Abstract
Allelic mutants exhibiting growth defects in Drosophila were isolated. Molecular cloning identified the responsible gene as a budding yeast Tim50 ortholog, and thus it was named tiny tim 50 (ttm50). The weak allele (ttm50Gp99) produced small flies due to reduced cell size and number, and growth terminated at the larval stage in the strong alleles (ttm50IE1 and ttm50IE2). Twin-spot analysis showed fewer cells in ttm50Gp99 clones, whereas ttm50IE1 clones did not proliferate, suggesting that the gene has an essential cellular function. Tim50 is known to maintain mitochondrial membrane potential (MMP) while facilitating inner-membrane protein transport. We found that tagged Ttm50 also localized to mitochondria and that mitochondrial morphology and MMP were affected in mutants, indicating that mitochondrial dysfunction causes the developmental phenotype. Conversely, ttm50 overexpression increased MMP and apoptosis. Co-expression of p35 suppressed this apoptosis, resulting in cell overproliferation. Interestingly, ttm50 transcription was tissue specific, corresponding to elevated MMP in the larval midgut, which was decreased in the mutant. The correlation of ttm50 expression levels with differences in MMP match its proposed role in mitochondrial permeability barrier maintenance. Thus a mitochondrial protein translocase component can play active roles in regulating metabolic levels, possibly for modulation of physiological function or growth in development.
SIZES of organs and organisms in each species are the result of the regulation of cell size and cell number (for reviews, see Neufeld and Edgar 1998; Polymenis and Schmidt 1999). Recent advances in the field of growth regulation have revealed the involvement of multiple signaling pathways, including the insulin/target of rapamycin (TOR) pathway (for a review, see Oldham and Hafen 2003), the Myc pathway (Johnston et al. 1999; Trumpp et al. 2001; de la Cova et al. 2004), and the Ras pathway (Prober and Edgar 2000). These findings suggest that nutrition levels are major input signals for this signaling network and that the output is the rate of protein synthesis causing changes in cell growth and proliferation. It would also be expected that intracellular energy levels affect cell growth and proliferation, although it is not yet proved whether such mechanisms are actually utilized in developmental growth regulation.
Intracellular energy levels are largely dependent on the synthesis of ATP in the mitochondria and their dysfunction causes severe developmental defects. In Drosophila melanogaster, mutants of nuclear-coded mitochondrial proteins, including DNA polymerases and ribosomal proteins, are reported to demonstrate growth retardation, indicating that they have at least a passive role in the regulation of development (Iyengar et al. 1999, 2002; Maier et al. 2001; Galloni 2003; Frei et al. 2005).
About 99% of mitochondrial proteins are encoded by the nuclear genome. After being synthesized by the cytoplasmic ribosomes, they are translocated into mitochondria by protein translocases (for reviews, see Endo et al. 2003; Rehling et al. 2003, 2004). In Caenorhabditis elegans, it has been shown that this transport of mitochondrial proteins is essential for mitochondrial biogenesis and that defects in the components of the translocases cause severe developmental aberrations (Curran et al. 2004). Mitochondrial architecture consists of the outer membrane, the intermembrane space, the inner membrane, and the matrix. Mitochondrial proteins are translocated into the intermembrane space or embedded into the outer membrane by the TOM complex. Many of the proteins translocated into the intermembrane space are further embedded in the inner membrane or tranlsocated into the matrix by the TIM22 or TIM23 complexes. The TIM23 complex consists of Tim23, Tim17, and Tim50 and is involved in the translocation of proteins with positively charged presequences (for reviews, see Endo et al. 2003; Rehling et al. 2003, 2004).
In this article, we show that Drosophila mutants of a gene similar to yeast Tim50 are defective in growth and cell cycle and exhibit reduced mitochondrial membrane potential (MMP). Overexpression of the protein induced increased MMP, apoptosis, and, in certain circumstances, extra cell proliferation.
MATERIALS AND METHODS
Genetics:
Transposon mutagenesis for X chromosomal recessive lethal mutants was performed by remobilizing the multiple incomplete P elements on the second chromosome of the Birmingham 2 strain with a supply of the transposase from PΔ2-3-32 (Robertson et al. 1988). Additional alleles were obtained by EMS mutagenesis. yellow (y) male flies were fed with 0.01 m EMS and mated with C(1)DX, y w/BS-Y, y+ w+ females, and the resultant male progeny were singly crossed with y ttm50Gp99/Binsc females to screen alleles.
Molecular techniques:
Standard molecular techniques were employed (Sambrook et al. 1989). The DNA isolated from y ttm50Gp99/Binsc female flies was partially digested with Sau3A and the fragments were ligated with EMBL3 arms. The ttm50Gp99 genomic library was screened with pπ25.1 (Rubin and Spradling 1983) as a probe to recover genomic DNA flanking the P element inserted in ttm50Gp99. The resultant clone was then used to probe a wild-type Drosophila genomic library (Tsuda et al. 1993). Fragments derived from the wild-type genomic clone were used as probes for cDNA library screening. A 12- to 24-hr embryonic cDNA library in pNB40 (Brown and Kafatos 1988) was screened and a full-length clone was obtained. The cDNA and genomic fragments were sequenced for both strands. Similarity searches were carried out using FASTA (Wisconsin GCG) and NCBI BLAST programs.
Transgene constructs:
The 3.5-kb SacII/EcoRI genomic fragment for mutant rescue was cloned into the P-element vector pW8 and used for transformation. The GMR-element-mediated expression construct was made by recloning the ttm50 containing the EcoRV/EcoRI fragment from the pNB40 cDNA clone into a pBST(SK+) vector and then again recloning the EcoRV/SmaI fragment into the pGMR expression vector (Hay et al. 1994). The hemagglutin (HA) epitope-tagged Ttm50 expression construct was produced by first PCR amplifying the pNB40 cDNA with an EcoRI site-incorporated 5′ primer (GAATTCGTGAACGGCTCTAGGACTC) and a XhoI site-incorporated 3′ primer (CTCGAGCGTGCCTCCCAATCCTGCT) and cloning it into the EcoRI/XhoI sites of the pUAST expression vector (Brand and Perrimon 1993) via a pGEM(TA) vector (Promega, Madison, WI). This was cut with Eco52I, and a NotI fragment containing a triple HA epitope was inserted, resulting in a C-terminal tagged construct that was confirmed by sequencing and used for transformation. Expression was induced using a heat-shock GAL4 transgene (Brand and Perrimon 1993), and expression of the correctly sized tagged protein was checked by Western blotting.
Clonal analysis:
X-ray clones were induced by irradiation at a dose of 1.5 kR using an Ohmic OM-100RE soft X-ray unit equipped with a 0.2-mm aluminum filter. Flp-recombinase clones were produced by recombining mutant alleles onto X chromosomes carrying the p[2πM]FRT construct at18A, crossing these with Ubi-GFP and p[2πM]FRT 18A, and inducing mitotic recombination with heat-shock-induced flp-recombinase expression (Golic 1991).
