 51.22 | 45.20–60.78 (MI62–BH.160L) | MI421 | II, 12.27 | 2.24–25.45 (MI320–THY_1) |
| E2 | MI390 | IV, 9.75 | 6.00–36.73 (G3843–PCITD23) | EMB514 | V, 110.74 | 104.35–110.74 (MI70–EMB514) | |
| E3 | M336 | II, 65.98 | 57.80–71.36 (VE017–MI79A) | M194 | V, 81.26 | NA | |
| E4 | G17311 | I, 131.83 | NA | RRS2 | II, 79.29 | NA | |
| E5 | AGP64 | I, 128 | 126.76–128 (VE011–AGP64) | MI138 | V, 33.14 | NA | |
| E6 | VE012 | II, 0 | NA | G4014 | III, 68.86 | NA | |
| E7 | PCITF3 | IV, 31.83 | NA | MI322 | V, 27.73 | NA | |
| E8 | M315 | II, 3.73 | NA | MI473 | II, 69.76 | 69.76–71.36 (MI473–MI79A) | |
| Lateral branch nodes | E1 | ATHFUS6 | III, 90.25 | 90.25–93.62 (ATHFUS6–NGA6) | MI174 | V, 23.96 | 0–27.73 (PATT80–MI322) |
| Short day environment | |||||||
| Lateral branches | E1 | NGA8 | IV, 25.31 | 13.06–33.64 (APP–M518) | H2A1 | V, 102.00 | 68.69–110.74 (M435–EMB514) |
| E2 | CDS7 | I, 51.22 | 14.65–51.22 (ATTS0477–CATTS039) | MI306 | IV, 22.11 | 22.11–50.88 (MI306–MI112) | |
| E3 | MI103 | I, 115.41 | NA | AG | IV, 55.18 | NA | |
| E4 | MI208 | I, 79.83 | NA | CA1 | III, 0 | NA | |
| Lateral branch nodes | E1 | M448A | IV, 21.59 | 2.75–36.73 (MI204–PCITD23) | H2A1 | V, 102.00 | 75.36–129.05 (G4028–CATHHAN) |
| E2 | MI139 | II, 28.11 | 28.11–29.38 (MI139–MI148) | M448A | IV, 21.59 | NA | |
| E3 | AGP64 | I, 128.00 | NA | G4028 | V, 75.36 | NA | |
IDs correspond to epistatic QTL in Figure 6.
TABLE 5.
Epistatic QTL detected in the Cvi × Ler RILs
| Trait | IDa | Marker 1 | Position | Range | Marker 2 | Position | Range |
|---|---|---|---|---|---|---|---|
| Long day environment | |||||||
| Lateral branches | E1 | AD.112L | III, 77.07 | 75.45–77.07 (AD.495L–AD.112L) | DF.231C | V, 26.53 | 20.19–31.80 (BH.107L–DF.184L) |
| E2 | EC.480C | I, 15.49 | NA | EG.75L | III, 7.25 | NA | |
| Lateral branch nodes | E1 | FD.167L | IV, 51.74 | 37.14–64.34 (CD.84C–GB.490C) | DF.231C | V, 26.53 | 15.39–49.32 (BH.325L–HH.480C) |
| E2 | CH.200C | I, 76.01 | 76.01–79.20 (CH.200C–DF.260L) | HH.158L | III, 26.55 | 22.45–26.89 (GH.390L–EC.83C) | |
| E3 | AXR1 | I, 7.70 | NA | HH.90L | III, 80.81 | NA | |
| E4 | GB.500C | I, 65.22 | NA | BH.92L | IV, 29.36 | NA | |
| Short day environment | |||||||
| Lateral branches | E1 | EG.129C | I, 57.56 | 49.42–60.96 (GB.112L–BH.162C) | AD.92L | III, 32.31 | 29.46–32.31 (GD.318C–AD.92L) |
| E2 | PVV4 | I, 0 | 0–7.70 (PVV4–AXR1) | HH.480C | V, 49.32 | 42.14–49.32 (GH.121L–HH.480C) | |
| E3 | PVV4 | I, 0 | NA | CC.318C | I, 108.57 | NA | |
| Lateral branch nodes | E1 | AD.121C | I, 39.46 | 39.46–40.44 (AD.121C–AD.106L) | FD.345C | V, 93.14 | 90.49–109.90 (CC.262C–GD.222C) |
| E2 | HH.159C | IV, 60.64 | 56.94–64.34 (CH.70L–GB.490C) | DF.231C | V, 26.53 | 24.55–42.14 (AD.114C–42.14) | |
| E3 | CH.200C | I, 76.01 | NA | BH.96L | V, 55.96 | NA | |
| E4 | DF.73L | I, 29.04 | NA | EC.83C | III, 26.89 | NA |
IDs correspond to epistatic QTL in Figure 6.
