Abstract
Decapping is a critical step in mRNA decay. In the 5′-to-3′ mRNA decay pathway conserved in all eukaryotes, decay is initiated by poly(A) shortening, and oligoadenylated mRNAs (but not polyadenylated mRNAs) are selectively decapped allowing their subsequent degradation by 5′ to 3′ exonucleolysis. The highly conserved heptameric Lsm1p-7p complex (made up of the seven Sm-like proteins, Lsm1p–Lsm7p) and its interacting partner Pat1p activate decapping by an unknown mechanism and localize with other decapping factors to the P-bodies in the cytoplasm. The Lsm1p-7p–Pat1p complex also protects the 3′-ends of mRNAs in vivo from trimming, presumably by binding to the 3′-ends. In order to determine the intrinsic RNA-binding properties of this complex, we have purified it from yeast and carried out in vitro analyses. Our studies revealed that it directly binds RNA at/near the 3′-end. Importantly, it possesses the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs such that the former are bound with much higher affinity than the latter. These results indicate that the intrinsic RNA-binding characteristics of this complex form a critical determinant of its in vivo interactions and functions.
Keywords: Lsm1, Sm-like, mRNA, decay, stability, decapping
INTRODUCTION
Turnover of mRNA is an important control point in gene expression as evident from the enormous number of genes known to be regulated at the level of mRNA decay (Hentze 1991; Tharun and Parker 2001b; Raghavan et al. 2002; Wang et al. 2002a; Frevel et al. 2003; Yang et al. 2003). Furthermore, it is a critical determinant of several crucial cellular processes (Harigaya et al. 2006; Hollien and Weissman 2006).
mRNA decay pathways and decay factors are well conserved in all eukaryotes from yeast to humans, wherein normal mRNAs are degraded via two major pathways of decay (Coller and Parker 2004; Meyer et al. 2004; Parker and Song 2004; Wilusz and Wilusz 2004; Fillman and Lykke-Andersen 2005). Decay is initiated in both pathways by poly(A) shortening (Wilson and Treisman 1988; Shyu et al. 1991; Decker and Parker 1993). In the 5′-to-3′ pathway, this is followed by decapping, which then permits the 5′-to-3′ exonucleolytic degradation of the message body (Muhlrad and Parker 1992; Decker and Parker 1993; Muhlrad et al. 1994, 1995; Couttet et al. 1997; Gao et al. 2001; Wilusz et al. 2001). In the 3′-to-5′ decay pathway, deadenylated mRNAs are degraded in a 3′-to-5′ exonucleolytic manner by the exosome followed by the hydrolysis of the resulting cap structure to m7GMP (Muhlrad et al. 1995; Anderson and Parker 1998; Chen et al. 2001; Wang and Kiledjian 2001; Liu et al. 2002; Mukherjee et al. 2002).
In the yeast Saccharomyces cerevisiae, poly(A) shortening is carried out by the Ccr4p complex (Tucker et al. 2001). Decapping, which in the 5′-to-3′ decay pathway occurs after the poly(A) tails of the mRNAs are shortened to oligo(A) length, is carried out by the decapping enzyme (Dcp1p–Dcp2p complex) (Beelman et al. 1996; Dunckley and Parker 1999). The 5′-to-3′ exonucleolytic digestion of the decapped mRNAs is carried out by the exonuclease Xrn1p (Hsu and Stevens 1993). Decapping is a crucial step controlled by several factors. While the translation initiation machinery and the poly(A) binding protein Pab1p are known to be inhibitory to decapping (Caponigro and Parker 1995; Coller et al. 1998; Schwartz and Parker 1999, 2000; Vilela et al. 2000; Wilusz et al. 2001; Ramirez et al. 2002; Khanna and Kiledjian 2004), several other factors are known to facilitate decapping. These include Pat1p, Dhh1p, Lsm1p-7p complex, Edc1p, Edc2p, and Edc3p (Hatfield et al. 1996; Boeck et al. 1998; Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Wyers et al. 2000; Coller et al. 2001; Dunckley et al. 2001; Fischer and Weis 2002; Schwartz et al. 2003; Kshirsagar and Parker 2004). All the decapping factors, the 5′-to-3′ exonuclease Xrn1p, the mRNA molecules targeted for decapping, and the mRNA degradation intermediates of the 5′-to-3′ pathway are localized in discrete cytoplasmic structures called P-bodies, suggesting that P-bodies are the cytoplasmic sites of decapping and 5′-to-3′ degradation (Sheth and Parker 2003; Teixeira et al. 2005). Importantly, P-bodies also are conserved in mammalian cells (Ingelfinger et al. 2002; Van Dijk et al. 2002).
The Lsm1p-7p complex, which is a heptameric complex of seven Sm-like proteins Lsm1p–Lsm7p, is a highly conserved component of eukaryotic mRNA decay machinery. The Like Sm (Lsm) proteins are homologous to Sm proteins, and together these proteins form a conserved family that is characterized by the presence of a bipartite sequence motif called the Sm domain (Cooper et al. 1995; Hermann et al. 1995; Seraphin 1995; Anantharaman and Aravind 2004). The tertiary structure of the Sm domain is also highly conserved among these proteins (Kambach et al. 1999; Collins et al. 2001; Mura et al. 2001; Toro et al. 2001; Schumacher et al. 2002). There are eight Lsm proteins (Lsm1p–Lsm8p) that are conserved in all eukaryotes, and they form two distinct heptameric complexes, Lsm1p-7p and Lsm2p-8p (Salgado-Garrido et al. 1999; Bouveret et al. 2000; Tharun et al. 2000), which are localized in the cytoplasm and nucleus, respectively (Ingelfinger et al. 2002; Sheth and Parker 2003; Stanek et al. 2003). The Lsm2p-8p complex, which has a ring-shaped structure similar to that of the Sm complex, binds to U6 snRNA, forms the core of that snRNP, and functions in splicing (Cooper et al. 1995; Pannone et al. 1998, 2001; Achsel et al. 1999; Mayes et al. 1999; Salgado-Garrido et al. 1999; Chan et al. 2003) but is not involved in cytoplasmic mRNA decay (Tharun et al. 2000). However, it is involved in the nuclear degradation of pre-mRNAs and nucleus-restricted mRNAs (Kufel et al. 2004). The Lsm1p-7p complex, on the other hand, is an activator of cytoplasmic mRNA decapping (Boeck et al. 1998; Bouveret et al. 2000; Tharun et al. 2000), and it is not involved in splicing (Boeck et al. 1998; Mayes et al. 1999). The human (recombinant) Lsm1p–Lsm7p proteins have been shown to associate into a ring-shaped heptamer in vitro (Zaric et al. 2005), suggesting that the Lsm1p-7p complex also has a quaternary structure similar to that of the Lsm2p-8p and Sm complexes.
The Lsm1p-7p complex physically interacts with several decay factors involved in the 5′-to-3′ decay pathway, namely, Pat1p, Dhh1p, and Xrn1p, as well as mRNPs (Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Coller et al. 2001). Furthermore, it also interacts with the RNA polymerase II subunit Rpb4p, which is implicated in the coordination of transcription and mRNA decay (Lotan et al. 2005). The variety of such interactions into which this complex engages indicates that it plays a key role in the control of mRNA decapping and decay. However, the mechanism by which this complex activates decapping in vivo is not clear. Our earlier studies suggested that in vivo this complex preferentially associates with mRNPs that are targeted for decay but not with mRNPs competent for translation (Tharun et al. 2000; Tharun and Parker 2001a). Such selective interaction of this complex with mRNPs is likely to be crucial for its function. In addition to decapping, the Lsm1p-7p–Pat1p complex also serves to protect the 3′-ends of deadenylated mRNAs in vivo from trimming (He and Parker 2001) presumably by binding to the 3′-ends. To gain insight into the factors that determine such interactions, it is important to elucidate the intrinsic RNA-binding properties of this complex. Such studies have not been done so far. In fact, the ability of this complex to directly bind RNA has not been demonstrated.
In the present study, we have purified the Lsm1p-7p–Pat1p complex from yeast and studied its RNA-binding properties in vitro. Our results show that this complex binds RNA directly, near the 3′-end, and that presence of a stretch of U-residues near the 3′-end facilitates the binding. Importantly, this complex binds to oligoadenylated RNA substrates with much higher affinity than polyadenylated RNA in vitro. These results are consistent with the known in vivo functions of this complex and suggest that the intrinsic RNA-binding characteristics of this complex form a key determinant of its in vivo functions.
RESULTS
Purification of the Lsm1p-7p–Pat1p complex from yeast
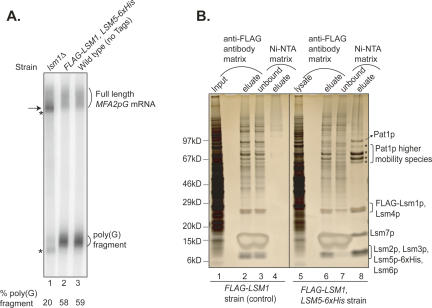
In order to determine the intrinsic RNA-binding properties of the Lsm1p-7p–Pat1p complex, we purified the Lsm1p-7p–Pat1p complex from yeast. Our strategy involved sequential affinity chromatography that exploited epitope tags attached to two different subunits of this complex. Specifically, we used a yeast strain that expressed FLAG-LSM1 (N-terminal tag) and LSM5–6xHis (C-terminal tag) as the only copies of these genes from their native promoters. Northern analyses (Fig. 1A) confirmed that the presence of these epitope tags did not affect mRNA decay and 3′-end protection functions in this strain based on the following observations. First, accumulation of the poly(G) decay intermediate of the MFA2pG mRNA (an indicator of the status of 5′-to-3′ mRNA decay) (Hatfield et al. 1996; Cao and Parker 2001; Tharun et al. 2005) was as efficient in this strain as in a wild-type strain not carrying any epitope tags. Second, full-length MFA2pG mRNA in this strain had a uniform poly(A) tail length distribution ranging from ∼10 to 70 A-residues similar to the wild-type strain without epitope tags. Thus, underaccumulation of poly(G) fragment and overaccumulation of deadenylated full-length mRNA, which are hallmarks of strains defective in the 5′-to-3′ decay pathway (Beelman et al. 1996; Dunckley and Parker 1999; Tharun et al. 2000), were not observed in our FLAG-LSM1, LSM5–6xHis strain. Finally, accumulation of 3′-trimmed forms of the full-length mRNA and poly(G) fragment, which is characteristic of strains defective in Lsm1p-7p–Pat1p complex function (Boeck et al. 1998; He and Parker 2001; Tharun et al. 2005), was also not observed in our strain (and the wild-type strain lacking epitope tags). Thus, all these observations indicate that this strain is indistinguishable from a wild-type strain lacking epitope tags with regard to mRNA decay and 3′-end protection functions.
FIGURE 1.