Histochemistry:
Mitotracker Red (chloromethyl-X-rosamine; Molecular Probes, Eugene, OR) was used at 10–100 nm, JC-1 (Molecular Probes) at 25–250 nm, and acridine orange (Wako Fine Chemicals) at 1 μm was used in PBS or Grace's medium with unfixed samples or before fixation. Concentrations were adjusted to obtain uniform staining in control sample tissues.
Fixation of heptane permeablized dechorionated eggs or dissected tissues were used with 4% paraformaldehyde in PBS. DAPI (Wako Fine Chemicals) was used at 1 ng/ml, rhodamine or Alexa Fluor 647-conjugated phalloidin (Molecular Probes) was used at 0.2 unit/ml in PBS containing 0.1% Triton-X. HA-tagged proteins were detected by indirect immunofluorescence using rabbit polyclonal anti-HA Y-11 serum (Santa Cruz) at 1:500 followed by Alexa Fluor 488-conjugated goat anti-rabbit IgG serum (Molecular Probes) at 1:1000 dilutions.
In situ hybridization was done according to the method of Tautz and Pfeifle (1989) with standard modifications for RNA probes and prestain washes in maleic acid buffer. The probes were transcribed from PCR products with T7 promoters incorporated into the primers during amplification from cDNA. Sense-strand probes were used as nonspecific staining controls. Bright-field and fluorescent images were taken on a Nikon Eclipse E800 microscope with a DXM 1200F digital camera, SEM images on a Phillips XL 20 scanning electron microscope, and confocal images on a Zeiss Axiovert 100M equipped with an LSM 510 module.
RESULTS
Isolation of the mutants:
Among a collection of newly induced P-insertional mutations, an X-linked semilethal line, Gp99, was obtained. Under standard culture conditions, few mutant flies appeared. When the mutant chromosome was marked with y and the y hemizygous male larvae were separately cultured in uncrowded conditions, 60–80% of the mutant larvae developed into adults but grew slowly (Figure 1D). Mutants took ∼3 more days to develop as compared to the normal 10-day developmental time. The mutant is considerably smaller than the control (Figure 2, A and B). The weight of the mutant male flies (0.65 ± 0.06 mg, n = 483) was ∼35% reduced as compared to the control y hemizygous flies (1.01 ± 0.04 mg, n = 347).
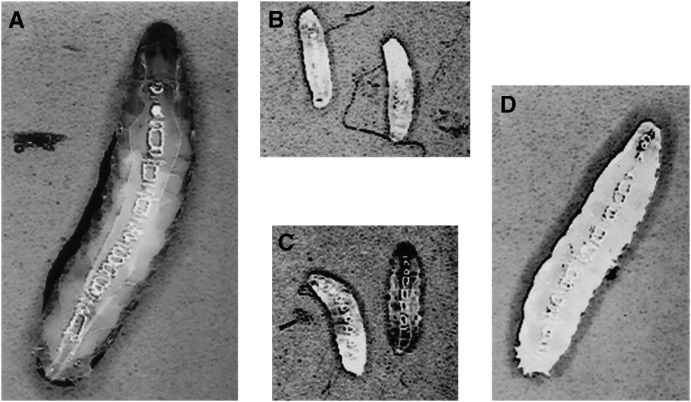
Figure 1.—
Growth defects observed in ttm50 mutant larvae. (A) A wild-type embryo at 4 days after hatching is fully grown and ready to pupate. (B) ttm50IE1 null allele and (C) ttm50IE2 strong allele larvae are much smaller at the same age and later die without showing significant increase in size. (D) A ttm50Gp99 hypomorphic allele larvae at the same age shows a smaller body size, requiring a few more days to pupate. All photos at same magnification.
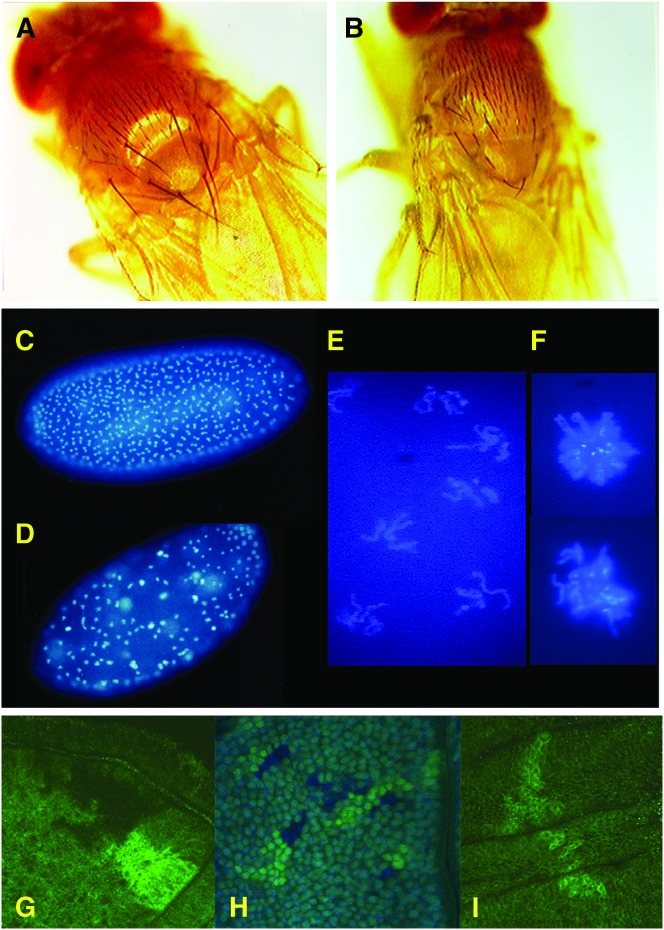
Figure 2.—
The ttm50Gp99 hypomorphic allele shows growth and cell cycle defects. (A) A yellow1 (y1) adult male control fly showing normal body size and bristle morphology. (B) A y1 ttm50Gp99 hemizygote male showing reduced body size and underdeveloped macrochaetae. (C) A DAPI-stained wild-type cleavage-stage embryo showing uniform distribution of nuclei in synchronous mitosis. (D) A DAPI-stained y1 ttm50Gp99 homozygous female-derived embryo showing irregular distribution of cleavage nuclei, with some showing stronger staining intensity. (E) Higher magnification image of typical nuclei in D shows condensed chromosomes in early mitosis. (F) Higher magnification image of nuclei with stronger staining intensity in D shows condensed polyploid chromosomes, which appear to have failed in nuclear division. (G) Mitotic sister (bright and dark) clones of wild-type cells are of similar size. (H) Mitotic clones of ttm50Gp99 homozygous cells (dark-blue nuclei) have fewer cells than their wild-type (ubi-GFP/ubi-GFP) sister clones (bright-green nuclei) when induced in a heterozygous (ttm50Gp99/ubi-GFP) background (pale green nuclei). (I) Only the control sister clones of ttm50IE1 null clones are observed, even after extensive growth.
