Abstract
Behaviors are complex traits influenced by multiple pleiotropic genes. Understanding the mechanisms that give rise to complex behaviors requires an understanding of how variation in transcriptional regulation shapes nervous system development and how variation in brain structure influences an organism's ability to respond to its environment. To begin to address this problem, we used olfactory behavior in Drosophila melanogaster as a model and showed that a hypomorphic transposon-mediated mutation of the early developmental gene Semaphorin-5c (Sema-5c) results in aberrant behavioral responses to the repellant odorant benzaldehyde. We fine mapped this effect to the Sema-5c locus using deficiency mapping, phenotypic reversion through P-element excision, and transgenic rescue. Morphometric analysis of this Sema-5c allele reveals subtle neuroanatomical changes in the brain with a reduction in the size of the ellipsoid body. High-density oligonucleotide expression microarrays identified 50 probe sets with altered transcriptional regulation in the Sema-5c background and quantitative complementation tests identified epistatic interactions between nine of these coregulated genes and the transposon-disrupted Sema-5c gene. Our results demonstrate how hypomorphic mutation of an early developmental gene results in genomewide transcriptional consequences and alterations in brain structure accompanied by profound impairment of adult behavior.
BEHAVIORS are complex traits influenced by the expression of multiple pleiotropic genes (Anholt 2004; Anholt and Mackay 2004). Understanding the mechanisms that give rise to complex behaviors requires an understanding of how variation in transcriptional regulation shapes nervous system development and how variation in brain structure influences the ability of an organism to respond appropriately to its environment. To begin to address this problem, we used olfactory behavior in Drosophila melanogaster as a model, since chemosensation is essential for survival, the olfactory system of Drosophila has been well characterized (Vosshall 2000; Dahanukar et al. 2005), and flies are readily amenable to powerful genetic, neuroanatomical, and behavioral analyses. Previously, we showed that the genetic architecture of odor-guided behavior in this system is composed of epistatic networks of pleiotropic genes (Fedorowicz et al. 1998; Anholt et al. 2003), that such networks are dynamic, and that genes functioning early in development feature prominently in genetic ensembles that mediate adult olfactory behavior (Sambandan et al. 2006). Here, we characterize the effects of hypomorphic disruption of the early developmental gene, Semaphorin-5c (Sema-5c), on the transcriptome, on development of the central nervous system, and on adult olfactory behavior.
Semaphorins are a family of secreted and membrane-associated proteins studied extensively for their role in nervous system development, especially in axon guidance (Kolodkin et al. 1992, 1993; Luo et al. 1993; Puschel et al. 1995). Semaphorins have also been implicated in the function of the immune system (Shi et al. 2000; Takegahara et al. 2005) and a variety of diseases, including cancer (Woodhouse et al. 2003; Neufeld et al. 2005; Basile et al. 2006), retinal degeneration (Rice et al. 2004; Abid et al. 2006), schizophrenia (Eastwood et al. 2003), and rheumatoid arthritis (Miller et al. 2004). Members of the semaphorin gene family are characterized by a conserved semaphorin domain ∼500 amino acids in length (Gherardi et al. 2004; Yazdani and Terman 2006). The family consists of eight classes differing in their sequence and primary structures. Class 1 and 2 semaphorins have been identified solely in invertebrates, classes 3–7 are present in vertebrates with class 5 also found in Drosophila, and class 5 semaphorins occur only in viruses (Semaphorin Nomenclature Committee 1999).
Of particular interest to olfactory behavior is the implication that class 3 soluble semaphorins and their receptors play a role in axon guidance in the mouse olfactory system (Renzi et al. 2000; Schwarting et al. 2000; Walz et al. 2002; Taniguchi et al. 2003; Cloutier et al. 2004). A null mutation in Sema3F resulted in defasciculation of the vomeronasal nerve and rerouting of axons from vomeronasal sensory neurons into the main olfactory bulb. Accurate innervation of the main olfactory bulb was also dependent on Sema3F signaling (Cloutier et al. 2004).
Of relevance to human disease are the class 5 semaphorins, which contain the characteristic semaphorin domain as well as seven thrombospondin repeat elements and a transmembrane domain (Semaphorin Nomenclature Committee 1999). A human class 5 semaphorin (Semaf) has been implicated in the cri-du-chat syndrome, a rare congenital neurological disorder (Simmons et al. 1998). The cri-du-chat critical region has been mapped to human chromosome 5 in which the human Semaf locus encompasses 10% of the interval. In Drosophila, a single class 5 semaphorin, Sema-5c, has been identified (Khare et al. 2000; Bahri et al. 2001) and is ubiquitously expressed in stage 2 embryos with a striped pattern emerging at later stages. Late-stage expression was observed at muscle attachment sites, the midgut, and the dorsal vessel (Khare et al. 2000; Bahri et al. 2001). Sema-5c expression patterns have not been characterized in the adult. Homozygous mutants of Sema-5c are viable with no detectable defects in embryonic development (Bahri et al. 2001). Furthermore, in a search for suppressors of the lethal giant larvae l(2)gl phenotype, which exhibits neoplastic growth of the larval brain and imaginal discs, a P-element insertion near Sema-5c disrupted tumorigenesis (Woodhouse et al. 2003). A well-defined role for Sema-5c in neural development, however, has not yet been documented.
Recently, a P-element insertional mutagenesis screen for mutations affecting Drosophila olfactory avoidance behavior identified four independent P-element insertion lines in two different genetic backgrounds in which the P element inserted near the Sema-5c locus. All four P-element insertion lines showed aberrant olfactory avoidance behavior, suggesting that Sema-5c may play a role in odor-guided behavior (Sambandan et al. 2006). Here, we characterize the role of Sema-5c in olfactory behavior by identifying and characterizing one of these P-element insertions in the Sema-5c gene region. We show that a P-element insertion upstream of the Sema-5c locus results in smell-impaired behavioral responses. We fine mapped this effect to the Sema-5c locus using deficiency mapping, phenotypic reversion through P-element excision, and transgenic rescue. We found that mutation of the Sema-5c gene results in subtle changes in brain morphology with a reduction in the size of the ellipsoid body. Furthermore, we provide insights into the function of Sema-5c in adults by characterizing the genomewide transcriptional effects of the P-element insertion at the Sema-5c locus. These experiments demonstrate how hypomorphic mutation of an early developmental gene results in genomewide transcriptional consequences and alterations in brain structure accompanied by profound impairment of adult behavior.