Purification of the Lsm1p-7p–Pat1p complex. (A) mRNA decay and mRNA 3′-end protection functions are normal in the FLAG-LSM1, LSM5–6xHis strain used for purification. RNA made from exponentially growing cultures of wild-type strain lacking epitope tags (yRP841, lane 3), the FLAG-LSM1, LSM5–6xHis strain (yST254, lane 2), and the lsm1Δ strain (yST247, lane 1) were subjected to Northern analysis for MFA2pG mRNA as described in Materials and Methods. The positions of the full-length MFA2pG mRNA and the poly(G) decay intermediate are indicated on the right. (Arrow on left) The position of the oligoadenylated full-length mRNA that accumulates in lsm1Δ cells due to block in decapping. (Upper and lower asterisks on the left) The positions of the 3′-trimmed forms of the full-length MFA2pG mRNA and the poly(G) decay intermediate, respectively, observed in lsm1Δ cells. The % poly(G) fragment was calculated by taking the sum of the signal from the full-length species (trimmed and normal) and poly(G) fragment (trimmed and normal) as 100%. (B) The purified Lsm1p-7p–Pat1p complex preparation contains the seven Lsm proteins and Pat1p. Lysate prepared from the FLAG-LSM1, LSM5–6xHis strain (lanes 5–8) or a control strain lacking the 6×His tag (lanes 1–4) was subjected to purification using anti-Flag antibody matrix followed by Ni-NTA matrix as described in Materials and Methods. The proteins present in the sample at different stages of purification (indicated on top) were revealed by SDS-PAGE analysis followed by silver staining. (Left) Positions of size markers. (Right) Identities of the bands observed with the final purified material obtained from the FLAG-LSM1, LSM5–6xHis strain (lane 8) as determined by mass spectrometry analysis of the corresponding gel slices. (Asterisks on the right) Pat1p bands of higher-than-expected mobility (see text).
Purification involved affinity chromatography using the anti-Flag antibody resin followed by Ni-NTA matrix. The band pattern of the purified material (Fig. 1B, lane 8) revealed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining closely matches the pattern expected based on the known sizes and mobilities of the component proteins and the pattern seen in another study where this complex was purified using a TAP tag fused to the Lsm protein (Bouveret et al. 2000). The identity of these protein bands (Fig. 1B) was confirmed by mass spectrometry of the gel slices containing them, based on the detection of tryptic peptides with unambiguous sequence match (seven for Pat1p, five for Lsm6p, three for Lsm1p, and one each for Lsm2p–Lsm5p and Lsm7p). Pat1p was not only present in a band of mobility corresponding to ∼97 kDa (in spite of its predicted size of 88 kDa), as observed by others earlier (Rodriguez-Cousino et al. 1995; Bouveret et al. 2000), but also in several faster-moving bands (Fig. 1B, labeled with asterisk) also observed by others earlier (Bonnerot et al. 2000; Bouveret et al. 2000). These analyses revealed the presence of all the previously observed components of the Lsm1p-7p–Pat1p complex (Bouveret et al. 2000) except Xrn1p in our preparation (Fig. 1B). Mass spectrometry analysis of the purified sample (without electrophoretic separation) was also unable to detect Xrn1p, indicating that Xrn1p is not present in significant amounts in our preparation. Absence of Xrn1p should help avoid undesirable degradation of the substrate RNA when RNA-binding assays are carried out with the purified complex. Similar analyses also failed to detect Lsm8p. Absence of Lsm8p in the preparation indicates that our preparation is not contaminated with the closely related Lsm2p-8p complex.
Purification of uncomplexed Lsm1p and partial complexes (lacking one or more subunits), if any, present in the cell lysate should be greatly reduced in our procedure since it involves use of two different epitope tags attached to two different subunits. Also, mRNA endogenously bound to the Lsm1p-7p–Pat1p complex is not likely to copurify with the complex because the purification procedure involves 300 mM salt wash (see Materials and Methods), while coimmunoprecipitation of mRNA with this complex from cell lysates is abolished with 100 mM salt wash (S. Tharun, unpubl.). Finally, while constructing the FLAG-LSM1, LSM5–6xHis strain, epitope tagging of LSM1 and LSM5 genes was done without changing any part of their native promoter and flanking sequences to ensure that native levels of expression of all the subunits is achieved.
The Lsm1p-7p–Pat1p complex directly binds to RNA
In order to determine the RNA-binding characteristics of the purified Lsm1p-7p–Pat1p complex, we carried out in vitro RNA-binding experiments. For our initial studies, we used an RNA substrate derived from the 3′-UTR of the yeast MFA2 mRNA, which is a commonly studied mRNA decay reporter in yeast (Muhlrad and Parker 1992; Decker and Parker 1993; Muhlrad et al. 1994) that is known to require the Lsm1p-7p–Pat1p complex for its normal rates of decay through the 5′-to-3′ decay pathway. The full-length MFA2pG mRNA and its decay intermediate (containing only its 3′-UTR sequences), the poly(G) fragment, are also known to coimmunoprecipitate with this complex (Tharun et al. 2000; Tharun and Parker 2001a). For our initial studies, we used a 47-nucleotide (nt)-long RNA prepared by run-on transcription with T7 polymerase using the plasmid pRP1039 as template (see Materials and Methods). This RNA contains the 3′-most 42 nt of the MFA2 mRNA 3′-UTR (Muhlrad and Parker 1992) attached to three additional U-residues (derived from the plasmid) at the 3′-end and two additional G-residues at the 5′-end, and it will be referred to as MFA2(u) RNA.
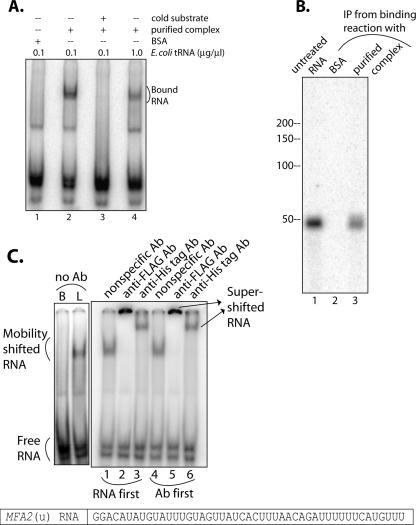
To test if the purified Lsm1p-7p–Pat1p complex can interact with RNA, we incubated the purified complex with uniformly radiolabeled MFA2(u) RNA (at final concentrations of 40.6 nM and 0.05 nM, respectively) and then analyzed the reaction mixture to check if ribonucleoprotein (RNP) complexes containing MFA2(u) RNA are formed. Separation of the binding reactions through a native polyacrylamide gel (Fig. 2A) revealed a clear gel mobility shift of the MFA2(u) RNA (Fig. 2A, lane 2), indicating that the purified Lsm1p-7p–Pat1p complex was able to interact with MFA2(u) RNA. The observed gel shift is not due to nonspecific interactions because of the following reasons: (1) The mobility shift was not observed (Fig. 2A, lane 1) if the MFA2(u) RNA was incubated with bovine serum albumin (BSA) instead of the purified complex, even though BSA was used at a higher molar concentration (147 nM) than the Lsm1p-7p–Pat1p complex. (2) The mobility shift was observed in spite of the presence of a large molar excess of Escherichia coli tRNA (100 ng/μL; ∼4 μM; see Materials and Methods) over the radiolabeled MFA2(u) RNA substrate in the RNA-binding reactions (Fig. 2A). (3) A further (10-fold) increase in the concentration of E. coli tRNA (to 1 μg/μL or ∼40 μM) in the binding reaction affected the mobility shift only moderately (Fig. 2A, lane 4), while including cold MFA2(u) RNA in the reaction at eightfold molar excess over the radiolabeled MFA2(u) RNA almost completely abolished the gel shift (Fig. 2A, lane 3).
FIGURE 2.
Purified Lsm1p-7p–Pat1p complex is capable of binding RNA. (A) Incubation of RNA with purified Lsm1p-7p–Pat1p complex results in gel mobility shift of the RNA. RNA-binding reactions containing MFA2(u) RNA (radiolabeled at U-residues) were carried out as described in Materials and Methods with the purified complex (at a final concentration of 40.6 nM; lanes 2–4) or BSA (lane 1) in the presence of E. coli tRNA at a final concentration of 0.1 μg/μL (lanes 1–3) or 1.0 μg/μL (lane 4) and in the presence (lane 3) or absence (lanes 1,2,4) of cold MFA2(u) RNA [used at eightfold molar excess over the hot MFA2(u) RNA]. Following incubation, samples were separated on a native polyacrylamide gel and visualized by Phosphorimaging as described in Materials and Methods. (Bottom) The nucleotide sequence of the RNA substrate. (B) Pull-down of the Lsm1p-7p–Pat1p complex from the RNA-binding reaction coprecipitates the RNA. RNA-binding reactions were carried out as described in Materials and Methods using radiolabeled MFA2(u) RNA and the purified complex (at a final concentration of 40.6 nM; lane 3) or BSA (lane 2). The reaction mix was then incubated with anti-Flag antibody matrix, and the RNA bound to the matrix was extracted and visualized by denaturing polyacrylamide gel electrophoresis and PhosphorImaging as described in Materials and Methods. (Lane 1) Untreated MFA2(u) RNA. (Left) Positions of size markers. About 59% of the input RNA was coprecipitated with the Lsm1p-7p–Pat1p complex in this experiment. (C) RNPs formed in the presence of the purified Lsm1p-7p–Pat1p complex can be supershifted using antibodies directed against the Lsm proteins. RNA-binding reactions were carried out as described in Materials and Methods using radiolabeled MFA2(u) RNA and the purified complex (at a final concentration of 40.6 nM), following which anti-Flag (lane 2), anti-His tag (lane 3), or nonspecific antibody (lane 1) was added to the reaction mix, and incubation was continued as described in Materials and Methods. The samples were finally visualized by native polyacrylamide gel electrophoresis and Phosphorimaging as described in Materials and Methods. (Lanes 4–6) Similar experiments (indicated on top of the lanes) in which the purified complex was incubated with the antibody before adding the RNA. (Left) Binding reactions carried out using the purified complex or BSA (labeled L and B, respectively, on top) in the absence of any antibody.
Two more analyses further confirmed that the observed gel shift of the MFA2(u) RNA is the result of its interaction with the Lsm1p-7p–Pat1p complex and not due to any nonspecific contaminant proteins present in the preparation. First, following incubation with radiolabeled MFA2(u) RNA, when the Lsm1p-7p–Pat1p complex was immunoprecipitated from the binding reaction using anti-Flag antibodies (targeting the Flag-Lsm1p subunit of the complex), the MFA2(u) RNA was coimmunoprecipitated as seen in Figure 2B, lane 3. Such coimmunoprecipitation failed to occur if the binding reactions contained BSA instead of the purified Lsm1p-7p–Pat1p complex, indicating that the MFA2(u) RNA is not being pulled down because of its nonspecific association with the antibody matrix (Fig. 2C, lane 2). Second, as seen in Figure 2C, incubation of the purified Lsm1p-7p–Pat1p complex with anti-Flag (Fig. 2C, lanes 2,5) or anti-His tag (Fig. 2C, lanes 3,6) antibodies (targeted against the Flag-Lsm1p or Lsm5p-6xHis subunit, respectively, of the purified complex), but not the unrelated anti-mouse IgG (Fig. 2C, lanes 1,4) either before (Fig. 2C, lanes 4–6) or after (Fig. 2C, lanes 1–3) the Lsm1p-7p–Pat1p complex is bound to the MFA2(u) RNA, results in a clear supershift of the RNPs formed. These results indicate that the gel-retarded RNP complexes observed in our experiments are, indeed, formed by the Lsm1p-7p–Pat1p complex. The fact that prior binding of the Lsm1p-7p–Pat1p complex to the anti-Flag or anti-His tag antibodies does not affect the ability of this complex to bind to the RNA suggests that the N-terminal region of Lsm1p and the C-terminal region of Lsm5p are not critical for RNA binding. This is consistent with our earlier observation that deletion of the N-terminal domain of Lsm1p does not affect mRNA decay or 3′-end protection (Tharun et al. 2005), and our results presented in Figure 1A, which indicate that these epitope tags do not affect the in vivo function of this complex.