To examine whether reduced body size might be due to reduced cell size, we compared the density of wing hairs (cells) within a fixed area with an arbitrary width just behind the posterior cross-vein. In the mutant, 125.5 ± 5.1 (n = 22) hairs were counted while 109.2 ± 5.1 (n = 15) hairs were counted in the control. This roughly corresponds to a 13% reduction in cell width and partially explains the reduction of body size. The number of hairs along the posterior cross-vein was also reduced in the mutant (30.0 ± 2.2, n = 21) as compared to the control (35.1 ± 2.6, n = 14), although the shape and vein pattern of the wing appeared normal. In addition, the macrochaetae (bristles) are considerably thinner and shorter than those of the control (Figure 2, A and B). Because they are secreted by a single polytene cell (shaft cell) that is much larger than the average diploid cell, the observed decrease in bristle size could be a result of defective cell growth or cell cycle progression.
Twin-spot analysis (mitotic clone analysis) utilizing flp recombinase showed that mutant clones induced in imaginal discs were consistently lower in cell number than their wild-type sister clones (Figure 2H), whereas wild-type control clones were the same size as their sister clones (Figure 2G). Acridine orange staining of the imaginal discs did not show increased cell death (data not shown). Thus, we conclude that the diminutive body size in the mutant is due to a reduction in both size and number of cells caused by a cell growth and proliferation defect.
The mutant males exhibited reduced fertility but crossings with heterozygous females produced homozygous females. These females laid eggs but none of the eggs hatched. Examination of the embryos revealed a maternal effect on cleavage divisions (Figure 2, D–F) although some embryos continued development past gastrulation. Those arrested in cleavage divisions seem to have arrested at around cycle 8 (Figure 2D), and most of the nuclei are at M-phase (Figure 2E). Polyploid nuclei were observed sporadically (Figure 2F), suggesting that some nuclei failed to complete nuclear division, but continued through DNA replication only to fail again in mitosis afterward. This could be because the G1-S checkpoint is not in effect at this developmental stage (Sibon et al. 1997).
As cleavage divisions consist of only S- and M-phases, we also examined the mitotic index in the brain squashes from third instar larvae (Table 1). In the larval brain lobes, cell cycles proceed through all four phases. If the mitotic defect were the major cause of cell cycle arrest, an increase of mitotic index would be expected. As shown in Table 1, the mitotic index was considerably reduced, suggesting that the cell cycle is affected during a phase(s) other than the M-phase. As aberrant mitotic figures were frequently observed in the brain squashes, mitosis is also affected. Thus, both mitosis and interphase are affected in this mutant.
TABLE 1.
Mitotic index in the larval brain lobe cells
| Genotype | % of mitotic cells | No. of cells counted |
|---|---|---|
| Control (y+) | 1.48 | 1356 |
| y ttm50Gp99/Y | 0.68 | 2217 |
Since Gp99 is a hypomorph (see below), we tried to obtain strong alleles by EMS mutagenesis. Two additional alleles, IE1 and IE2, were isolated. IE1 is thought to be null (see below). The growth of the IE1 mutant larvae was severely affected, and the larvae died as first instar larvae after surviving for ∼1 week (Figure 1B). The molecular defect of IE2 was not identified, but the growth of the mutant larvae was also severely affected and IE2 mutant larvae died as larvae after surviving a considerable period (Figure 1C). These phenotypes resemble those of typical growth-defect mutants such as peter pan (Migeon et al. 1999), bonsaï (Galloni 2003), and the mutants in the insulin/TOR network (Chen et al. 1996; Böhni et al. 1999; Montagne et al. 1999; Weinkove et al. 1999; Oldham et al. 2000, 2002; Zhang et al. 2000; Rintelen et al. 2001; Saucedo et al. 2003; Stocker et al. 2003).
Clonal analyses of IE1 produced clones of one or two cells in contrast to the control twin spots with large numbers of cells in wings (Figure 2I), indicating a loss of cell proliferation ability in the null mutant. Germline clones of IE1 could not be obtained.
Cloning and identification of the gene:
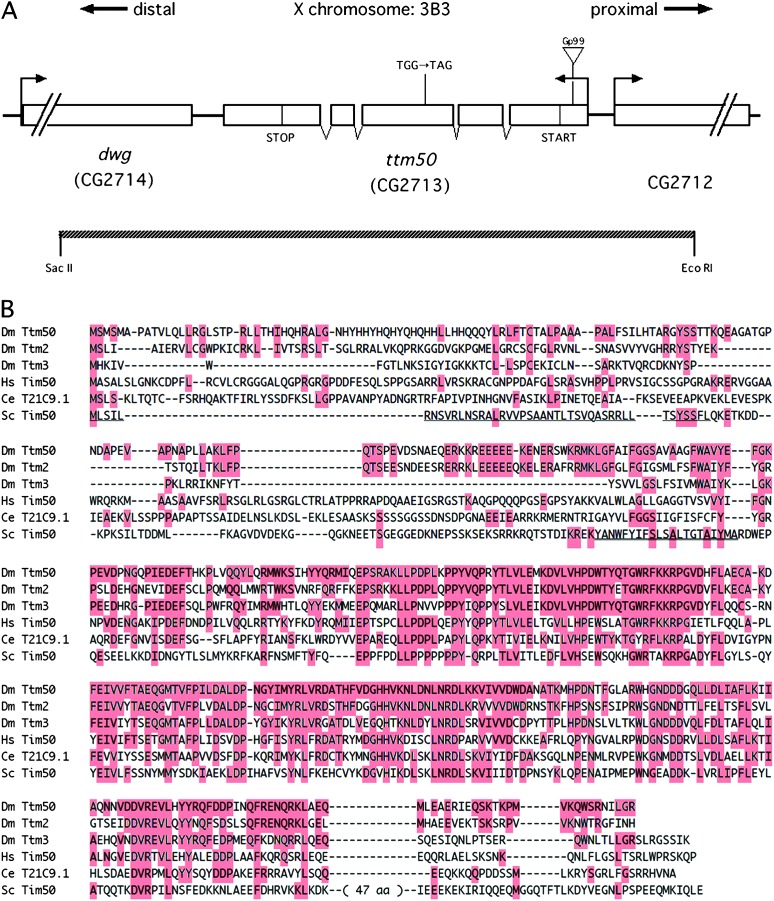
Using the P element as a probe, we cloned the DNA fragment flanking the insertion site in Gp99, and the wild-type locus was also cloned (see materials and methods). The P element was inserted in the 5′-UTR of the transcription unit, CG2713 (Figure 3A), mapping to 3B3 on the X chromosome. Excision of the P element using transposase reverted the mutant phenotypes of Gp99. Furthermore, a 3.5-kb SacII/EcoRI genomic fragment containing the intact gene but only portions of the neighboring genes (Figure 3A) completely rescued the bristle phenotype of Gp99 and the viability of all three alleles, showing that CG2713 is the responsible gene. A cDNA clone was isolated and sequenced. Comparison with the genomic sequence revealed the presence of four short introns and an ORF encoding a protein of 428 amino acid residues. These sequences did not differ significantly from those published by the Berkeley Drosophila Genome Project. Two more related sequences are present in the Drosophila genome (Figure 3B). A search of the Genome Databases revealed that the sequence at the carboxyl-terminal portion is highly conserved in human and nematode (Figure 3B). The sequence showed a low but significant similarity to the phosphatase for the C-terminal domain of RNA polymerase II. Similar sequences were also found in budding and fission yeasts. The budding-yeast sequence has been reported to encode Tim50, which is located in the inner membrane of mitochondria and is involved in protein translocation (Geissler et al. 2002; Yamamoto et al. 2002) while maintaining MMP (Meinecke et al. 2006). Drosophila sequences contain a putative, positively charged presequence at the N-terminal ends and a putative transmembrane domain, suggesting that these gene products themselves also translocate into mitochondria and are embedded in the inner membrane. From the phenotype and sequence homology we named the genes tiny tim50 (ttm50), tiny tim2 (ttm2: CG12313), and tiny tim3 (ttm3: CG6691). The ttm50 mutant lines are indicated as ttm50Gp99, ttm50IE1, and ttm50IE2 hereafter.