MATERIALS AND METHODS
Drosophila stocks:
The BG01245 line was generated and donated by Hugo Bellen as part of the Berkeley Drosophila Gene Disruption Project and has a p[GT1]-element insertion at cytological position 68F in the isogenic Canton-S (B) background (Lukacsovich et al. 2001; Bellen et al. 2004). The BG01245 revertant line was generated as described below. The E215 and E77 excision lines and a UAS-Sema-5c transgenic line were generously made available by Sami M. Bahri (Bahri et al. 2001). The transgenic construct was mobilized and placed into the Canton-S (B) background, as described below. The following mutants used for quantitative complementation tests for epistasis were obtained from the Bloomington Stock Center (stock number in parentheses): no mitochondrial derivative (BL-10715), eukaryotic release factor 1 (BL-11488; BL-10266), Sec61beta (BL-12339; BL-10376), CG3168 (BL-12516), CG2994 (BL-12657), CG17259 (BL-12894), CG8533 (BL-13060), laminin B1 (BL-13957), CG4607 (BL-14408), laminin A (BL-14568), CG4769 (BL-14909), CG7800 (BL-14958), CG8545 (BL-14964), between CG5579/CG33177/CG33178 and lipid storage droplet 2 (BL-15119), lipid storage droplet 2 (BL-12398), CG15557-9 (BL-15221), CG8386 (BL-13815), heterogeneous nuclear ribonucleoprotein at 27C (BL-10375), protein phosphatase 2A regulatory B subunit (BL-12974), signal sequence receptor beta (BL-12094), CG1383 (BL-15240), and homothorax (BL-11670). All flies were reared on standard agar–cornmeal–molasses medium in vials maintained at 25° and under a 12-hr light/dark cycle.
Benzaldehyde avoidance assay:
All behavioral assays were conducted as described previously (Anholt et al. 1996). Briefly, one replicate assay consisted of a single sex group of five individuals, 5–7 days post-eclosion, removed from their food source ∼1–2 hr prior to the assay. Test vials were demarcated at 3 and 6 cm from the bottom of the vial. Benzaldehyde was introduced on a saturated cotton swab wedged between the cotton plug and the vial wall at the 6-cm mark. Flies were allowed to acclimate to the vial for 15 sec after the introduction of the odor source. The number of flies present in the distal compartment of the vial was recorded every 5 sec for 1 min. The “avoidance score” for the replicate is the average of the 10 counts. All behavioral assays were conducted in an environmental chamber at 25° and multiple replicate assays were conducted for each genotype. Dose-response curves were determined using concentrations of 0.03, 0.06, 0.1, 0.3, 0.6, 1, and 3% (v/v) benzaldehyde, with subsequent behavioral assays conducted at 0.3% (v/v) benzaldehyde. Benzaldehyde avoidance responses of Canton-S (B) and BG01245 at different benzaldehyde concentrations were analyzed by a two-way fixed-effects ANOVA according to the model y = μ + L + S + (L × S) + E, where L denotes line, S designates sex, and E the environmental variation within sex and genotype. Ten replicates per sex and genotype were measured at each benzaldehyde concentration.
Quantitative complementation test:
The P-element insertion line (BG01245) and its co-isogenic control [Canton-S (B)] were crossed to excision lines E215 and E77 (Bahri et al. 2001). Progeny from each cross were assayed for avoidance behavior to 0.3% (v/v) benzaldehyde and significant differences between phenotypic values were assessed between excision lines crossed to BG01245 and Canton-S (B). Forty replicates per genotype and sex were assayed over three blocks. For each complementation test, the data were analyzed by a three-way fixed-effects ANOVA with the model y = μ + G + S + B + (G × S) + (G × B) + (S × B) + (G × S × B) + E, with genotype (G), block (B), and sex (S) as fixed effects, and E indicating environmental variance. Quantitative failure to complement was inferred if the genotype or genotype-by-sex interaction (G × S) terms were significant.
Phenotypic reversion through P-element excision:
The p[GT1] construct was mobilized by crossing BG01245 females to w;Cy/Sp;SbΔ2-3/TM6,Tb males. Male offspring of the genotype w;Cy/CS(B);BG01245/SbΔ2-3 were subsequently mated to w;CS(B);H/TM3,Sb females, and single male offspring (w;CS(B); P-/H) were crossed to w;CS(B);H/TM3,Sb females. Male and female w;CS(B); P-/TM3,Sb in which the P element has been excised were mated inter se to make a homozygous P-element excision line in the isogenic Canton-S (B) background. Precise excision of the construct was characterized by PCR amplification using primers flanking the original P-element insertion site. PCR products were subsequently sequenced using ABI big dye terminator cycle sequencing chemistry (Applied Biosystems, Foster City, CA). Sequences were analyzed using Vector NTI Suite 9 software (Informax, Frederick, MD). Avoidance responses to benzaldehyde of the precise P-element excision line, Canton-S (B) control, and the BG01245 P-element insertion line were measured. Ten replicates per sex and line were scored and data were analyzed by a two-way fixed-effects ANOVA according to the model y = μ + L + S + (L × S) + E, where L denotes line, S designates sex, and E environmental variance. Significant differences among lines were determined by post-hoc Tukey's test.
Transgenic rescue:
The UAS-Sema-5c transgene was mobilized and inserted into an isogenic Canton-S (B) background by crossing +;+;UAS-Sema-5c males to w;Cy/Sp;TM3,Sb/H females. Male offspring of the genotype w; Cy/+;TM3/UAS-Sema-5c were subsequently mated to w;CS(B);TM3/H virgin females and w; Cy/CS(B);UAS-Sema-5c/TM3 female offspring were crossed to w;Cy/Sp;SbΔ2-3/TM6,Tb males. Male progeny, w;Cy/CS(B);UAS-Sema-5c/SbΔ2-3, were then crossed to w;Cy/Sp;TM3/H virgin females. Single males of the genotype w;UAS-Sema-5c/Cy;SbΔ2-3/H were subsequently mated to w;Cy/Sp;TM3/H females. Male progeny of the genotype w;Cy/UAS-Sema-5c;TM3/H were crossed to w;Cy/Sp;TM3/H females and w;Cy/UAS-Sema-5c;TM3/H offspring mated inter se. Male offspring, w;UAS-Sema-5c;TM3/H, were crossed to w;Cy/Sp;BG01245 virgin females. Finally, males and females of the genotype w;UAS-Sema-5c/Cy;BG01245/TM3 were mated inter se. Olfactory avoidance behavior to benzaldehyde was tested contemporaneously for the BG01245 P-element insertion line (N = 30 replicates/sex), Canton-S (B) control (N = 30 replicates/sex), and transgenic rescue line w;UAS-Sema-5c/CS(B);BG01245 (N = 20 replicates/sex). Avoidance responses were analyzed by a two-way fixed-effects ANOVA according to the model y = μ + L + S + (L × S) + E, where L denotes line, S designates sex, and E environmental variance. Significant differences among lines were determined by post-hoc Tukey's test.