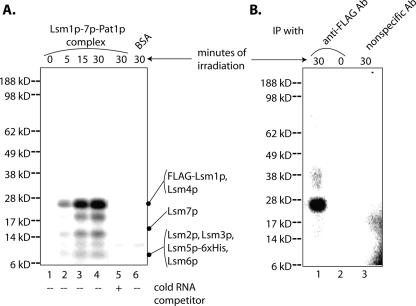
In order to confirm that the Lsm1p-7p–Pat1p complex makes direct contacts with the RNA substrate, we carried out UV cross-linking experiments. For this, binding reactions with MFA2(u) RNA (radiolabeled with 32P-UTP) were carried out using the purified complex or BSA as described above followed by UV irradiation for various lengths of time. The irradiated samples were then subjected to ribonuclease digestion (to digest unprotected RNA) before visualizing the cross-linked proteins by SDS-PAGE and autoradiography (see Materials and Methods). As seen in Figure 3A, a protein(s) with a mobility similar to that of Lsm4p and/or Flag-Lsm1p (see Fig. 1B, lane 8) corresponding to ∼23 kDa was strongly radiolabeled in a UV-irradiation-dependent manner in the binding reaction containing the purified complex (Fig. 3A, lanes 1–4), suggesting that Lsm1p and/or Lsm4p subunits of the Lsm1p-7p–Pat1p complex directly contact the RNA. Additional weaker bands of lower mobilities were also observed, suggesting possible RNA contacts of other Lsm subunits. Importantly, neither the 23-kDa band nor most of the weaker bands were observed if cold MFA2(u) RNA was included in the reaction at eightfold molar excess over the hot MFA2(u) RNA (Fig. 3A, lane 5) or if the binding reaction was carried out with BSA instead of the purified complex (Fig. 3A, lane 6). As shown in Figure 3B, when the UV cross-linking reaction (carried out with the purified complex) was subjected to detergent treatment (to disrupt the protein complexes) after ribonuclease treatment and then subjected to immunoprecipitation using anti-Flag antibody matrix (Fig. 3B, lane 1), it led to the specific pull-down of a 23-kDa protein(s). This did not occur if unrelated anti-HA antibody matrix (Fig. 3B, lane 3) was used for immunoprecipitation or if the binding reactions were not UV-irradiated (Fig. 3B, lane 2). This observation indicates that the 23-kDa band revealed by the cross-linking experiment shown in Figure 3A, indeed, contains Flag-Lsm1p. These results strongly suggest that the purified Lsm1p-7p–Pat1p complex is capable of binding RNA directly, with the Lsm1p subunit being at least one of the subunits in the complex making such direct RNA contacts.
FIGURE 3.
Lsm1p-7p–Pat1p complex makes direct contacts with RNA. (A) Proteins present in the purified Lsm1p-7p–Pat1p complex can be UV cross-linked to RNA. RNA-binding reactions containing MFA2(u) RNA (radiolabeled at U-residues) were carried out as described in Materials and Methods with the purified complex (at a final concentration of 40.6 nM; lanes 1–5) or BSA (lane 6) in the presence (lane 5) or absence (lanes 1–4 and 6) of cold MFA2(u) RNA [used at eightfold molar excess over the hot MFA2(u) RNA]. Aliquots of the reaction mix were then exposed to UV irradiation for varying lengths of time (indicated above the lanes) followed by ribonuclease treatment, SDS-PAGE separation, and Phosphorimaging as described in Materials and Methods. (Left) Positions of size markers. (Right) Expected positions of the Lsm proteins. (B) The 23-kDa band that gets UV cross-linked to RNA contains Flag-Lsm1p. Purified Lsm1p-7p–Pat1p complex was bound to radiolabeled MFA2(u) RNA and exposed to UV for 0 (lane 2) or 30 min (lanes 1,3). After ribonuclease treatment, the samples were treated with detergent (to disrupt the protein complexes) and then immunoprecipitated with either anti-Flag antibody matrix (lanes 1,2) or anti-HA antibody matrix (lane 3) before separation of the proteins pulled down by SDS-PAGE and their visualization by Phosphorimaging. (Left) Positions of molecular weight markers.
Comparison of the binding affinities of MFA2(u) RNAs carrying or lacking an m7G cap structure at their 5′-ends (using gel shift assays) revealed that the 5′-cap has no significant effect on the binding of the Lsm1p-7p–Pat1p complex (data not shown). This is consistent with the following earlier observations: (1) The poly(G) decay intermediate of the MFA2pG mRNA (which is generated by decapping followed by Xrn1p-mediated 5′-to-3′ exonucleolysis up to the 5′-end of the poly[G] insertion and hence lacks all of the coding region and 5′-UTR of the mRNA) efficiently coimmunoprecipitates with this complex from cell lysates (Tharun and Parker 2001a) just like the full-length mRNA. (2) mRNA coimmunoprecipitates with this complex from wild-type cells as well as xrn1Δ (which accumulate uncapped mRNAs) and dcp1Δ cells (which accumulate capped mRNAs) (Tharun and Parker 2001a).
Lsm1p-7p–Pat1p complex binds near the 3′-end of RNA
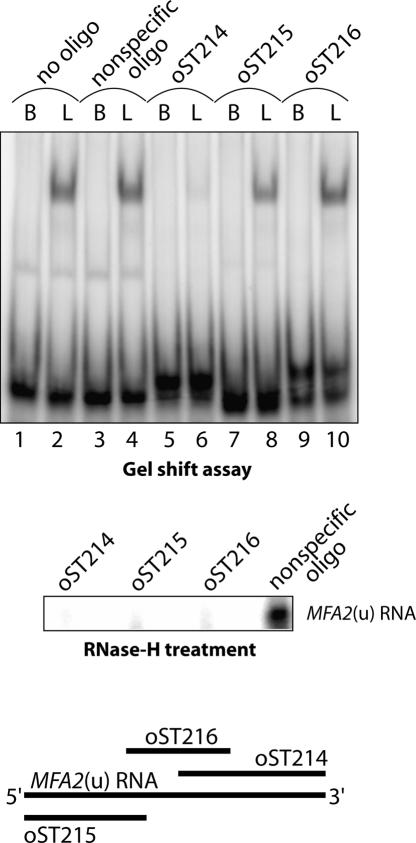
In order to determine which part of the RNA is bound by the Lsm1p-7p–Pat1p complex, we carried out oligonucleotide inhibition experiments. We designed DNA oligonucleotides complementary to the 5′, middle, and 3′ thirds of the MFA2(u) RNA and asked if they inhibited the binding of the purified Lsm1p-7p–Pat1p complex to the MFA2(u) RNA when they were annealed to that RNA prior to the binding reaction. As seen in Figure 4, while the oligonucleotide complementary to the 3′ portion of the MFA2(u) RNA almost completely abolished the binding (as revealed by gel shift analysis), the oligos complementary to the 5′ or middle segments of the MFA2(u) RNA, or a nonspecific oligo, did not have any significant effect on the binding. The differential effects of the oligos complementary to different regions of the MFA2(u) RNA are not due to differences in their efficiency of annealing because RNase-H treatment of the MFA2(u) RNA after annealing to each of these oligonucleotides (but not the nonspecific oligonucleotide) led to a complete loss of the full-length substrate RNA. These results therefore suggest that the Lsm1p-7p–Pat1p complex binds at or near the 3′-end of this RNA.
FIGURE 4.
Lsm1p-7p–Pat1p complex binds near the 3′-end of RNA. Radiolabeled MFA2(u) RNA was annealed to DNA oligonucleotides oST214, oST215, oST216, a nonspecific DNA oligonucleotide, or no oligonucleotide (indicated above the lanes in upper and middle panels). The annealed RNA was then subjected either to RNA-binding reactions with the purified complex (at a final concentration of 40.6 nM; lanes labeled with L on top) or BSA (lanes labeled with B on top), followed by visualization of gel-shifted RNA as described in Materials and Methods (upper panel) or RNase-H treatment followed by separation on denaturing gels and autoradiography (middle panel). (Lower panel) Regions of MFA2(u) RNA spanned by the oligonucleotides oST214, oST215, and oST216 are shown schematically.
Presence of a stretch of U-residues near the 3′-end facilitates Lsm1p-7p–Pat1p complex binding
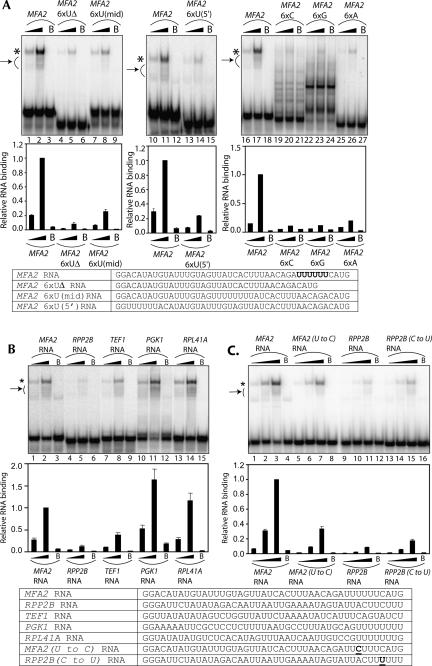
Given that the Lsm1p-7p–Pat1p complex binds at or near the 3′-end of the MFA2(u) RNA, we asked if there are any specific sequence features in that region of the MFA2(u) RNA that are important for the binding of this complex. A stretch of six U-residues is present very close to the 3′-end of the MFA2 mRNA (Muhlrad and Parker 1992) and hence the MFA2(u) RNA. Similar stretches of U-residues form the binding sites of the Sm complex and the Lsm2p-8p complex (Branlant et al. 1982; Achsel et al. 1999; Raker et al. 1999; Vidal et al. 1999). Therefore we tested if the 6×U-stretch near the 3′-end of the MFA2(u) RNA is important for binding to the Lsm1p-7p–Pat1p complex.