Figure 3.—
Molecular characterization of the ttm50 loci and its conservation through evolution. (A) The ttm50 locus and the intron/exon organization of its transcript. The ORF is indicated by START and STOP. The ttm50Gp99 allele is caused by a P-element-insertion in the 5′-UTR. The ttm50IE1 allele is caused by a nucleotide alteration of Trp197 to a TAG termination codon. The extent of the 3.5-kb SacII/EcoRI fragment used for rescue transgene constructs is indicated. (B) Amino acid sequence comparison of genes sharing homology to ttm50. Amino acid residues identical to those of Dm Ttm50 are highlighted and gaps are indicated by dashes. Dm Ttm50, D. melanogaster Tiny tim50; Dm Ttm2, D. melanogaster Tiny tim2; Dm Ttm3, D. melanogaster Tiny tim3; HsTim50, Homo sapiens Tim50; Ce T21C9.1; C. elegans T21C9.1; Sc Tim50, Saccharomyces cerevisiae Tim50. The thin underline in S. cerevisiae Tim50 indicates the mitochondria-targeting presequence and the thick underline indicates the transmembrane domain.
Sequencing of the PCR-amplified ORF fragments from genomic DNA of ttm50IE1 revealed a point mutation altering guanine to adenine, which causes a change of Trp197 to a TAG termination codon (Figure 3A). This causes a truncation before the conserved region, and the mutation is interpreted to be null. We could not find any nonsilent alterations in the ORF of ttm50IE2, and the mutation may be in the regulatory sequence.
Ttm50 is a mitochondrial protein:
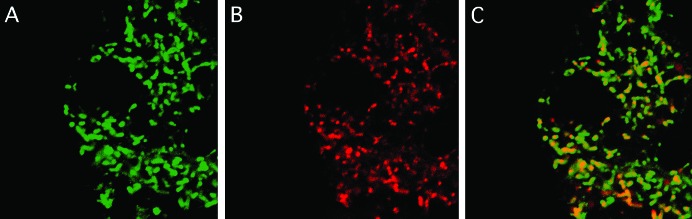
As Ttm50 is similar to yeast Tim50, we examined whether Ttm50 is also localized in mitochondria by expressing a HA-tagged Ttm50 protein using the GAL4/UAS system (Brand and Perrimon 1993). This transgene was able to rescue the ttm50IE1 mutant lethality, indicating that it is functional. When the Ttm50-HA protein was expressed using various GAL4 drivers, the presence of Ttm50-HA was observed in a reticulated pattern in the cytoplasm in tissues such as those of the central nervous system, imaginal discs, salivary glands, and fat bodies of larvae (Figure 4A). Costaining with Mitotracker Red, which stains mitochondria specifically, showed an overlapping pattern, demonstrating that Ttm50-HA is localized to mitochondria (Figure 4C) as anticipated from its sequence homology.
Figure 4.—
HA-tagged Ttm50 localizes to the mitochondria. (A) Highly magnified confocal image of a fat-body cell. The open area in the center corresponds to the nuclei. The HA-tagged Ttm50 protein staining appears as reticulated structures in the cytoplasm. (B) Mitochondria stained by Mitotracker Red. (C) Merged image shows a good overlap (yellow), although the distribution of intensity within the mitrochondria differs.
Northern blot and real time RT–PCR analyses demonstrated that ttm50 is expressed throughout development (data not shown). In situ hybridization in embryos demonstrated a large amount of mRNA in the cleavage-division-stage embryos (Figure 5A), suggesting a contribution by maternal mRNA. After gastrulation, ttm50 is highly expressed in the developing midgut (Figure 5, C–H). In adult flies, significant expression of ttm50 was detected in isolated testis as well as in the remaining carcass, whereas ttm2 and ttm3 are expressed exclusively in the testis when analyzed by real time RT–PCR (M. Nishimura, T. Yamamoto, S. Sugiyama and Y. Nishida, unpublished observations).
Figure 5.—
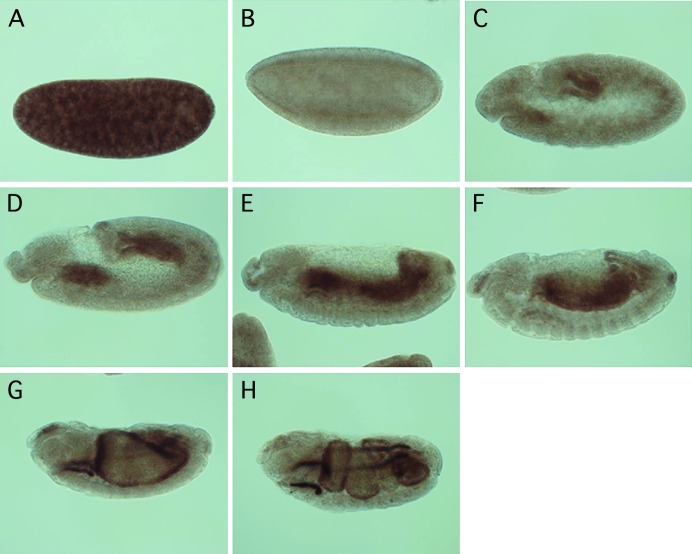
Expression of ttm50 transcripts is developmentally regulated during embryogenesis. In situ hybridization with antisense RNA probes was used to study the tissue specificity of ttm50 transcription at various embryonic stages. (A) Transcripts were ubiquitously distributed during the cleavage stage and are presumably maternally derived. (B) Transcripts temporarily disappeared at the cellular blastoderm stage. (C and D) Expression reappeared in the midgut primordial and weakly in the mesoderm of gastrulas. (E and F) Additional transcripts appeared in the hindgut and Malpighian tubules during germband retraction. The mesodermal expression could be seen in the differentiating skeletal and visceral muscles. (G and H) This expression pattern was retained throughout the duration of organogenesis. In all photos, anterior is to the left and dorsal to the top.