Whole-mount immunohistochemistry:
Brains were dissected in ice-cold phosphate buffered saline (PBS) for up to 1 hr and collected in PBS in a microcentrifuge tube on ice. All the following steps were done on a rotating platform. After removal of PBS, the brains were fixed in PBS with 3.7% formaldehyde for 15 min at room temperature followed with three 10-min washes in PBS. The tissues were then preincubated with PAXD (PBS + 5% bovine serum albumin, 0.3% Triton-X100, and 0.3% sodium deoxycholate) for 10 min. Subsequently, the tissues were incubated overnight at 4° with PAXD containing the primary antibody at the appropriate dilution. Tissues were then washed for 4–6 hr with several changes of PAXD at room temperature and incubated overnight at 4° with PAXD with secondary antibody at the appropriate dilution. Tissues were washed for at least 2 hr with several changes prior to mounting in Vectashield medium (Vector Laboratories, Burlingame, CA).
Antibody:
The monoclonal antibody 1D4 (antifasciclin2) was obtained from the Developmental Studies Hybridoma Bank (under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242) and used at a dilution of 1:100. Cy3 and FITC-coupled secondary antibodies (Jackson Immunoresearch, Westgrove, PA) were used at 1:200 and 1:100 dilutions.
Microscopy:
The antifasciclin2 antibody staining used for morphometric analysis was documented using an Olympus BX61 epifluorescence microscope equipped with a DP70 digital camera controlled with analySIS software.
Morphometric analysis:
For morphometric analyses, images were sampled using the analySIS software and the DP70 digital camera. Relevant dimensions (length and diameter of α-and β-lobes and radii of ellipsoid body; see Figure 2D) were measured on the computer screen using a normal ruler and subsequently converted to values (expressed as percentages) relative to the distance between the two mushroom body heels. Statistical analyses used two-way ANOVA with post-hoc Tukey's tests.
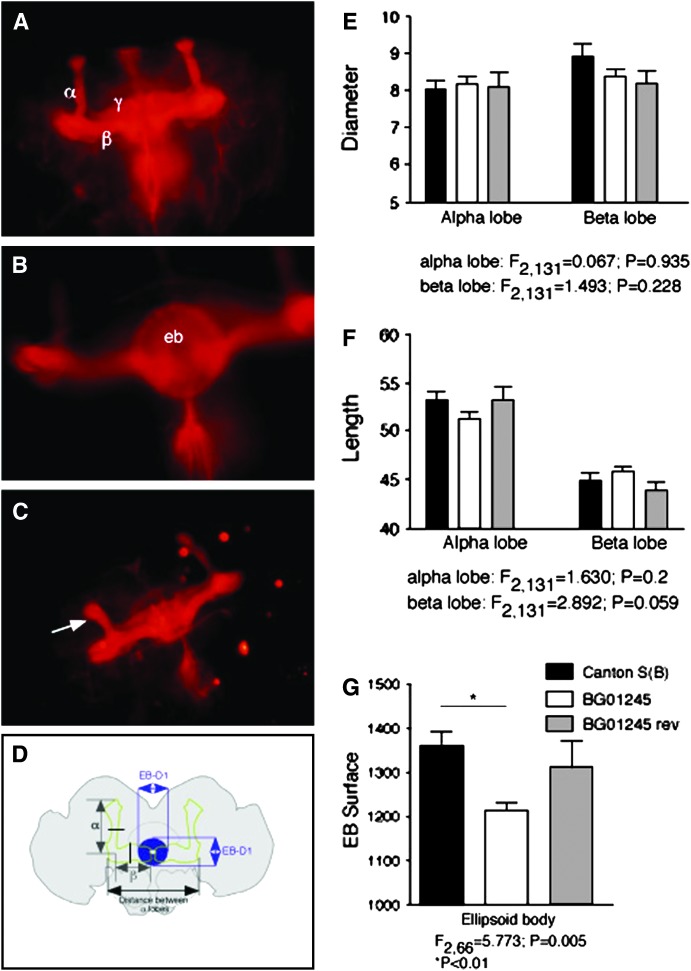
Figure 2.—
Morphometric analysis of antifasciclin2 (1D4) antibody-stained adult brains of Canton S (B) control flies and BG01245 Sema-5c mutants. (A) Antifasciclin2 labels α-, β-, and gamma (γ) lobes as well as the ellipsoid body (eb) (B). (C) Short α-lobe phenotype that was observed in 7 of 40 brains analyzed. Note that in the morphometric analysis brains were scored per scorable hemisphere (i.e., without mechanical defects caused by dissections). Expressed per hemisphere, a total of 77 hemispheres were scorable, of which 7 displayed a readily identifiable short α-lobe (arrow). This phenotype is never seen in wild-type flies. (D) Schematic of the adult Drosophila brain. For α- and β-lobes, the lengths were measured at the positions indicated (double arrows marked with α and β, respectively). The widths of the α- and β-lobes were measured at the positions indicated by the black lines perpendicular to the lobes. For the ellipsoid body (in blue), two diameters were measured, EB-D1 and EB-D2, respectively. From these, the radii R1 and R2 were derived and the product R1 × R2 was used as a measure to compare surfaces of the ellipsoid body. All measurements were expressed as percentages relative to the distance between the heels (marked as the distance between α-lobes). (E) No significant differences were observed between α- or β-lobe diameters of Canton S(B) (N = 25, solid bars), Sema-5c (BG01245) mutants (N = 77, open bars), and Sema-5C revertants (BG01245 rev) (N = 32, shaded bars). (F) No significant differences were observed between α- or β-lobe lengths of Canton S(B) (N = 25, solid bars), Sema-5c (BG01245) mutants (N = 77, open bars), and Sema-5C revertants (BG01245 rev) (N = 32, shaded bars). (G) A significant reduction (P < 0.01) in ellipsoid body surface was found when control (N = 13, solid bar) and Sema-5c mutant (BG01245) (N = 40, open bar) ellipsoid body surfaces were compared. This phenotype is restored to wild type in the Sema-5c revertant (BG01245 rev) (N = 16, shaded bar).
Transcriptional profiles:
For each genotype and sex, total RNA was isolated from two independent replicate groups of Drosophila heads (5–7 days post-eclosion) using Trizol (GIBCO-BRL, Gaithersburg, MD). RNA samples were purified through RNeasy columns (QIAGEN, Valencia, CA) and cDNA was generated from 5 μg of total RNA. Biotinylated cRNA probes were generated and hybridized to Drosophila high-density oligonucleotide microarrays (GeneChip Drosophila Genome Array based on FlyBase version 1.0, Affymetrix) and visualized with a streptavidin–phycoerythrin conjugate according to Affymetrix protocols. The data were analyzed with Microarray Suite version 5.0 (MAS 5.0) using Affymetrix default analysis settings and global scaling as the normalization method. The signal values of any hybridization were multiplied by a scaling factor to make their mean intensity equal to 500. Differences in signal values between BG01245 and its control Canton-S (B) were analyzed by two-way factorial ANOVA according to the model y = μ + L + S + (L × S) + E, where L denotes line, S designates sex, and E the variance between replicate arrays. A significance threshold of P < 0.001 was used to identify probe sets that differed between BG01245 and the Canton-S (B) control. Analysis of overrepresentation of molecular function and gene ontology categories among transcripts with altered transcriptional regulation was implemented with the DAVID program (Dennis et al. 2003).