To this end, different RNA substrates were made (as described in Materials and Methods) by T7 transcription using oligonucleotide templates. As a control, we used the MFA2 RNA, which is the same as the MFA2(u) RNA except that it does not contain the three additional U-residues at the 3′-end (Figs. 2, 5, cf. sequences). The effect of deleting, mutating, or relocating the 6×U-stretch near the 3′-end of this RNA was determined by comparing the MFA2 RNA and its various modified versions (in which the 6×U-stretch is subjected to such changes) for their ability to bind to the purified Lsm1p-7p–Pat1p complex in gel shift assays. Figure 5A shows that, as expected, the MFA2 RNA was efficiently bound by the Lsm1p-7p–Pat1p complex (Fig. 5A, lanes labeled MFA2 on top). However, deletion of the 6×U-stretch from this RNA significantly weakened the binding such that the fraction of bound RNA dropped by ∼10-fold (Fig. 5A, lanes labeled MFA2 6×UΔ). Moreover, moving the 6×U-stretch away from the 3′-end to the middle or near the 5′-end of the RNA also led to about fourfold weakening of binding [Fig. 5A, lanes labeled MFA2 6×U(mid) and MFA2 6×U(5′), respectively]. Finally, replacing the 6×U-stretch with a stretch of six As, Cs, or Gs (Fig. 5A, lanes labeled MFA2 6×A, MFA2 6×C, and MFA2 6×G, respectively) also weakened the binding by at least fivefold. These results indicate that the presence of the 6×U-stretch near the 3′-end of the MFA2 RNA facilitates the binding of that RNA to the Lsm1p-7p–Pat1p complex. Furthermore, together with the results of the oligonucleotide inhibition experiments (Fig. 4), they suggest that this 6×U-stretch forms at least part of the binding site on that RNA for the Lsm1p-7p–Pat1p complex. In any case, the band pattern of the MFA2 6×G RNA (Fig. 5A) suggests that its decreased binding may at least partly be due to its ability to form a more stable secondary structure. Secondary structure prediction using the RNAfold program (Mathews et al. 1999) revealed that the MFA2 6×G RNA could form a slightly stronger structure (−6.8 kcal/mol) than the MFA2, MFA2 6×C, and MFA2 6×A RNAs (−4.85 kcal/mol, −4.36 kcal/mol, and −6.12 kcal/mol, respectively), although in general all these RNAs formed only weak structures.
FIGURE 5.
Presence of a stretch of U-residues near the 3′-end of the RNA facilitates the Lsm1p-7p–Pat1p complex binding. (A) Mutation, deletion, or relocation of the 6×U-stretch near the 3′-end of the MFA2 RNA impairs Lsm1p-7p–Pat1p complex binding. (B) RNA substrates that carry an uninterrupted stretch of six or more U-residues near their 3′-ends bind to the Lsm1p-7p–Pat1p complex better than RNAs that do not. (C) A single U-to-C change within the U-stretch could significantly affect binding. (Top panels) RNA-binding reactions were carried out as described in Materials and Methods using various radiolabeled RNAs (indicated above the lanes) in the presence of the purified Lsm1p-7p–Pat1p complex at a final concentration of 2.8 nM (lanes 1,5,9,13 in C), 8.12 nM (lanes 16,19,22,25 in A; lanes 1,4,7,10,13 in A,B), 14 nM (lanes 2,6,10,14 in C), 40.6 nM (lanes 17,20,23,26 in A; lanes 2,5,8,11,14 in A,B), or 56 nM (lanes 3,7,11,15 in C) or in the presence of BSA (lanes labeled with B on top). After the reaction, gel shift of the RNA was visualized as described in Materials and Methods. (*) Position of gel-shifted RNA. (Arrow on the left of the gel) The smear observed below the band of gel-shifted RNA resulting from destabilization of the RNPs during the gel run. (Middle panels) Fraction of the RNA bound in each reaction (quantitated from the gel using a PhosphorImager) normalized to the value obtained with MFA2 RNA at 40.6 nM (in A,B) or 56 nM (in C) concentration of the purified complex is shown as a bar diagram directly under the corresponding lane of the gel picture. (Bottom panels) Sequences of the various RNA substrates used for the gel shift assays. The 6×U-stretch of MFA2 RNA, which is replaced with 6×A or 6×C or 6×G in MFA2 6×A, MFA2 6×C and MFA2 6×G RNAs, respectively, is shown in bold. The U-to-C and C-to-U changes introduced in MFA2 (U-to-C) RNA and RPP2B (C-to-U) RNA are underlined.
We then asked if the observed influence of the stretch of Us near the 3′-end of the RNA on the Lsm1p-7p–Pat1p complex binding is specific to the MFA2 mRNA or if this complex has a general binding preference for RNAs that carry such sequences near their 3′-ends. To this end, we made RNA substrates derived from the 3′-UTRs of different mRNAs that do or do not carry an uninterrupted stretch of six or more Us near their 3′-ends. This included the PGK1 and RPL41A mRNAs, which carried stretches of eight and seven U-residues, respectively, near their 3′-ends and the TEF1 and RPP2B mRNAs whose 3′-ends, although rich in Us, were interrupted with other nucleotides (Cottrelle et al. 1985; Remacha et al. 1988; Muhlrad and Parker 1994; Muhlrad et al. 1995; Yu and Warner 2001). RNA substrates carrying the 3′-most 42 nt of each of these mRNAs (attached to two additional G-residues at the 5′-end) were made by T7 transcription using oligonucleotide templates, and their ability to bind to the purified Lsm1p-7p–Pat1p complex was compared to that of the MFA2 RNA by gel shift assays. The results shown in Figure 5B reveal that the PGK1 and RPL41A RNAs (which carry 8×U and 7×U-stretches near their 3′-ends) bind more strongly than the MFA2 RNA to the purified complex. On the other hand, the TEF1 and RPP2B RNAs (which lack uninterrupted stretches of six or more Us) show much weaker binding. These results reveal a correlation between the presence of an uninterrupted stretch of six or more U-residues near the 3′-end of the RNA and the efficiency of the RNA to bind to the Lsm1p-7p–Pat1p complex and, therefore, suggest that this complex has a general binding preference for RNAs carrying a stretch of U-residues near their 3′-ends. Nevertheless, these results also suggest that in addition to the U-rich sequences near the 3′-end, other sequences of the RNA also affect binding by this complex, because the TEF1 RNA carrying the most disrupted U-rich sequence binds only twofold weaker than the MFA2 RNA, and the RPP2B RNA shows very poor binding in spite of bearing a U-rich element that differs from an uninterrupted 6×U-stretch only by a single base change (Fig. 5B). Consistent with this idea, the MFA2 (U-to-C) RNA (in which a single U-to-C change is introduced in the 6×U-stretch of MFA2 RNA so that it resembles the U-rich element of RPP2B RNA) binds only threefold weaker than MFA2 RNA (Fig. 5C), while RPP2B RNA binds almost eightfold to 10-fold weaker than MFA2 RNA (Fig. 5B). Similarly, the RPP2B (C-to-U) RNA (carrying a single C-to-U change in its U-rich element that converts it into an uninterrupted 6×U-stretch) binds only twofold better than the RPP2B RNA (Fig. 5C).
The binding preference of this complex for U-rich sequences is also supported by the fact that when synthetic homo-oligoribonucleotides (20-mers) were used in gel shift assays, detectable binding was observed only with oligo(U) but not with the other oligomers (data not shown).
Oligoadenylated RNAs interact stronger than polyadenylated RNAs with the Lsm1p-7p–Pat1p complex
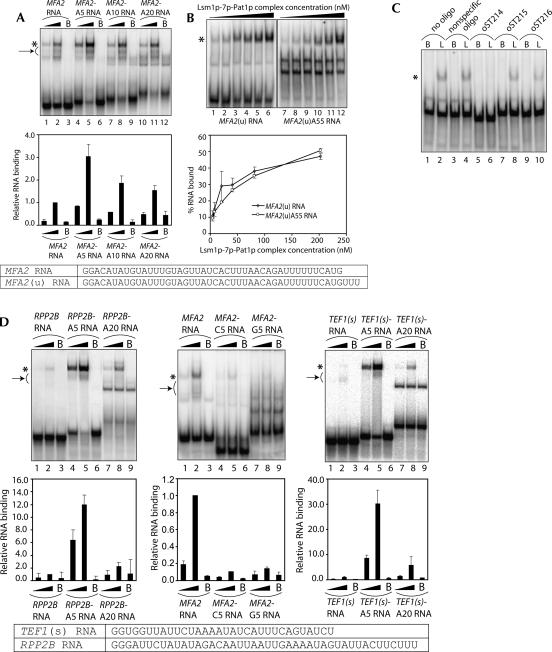
Oligoadenylated (but not polyadenylated) mRNAs are selectively decapped in the 5′-to-3′ decay pathway, and the ability of the decapping activator Lsm1p-7p–Pat1p complex to interact with RNA is crucial for this pathway to proceed at normal rates in vivo (Tharun et al. 2005). Therefore, in order to determine if this complex has an intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs, we tested the effect of adding varying number of adenylate residues to the 3′-end of the RNA on the binding of the RNA to the purified complex. Using oligonucleotide templates, we made MFA2 RNA carrying five, 10, or 20 residues-long 3′-A-tails (referred to as MFA2-A5, MFA2-A10, and MFA2-A20 RNAs, respectively). Gel shift assays revealed that presence of an A5 tail at the 3′-end enhanced binding about threefold; however, further increase in the length of the A-tail to 10 and 20 A-residues progressively weakened the binding so that the MFA2-A20 RNA was only moderately better than the MFA2 RNA in binding to this complex (Fig. 6A). In order to test the effect of a long poly(A) tail, we made the MFA2(u)A55 RNA (which is the MFA2[u] RNA carrying a 55-residues-long 3′-poly[A] tail) and compared it with the MFA2(u) RNA for its affinity to bind to the Lsm1p-7p–Pat1p complex. As seen in Figure 6B, both the MFA2(u) and MFA2(u)A55 RNAs bound to this complex with similar affinities, giving an apparent K D of ∼200 nM. Consistent with this, oligonucleotide inhibition assays (similar to those shown in Fig. 4) suggested that this complex binds to the same region in both MFA2[u] and MFA2[u]-A55 RNAs [Figs. 4, 6C]). Thus, these results indicate that oligoadenylated RNA binds to this complex more strongly than polyadenylated RNA. Such influence of the 3′-oligoadenylate tail on the binding of this complex is not specific for the MFA2 RNA, because similar results were obtained with RPP2B RNA also. As seen in Figure 6D (left panel), the Lsm1p-7p–Pat1p complex binds RPP2B-A5 RNA with much greater affinity than RPP2B-A20 RNA. Importantly, the enhancement of RNA binding caused by the 3′-tail is specific to an oligoadenylate tail as shown by the observation (Fig. 6D, middle panel) that attaching a C5 or a G5 tail at the 3′-end of the MFA2 RNA fails to enhance the binding of this complex and, in fact, results in weakening of binding. Together, these results suggest that this complex has a general intrinsic binding preference for oligoadenylated RNAs over polyadenylated RNAs.
FIGURE 6.