To evaluate the effect of the ttm50 mutations on mitochondrial activity, either the body wall of control and second instar ttm50IE2 larvae was ripped to allow the gut to spill out (Figure 6A) or the entire larva was completely inverted and then stained with Mitotracker Red. Mitotracker Red is a lipophilic cationic dye that accumulates in mitochondria due to their negative membrane potential and is a sensitive nontoxic relative indicator of MMP, although it also reflects changes in mitochondrial mass (Pendergrass et al. 2004). We found that mitochondrial activity was strongest in the gut, corresponding to the transcription pattern observed in the late embryo. Gut staining in the mutant was weaker than in wild type (Figure 6B). Results were similar when the larvae were fed Mitotracker Red and the anterior midgut epithelia were compared (Figure 6, C–F) as described by Galloni (2003). Since Mitotracker Red staining is dependent on MMP, this indicates that respiratory activity is reduced in the mutant midgut.
Figure 6.—
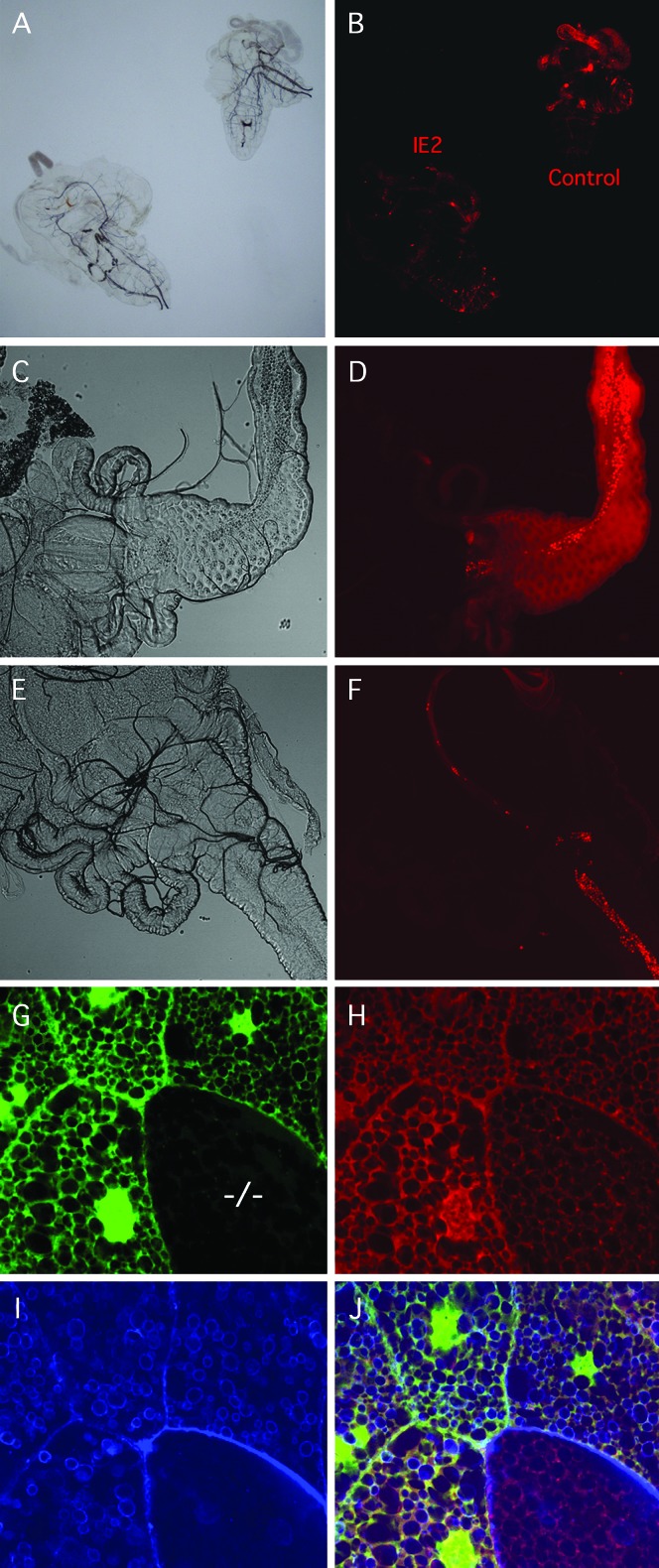
Strong alleles of ttm50 show reduced mitochondrial activity. (A) Bright-field image of ttm50IE2 (left) and wild-type (right) larvae partially dissected and incubated in PBS containing 10 ng/ml Mitotracker Red. (B) Fluorescent image of same sample shows weaker respiration-dependent staining in the mutant. Staining is strongest in the wild-type gut. (C) Bright-field image of proventriculus and anterior midgut of wild-type second instar larva. (D) Mitotracker-Red-stained fluorescent image of same sample. Bright red spots are stained yeast in the alimentary tract. (E) Bright-field image of the same organs in a ttm50IE2 larva. (F) Mitotracker Red staining is reduced in mutant. (G) Homozygous ttm50IE1 mutant clone cells were induced in heterozygous embryos by X-ray irradiation and examined in third instar larva. Mutant salivary gland cell (−/−) is identified by reduced GFP fluorescence. (H) Mitotracker Red staining in mutant cell is reduced in both intensity and area. (I) Alexa Fluor 647-conjugated phalloidin staining reveals accumulation of actin at cell periphery and reduction in actin around vesicle-like structures of mutant cell. (J) Merged image of G–I.
When mitotic clones were X-ray induced in ttm50IE1 heterozygous embryos and observed at larval stages, polyploid clone cells identified in the salivary gland by reduced GFP fluorescence showed weakened Mitotracker Red staining compared to surrounding cells (Figure 6, G–J). The network of mitochondria appeared less developed and the staining intensity of individual components was reduced. Thus it appears that there was a decrease in overall volume of mitochondria in the cytoplasm in addition to a drop in MMP, leading to a combined decline in mitochondrial activity.
Overexpression of ttm50 can cause increased mitochondrial membrane potential, apoptosis, and an increase in cell proliferation:
When the Act5C-GAL4 driver was used to express ttm50 cDNA ubiquitously, some of the transgenic lines showed lethality at different stages. UAS-ttm50#48 was lethal at an early larval stage, UAS-ttm50#2 died as pharate adults, and UAS ttm50#32, viable with no visible phenotypes, was still able to rescue the lethality of ttm50IE1. These differences in phenotype probably reflect differences in strength of expression due to a “position effect” of the chromosomal sites in which the transgene was inserted. Thus overexpression of ttm50 appears to be lethal and indicates that a moderate level of expression needs to be maintained for proper function and viability.