Epistasis:
The Sema-5c P-element mutation is recessive. To assess epistatic interactions between genes with altered transcriptional regulation in the mutant background, trans-heterozygous flies were generated by crossing mutant lines from the Bloomington Stock Center to BG01245 and Canton-S (B). Ten replicates of five flies/sex and genotype were assayed for avoidance behavior to 0.3 and 0.6% (v/v) benzaldehyde as described above. Epistatic interactions were determined by ANOVA according to the model y = μ + G + S + C + (G × S) + (G × C) + (S × C) + (G × S × C) + E, where G (genotype), sex (S), and benzaldehyde concentration (C) are fixed main effects and E indicates environmental variance.
RESULTS
A P-element insertion near the Sema-5c gene results in aberrant olfactory behavior:
Previously, in a screen for genes influencing olfactory avoidance behavior in Drosophila, four independent p[GT1]-element insertion lines adjacent to the Sema-5c locus showed aberrant behavioral responses to benzaldehyde (Sambandan et al. 2006). The insertion BG01245 resulted in significantly reduced transcription levels of Sema-5c in larval and pupal development, whereas in the adult no such differences were observed (Sambandan et al. 2006). We conducted a comprehensive characterization of the effects of this P-element insertion line on olfactory avoidance behavior (Figure 1). The P-element insertion in the BG01245 mutant line inserted 197 bp upstream of the first exon of Sema-5c (Figure 1A; Sambandan et al. 2006). Sema-5c contains 12 exons and is ∼14 kb in length with two predicted alternatively spliced transcripts that differ in the number of thrombospondin type 1 repeats (Khare et al. 2000; Bahri et al. 2001). Two predicted translation initiation sites have been identified in the first and second exons (Bahri et al. 2001). In the BG01245 line, insertion of the p[GT1]-element into an isogenic Canton-S (B) background resulted in reduced avoidance responses to the standard test odorant benzaldehyde (Figure 1B). Reduced behavioral responses were similar for both sexes; therefore, measurements for males and females were pooled (Figure 1B). Dose responses to benzaldehyde for the mutant line compared to the control showed a decrease in avoidance behavior with a shift of the dose-response profile toward higher stimulus concentrations in the BG01245 mutant at concentrations of 0.1–3% (v/v) benzaldehyde (Figure 1B). Subsequent comparisons of avoidance responses between control and mutant flies were conducted at an intermediate concentration of 0.3% (v/v) benzaldehyde, which is optimal for resolving behavioral differences between the lines.
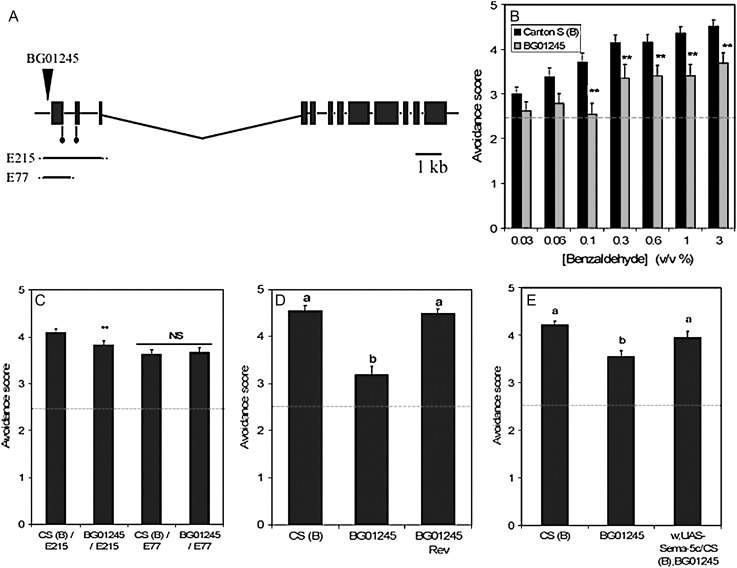
Figure 1.—
Phenotypic characterization of disruption of the Sema-5c gene by p[GT1] insertion. (A) P-element insertion site and deletions in the Sema-5c gene region. The horizontal line represents genomic DNA on the third chromosome at cytological position 68F2. Exons of the Sema-5c gene are represented by solid boxes. The location of the p[GT1] insertion site is indicated with an arrowhead and putative translation initiation sites are denoted by lines ending in circles. Excision lines E215 and E77 are represented as horizontal bars denoting the extent of the deletions of the Sema-5c gene region in these lines (Bahri et al. 2001). (B) Dependence of olfactory avoidance behavior on the concentration of benzaldehyde. P-element insertion line BG01245 (shaded bars) and its control, Canton S (B) (solid bars), were measured at different benzaldehyde concentrations. The dashed red line denotes the expected baseline avoidance behavior score at which flies are indifferent to the odorant. Double asterisks denote P < 0.01 at different concentrations of benzaldehyde (v/v) (0.1% F1,36 = 10.42, P < 0.0027; 0.3% F1,36 = 6.93, P < 0.0124; 0.6% F1,36 = 6.82, P < 0.0131; 1% F1,36 = 11.39, P < 0.0018; 3% F1,36 = 10.10, P < 0.003). (C) Fine mapping of aberrant olfactory avoidance responses in the BG01245 mutant to the Sema-5c gene region. Quantitative complementation tests were conducted with deletion lines E215 and E77 (A). Complementation is observed for deletion line E77. However, failure to complement is observed between progeny of crosses of Canton S (B) and BG01245 to E215, which spans both putative translation initiation sites of Sema-5c. The dashed red line denotes the expected baseline avoidance behavior score at which flies are indifferent to the odorant. No difference between the sexes was observed. Double asterisks denote P < 0.01. (D) Phenotypic reversion of avoidance responses to benzaldehyde by P-element excision. Precise excision of the p[GT1] construct restores the wild-type phenotype. Whereas P-element insertion line BG01245 shows reduced avoidance responses to 0.3% benzaldehyde compared with the control line Canton S (B), no significant difference is observed between Canton S (B) and the revertant line BG01245 Rev. Letters above bars denote results of the post-hoc Tukey's test with means designated by the same letter (a, b) not significantly statistically different from one another. The dashed red line denotes the expected baseline avoidance behavior score at which flies are indifferent to the odorant. (E) Transgenic rescue of the BG01245 aberrant behavioral phenotype. A full-length copy of the UAS-Sema-5c transgene, provided by Sami Bahri (Bahri et al. 2001), was hopped to the second chromosome and placed into the co-isogenic Canton S (B) background (see materials and methods). Canton S (B), BG01245, and the transgenic line w;UAS-Sema-5c/CS(B);BG01245 were measured contemporaneously for avoidance behavior to 0.3% benzaldehyde. The dashed red line denotes the expected baseline avoidance behavior score at which flies are indifferent to the odorant. Letters above bars denote results of the post-hoc Tukey's test with different letters (a, b) denoting statistically significant mean line differences.