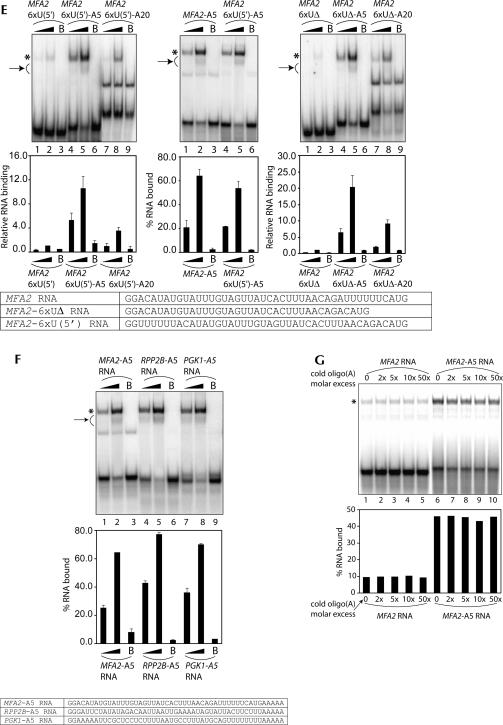
Lsm1p-7p–Pat1p complex has a higher affinity for oligoadenylated RNA than polyadenylated RNA. (A) Lengthening the oligoadenylate tail of the MFA2 RNA leads to a progressive weakening of Lsm1p-7p–Pat1p complex binding. (Upper panel) RNA-binding reactions were carried out as described in Materials and Methods using various radiolabeled RNAs (indicated above the lanes) in the presence of the purified Lsm1p-7p–Pat1p complex at a final concentration of 8.12 nM (lanes 1,4,7,10) or 40.6 nM (lanes 2,5,8,11) or in the presence of BSA (lanes labeled with B on top). After the reaction, gel shift of the RNA was visualized as described in Materials and Methods. (Lower panel) Fraction of the RNA bound in each reaction (quantitated from the gel using a PhosphorImager) normalized to the value obtained with MFA2 RNA at 40.6 nM concentration of the purified complex is shown as a bar diagram directly under the corresponding lane of the gel picture. (B) MFA2(u) RNA carrying a 55-residue-long 3′-poly(A) tail [MFA2(u)A55 RNA] binds to the Lsm1p-7p–Pat1p complex with an affinity similar to that of the MFA2(u) RNA. (Upper panel) RNA-binding reactions were carried out as described in Materials and Methods using the radiolabeled RNAs indicated below the lanes in the presence of the purified Lsm1p-7p–Pat1p complex at a final concentration of 4.06 nM (lanes 1,7), 8.12 nM (lanes 2,8), 20.3 nM (lanes 3,9), 40.6 nM (lanes 4,10), 81.2 nM (lanes 5,11), or 203 nM (lanes 6,12). After the reaction, gel shift of the RNA was visualized as described in Materials and Methods. (Lower panel) Percentage of RNA bound (quantitated from the gel using a PhosphorImager) is plotted against the concentration of the Lsm1p-7p–Pat1p complex used in the reaction. Both MFA2(u) and MFA2(u)A55 RNAs bound to the purified complex with an apparent K D of ∼200 nM. Sequences of MFA2 and MFA2(u) RNA substrates used in the experiments are shown at the bottom of A and B. (C) The binding site of the Lsm1p-7p–Pat1p complex is the same in both MFA2(u) and MFA2(u)A55 RNAs. Experiments were carried out as described for Figure 4 except that MFA2(u)A55 RNA was used as the substrate in the RNA-binding reactions. (D) The 3′-tail length effect (on the binding of the Lsm1p-7p–Pat1p complex) is specific for oligo(A) tail (middle panel) and is also observed with RPP2B RNA (left panel), and TEF1(s) RNA (right panel) as with the MFA2 RNA. (Upper panels) RNA-binding reactions (using various radiolabeled RNA substrates indicated above the lanes) and visualization of results were carried out as described in A. Reactions carried out with purified Lsm1p-7p–Pat1p complex at a final concentration of 8.12 nM (lanes 1,4,7) or 40.6 nM (lanes 2,5,8) or in the presence of BSA (lanes labeled with B on top) are shown. (Lower panels) Fraction of the RNA bound in each reaction (quantitated from the gel using a PhosphorImager) normalized to the value obtained with RNA lacking a 3′-tail (at 40.6 nM concentration of the purified complex) in each set is shown as a bar diagram directly under the corresponding lane of the gel picture. Sequences of the TEF1(s) and RPP2B RNAs are shown at the bottom. (E) Influence of the 3′-oligoadenylate tail length on the binding of the Lsm1p-7p–Pat1p complex to the RNA is not directly related to the U-rich sequence near the 3′-end of the body of the RNA. Binding reactions were carried out and results were visualized as described for D. The upper and lower panels are presented as in D except that in the case of comparison of MFA2-A5 and MFA2-6xU(5′)-A5 RNAs, the RNA binding quantitated after the gel shift assays is shown as a percentage of RNA bound in each reaction (without normalization) in the bar diagram. Sequences of the relevant RNA substrates are shown at the bottom. (F) The difference in the binding affinity for the Lsm1p-7p–Pat1p complex of RNAs that bear or lack an uninterrupted stretch of Us near their 3′-ends disappears when they are oligoadenylated. The binding reactions were carried out and results were visualized as described for D. The upper and lower panels are presented as in D except that the bar diagram in the lower panel shows the percentage of RNA bound in each reaction without normalization. (Bottom) Sequences of the RNA substrates used for gel shifts. (G) Binding of the Lsm1p-7p–Pat1p complex to the oligoadenylated RNA cannot be competitively inhibited by oligo(A). (Upper panel) RNA-binding reactions were carried out as described in Materials and Methods using radiolabeled MFA2 RNA (lanes 1–5) or MFA2-A5 RNA (lanes 6–10) with the purified Lsm1p-7p–Pat1p complex (at a final concentration of 20.3 nM) that had been preincubated for 10 min at 30°C (before adding the radiolabeled RNA) in the presence of varying folds of molar excess (over the radiolabeled RNA) of oligo(A) (indicated above the lanes). After the reaction, gel shift of the RNA was visualized as described in Materials and Methods. (Lower panel) The percentage of the RNA bound in each reaction (quantitated from the gel using a PhosphorImager) is shown as a bar diagram directly under the corresponding lane of the gel picture. (* on the left in each figure) Positions of gel-shifted RNAs. (A,D,E,F, arrow on left) The smear observed below the band of gel-shifted RNA resulting from destabilization of the RNPs during the gel run.
The enhancement of Lsm1p-7p–Pat1p complex binding observed upon attaching an A5 tail to the 3′-end of the RNA is unlikely to be the result of an increase in the length of the RNA because further lengthening of the A-tail leads to weakening of binding (Fig. 6A,B,D). In order to test if the MFA2-A5 and RPP2B-A5 RNAs bind to this complex with greater affinity because they have the length (49 nt long) that is optimal for binding to this complex, we studied the effect of 3′-oligoadenylate tail length on the binding of a shorter RNA substrate, the TEF1(s) RNA. This RNA is the same as the TEF1 RNA (used in the experiment shown in Fig. 5B), except that it carries only the 3′-most 30 nt (not 42 nt) of TEF1 mRNA. As shown in Figure 6D (right panel), attaching a 3′-A5 tail to this RNA [yielding the 37-nt-long TEF1(s)-A5 RNA] results in strong enhancement of binding while lengthening the tail further to 20 As (TEF1[s]-A20 RNA; 52 nt-long]) results in weakening of binding. Similar results were also obtained with MFA2-25 RNA (which carries the 3′-most 25 nt of MFA2 mRNA preceded by two Gs at the 5′-end) (see Fig. 7 for sequence) carrying A5 or A20 tails (data not shown). These results indicate that the factor determining the affinity of the Lsm1p-7p–Pat1p complex for the oligoadenylated RNAs is the length of the oligoadenylate tail rather than the overall length of the RNA substrate. The binding of TEF1(s) RNA (Fig. 6D, right panel) is weaker than that of TEF1 RNA (Fig. 5B, top panel) because the former is shorter in length (see below and Fig. 7).
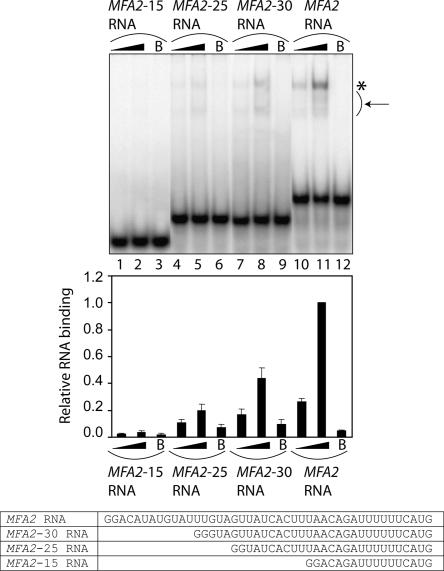
FIGURE 7.
The shorter the RNA, the weaker is its binding to the Lsm1p-7p–Pat1p complex. (Top panel) RNA-binding reactions were carried out as described in Materials and Methods using various radiolabeled RNAs (indicated above the lanes) in the presence of the purified Lsm1p-7p–Pat1p complex at a final concentration of 8.12 nM (lanes 1,4,7,10) or 40.6 nM (lanes 2,5,8,11) or in the presence of BSA (lanes labeled with B on top). After the reaction, gel shift of the RNA was visualized as described in Materials and Methods. (*) Position of gel-shifted RNA. (Arrow) The smear observed below the band of gel-shifted RNA due to the destabilization of the RNPs during the gel run. (Middle panel) The fraction of the RNA bound in each reaction (quantitated from the gel using PhosphorImager) normalized to the value obtained with MFA2 RNA at 40.6 nM concentration of the purified complex is shown as a bar diagram directly under the corresponding lane of the gel picture. (Bottom panel) Sequences of the RNA substrates used for the gel shift assays.
As discussed above, U-rich sequences when present near the 3′-end of the RNA promote the binding of the Lsm1p-7p–Pat1p complex (Figs. 4, 5). Therefore, it is possible that this complex distinguishes between RNAs carrying 3′-A5 and 3′-A20 tails based on the difference in the distance between the 3′-end of the A-tail and the U-rich sequence. For example, such a distance of ∼9 nt could be optimal for binding (as in MFA2-A5 RNA), while a longer distance of 24 nt or more may not be optimal (as in MFA2-A20 and MFA2[u]-A55 RNAs). If this is true, MFA2-6×U(5′) RNA (in which the 6×U-stretch near the 3′-end is moved to an upstream location) (see Fig. 5A) carrying a 3′-A5 tail should not be an optimal substrate for binding since in the MFA2-6×U(5′)-A5 RNA the 3′-end of the A-tail will be 41 nt away from the U-rich sequence. However, Figure 6E (left panel) shows that the effect of 3′-oligoadenylate tail length on the binding observed with the MFA2-6×U(5′) RNA is similar to that seen with the MFA2 RNA. Furthermore, the binding of MFA2-6×U(5′)-A5 RNA to the purified complex was only slightly weaker compared to that of the MFA2-A5 RNA (Fig. 6E, middle panel) although the MFA2-6×U(5′) RNA was about fourfold weaker than the MFA2 RNA in binding to the purified complex (Fig. 5A). Finally, even in the case of the MFA2-6×UΔ RNA (in which the 6×U-stretch near the 3′-end of the RNA body has been deleted) (see Fig. 5A), the influence of the 3′-oligoadenylate tail length on the binding was similar to that observed with MFA2 RNA (Fig. 6E, right panel). These results suggest that the effect of the 3′-oligoadenylate tail length on the binding of the RNA to the Lsm1p-7p–Pat1p complex is not related to the presence of the U-rich sequences near the 3′-end of the RNA body. This idea was further supported by the observation that the large difference between the binding affinities of RNAs that bear (MFA2 and PGK1 RNAs) or lack (RPP2B RNA) an uninterrupted stretch of U-residues near their 3′-ends mostly disappears when such RNAs carry an A5 tail at their 3′-ends (Fig. 6F).