The intermediate strength UAS-ttm50#2 transgene was next expressed in the eye under the control of the GMR-GAL4 driver, which expresses target sequences posterior to the morphogenetic furrow in the developing eye disc by utilizing the glass-mediated response element (Hay et al. 1994). This caused a mild rough-eye phenotype, indicating disruption of ommatidial cell organization (Figure 7B and data not shown). To check whether this rough-eye phenotype was caused by cell death, the deficiency Df(3R)H99, which uncovers a set of the proapoptotic genes reaper (rpr), head involution defective (hid), and grim, was used. The gene dosage of these genes was reduced by half in flies expressing UAS-ttm50#2 under the control of the GMR-GAL4. The deficiency suppressed the rough-eye phenotype, suggesting that overexpression of ttm50 induced apoptosis (Figure 7C). It was then found that a half reduction of the hid gene dosage alone is sufficient for suppression (Figure 7D), suggesting that hid is the main factor responsible. On the other hand, a half reduction of the gene dosage of the inhibitor of the apoptosis protein gene DIAP1 enhanced the rough-eye phenotype (Figure 7E). This was expected because DIAP1 is negatively regulated by hid. Staining of the larval eye discs with acridine orange demonstrated increased cell death, supporting the above results (Figure 8B).
Figure 7.—
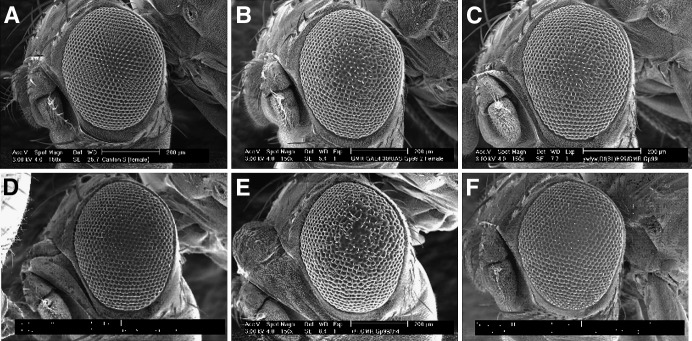
Overexpression of ttm50 in the compound eye causes extra proliferation of cells. (A) Scanning electron micrograph of wild-type eye. The number of ommatidia was 752.5 ± 23.5 (n = 13). (B) Rough-eye phenotype caused by overexpression of ttm50 under the control of the glass-mediated response element. (C) Suppression of rough-eye phenotype by a reduction by half in the gene dosage of hid grim reaper using the Df(3R)H99. (D) Suppression of rough-eye phenotype by a reduction by half in the gene dosage of hid alone. (E) Enhancement of rough-eye phenotype by a reduction by half in the gene dosage of DIAP. (F) Suppression of rough-eye phenotype by the co-expression of the caspase inhibitor p35. The number of ommatidia increased (798.5 ± 11.6, n = 13) significantly compared to wild type. Expression of p35 alone did not affect ommatidial organization or number (754.0 ± 10.8, n = 6) compared to wild type.
Figure 8.—
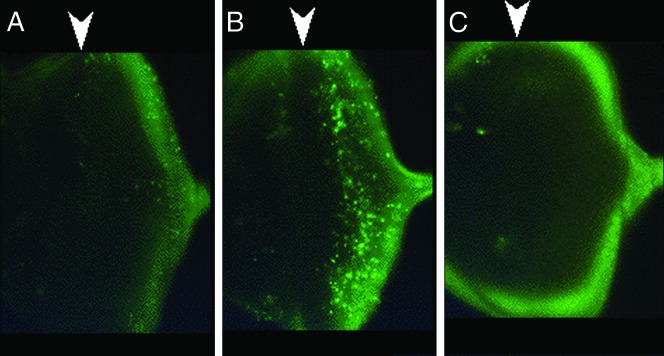
Cell death is induced by overexpression of ttm50. (A) Normal levels of apoptosis detected in the eye imaginal disc by acridine orange. (B) Increased cell death induced by the GMR element mediated expression of ttm50 in the eye imaginal disc. Note that the induction occurs after the passage of (and to the right of) the morphogenetic furrow. (C) Almost complete suppression of apoptosis by the co-expression of p35. Arrowheads above each image indicate the position of the morphogenetic furrow.
In contrast to overexpression of ttm50, imaginal discs from the loss-of-function mutant, the hypomorph ttm50Gp99, demonstrated no evidence of increase of cell death by the TUNEL assay (data not shown). This differs from observations with human Tim50, whose downregulation of expression increased the sensitivity to death stimuli in cultured cells (Guo et al. 2004).
When the baculovirus P35 caspase inhibitor was co-expressed with ttm50 in the compound eyes to suppress cell death, the rough-eye phenotype was almost completely suppressed (Figure 8C). At the same time, the number of ommatidia increased (798.5 ± 11.6, n = 13; Figure 7F) as compared to the normal eyes (752.5 ± 23.5, n = 13; Figure 7A). Expression of p35 alone did not increase the number of ommatidia (754.0 ± 10.8, n = 6). The results indicate that overexpression of ttm50 induces cell proliferation when cell death is inhibited by p35 co-expression.
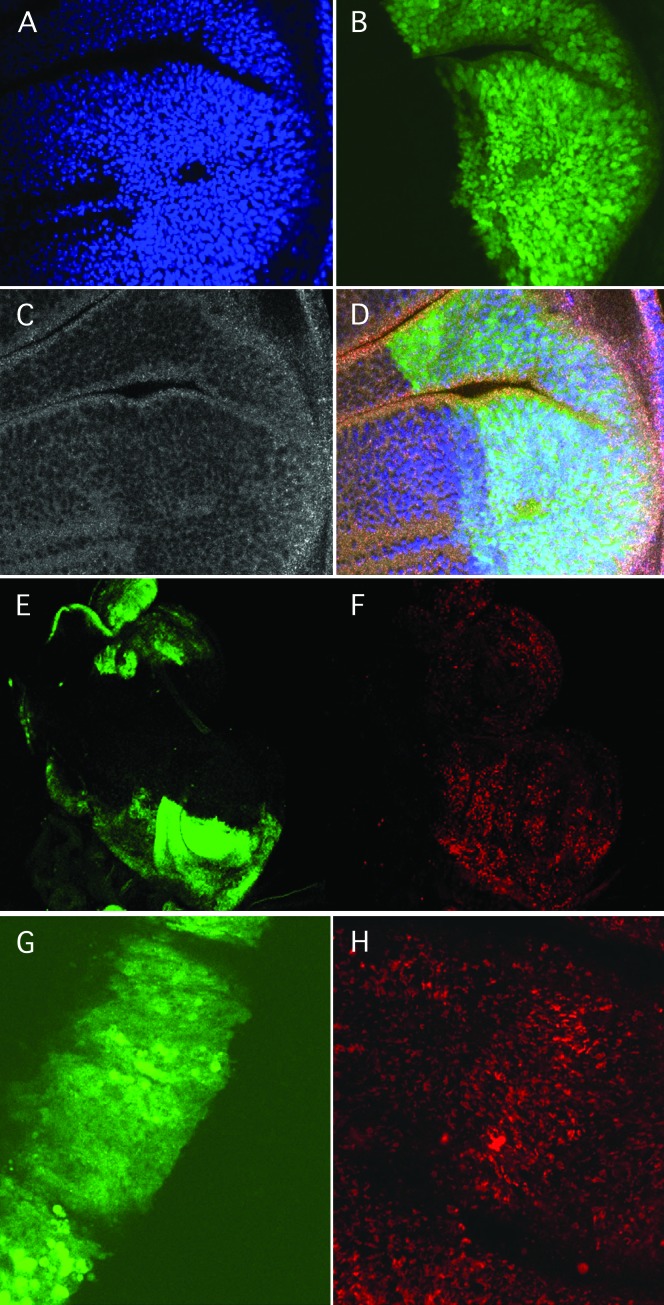
This was confirmed by localized overexpression in the larval wing imaginal disc using the engrailed (en)-GAL4 expression driver construct. The localized overexpression of UAS-ttm50#48 with p35 co-expression caused an increase in nuclear density (Figure 9A) and an increase in cell proliferation as determined by the ratio of S-phase cells detected by BrdU incorporation (Figure 9F). It also resulted in an increase in MMP as judged by the staining intensity of Mitotracker Red (Figure 9H) and JC-1 (not shown). Thus, raising the level of ttm50 expression causes an increase in respiratory activity and in addition stimulates cell proliferation when P35 is present to inhibit apoptosis.