p[GT1]-element disruption of Sema-5c accounts for the smell-impaired phenotype:
To demonstrate that the smell-impaired phenotype is due to the p[GT1]-element insertion, we fine mapped the phenotype by quantitative complementation testing (Pasyukova et al. 2000; Fanara et al. 2002) using excision lines with small deletions in the Sema-5c gene, derived from an independent EP insertion line (Figure 1A; Bahri et al. 2001). Results from these experiments allowed us to uncover genomic regions that fail to complement the mutant phenotype. The E77 and E215 excision lines differ in the sizes of their deletions with the E215 deletion spanning a portion of the 5′-untranslated region and first three exons of Sema-5c, whereas the E77 deletion removed a smaller portion of the coding region (see Bahri et al. 2001; Figure 1A). Olfactory avoidance behavior of E77/Canton-S (B) flies was not significantly different from E77/BG01245 flies, indicating complementation of the mutant phenotype (Figure 1C). However, the E215/BG01245 genotype showed smell-impaired responses relative to the E215/Canton-S (B) genotype (Figure 1C; F1,148 = 6.84, P < 0.0098). Quantitative failure of the E215 excision to complement the BG01245 P-element insertion directly maps the effects of the mutation to the nonoverlapping region of the E77 and E215 deletions of the Sema-5c gene.
To further verify that aberrant olfactory avoidance responses are due to the p[GT1]-element insertion rather than to an independent mutation, we mobilized the p[GT1] transposon to obtain a precise P-element excision. Precise excision was verified by DNA sequencing the region around the original P-element insertion site. We observed significant differences in avoidance response between the control and the mutant, but not between the precise revertant and the control (Figure 1D; F2,54 = 32.53, P < 0.0001). Thus, precise excision of the p[GT1] construct resulted in restoration of avoidance responses to wild-type levels (Figure 1D).
Conclusive evidence that mutation of the Sema-5c gene is causal for the reduction in olfactory avoidance behavior was obtained by introducing an UAS-Sema-5c transgene into the BG01245 mutant background. The p[GT1] construct in the Sema-5c gene contains a GAL4 cassette (Lukacsovich et al. 2001) that can be driven by the Sema-5c promoter, allowing transgenes cloned behind a UAS promoter to be expressed in cells that normally express the disrupted gene. The Canton-S (B) control, the BG01245 mutant, and the transgenic rescue line w;UAS-Sema-5c/CS(B);BG01245 were tested contemporaneously. As expected, avoidance responses of Canton-S (B) differed significantly from those of the mutant (Figure 1E; F2,154 = 9.52, P < 0.0001). However, introduction of the UAS-Sema-5c transgene into the BG01245 genetic background resulted in rescue of avoidance behavior to control levels with no significant difference between the transgenic line and the Canton-S (B) control (Figure 1E). Quantitative complementation testing with deletions, phenotypic reversion through P-element excision, and transgenic rescue of avoidance responses to benzaldehyde show that disruption of the Sema-5c gene caused the observed smell-impaired phenotype.
Neuroanatomical consequences of disruption of the Sema-5c gene:
In third instar Canton-S (B) larvae, we observed expression of Sema-5c in scattered cells in the brain and ventral ganglion and a broad pattern of expression at the level of the thoracic ganglion and in the midline glia (supplemental Figure 1 at http://www.genetics.org/supplemental/). However, we were not able to detect expression of Sema-5c in adult brain by in situ hybridization.
It was previously suggested that hypomorphic mutations due to insertion of p[GT1] might lead to subtle morphological defects (Sambandan et al. 2006). To examine whether disruption of the Sema-5c gene during development impacts the neuroanatomical organization of the adult brain, we performed whole-mount antibody labeling using the 1D4 monoclonal antibody against fasciclin 2 (fas2). Fasciclin 2 is strongly expressed in the ellipsoid body of the central complex, in the α- and β-lobes of the mushroom bodies, and at somewhat lower levels in the gamma lobes of the mushroom bodies (Figure 2, A and B). This antibody staining allows good resolution for the identification of morphological defects in these neuropils. The gross anatomy of the fas2-positive neuropils was largely intact. We did, however, observe α-lobe defects in ∼10% of the Sema-5c mutant brains (Figure 2C). These defects were never seen in the Canton-S (B) wild-type controls.
We measured the diameters and lengths of the α- and β-lobes of the mushroom bodies and the surface of the ellipsoid body (Figure 2D). Morphometric analyses revealed no significant differences in the diameters and lengths of the α- and β-lobes of the mushroom bodies between controls, Sema-5c mutants (BG01245), and Sema-5c revertants (BG01245 rev) (Figure 2, E and F). We did, however, observe a significant reduction in surface area of the ellipsoid body, a phenotype that was completely reverted to wild type in the P-element excision revertant (BG01245 rev) (Figure 2G). A similar reversion of the phenotype was also seen in a transgenic rescue line, w;UAS-Sema-5c/CS(B);BG01245 (results not shown). These experiments demonstrate that substantial impairments in adult behavior are associated with subtle alterations in brain structure due to disruption of the early developmental gene Sema-5c.
Disruption of the Sema-5c gene alters genomewide expression levels:
Previous studies have shown that the insertion of single P elements that affect olfactory avoidance behavior have genomewide transcriptional consequences (Anholt et al. 2003). Therefore, we evaluated to what extent disruption of the Sema-5c locus by the P element results in alterations in expression levels of transregulated genes. We assessed whole-genome transcriptional profiles of the BG01245 and Canton-S (B) lines by RNA hybridization to high-density oligonucleotide Affymetrix expression microarrays and used two-way ANOVA to partition variation in transcript abundance between main effects of sex and line (mutant vs. control genotypes) and the sex-by-line interaction term. We observed a total of 64 probe sets with significant difference in transcript abundance between genotypes and/or significant line-by-sex interaction terms at a significance threshold of P < 0.001 (supplemental Table 1 at http://www.genetics.org/supplemental/; the conservative Bonferroni correction for multiple testing predicts that 14 of these coregulated genes may be false positives). A total of 43 probe sets were significant only for the line term, with 20 upregulated and 23 downregulated in the Sema-5c mutant background. Seven probe sets were significant for both line and sex-by-line interaction terms. For three of these probe sets, the direction of the effects was the same in males and females, and the interaction term was attributable to a change in relative magnitude of transcript between males and females. However, in four of these probe sets, and in the remaining 14 probe sets that were significant only for the sex-by-line interaction term, the changes in transcript abundance in the Sema-5c mutant were in opposite directions in males and females (supplemental Table 1 at http://www.genetics.org/supplemental/).