Recent studies (Mikulecky et al. 2004) on the Sm-like protein Hfq suggest that the Hfq hexamer has two RNA-binding sites, with one of them involved in binding poly(A). This raises the possibility of a similar scenario with regard to the Lsm1p-7p–Pat1p complex given that the length of the 3′-oligoadenylate tail of the RNA can influence the binding of this complex. However, as mentioned above, oligo(U) was the only homo-oligoribonucleotide that was able to bind to this complex, and the other homo-oligomers including oligo(A) were not able to bind to this complex. To confirm this further, we tested if binding of this complex to oligoadenylated substrate RNA can be inhibited by preincubation of the purified complex with cold oligo(A) before starting the binding reaction. As shown in Figure 6G, preincubation of the purified complex with oligo(A) (20-mer) had no effect on its ability to bind to either the MFA2 or the MFA2-A5 RNA, even when oligo(A) was used at 50-fold molar excess over those RNAs. These results indicate that the Hfq complex and the Lsm1p-7p–Pat1p complex probably recognize A-tails by different mechanisms. Nevertheless, they do not completely rule out the presence of a second RNA-binding site in the Lsm1p-7p–Pat1p complex (see Discussion).
Shorter RNA substrates bind more weakly than the longer RNAs to the Lsm1p-7p–Pat1p complex
In order to test the effect of RNA length on Lsm1p-7p–Pat1p complex binding, we made the MFA2-15, MFA2-25, and MFA2-30 RNAs, which carry the 3′-most 15, 25, and 30 nt of MFA2 mRNA sequence, respectively, preceded by two G-residues at the 5′-end (see Fig. 7). When these RNAs were studied for their ability to bind to the purified Lsm1p-7p–Pat1p complex by gel shift assays, it was observed that a decrease in RNA length led to a progressive weakening of the binding such that with the shortest RNA (MFA2-15), the binding was almost undetectable (Fig. 7). Thus, longer RNAs bind more strongly than shorter RNAs to this complex. Nevertheless, as shown by the results with TEF1(s) RNA (Fig. 6D), it is clear that the binding of shorter RNAs could be enhanced when they carry a 3′-oligo(A) tail. Therefore, the minimum RNA length needed for detectable binding could be lower in the case of oligoadenylated RNAs.
DISCUSSION
The Lsm1p-7p–Pat1p complex plays a key role in mRNA decay, and its interaction with mRNA is an important event in this process. Mutations disrupting the predicted RNA-binding residues of the Lsm1p subunit lead to strong inhibition of mRNA decay (Tharun et al. 2005). In order to understand how the intrinsic RNA-binding properties of this complex are related to its in vivo interactions and functions, we have purified this complex from yeast and determined its RNA-binding characteristics. Our studies reveal that the intrinsic RNA-binding characteristics of this complex are consistent with its in vivo functions.
Electrophoretic analysis of our purified preparation of the Lsm1p-7p–Pat1p complex reveals the presence of Pat1p not only in a band of expected mobility but also in additional faster-moving bands. Such species were also observed by other investigators (Bonnerot et al. 2000; Bouveret et al. 2000), and it is not clear if they are present in vivo or are degradation products generated upon cell lysis. Given the fact that Pat1p exhibits abnormal mobility during electrophoresis (Fig. 1B; Rodriguez-Cousino et al. 1995; Bouveret et al. 2000), it is also possible that some of these faster-moving bands contain full-length Pat1p. In fact, mass spectrometry analysis of the top-most band among the higher mobility species (Fig. 1B, labeled with asterisks) identified the peptides DGPLDFEESYK and SQELDTELK, which are very close to the N and C termini in the Pat1p sequence (13 and 33 residues away from the N and C terminus, respectively), suggesting that this band may contain full-length Pat1p. In any case, the RNA-binding properties of the purified complex observed here are unlikely to be unphysiological anomalous activities resulting from the possible presence of truncated forms of Pat1p for the following reasons. First, the ability to bind preferentially to U-rich sequences is a property shared by several Sm-like protein complexes and not just the Lsm1p-7pPat1p complex. Second, our earlier studies also showed that the Lsm proteins preferentially associate with oligoadenylated mRNAs while exhibiting very little association, if any, with polyadenylated mRNAs in vivo (Tharun and Parker 2001a) and that the mRNPs associated with the Lsm1p are not bound to the poly(A) binding protein, Pab1p. Future work will reveal the contribution of Pat1p to the various RNA-binding properties of the Lsm1p-7p–Pat1p complex.
Although the association of the Lsm1p-7p–Pat1p complex with mRNA was revealed by immunoprecipitation studies earlier, it was not clear if it is the result of a direct interaction (Tharun and Parker 2001a). Our studies presented here not only show that the purified Lsm1p-7p–Pat1p complex can bind to RNA in vitro (as revealed by gel shift assays and pull-down of the RNPs using anti-Flag antibody matrix) (Fig. 2) but also that Lsm1p and possibly a few other subunits in the complex can be cross-linked to RNA (as revealed by UV cross-linking experiments and immunoprecipitation analyses after UV cross-linking) (Fig. 3), supporting the idea that this complex directly binds RNA. These results suggest that the in vivo association of this complex with mRNAs is by virtue of its ability to directly bind RNA. This is consistent with inhibition of mRNA decay resulting from mutations targeting the predicted RNA-binding residues of Lsm1p (Tharun et al. 2005). Nevertheless, it is possible that other factors in vivo modulate the interaction of this complex with mRNA.
The binding affinity of the purified Lsm1p-7p–Pat1p complex for various RNA substrates is likely an underestimate because our purified complex is not 100% active. Binding reactions carried out using various concentrations of MFA2 RNA in the presence of a constant amount of the purified complex (such that RNA:Lsm1p-7p–Pat1p complex molar ratios ranging from 0.012 to 12.3 are attained) revealed that our preparation is 50% active (data not shown).
Our studies show that Lsm1p-7p–Pat1p complex binding is weaker with shorter RNAs such that the binding is undetectable with a 17-nt-long RNA (Fig. 7). In this regard, this complex differs from many other Sm-like protein complexes like the Sm complex (Raker et al. 1999; Urlaub et al. 2001), Lsm2p-8p complex (Achsel et al. 1999), the archaebacterial Sm1 (Toro et al. 2001) and Sm2 homoheptamers (Achsel et al. 2001), and the Hfq hexamer (Schumacher et al. 2002), which have been shown to bind efficiently very short (5–10 residues long) RNA substrates. In our gel shift assays using the RNA substrates discussed here, we did not observe multiple gel-shifted bands indicative of ternary or higher-order complexes, even though the Lsm1p-7p–Pat1p complex is used at several-fold molar excess over the radiolabeled RNA substrate in these experiments (see Materials and Methods). Furthermore, when assays were carried out using various concentrations of MFA2 RNA in the presence of a constant amount of the purified complex such that RNA:Lsm1p-7p–Pat1p complex molar ratios ranging from 0.012 to 12.3 are attained, multiple gel-shifted bands were not observed and the mobility of the gel-shifted band was not altered (data not shown). Thus, our results are consistent with a 1:1 molar complex being formed between RNA and the Lsm1p-7p–Pat1p complex under our assay conditions. However, it is possible that higher-order complexes may be formed when substrates with an RNA body longer than 42 nt are used.
Our results support the idea that the Lsm1p-7p–Pat1p complex binds at or near the 3′-end of the RNA. First, an oligonucleotide that anneals to the 3′-third of the MFA2(u) RNA impairs the binding of the Lsm1p-7p–Pat1p complex to this RNA, while oligonucleotides annealing to other regions of this RNA have no effect on binding (Fig. 4). Second, several experiments indicate that changes introduced into the RNA substrate near its 3′-end affect binding of this complex to the RNA. This includes mutation or deletion of the 6×U-stretch (which is located very close to the 3′-end of the MFA2 RNA) and addition of an oligo(A) tail at the 3′-end of the RNA (Figs. 5, 6, respectively). Therefore, these results suggest that the protection provided by this complex to the 3′-ends of deadenylated mRNAs in vivo is a result of the direct interaction of this complex with mRNA 3′-ends. Consistent with such an idea, mutations disrupting the predicted RNA-binding residues of Lsm1p not only lead to inhibition of mRNA decay but also to loss of mRNA 3′-end protection (Tharun et al. 2005).
An important observation made in our studies is that the purified Lsm1p-7p–Pat1p complex shows a significantly stronger interaction with oligoadenylated RNA than with polyadenylated RNA. Strongest binding was observed with RNA carrying a 3′-A5 tail, while further increase in the tail length led to weakening of binding such that RNA with a poly(A) tail (55 residues) has similar affinity as an RNA lacking a tail (Fig. 6A,B). The ability of this complex to distinguish between oligoadenylated and polyadenylated forms of RNA was not specific to a particular RNA or dependent on the presence of U-rich sequences near the 3′-end of the RNA body and is determined by the length of the A-tail rather than the overall length of the RNA (Fig. 6D,E). Importantly, only an oligo(A) tail but not an oligo(C) or oligo(G) tail at the 3′-end could enhance the binding of RNA to this complex (Fig. 6D). These results suggest that a selective in vivo association of this complex with oligoadenylated mRNAs driven by the intrinsic ability of this complex to distinguish between polyadenylated and oligoadenylated RNAs is crucial for the 5′-to-3′ mRNA decay pathway since a key aspect of this pathway is that decapping is allowed only after poly(A) shortening of the mRNA is completed such that only oligoadenylated mRNAs, but not polyadenylated mRNAs, form the substrates for decapping. It is known that Pab1p bound to the poly(A) tail is involved in the inhibition of decapping of polyadenylated mRNAs (Caponigro and Parker 1995; Coller et al. 1998; Wilusz et al. 2001; Khanna and Kiledjian 2004). Our results presented here support the idea that the selective binding (and decapping activation) of oligoadenylated mRNAs by the Lsm1p-7p–Pat1p complex could be an additional mechanism that contributes to the deadenylation dependence of decapping in the 5′-to-3′ pathway.