Figure 9.—
Co-expression of UAS-ttm50 and UAS-p35 driven by en-GAL4 increases cell density (A–D) and proliferation (E and F) in the imaginal discs. (A) Wing disc nuclei stained by DAPI. (B) en-GAL4-driven gene expression in the posterior compartment marked by UAS-GFP. (C) Tissue morphology visualized by actin stained with Alexa Fluor 647-conjugated phalloidin shows a change in the posterior compartment. (D) Merged image of A–C. (E) en-GAL4-driven gene expression in the posterior compartments of two imaginal discs marked by UAS-GFP. (F) S-phase cells visualized by Brd-U incorporation increased in the posterior compartments. (G) Expression of UAS-GFP driven by ptc-GAL along the anterior posterior boundry of the wing imaginal disc pouch. (H) Increase in Mitotracker Red staining induced by overexpression of UAS-ttm50 driven by ptc-GAL in the same sample.
The increase in Mitotracker Red staining was also observed without p35 co-expression and was not seen with p35 expression alone (data not shown). This further suggests that controlling the selective permeability of the mitochondrial inner membrane through the expression level of ttm50 results in corresponding changes in MMP and presumably in the resulting cellular energy output.
DISCUSSION
Mutants of ttm50, which encodes a Drosophila ortholog of Tim50, showed systemic growth defects, such as delayed development and reduced cell size and cell proliferation, whereas overexpression of ttm50 induced cell death and extra cell proliferation when that cell death was artificially inhibited. These phenotypes correlated well with the presumed roles of Ttm50 in mitochondrial physiology, with the loss of Ttm50 affecting mitochondrial morphology, and with causing a drop in MMP, while its overexpression caused an increase in MMP. Expression of ttm50 during development was found to be tissue specifically regulated, and its expression levels matched the MMP observed in tissues; thus ttm50 also may have specific roles in tissue development and physiology.
Ttm 50 function in mitochondria and its relationship to observed phenotypes:
Whereas loss of ttm50 in mutants reduced MMP, its overexpression elevated MMP. The maintenance of MMP is essential for ATP synthesis by the mitochondria, and thus transport through the inner membrane is highly selective to maintain the proton gradient. The Tim23 complex serves as the channel for preproteins, and the presequences in preproteins induce rapid gating of the channel by Tim50 (Meinecke et al. 2006). The function of Tim50 is to keep the channel closed in the absence of presequences, so the loss or reduction of its homolog, Ttm50, would be expected to result in a “leaky” membrane with a decrease in MMP. Such a decrease was observed in mutants in this study and has been reported for Tim50 mutants in budding yeast (Meinecke et al. 2006). Extending this logic, too much Ttm50 could cause an overly tight permeability barrier with an increase in MMP, as was observed in our overexpression experiments. Alternatively, reduced protein import by loss of Ttm50 could cause a general dysfunction in mitochondria, leading to an MMP decrease, whereas overexpression could cause increased protein import, resulting in increased MMP due to a rise in overall mitochondrial metabolism.
Both M-phase and interphase cell cycle defects were found in ttmGp99 mutants, although the M-phase defects were prominent only during the cleavage stage when the G1/S phase checkpoint was not in place. Malnutrition usually affects the G1 phase, and mutants of the tenured gene, which encodes the cytochrome oxidase subunit Va, have been shown to cause drops in intracellular ATP levels that result in cell cycle arrest at the G1-S cell cycle checkpoint. This occurs through a pathway involving AMP-activated protein kinase and p53 activation that causes elimination of cyclin E (Mandal et al. 2005). Furthermore, cleavage-stage embryos are known to be sensitive to hypoxia that induces declines in ATP levels and results in mitotic arrest (DiGregorio et al. 2001); thus a similar phenomenon may be occurring in ttmGp99 mutants.
Induction of cell death by excess Ttm50:
Overproduction of Ttm50 induced cell death. Apoptosis is generally accompanied by mitochondrial permeability transition (MPT) in which the inner mitochondrial membrane increases in permeability, resulting in loss of membrane potential, swelling, and eventual rupture of the outer membrane (Zoratti and Szabo 1995). A scenario in which the integration of too much Ttm50 into the inner mitochondrial membrane disrupts its integrity and induces MPT may explain the apoptosis observed. Other possibilities are that abnormally elevated MMP or abnormal protein transport caused by excess Ttm50 causes the apoptosis observed.
When the induced apoptosis was inhibited by co-expression of p35, overproduced Ttm50 induced excessive proliferation. This could be explained by increased MMP that would result in upregulation of ATP synthesis and may have stimulated cellular growth and, subsequently, cell proliferation. It has been shown that cell cycle progression could be regulated via the mitochondria in the case of mRpL12 required for cell growth driven by cyclin D/Cdk4 (Frei et al. 2005).
An alternative interpretation is that when cell death induced by overexpressing ttm50 is artificially prevented through p35 co-expression, it results in overgrowth through a phenomenon known as compensatory cell proliferation. Apoptotic cells induce the proliferation of surrounding cells to compensate for their death by secreting growth-promoting signals such as Decapentaplegic (Dpp) and Wingless (Wg). Thus, when apoptosis is induced but at the same time inhibited by P35, the “nondying” apoptotic cells can bring on excessive compensatory division (Huh et al. 2004; Ryoo et al. 2004).
Tissue-specific roles of ttm50:
Ttm50 is probably essential for mitochondrial function since its two homologs, ttm2 and ttm3, appear to be testes specific and probably cannot complement its function in other tissues. Thus the tissue-specific transcription seen by in situ hybridization in embryos after gastrulation is interesting. The lethal phase for the null ttm50IE1 allele was at the early larval stage, so embryonic development of tissues not transcribing ttm50 is presumably sustained by maternally provided transcripts or translation products. Accordingly, ubiquitously distributed transcripts were seen in early cleavage-stage embryos. They are thought to be of maternal origin because expression of zygotic genes generally does not occur at this early stage. From gastrulation onward in embryogenesis ttm50 transcripts are expressed strongly in the midgut, and also in the hindgut, Malpighian tubules, and mesoderm. While we did not detect ttm50 transcription in the nervous system, zebrafish Tim50 mRNA expression is reported to be prominent in the brain of the embryo (Guo et al. 2004). These differences imply that the tissue-specific roles of Tim50/ttm50 differ between the two organisms, but nonetheless the control of both genes during development seems to be critical.