Figure 6.—
Genomic map of candidate gene associations and QTL in the Ler × Col (A) and Cvi × Ler (B) RILs. Environment–trait combinations are colored as red, green, blue, and purple for lateral branches in long day, lateral branch nodes in long day, lateral branches in short day, and lateral branch nodes in short day, respectively. Each epistatic QTL is referenced to the table of epistatic QTL by number. Additive QTL are included on the map as colored rectangles at the marker location reported in Ungerer et al. (2002, 2003), using the same color scheme as for the epistatic QTL.
It is thus possible that what we are observing is the effect of population stratification in our association studies, but in this context the population structure works to maintain epistatic interactions in a large, genetically diverse sample. Indeed, it is well known that epistatic effects can manifest themselves as additive genetic variation, particularly in species that are highly inbred and/or highly structured at the population level (as reviewed in Neiman and Linksvayer 2006). If association-mapping populations contain inbred genotypes and/or individuals from different subpopulations, alleles involved in epistatic interactions may prove detectable through conventional association-mapping techniques, but undetectable as additive QTL through QTL mapping. The results presented here support those from recent studies that point to the prevalence of epistasis on the genetic architecture of various traits in A. thaliana (Malmberg et al. 2005).
Candidate gene association mapping of branch variation in Arabidopsis:
Shoot architecture, in particular the organization of axillary branches, is one of the most visible features that differentiate plant species (Sussex and Kerk 2001). Characterizing the molecular basis of microevolutionary variation in branching in tractable plant models, such as A. thaliana, may provide clues to the molecular mechanisms underlying shoot macroevolution. This study is the first report of a large-scale candidate gene association mapping in A. thaliana, which evaluates many of the genes found in the genetic network that underlies shoot branching in this plant species. By screening 36 genes, we have identified at least one strong candidate gene for branching variation in this species in SPS1 and two weaker candidates in MAX2 and MAX3. Additionally, we have localized the MAX2 and MAX3 associations to the genes themselves.
The associations described in this study provide evidence that variation in phytohormone signaling pathways for auxin and cytokinin may play important roles in generating branching diversity in A. thaliana. Both auxin and cytokinin have long been known to play central roles in apical dominance and shoot branch development, so it is plausible that these signals could contribute to branching variation. This study also emphasizes the strong influence the environment exerts over quantitative variation in the shoot and its genetic architecture. The nominally significant candidate gene associations detected in each environment were typically very different and are comparable to those from QTL-mapping experiments that have shown a large number of environment-specific QTL for shoot architectural traits (e.g., Ungerer et al. 2003). This environmental sensitivity provides support for the importance of genotype–environment interactions in shaping shoot morphology, and it is likely that such environment-specific control of branching exists and may be of adaptive value as plants modulate their architecture to various ecological signals such as photoperiod length, available nutrients, and herbivory (Bonser and Aarssen 1996).
Our results demonstrate the utility and difficulty of association mapping, which is increasingly being applied to a large number of trait-mapping studies. This study shows that in A. thaliana one can conceivably obtain high resolution and localize haplogroup–phenotype associations to single genes with this approach. Detailed studies are necessary to validate the associations reported in this study at the causal level, and these studies are currently underway. Our results suggest, however, that candidate gene association studies can provide strong candidate quantitative trait genes, although certainly this approach is not as comprehensive as genomewide analyses that are not limited to characterized genes. Nevertheless, the challenge is to determine the biological significance of associations detected by association mapping, and the possibility that epistatic interactions may underlie the effects of many of these genes poses challenges on how association studies are replicated and validated in the future.
Acknowledgments
We are grateful to past and present members of the Purugganan lab for assistance in performing experiments and analyses in this article, as well as for thoughtful discussion regarding this manuscript. Also, we thank Tonia Korves, Ottoline Leyser, and Magnus Nordborg for comments on a previous version of this manuscript. This work was supported by a Department of Education Graduate Assistance in Areas of National Need Fellowship and a National Science Foundation (NSF) Graduate Research Fellowship to I.M.E. and by grants from the NSF's Frontiers in Integrated Biological Research and Plant Genome Research Programs to M.D.P.