Our results indicate that the difference in the binding affinities (for the Lsm1p-7p–Pat1p complex) of RNAs bearing or lacking an uninterrupted stretch of Us at their 3′-ends disappears considerably when an A5 tail is attached to their 3′-ends. While this supports the idea that this complex plays a general role in targeting mRNAs for decapping in vivo (rather than selectively targeting certain groups of mRNAs), it also raises the possibility that binding of this complex to the oligoadenylated mRNAs could affect their further deadenylation and thereby influence the endpoint of deadenylation. However, in this regard, it is important to note that accumulation of fully deadenylated mRNAs occurs not only in lsm and pat1 mutants but also in dcp1 or dcp2 mutants, where the integrity of the Lsm1p-7p–Pat1p complex is unaffected. This suggests that the further shortening of the oligo(A) tail of oligoadenylated mRNAs in vivo is mainly determined by decapping rates. Nevertheless, it is possible that when the decapping rate of an mRNP is impaired in vivo (as in a dcp1 or dcp2 mutant) that weakens or alters the association of the Lsm1p-7p–Pat1p complex with the mRNP, allowing further shortening of the tail by the deadenylase.
Our experiments indicate that although polyadenylated RNA and RNA lacking a poly(A) tail have a lower affinity (than the oligoadenylated RNA) for the purified Lsm1p-7p–Pat1p complex in vitro, they do bind to this complex. However, polyadenylated mRNA does not coprecipitate at detectable amounts with this complex when this complex is pulled down from cell lysates (Tharun and Parker 2001a). This suggests that other factors may interfere with the interaction of this complex with polyadenylated mRNA in vivo. Polyadenylated mRNA is not only bound to Pab1p in vivo, but also to other translation factors depending on its translation status. Furthermore, translating polyadenylated mRNA has a distinct mRNP structure due to the interactions between Pab1p and eIF4G (Wells et al. 1998) and is likely to be localized outside the P-bodies in the cytoplasm (Kedersha et al. 2005; Teixeira et al. 2005), while the Lsm1p-7p–Pat1p complex (like other proteins involved in decay) is concentrated in the P-bodies. On the other hand, mRNPs that are targeted for decapping and not efficient substrates for translation (like the oligoadenylated mRNPs) are likely to be localized in the P-bodies (Sheth and Parker 2003; Cougot et al. 2004; Teixeira et al. 2005). Due to these reasons, the difference in the abilities of polyadenylated and oligoadenylated mRNAs to associate with this complex could be higher in vivo compared to what is observed in vitro.
The enhancement of RNA binding of the Lsm1p-7p–Pat1p complex by the 3′-oligo(A) tail observed in our studies is reminiscent of observations on another Sm-like protein complex, the Hfq homohexamer. However, there are several differences between the Hfq complex and the Lsm1p-7p–Pat1p complex. First, while attaching a 3′-oligo(A) tail to the RNA enhances binding in the case of both the Lsm1p-7p–Pat1p complex and the Hfq complex, further increase in the length of the 3′-A-tail results in weakening of binding only in the case of the Lsm1p-7p–Pat1p complex [such that polyadenylated RNA has the same affinity as the RNA lacking a poly(A) tail] (Fig. 6A,B) but not the Hfq complex (Hajnsdorf and Regnier 2000). In fact, lengthening the oligo(A) tail to a poly(A) tail seems to enhance binding in the case of the Hfq complex (Hajnsdorf and Regnier 2000). Second, the Hfq complex is capable of binding to oligo(A) as shown by direct binding and competition experiments (Hajnsdorf and Regnier 2000; Schumacher et al. 2002; Zhang et al. 2002; Mikulecky et al. 2004). On the other hand, our experiments (data not shown) reveal that only oligo(U), but not oligo(A) or oligo(C) or oligo(G), is able to bind to the purified Lsm1p-7p–Pat1p complex. Furthermore, preincubation with cold oligo(A) had absolutely no effect on the interaction of this complex with MFA2 or MFA2-A5 RNA (Fig. 6G). Finally, while the presence of a 3′-terminal A5 tail on the RNA promotes binding of the Lsm1p-7p–Pat1p complex, a similar stretch of A-residues located internally (4 nt upstream of the 3′-end) fails to have such an effect as shown by the poor binding of the MFA2-6×A RNA (Fig. 5A) (this RNA binds threefold to fourfold weaker than MFA2 RNA). However, the Hfq complex is able to bind A-rich sequences located internally (Senear and Steitz 1976; Zhang et al. 1998, 2002). These observations indicate that the Hfq and Lsm1p-7p–Pat1p complexes recognize stretches of A-residues in distinct ways. Recent studies on Hfq have suggested that it has two RNA-binding sites, with one of them capable of binding to poly(A) sequences (Mikulecky et al. 2004). At present, we do not know if a similar scenario exists in the case of the Lsm1p-7p–Pat1p complex also. In any case, the fact that the Lsm1p-7p–Pat1p complex is able to recognize the oligo(A) tail attached to the 3′-end of the RNA but not able to bind to free oligo(A) is analogous to the decapping enzyme, which is able to recognize and cleave the cap when the cap structure is part of the capped RNA molecule but not when the cap structure is in the form of free cap analog (LaGrandeur and Parker 1998; Van Dijk et al. 2002; Wang et al. 2002b; Piccirillo et al. 2003; Steiger et al. 2003). Similarly, the mammalian poly(A) binding protein (PABP) and the decapping inhibitor VCX-A are also able to bind to capped RNA but not free cap (Khanna and Kiledjian 2004; Jiao et al. 2006).
Sm-like protein complexes in general prefer to bind U-rich sequences (Achsel et al. 1999, 2001; Raker et al. 1999; Toro et al. 2001; Urlaub et al. 2001; Schumacher et al. 2002), and our results (Figs. 4, 5) indicate that the Lsm1p-7p–Pat1p complex also shares that property. Importantly, we also find that in order to facilitate the binding of the Lsm1p-7p–Pat1p complex, the U-stretch needs to be located near the 3′-end (Fig. 5A). In any case, this complex is also capable of binding RNAs with disrupted stretches of Us, albeit with lower affinity (Fig. 5B). Such relatively less stringent binding specificity of this complex may permit its binding to multiple mRNAs such that it plays a general role in mRNA decay (see below).
The Lsm1p-7p–Pat1p complex plays a very general role in mRNA decay and 3′-end protection as shown by the observation that decay and 3′-end protection of multiple mRNAs get affected in lsm and pat1 loss-of-function mutants, and that multiple mRNAs associate with this complex in vivo (Tharun and Parker 2001a). These observations imply that the sequence elements recognized by this complex must be relatively common among cellular mRNAs, and our results presented here indicate that this is, indeed, true; the oligo(A) tail is a structural feature common to the mRNAs subjected to decapping in vivo, and it is also known that presence of U-rich sequences near the 3′-end is a feature shared by a significant fraction of yeast mRNAs (Graber et al. 1999a,b).
The closely related Lsm2p-8p complex has been shown to interact with both U6 snRNA and mRNA (Achsel et al. 1999; Kufel et al. 2004), and an Lsm8p-containing complex also associates with U8 snoRNA (Tomasevic and Peculis 2002). While the Lsm2p-8p complex binds to the U-rich region at the 3′-end (of U6 snRNA) similarly to the Lsm1p-7p–Pat1p and other Sm-like protein complexes, it also binds to other internal sites in U6 snRNA (Achsel et al. 1999; Vidal et al. 1999). Thus, just like the Lsm1p-7p–Pat1p complex (as shown above), the Lsm2p-8p complex is also likely to have the intrinsic ability to bind to a variety of different RNAs. On the other hand, in vivo, the binding of these complexes with RNAs could be made more selective by their compartmentation and/or by the influence of other factors associating with them.
The exact mechanism by which the Lsm1p-7p–Pat1p complex promotes decapping in vivo is not known. The observation that this complex selectively associates with oligoadenylated mRNPs targeted for decay (but not with polyadenylated mRNPs bound to translation factors) and that it promotes the interaction of the decapping enzyme with mRNA in vivo supported the model that it activates decapping by promoting a transition in the mRNP organization from a translation-competent state to a decay-competent state (Tharun and Parker 2001a). Several studies have shown that while translating mRNPs and translation factors are localized outside P-bodies, decapping factors and mRNPs targeted for decapping are concentrated in P-bodies, suggesting that translation and decapping occur in different compartments (Sheth and Parker 2003; Cougot et al. 2004; Fenger-Gron et al. 2005; Ferraiuolo et al. 2005; Kedersha et al. 2005; Teixeira et al. 2005; Yu et al. 2005). Therefore, these observations together support the idea that the Lsm1p-7p–Pat1p complex could act by promoting the transfer of mRNPs into P-bodies. The intrinsic binding preference of this complex for oligoadenylated mRNAs could drive the association of this complex to mRNPs as soon as poly(A) shortening is completed. This association could then facilitate the interaction of other decay factors such that the mRNP is fully reorganized into a decay-competent state and transported into the P-bodies. Given that mRNA is needed for the formation of the P-bodies (Sheth and Parker 2003; Cougot et al. 2004; Kedersha et al. 2005; Teixeira et al. 2005; Yu et al. 2005), the studies in mammalian cells indicating that down-regulation of Lsm1p expression affects the localization of other decay factors into the P-bodies (Andrei et al. 2005) is consistent with the above model. However, since these studies also show that localization of Lsm1p to P-bodies is affected upon down-regulation of rck/p54 (homolog of Dhh1p) expression (Andrei et al. 2005), it is likely that the Lsm1p-7p–Pat1p complex is not sufficient for the transport for mRNPs into P-bodies and needs the help of other factors for this function.
MATERIALS AND METHODS
Yeast strains
Strains used in this study are in the genetic background of yRP841 (MATα, leu2, ura3, lys2, trp1, cup1Δ∷LEU2[PM]) (Hatfield et al. 1996). The Lsm1p-7p–Pat1p complex was purified from the strain yST254, which expresses Flag-tagged Lsm1p and His-tagged Lsm5p. This strain was made by transforming the strain yST247 (MATα, leu2, ura3, lys2, trp1, cup1Δ∷LEU2[PM], lsm1Δ∷TRP1, LSM5–6xHis-NEOr) with the plasmid pST17, which expresses Flag-Lsm1p from the native promoter. pST17 was made by the Quikchange mutagenesis procedure (Stratagene) using pST11 (Tharun et al. 2005) as the template. C-terminal tagging of LSM5 with the His-tag was carried out following the PCR-based method of gene modification (Longtine et al. 1998) using yRP1365 (MATα, leu2, ura3, lys2, trp1, cup1Δ∷LEU2[PM], lsm1Δ∷TRP1) as the host strain (Tharun et al. 2000) to generate yST247.