In these probe sets, genes that contribute directly to chemosensation included antdh (Wang et al. 1999), homothorax (Casares and Mann 1998; Dong et al. 2000), defective proboscis extension response 20 (Nakamura et al. 2002), and odorant binding proteins (Obps) Obp56d, Obp99a, Obp99c, and Obp99d (Galindo and Smith 2001; Hekmat-Scafe et al. 2002). Notably, the expression of Obp99a and Obp99c was downregulated in Sema-5c mutant males and upregulated in Sema-5c mutant females. Seven of the probe sets that differed in expression between lines represent predicted transcripts of unknown function. Gene ontology analysis identified overrepresented probe sets with altered transcript abundance in molecular function categories related to protein biosynthesis and metabolism, including purine nucleotide binding (P = 3.9E-2), nucleotide binding (P = 7.4E-2), oxidoreductase activity (P = 7.0E-2), adenyl nucleotide binding (P = 8.3E-2), signal sequence binding (P = 4.7E-2), elastase activity (P = 6.2E-2), translation regulator activity (P = 6.7E-2), translation factor activity/nucleic acid binding (P = 6.2E-2), ATP binding (P = 7.0E-2), as well as odorant binding (P = 1.4E-2).
To investigate the extent to which genes with altered transcriptional regulation in the BG01245 mutant background interact epistatically with the transposon-tagged Sema-5c gene, we conducted quantitative complementation tests for epistasis (Pasyukova et al. 2000; Fanara et al. 2002; Anholt et al. 2003). We crossed the BG01245 mutant and its co-isogenic control to 24 available mutations (m) and tested the trans-heterozygous flies for avoidance behavior to 0.3 and 0.6% (v/v) benzaldehyde. We identified nine loci (37.5%) in which mutant alleles interacted epistatically with the Sema-5c P-element-tagged gene (Table 1). We found a significant reduction in avoidance behavior at 0.6% (v/v) benzaldehyde between mutants of coregulated genes crossed to the BG01245 mutant compared to the control for heterogeneous nuclear ribonucleoprotein at 27C, lipid storage droplet 2, protein phosphatase 2A regulatory B subunit, CG1383, and homothorax. Eukaryotic release factor 1 and Sec61beta double heterozygotes were smell impaired at 0.3% (v/v) benzaldehyde relative to the control and double heterozygotes for signal sequence receptor beta showed reduced avoidance responses at both concentrations relative to the control genotype. Furthermore, the CG8386 mutant showed a suppressor effect as a double heterozygote with the Sema-5c mutant when tested at 0.3% (v/v) benzaldehyde, but an enhancer effect at 0.6% (v/v) benzaldehyde (Table 1). Such plasticity of epistatic interactions and dependence on stimulus concentrations are consistent with previous observations (Sambandan et al. 2006).
TABLE 1.
Significant epistatic interactions for olfactory avoidance behavior to benzaldehyde between BG01245 and loci with altered transcriptional regulation in the BG01245 mutant background
| Average avoidance score |
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene namea | Benzaldehyde (% v/v) | m × Canton S (B) | m × BG01245 | FG statisticb | PGc | PG×Cc | PG(0.3%)d | PG(0.6%)d |
| CG8386 (BL-13815) | 0.3 | 2.77 ± 0.15; | 3.19 ± 0.14; | 0.06 | 0.8082 | 0.0046 | 0.0516 | 0.0403 |
| 0.6 | 3.6 ± 0.15 | 3.11 ± 0.18 | ||||||
| Eukaryotic release factor 1 (BL-10266) | 0.3 | 3.93 ± 0.14; | 3.53 ± 0.14; | 5.47 | 0.0222 | 0.4806 | 0.0311 | 0.2747 |
| 0.6 | 4.29 ± 0.15 | 4.07 ± 0.14 | ||||||
| Heterogeneous nuclear ribonucleoprotein at 27C (BL-10375) | 0.3 | 3.3 ± 0.16; | 2.87 ± 0.22; | 9.98 | 0.0023 | 0.5993 | 0.1159 | 0.0029 |
| 0.6 | 3.78 ± 0.15 | 3.2 ± 0.11 | ||||||
| Lipid storage droplet 2 (BL-12398) | 0.3 | 3.79 ± 0.18; | 3.39 ± 0.22; | 9.69 | 0.0027 | 0.303 | 0.1788 | 0.0031 |
| 0.6 | 4.01 ± 0.13 | 3.21 ± 0.21 | ||||||
| Protein phosphatase 2A regulatory B subunit (BL-12974) | 0.3 | 3.19 ± 0.13; | 2.86 ± 0.13; | 7.91 | 0.0063 | 0.6738 | 0.091 | 0.0322 |
| 0.6 | 3.88 ± 0.14 | 3.44 ± 0.14 | ||||||
| sec61beta (BL-10376) | 0.3 | 3.95 ± 0.11; | 3.32 ± 0.17; | 13.39 | 0.0005 | 0.1636 | 0.0036 | 0.0608 |
| 0.6 | 4.2 ± 0.10 | 3.92 ± 0.10 | ||||||
| Signal sequence receptor beta (BL-12094) | 0.3 | 3.56 ± 0.16; | 2.75 ± 0.12; | 24.25 | <0.0001 | 0.5401 | 0.0004 | 0.0041 |
| 0.6 | 3.76 ± 0.14 | 3.13 ± 0.14 | ||||||
| CG1383 (BL-15240) | 0.3 | 3.04 ± 0.17; | 2.84 ± 0.15; | 5.34 | 0.0237 | 0.336 | 0.3814 | 0.0167 |
| 0.6 | 3.7 ± 0.11 | 3.21 ± 0.16 | ||||||
| Homothorax (BL-11670) | 0.3 | 3.6 ± 0.13; | 3.39 ± 0.14; | 5.1 | 0.027 | 0.3948 | 0.2713 | 0.0532 |
| 0.6 | 3.7 ± 0.16 | 3.24 ± 0.14 | ||||||
m, mutant background.
Bloomington Drosophila stock numbers are in parentheses.
F-statistic for genotype.
P-values for genotype and genotype-by-concentration interactions.
P-values for genotype when concentrations of benzaldehyde are analyzed separately. Lines BL-10715, 11488, 12339, 12516, 12657, 12894, 13060, 13957, 14408, 14568, 14909, 14958, 14964, 15119, and 15221 did not show epistatic interactions with BG01245.
Trans-heterozygotes were also assayed for locomotor activity. Individual flies were placed into empty vials, given a gentle tap, and after acclimating for 15 sec were scored for locomotor activity over a 30-sec period. Nominally significant differences (P < 0.03) were observed only for trans-heterozygotes of CG8386 [27.96 ± 0.34 sec for m/Canton-S (B) and 26.55 ± 0.55 sec for m/BG01245] and for protein phosphatase 2A regulatory B subunit [27.53 ± 0.28 sec for m/Canton-S (B) and 26.24 ± 0.51 sec for m/BG01245]. No data were obtained for CG1383 (BL-15240) and homothorax (BL-11670).
DISCUSSION
We showed that P-element disruption of the early developmental gene Sema-5c results in smell-impaired behavioral responses to benzaldehyde. We mapped these behavioral effects to the Sema-5c gene using quantitative complementation tests to deletions, a precise P-element excision line, and transgenic rescue. Sema-5c mutants are homozygous viable and embryos do not show overt morphological defects (Bahri et al. 2001).