References
- Aranzana, M. J., S. Kim, K. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1 e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, T., T. Sieberer, B. Willett, J. Booker, C. Luschnig et al., 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16 553–563. [DOI] [PubMed] [Google Scholar]
- Bonser, S., and L. Aarssen, 1996. Meristem allocation: a new classification theory for adaptive strategies in herbaceous plants. Oikos 77 347–352. [Google Scholar]
- Booker, J., T. Sieberer, W. Wright, L. Williamson, B. Willett et al., 2005. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8 443–449. [DOI] [PubMed] [Google Scholar]
- Bradley, D., O. Ratcliffe, C. Vincent, R. Carpenter and E. Coen, 1997. Inflorescence commitment and architecture in Arabidopsis. Science 275 80–83. [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., J. R. Stinchcombe, K. M. Olsen, J. Schmitt and M. D. Purugganan, 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon, L. R., and G. R. Abecasis, 2003. Using haplotype blocks to map human complex trait loci. Trends Genet. 19 135–140. [DOI] [PubMed] [Google Scholar]
- Clark, R. M., E. Linton, J. Messing and J. F. Doebley, 2004. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. USA 101 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. M., T. N. Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38 594–597. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386 485–488. [DOI] [PubMed] [Google Scholar]
- Gallavotti, A., Q. Zhao, J. Kyozuka, R. B. Meeley, M. K. Ritter et al., 2004. The role of barren stalk1 in the architecture of maize. Nature 432 630–635. [DOI] [PubMed] [Google Scholar]
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb, T., O. Clarenz, E. Schafer, D. Muller, R. Herrero et al., 2003. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., and M. Nordborg, 2002. Sequence variation and haplotype structure surrounding the flowering time locus FRI in Arabidopsis thaliana. Genetics 161 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., C. Tang, J. Molitor, J. Werner, K. Zhao et al., 2004. Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics 168 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, O., and X. Vekemans, 2002. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2 618–620. [Google Scholar]
- Hill, W. G., 1974. Estimation of linkage disequilibrium in randomly mating populations. Heredity 33 229–239. [DOI] [PubMed] [Google Scholar]
- Holland, J. B., 1998. EPISTACY: a SAS program for detecting two-locus epistatic interactions using genetic marker information. J. Hered. 89 374–375. [Google Scholar]
- Keller, T., J. Abbott, T. Moritz and P. Doerner, 2006. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves, T., K. Schmid, A. Caicedo, C. Mays, J. Stinchcombe et al., 2007. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169 E141–157. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Leyser, H. M., C. A. Lincoln, C. Timpte, D. Lammer, J. Turner et al., 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., J. H. Britton and M. Estelle, 1990. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J. A., E. I. Moan, J. I. Medford and M. K. Barton, 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mackay, T. F., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14 253–257. [DOI] [PubMed] [Google Scholar]
- Malmberg, R. L., S. Held, A. Waits and R. Mauricio, 2005. Epistasis for fitness-related quantitative traits in Arabidopsis thaliana grown in the field and in the greenhouse. Genetics 171 2013–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen, P., and O. Leyser, 2005. Shoot branching. Annu. Rev. Plant Biol. 56 353–374. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and J. Schmitt, 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441 947–952. [DOI] [PubMed] [Google Scholar]
- Muller, D., G. Schmitz and K. Theres, 2006. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, M., and T. A. Linksvayer, 2006. The conversion of variance and the evolutionary potential of restricted recombination. Heredity 96 111–121. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. L., N. J. Patterson, R. M. Plenge, M. E. Weinblatt, N. A. Shadick et al., 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. a Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens, N. A. Rosenberg and P. Donnelly, 2000. b Association mapping in structured populations. Am. J. Hum. Genet. 67 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D., and J. I. Suddith, 1998. Molecular population genetics of the Arabidopsis CAULIFLOWER regulatory gene: nonneutral evolution and naturally occurring variation in floral homeotic function. Proc. Natl. Acad. Sci. USA 95 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D., A. L. Boyles and J. I. Suddith, 2000. Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics 155 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., O. Torjek, R. Meyer, H. Schmuths, M. H. Hoffmann et al., 2006. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 112 1104–1114. [DOI] [PubMed] [Google Scholar]
- Sharbel, T. F., B. Haubold and T. Mitchell-Olds, 2000. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol. Ecol. 9 2109–2118. [DOI] [PubMed] [Google Scholar]
- Sorefan, K., J. Booker, K. Haurogne, M. Goussot, K. Bainbridge et al., 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., S. P. Chatfield and H. M. Leyser, 1999. AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., K. van De Sande and H. M. Leyser, 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sussex, I. M., and N. M. Kerk, 2001. The evolution of plant architecture. Curr. Opin. Plant Biol. 4 33–37. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P. B., H. T. Adler, D. W. Parks and L. Comai, 1995. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735. [DOI] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, J. L. Modliszewski, T. F. Mackay and M. D. Purugganan, 2002. Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160 1133–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, M. D. Purugganan and T. F. Mackay, 2003. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., P. S. Springer, L. Goh, E. S. Buckler, IV and R. Martienssen, 2005. Architecture of floral branch systems in maize and related grasses. Nature 436 1119–1126. [DOI] [PubMed] [Google Scholar]
- Wang, R. L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398 236–239. [DOI] [PubMed] [Google Scholar]
- Ward, S. P., and O. Leyser, 2004. Shoot branching. Curr. Opin. Plant Biol. 7 73–78. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Weigel, D., J. Alvarez, D. R. Smyth, M. F. Yanofsky and E. M. Meyerowitz, 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Yoon, H. S., and D. A. Baum, 2004. Transgenic study of parallelism in plant morphological evolution. Proc. Natl. Acad. Sci. USA 101 6524–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. [DOI] [PubMed] [Google Scholar]
- Zhao, K., M. J. Aranzana, S. Kim, C. Lister, C. Shindo et al., 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]