The midgut expression pattern of ttm50 is similar to that of Drosophila bonsaï, which encodes the mitochondrial ribosomal protein S15 (Galloni 2003). In bonsaï mutant larvae, a gut-specific respiration defect similar to the one that we report has been observed. The larval midgut is known to show high-cytochrome-C-oxidase activity (Galloni 2003) and in this study we find elevated MMP. So it appears that bonsaï as well as ttm50 are responsible for elevated respiratory activity in this organ that could be required for its proper development or physiological function.
Acknowledgments
We thank J. M. Abrams and M. Yamaguchi for fly strains and N. Brown for cDNA libraries. We are indebted to M. Wada, M. Motegi, and Y. Tanaka for technical assistance. This work is supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Science and Technology Agency.
References
- Böhni, R., J. Riesgo-Escovar, S. Oldham, W. Brogiolo, H. Stocker et al., 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97 865–875. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Brown, N. H., and F. C. Kafatos, 1988. Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203 425–437. [DOI] [PubMed] [Google Scholar]
- Chen, C., J. Joseph and R. S. Garofalo, 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137 846–856. [DOI] [PubMed] [Google Scholar]
- Curran, S, P., E. P. Leverich, C. M. Koehler and P. L. Larsen, 2004. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J. Biol. Chem. 279 54655–54662. [DOI] [PubMed] [Google Scholar]
- de la Cova, C., M. Abril, P. Bellosta, P. Gallant and L. A. Johnston, 2004. Drosophila myc regulates organ size by inducing cell competition. Cell 117 107–116. [DOI] [PubMed] [Google Scholar]
- DiGregorio, P. J., J. A. Ubersax and P. H. O'Farrell, 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 19 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T., H. Yamamoto and M. Esaki, 2003. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 116 3259–3267. [DOI] [PubMed] [Google Scholar]
- Frei, C., M. Galloni, E. Hafen and B. A. Edgar, 2005. The Drosophila mitochondrial ribosomal protein mRpL12 is required for cyclin D/Cdk4-driven growth. EMBO J. 24 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni, M., 2003. bonsaï, a ribosomal protein S15 homolog, involved in gut mitochondrial activity and systemic growth. Dev. Biol. 264 482–494. [DOI] [PubMed] [Google Scholar]
- Geissler, A., A. Chacinska, K. N. Truscott, N. Wiedemann, K. Brandner et al., 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111 507–518. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., 1991. Site-specific recombination between homologous chromosomes in Drosophila. Science 5008 958–961. [DOI] [PubMed] [Google Scholar]
- Guo, Y., N. Cheong, Z. Zhang, R. De Rose, Y. Deng et al., 2004. Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J. Biol. Chem. 279 24813–24825. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120 2121–2129. [DOI] [PubMed] [Google Scholar]
- Huh, J. R., M. Guo and B. A. Hay, 2004. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 14 1262–1266. [DOI] [PubMed] [Google Scholar]
- Iyengar, B., J. Roote and A. R. Campos, 1999. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-125). Genetics 153 1809–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar, B., N. Luo, C. L. Farr, L. S. Kaguni and A. R. Campos, 2002. The accessory subunit of DNA polymerase is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman and P. Gallant, 1999. Drosophila myc regulates cellular growth during development. Cell 98 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, D., C. L. Farr, B. Poeck, A. Alahari, M. Vogel et al., 2001. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell 12 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S., P. Guptan, E. Owusu-Ansah and U. Banerjee, 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9 843–854. [DOI] [PubMed] [Google Scholar]
- Meinecke, M., R. Wagner, P. Kovermann, B. Guiard, D. U. Mick et al., 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312 1523–1526. [DOI] [PubMed] [Google Scholar]
- Migeon, J. C., M. S. Garfinkel and B. A. Edgar, 1999. Cloning and characterization of peter pan, a novel Drosophila gene required for larval growth. Mol. Biol. Cell 10 1733–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma et al., 1999. Drosophila S6 kinase: a regulator of cell size. Science 285 2126–2129. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., and B. A. Edgar, 1998. Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 10 784–790. [DOI] [PubMed] [Google Scholar]
- Oldham, S., and E. Hafen, 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13 79–85. [DOI] [PubMed] [Google Scholar]
- Oldham, S., J. Montagne, T. Radimerski, G. Thomas and E. Hafen, 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham, S., H. Stocker, M. Laffargue, F. Wittwer, M. Wymann et al., 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP3 levels. Development 129 4103–4109. [DOI] [PubMed] [Google Scholar]
- Pendergrass, W., N. Wolf and M. Poot, 2004. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A 61 162–169. [DOI] [PubMed] [Google Scholar]
- Polymenis, M., and E. V. Schmidt, 1999. Coordination of cell growth with cell division. Curr. Opin. Genet. Dev. 9 76–80. [DOI] [PubMed] [Google Scholar]
- Prober, D. A., and B. A. Edgar, 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100 435–446. [DOI] [PubMed] [Google Scholar]
- Rehling, P., N. Pfanner and C. Meisinger, 2003. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane: a guided tour. J. Mol. Biol. 326 639–657. [DOI] [PubMed] [Google Scholar]
- Rehling, P., K. Brandner and N. Pfanner, 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5 519–530. [DOI] [PubMed] [Google Scholar]
- Rintelen, F., H. Stocker, G. Thomas and E. Hafen, 2001. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. Johnson-Shlitz, W. Benz et al., 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1983. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 11 6341–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, H. D., T. Gorenc and H. Steller, 2004. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 7 491–501. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan et al., 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5 566–571. [DOI] [PubMed] [Google Scholar]
- Sibon, O. C., V. A. Stevenson and W. E. Theurkauf, 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388 93–97. [DOI] [PubMed] [Google Scholar]
- Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat et al., 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5 559–566. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals a translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy et al., 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414 768–773. [DOI] [PubMed] [Google Scholar]
- Tsuda, L., Y. H. Inoue, M.-A. Yoo, M. Mizuno, M. Hata et al., 1993. A protein kinase similar to MAP kinase activator acts downstream of the raf kinase in Drosophila. Cell 72 407–414. [DOI] [PubMed] [Google Scholar]
- Weinkove, D., T. P. Neufeld, T. Twardzik, M. D. Waterfield and S. J. Leevers, 1999. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class IA phosphoinositide 3-kinase and its adaptor. Curr. Biol. 9 1019–1029. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., M. Esaki, T. Kanamori, Y. Tamura, S. Nishikawa et al., 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111 519–528. [DOI] [PubMed] [Google Scholar]
- Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard and T. P. Neufeld, 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoratti, M., and I. Szabo, 1995. The mitochondrial permeability transition. Biochim. Biophys. Acta 1241 139–176. [DOI] [PubMed] [Google Scholar]