To assess whether disruption of Sema-5c affects the structure of the adult brain, we performed morphometric measurements on the mushroom bodies and ellipsoid body, integrative centers in the central brain that could be readily visualized by staining with an antibody to fasciclin 2. We found neuroanatomical differences in the size of the ellipsoid body and in some cases the α-lobes of the mushroom bodies. The observation that Sema-5c is expressed in numerous structures in the third instar larval brain (supplemental Figure 1 at http://www.genetics.org/supplemental/) and that mutations in Sema-5c are associated with mild neuroanatomical defects (Figure 2) suggests that Sema-5c is required for normal Drosophila brain development and function. Intriguing in this regard is the demonstration that Sema-5c is linked to activation of the Dpp/Mothers against Dpp (Mad) signal transduction pathway (Woodhouse et al. 2003). This suggests a possible developmental mechanism by which disruption of Sema-5c may lead to the neuroanatomical and behavioral phenotypes that we describe here. The Dpp pathway is also important for synaptic development (Sweeney and Davis 2002; Rawson et al. 2003; Dudu et al. 2006). Thus, the observed neuroanatomical differences and the behavioral defects could in part be due to abnormal synapse structure and function. Whereas we were not able to detect expression of Sema-5c in adult brains by in situ hybridization, a previous study reported expression of Sema-5c in adult heads by real-time quantitative PCR (Sambandan et al. 2006). Thus, expression of Sema-5c in the antennae cannot be excluded and would be consistent with the observed altered regulation of expression of the odorant-binding proteins Obp56d, Obp99a, and Obp99d (supplemental Table 1 at http://www.genetics.org/supplemental/).
A low level or absence of Sema-5c expression in the adult brain suggests that the altered neuroanatomical features in the central brain arise as a consequence of earlier developmental impairments. This would also be consistent with the observation by Sambandan et al. (2006) that the biggest differences in Sema-5c expression levels between the control Canton-S(B) line and the Sema-5c transposon-induced mutation are observed in earlier developmental stages. Furthermore, the neuroanatomical defects that we documented are more likely glial than neuronal in origin since the pattern of expression of Sema-5c in larvae is consistent with expression in glia.
The genetic architecture of olfactory behavior in adult Drosophila depends on a dynamic epistatic genetic network (Anholt et al. 2003). Developmental genes, including Sema-5c, form part of this network (Sambandan et al. 2006). To assess to what extent transposon-mediated disruption of Sema-5c affects the expression of coregulated genes, we performed transcriptional profiling. We found 50 probe sets with altered regulation in the Sema-5c mutant background (supplemental Table 1 at http://www.genetics.org/supplemental/). This is in accordance with previous studies that have shown that the insertion of a single P element affecting olfactory avoidance behavior results in a similar number of genes with altered transcription (Anholt et al. 2003). Quantitative complementation tests revealed significant concordance between transcriptional coregulation and enhancer or suppressor effects on phenotypic values in trans-heterozygotes (Table 1), in line with previous observations (Anholt et al. 2003).
Gene ontology analysis of the altered transcriptional profile in the Sema-5c mutant implicates genes involved in protein biosynthesis, transport, and secretion. The genes listed in supplemental Table 1 at http://www.genetics.org/supplemental/ can be organized in a cellular pathway that directs protein synthesis and secretion of, among others, odorant-binding proteins, in line with a possible role of Sema-5c in support cells of the adult antenna. It is important, however, to note that the observed transcriptional profile is not necessarily due to the effect of Sema-5c disruption on a homogeneous population of cells, but may arise from a heterogeneous spatial and temporal pattern of expression. In addition, although changes in the size of the ellipsoid body and occasionally in the structure of the mushroom bodies accompany observed effects on olfactory behavior, we cannot be certain that aberrant olfactory behavior in Sema-5c flies is entirely due to effects of the mutation on the central brain. The implication of the ellipsoid body in altered olfactory behavior suggests the intriguing possibility that the ellipsoid body might play a direct role in processing olfactory information. On the other hand, the association of alterations in the ellipsoid body with altered olfactory behavior may reflect the previously demonstrated role of the ellipsoid body and other central complex structures in the control of locomotion (Martin et al. 1999; Strauss 2002).
Our results demonstrate how hypomorphic disruption of an early developmental gene can alter adult brain structure and transcriptional networks that contribute to adult behavior.
Acknowledgments
We thank Sami Bahri for kindly providing Drosophila stocks and Chao-Qiang Lai (Tufts University, Boston) for hybridization of the Affymetrix microarray GeneChips. This work was supported by National Institutes of Health grants GM059469 (to R.R.H.A.) and GM045146 (to T.F.C.M.). K.N. is a Senior Clinical Investigator of the Foundation for Scientific Research-Flanders and L.Z. and T.G. are supported by the Flanders Interuniversity Institute for Biotechnology (VIB). This work was partially supported by a Federal Science Policy Return Grant (K.N.), by a Marie-Curie International Reintegration Grant (K.N.), and VIB. This is a publication of the W. M. Keck Center for Behavioral Biology at North Carolina State University.
References
- Abid, A., M. Ismail, S. Q. Mehdi and S. Khaliq, 2006. Identification of novel mutations in the SEMA4A gene associated with retinal degenerative diseases. J. Med. Genet. 43 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt, R. R. H., 2004. Genetic modules and networks for behavior: lessons from Drosophila. BioEssays 26 1299–1306. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., and T. F. C. Mackay, 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 5 838–849. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., R. F. Lyman and T. F. C. Mackay, 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt, R. R. H., C. L. Dilda, S. Chang, J. J. Fanara, N. H. Kulkarni et al., 2003. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat. Genet. 35 180–184. [DOI] [PubMed] [Google Scholar]
- Bahri, S. M., W. Chia and X. Yang, 2001. Characterization and mutant analysis of the Drosophila sema 5c gene. Dev. Dyn. 221 322–330. [DOI] [PubMed] [Google Scholar]
- Basile, J. R., R. M. Castilho, V. P. Williams and J. S. Gutkind, 2006. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proc. Natl. Acad. Sci. USA 103 9017–9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares, F., and R. S. Mann, 1998. Control of antennal versus leg development in Drosophila. Nature 392 723–726. [DOI] [PubMed] [Google Scholar]
- Cloutier, J. F., A. Sahay, E. C. Chang, M. Tessier-Lavigne, C. Dulac et al., 2004. Differential requirements for semaphorin 3F and Slit-1 in axonal targeting, fasciculation, and segregation of olfactory sensory neuron projections. J. Neurosci. 24 9087–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar, A., E. A. Hallem and J. R. Carlson, 2005. Insect chemoreception. Curr. Opin. Neurobiol. 15 423–430. [DOI] [PubMed] [Google Scholar]
- Dennis, G., Jr., B. J. Sherman, D. A. Hosack, J. Yang, W. Gao et al., 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 P3. [PubMed] [Google Scholar]
- Dong, P. D., J. Chu and G. Panganiban, 2000. Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development 127 209–216. [DOI] [PubMed] [Google Scholar]
- Dudu, V., T. Bittig, E. Entchev, A. Kicheva, F. Julicher et al., 2006. Postsynaptic mad signaling at the Drosophila neuromuscular junction. Curr. Biol. 16 625–635. [DOI] [PubMed] [Google Scholar]
- Eastwood, S. L., A. J. Law, I. P. Everall and P. J. Harrison, 2003. The axonal chemorepellant semaphorin 3A is increased in the cerebellum in schizophrenia and may contribute to its synaptic pathology. Mol. Psychiatry 8 148–155. [DOI] [PubMed] [Google Scholar]
- Fanara, J. J., K. O. Robinson, S. M. Rollmann, R. R. H. Anholt and T. F. C. Mackay, 2002. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorowicz, G. M., J. D. Fry, R. R. H. Anholt and T. F. C. Mackay, 1998. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics 148 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, K., and D. P. Smith, 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi, E., C. A. Love, R. M. Esnouf and E. Y. Jones, 2004. The sema domain. Curr. Opin. Struct. Biol. 14 669–678. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe, D. S., C. R. Scafe, A. J. McKinney and M. A. Tanouye, 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare, N., N. Fascetti, S. DaRocha, R. Chiquet-Ehrismann and S. Baumgartner, 2000. Expression patterns of two new members of the semaphorin family in Drosophila suggest early functions during embryogenesis. Mech. Dev. 91 393–397. [DOI] [PubMed] [Google Scholar]
- Kolodkin, A. L., D. J. Matthes, T. P. O'Connor, N. H. Patel, A. Admon et al., 1992. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron 9 831–845. [DOI] [PubMed] [Google Scholar]
- Kolodkin, A. L., D. J. Matthes and C. S. Goodman, 1993. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 75 1389–1399. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., Z. Asztalos, W. Awano, K. Baba, S. Kondo et al., 2001. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics 157 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y., D. Raible and J. A. Raper, 1993. Collapsin: a protein in the brain that induces the collapse and paralysis of neuronal growth cones. Cell 75 217–227. [DOI] [PubMed] [Google Scholar]
- Martin, J. R., T. Raabe and M. Heisenberg, 1999. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J. Comp. Physiol. A 185 277–288. [DOI] [PubMed] [Google Scholar]
- Miller, L. E., C. Weidler, W. Falk, P. Angele, J. Schaumburger et al., 2004. Increased prevalence of semaphorin 3C, a repellent of sympathetic nerve fibers, in the synovial tissue of patients with rheumatoid arthritis. Arthritis Rheum. 50 1156–1163. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., D. Baldwin, S. Hannaford, J. Palka and C. Montell, 2002. Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J. Neurosci. 22 3463–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, G., N. Shraga-Heled, T. Lange, N. Guttmann-Raviv, Y. Herzog et al., 2005. Semaphorins in cancer. Front. Biosci. 10 751–760. [DOI] [PubMed] [Google Scholar]
- Pasyukova, E. G., C. Vieira and T. F. C. Mackay, 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschel, A. W., R. H. Adams and H. Betz, 1995. Murine semaphorin D/collapsin is a member of a diverse gene family and creates domains inhibitory for axonal extension. Neuron 14 941–948. [DOI] [PubMed] [Google Scholar]
- Rawson, J. M., M. Lee, E. L. Kennedy and S. B. Selleck, 2003. Drosophila neuromuscular synapse assembly and function require the TGF-beta type I receptor saxophone and the transcription factor Mad. J. Neurobiol. 55 134–150. [DOI] [PubMed] [Google Scholar]
- Renzi, M. J., T. L. Wexler and J. A. Raper, 2000. Olfactory sensory axons expressing a dominant-negative semaphorin receptor enter the CNS early and overshoot their target. Neuron 28 437–447. [DOI] [PubMed] [Google Scholar]
- Rice, D. S., W. Huang, H. A. Jones, G. Hansen, G.-L. Ye et al., 2004. Severe retinal degeneration associated with disruption of semaphorin 4A. Invest. Ophthalmol. Vis. Sci. 45 2767–2777. [DOI] [PubMed] [Google Scholar]
- Sambandan, D., A. Yamamoto, J. J. Fanara, T. F. C. Mackay and R. R. H. Anholt, 2006. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics 174 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting, G. A., C. Kostek, N. Ahmad, C. Dibble, L. Pays et al., 2000. Semaphorin 3A is required for guidance of olfactory axons in mice. J. Neurosci. 20 7691–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaphorin Nomenclature Committee, 1999. Unified nomenclature for the semaphorins/collapsins. Cell 97 551–552. [DOI] [PubMed] [Google Scholar]
- Shi, W., A. Kumanogoh, C. Watanabe, J. Uchida, X. Wang et al., 2000. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity 13 633–642. [DOI] [PubMed] [Google Scholar]
- Simmons, A. D., A. W. Puschel, J. D. McPherson, J. Overhauser and M. Lovett, 1998. Molecular cloning and mapping of human semaphorin F from the Cri-du-chat candidate interval. Biochem. Biophys. Res. Commun. 242 685–691. [DOI] [PubMed] [Google Scholar]
- Strauss, R., 2002. The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 12 633–638. [DOI] [PubMed] [Google Scholar]
- Sweeney, S. T., and G. W. Davis, 2002. Unrestricted synaptic growth in spinster, a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron 36 403–416. [DOI] [PubMed] [Google Scholar]
- Takegahara, N., A. Kumanogoh and H. Kikutani, 2005. Semaphorins: a new class of immunoregulatory molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360 1673–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, M., H. Nagao, Y. K. Takahashi, M. Yamaguchi, S. Mitsui et al., 2003. Distorted odor maps in the olfactory bulb of semaphorin 3A-deficient mice. J. Neurosci. 23 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall, L. B., 2000. Olfaction in Drosophila. Curr. Opin. Neurobiol. 10 498–503. [DOI] [PubMed] [Google Scholar]
- Walz, A., I. Rodriguez and P. Mombaerts, 2002. Aberrant sensory innervation of the olfactory bulb in neuropilin-2 mutant mice. J. Neurosci. 22 4025–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., G. Hasan and C. W. Pikielny, 1999. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 274 10309–10315. [DOI] [PubMed] [Google Scholar]
- Woodhouse, E. C., A. Fisher, R. W. Bandle, B. Bryant-Greenwood, L. Charboneau et al., 2003. Drosophila screening model for metastasis: Semaphorin 5c is required for l(2)gl cancer phenotype. Proc. Natl. Acad. Sci. USA 100 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani, U., and J. R. Terman, 2006. The semaphorins. Genome Biol. 7 211. [DOI] [PMC free article] [PubMed] [Google Scholar]