Purification of Lsm1p-7p–Pat1p complex
yST254 cells were grown in −ura drop out medium at 25°C, collected, and lysed as described earlier (Tharun and Parker 1999). The lysate was first incubated with the anti-Flag antibody matrix (Sigma). After washing of the matrix to remove unbound proteins, the bound proteins were eluted. The eluted proteins were then incubated with the Ni-NTA matrix (Invitrogen), followed by washing of the Ni-NTA matrix and elution of the final purified material. Purification steps using these two matrices were carried out following the manufacturer's protocols except for the differences noted below. All the steps were carried out at 4°C in 50 mM phosphate buffer (pH 7.5) with 0.1% NP-40 containing various other components as required for the different steps as indicated below. The buffers used for cell lysis and anti-Flag antibody matrix binding contained 100 mM NaCl, while the lysis buffer also had 1× COMPLETE protease inhibitor mix (Roche). The buffers used for washing of unbound proteins and elution of bound proteins from this matrix and all the steps of purification over the Ni-NTA matrix had 300 mM NaCl. The elution buffers used with the anti-Flag antibody matrix and the Ni-NTA matrix also had 150 ng/μL Flag peptide and 250 mM Imidazole, respectively. Determination of protein concentration, SDS-PAGE analysis, and silver staining of gels were carried out using standard procedures. Mass spectrometry analysis of the proteins was done by Prottech, Inc.
RNA analyses
Both MFA2(u) and MFA2(u)A55 RNAs were made by in vitro transcription using T7 RNA polymerase (Roche) and [α-32P]-labeled UTP following the manufacturer's protocol. The plasmid pRP1039 codes for MFA2(u) RNA [with or without poly(A) tail] under the bacteriophage T7 promoter control (D. Muhlrad and R. Parker, unpubl.). pRP1039 linearized with DraI formed the template for making MFA2(u) RNA, while pRP1039 linearized with HindIII and then treated with mung bean nuclease served as the template for making MFA2(u)A55 RNA. All the other RNA substrates used for gel shifts were made using the mirvana kit (Ambion Inc.) with [α-32P]-labeled UTP or CTP and oligonucleotide templates. The template for each RNA was made by annealing the oligonucleotide carrying T7 promoter sequence to a bottom-strand oligonucleotide that codes for that RNA. After transcription, reactions were treated with DNase I and cleaned up using a spin column. The radiolabeled RNAs were gel purified before they were used in gel shift assays. The integrity of purified RNAs was confirmed by denaturing polyacrylamide gel electrophoresis.
RNA-binding reactions (10 μL) were carried out by incubating purified Lsm1p-7p–Pat1p complex (at the final concentrations indicated in the figure legends) or BSA (at final 147 nM concentration) with the radiolabeled RNA substrate in the presence of 0.1 μg/μL E. coli tRNA and 4 u/μL ribonuclease inhibitor in 50 mM Tris (pH 7.5), 50 mM Nacl, 0.5 mM MgCl2, 10% (v/v) glycerol, and 0.1% NP-40 for 45 min at 30°C. For gel shift assays, the reactions were then separated on a 6% native polyacrylamide gel. The gels were then dried and exposed to a PhosphorImager. Concentration of the radiolabeled RNA substrate in the binding reaction was 46 nM for experiment shown in Figure 2B and 6.7 nM for the experiment in Figure 3. In all the other experiments, it was 0.05 nM so that the molar concentration of the radiolabeled RNA substrate is several-fold lower than that of the lowest concentration of the purified complex used in the experiment. In all the experiments, substrate RNA was heat-denatured before it was included in the binding reaction. RNA substrates whose binding affinities are compared in a given experiment were prepared and tested together. Typically ∼25%–30% of the MFA2 RNA got gel shifted when the Lsm1p-7p–Pat1p complex was used at 40.6 nM concentration in the binding reaction. For oligonucleotide inhibition experiments, radiolabeled RNA substrate was annealed to an excess of DNA oligonucleotide in 50 mM Tris (pH 7.5), 50 mM Nacl, and 0.5 mM MgCl2, by heat denaturation followed by slow cooling to room temperature. An appropriate amount of this annealed RNA was then used for setting up the RNA-binding reaction. For RNase-H reactions, the same amount of annealed RNA was digested with 1.5 units of RNase-H followed by phenol:chloroform treatment and ethanol precipitation of the digested RNA and visualization of the digested RNA by denaturing polyacrylamide gel electrophoresis and autoradiography. For supershift assays, 0.9 μg of nonspecific antibody (anti-mouse IgG) or 0.6 μg of anti-His tag or anti-Flag antibody was used. Binding of the antibody to the purified complex (before or after it is bound to the RNA) was done by incubation for 30 min at 30°C. UV irradiation and ribonuclease treatment of the RNA-binding reactions and immunoprecipitation analysis of the UV cross-linked proteins were carried out as described (Ausubel et al. 2003). For anti-Flag antibody pull-down experiments (Fig. 2B), 50-μL binding reactions carried out as described above were incubated with anti-Flag antibody matrix at 4°C. After washing the matrix with 50 mM Tris (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 0.1% NP-40 to remove unbound RNA, the bound RNA was extracted by phenol:chloroform treatment of the washed matrix and analyzed by separation on denaturing gels and autoradiography.
Isolation of RNA from yeast and Northern analyses (to visualize MFA2pG mRNA and the poly[G] fragment) were performed as described (Tharun and Parker 1999).
ACKNOWLEDGMENTS
We thank Dr. Roy Parker and Denise Muhlrad for providing us the plasmid pRP1039 and Drs. David Horowitz, Rich Maraia, and Mordechai Choder for providing helpful comments on this manuscript. This work was supported by USUHS intramural grant C071HJ and NIH grant GM072718 to S.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.502507.
REFERENCES
- Achsel, T., Brahms, H., Kastner, B., Bachi, A., Wilm, M., Luhrmann, R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel, T., Stark, H., Luhrmann, R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V., Aravind, L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.S.J., Parker, R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., Luhrmann, R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. Current protocols in molecular biology. Wiley; New York: 2003. [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., Parker, R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Boeck, R., Lapeyre, B., Brown, C.E., Sachs, A.B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot, C., Boeck, R., Lapeyre, B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., Seraphin, B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant, C., Krol, A., Ebel, J.P., Lazar, E., Haendler, B., Jacob, M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, D., Parker, R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., Parker, R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chan, S.-P., Kao, D.-I., Tsai, W.-Y., Cheng, S.-C. The Prp 19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Coller, J., Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller, J.M., Gray, N.K., Wickens, M.P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B.M., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M., Mabbutt, B.C. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- Cooper, M., Johnston, L.H., Beggs, J.D. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrelle, P., Thiele, D., Price, V.L., Memet, S., Micouin, J.Y., Marck, C., Buhler, J.M., Sentenac, A., Fromageot, P. Cloning, nucleotide sequence, and expression of one of two genes coding for yeast elongation factor 1 α. J. Biol. Chem. 1985;260:3090–3096. [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R., Grange, T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T., Tucker, M., Parker, R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae . Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron, M., Fillman, C., Norrild, B., Lykke-Andersen, J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo, M.A., Basak, S., Dostie, J., Murray, E.L., Schoenberg, D.R., Sonenberg, N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman, C., Lykke-Andersen, J. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Fischer, N., Weis, K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevel, M.A., Bakheet, T., Silva, A.M., Hissong, J.G., Khabar, K.S., Williams, B.R. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Wilusz, C.J., Peltz, S.W., Wilusz, J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, J.H., Cantor, C.R., Mohr, S.C., Smith, T.F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999a;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, J.H., Cantor, C.R., Mohr, S.C., Smith, T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. 1999b;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf, E., Regnier, P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya, Y., Tanaka, H., Yamanaka, S., Tanaka, K., Watanabe, Y., Tsutsumi, C., Chikashige, Y., Hiraoka, Y., Yamashita, A., Yamamoto, M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., Parker, R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, W., Parker, R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W. Determinants and regulation of cytoplasmic mRNA stability in eukaryotic cells. Biochim. Biophys. Acta. 1991;1090:281–292. doi: 10.1016/0167-4781(91)90191-n. [DOI] [PubMed] [Google Scholar]
- Hermann, H., Fabrizio, P., Raker, V.A., Foulaki, K., Hornig, H., Brahms, H., Luhrmann, R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien, J., Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L., Stevens, A. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., Achsel, T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jiao, X., Wang, Z., Kiledjian, M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Luhrmann, R., Li, J., Nagai, K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, R., Kiledjian, M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar, M., Parker, R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae . Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Bousquet-Antonelli, C., Beggs, J.D., Tollervey, D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T.E., Parker, R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Rodgers, N.D., Jiao, X., Kiledjian, M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A., III, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lotan, R., Bar-On, V.G., Harel-Sharvit, L., Duek, L., Melamed, D., Choder, M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes & Dev. 2005;19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mayes, A.E., Verdone, L., Legrain, P., Beggs, J.D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Mikulecky, P.J., Kaw, M.K., Brescia, C.C., Takach, J.C., Sledjeski, D.D., Feig, A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS, and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes & Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., Parker, R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., Parker, R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O'Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., Wilusz, J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura, C., Cascio, D., Sawaya, M.R., Eisenberg, D.S. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone, B.K., Xue, D., Wolin, S.L. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone, B.K., Kim, S.D., Noe, D.A., Wolin, S.L. Multiple functional interactions between components of the Lsm2–Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158:187–196. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Piccirillo, C., Khanna, R., Kiledjian, M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan, A., Ogilvie, R.L., Reilly, C., Abelson, M.L., Raghavan, S., Vasdewani, J., Krathwohl, M., Bohjanen, P.R. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker, V.A., Hartmuth, K., Kastner, B., Luhrmann, R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, C.V., Vilela, C., Berthelot, K., McCarthy, J.E. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 2002;318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- Remacha, M., Saenz-Robles, M.T., Vilella, M.D., Ballesta, J.P. Independent genes coding for three acidic proteins of the large ribosomal subunit from Saccharomyces cerevisiae . J. Biol. Chem. 1988;263:9094–9101. [PubMed] [Google Scholar]
- Rodriguez-Cousino, N., Lill, R., Neupert, W., Court, D.A. Identification and initial characterization of the cytosolic protein Ycr77p. Yeast. 1995;11:581–585. doi: 10.1002/yea.320110608. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido, J., Bragado-Nilsson, E., Kandels-Lewis, S., Seraphin, B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., Parker, R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., Parker, R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D., Decker, C.J., Parker, R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear, A.W., Steitz, J.A. Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J. Biol. Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- Seraphin, B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu, A.B., Belasco, J.G., Greenberg, M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Stanek, D., Rader, S.D., Klingauf, M., Neugebauer, K.M. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., Parker, R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., Parker, R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., Parker, R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001a;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun, S., Parker, R. Turnover of mRNA in eukaryotic cells. In: Soll D., et al., editors. RNA. Pergamon; New York: 2001b. pp. 245–257. [Google Scholar]
- Tharun, S., He, W., Mayes, A.E., Lennertz, P., Beggs, J.D., Parker, R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun, S., Muhlrad, D., Chowdhury, A., Parker, R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasevic, N., Peculis, B.A. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 2002;22:4101–4112. doi: 10.1128/MCB.22.12.4101-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, I., Thore, S., Mayer, C., Basquin, J., Seraphin, B., Suck, D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Urlaub, H., Raker, V.A., Kostka, S., Luhrmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., Seraphin, B. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, V.P., Verdone, L., Mayes, A.E., Beggs, J.D. Characterization of U6 snRNA–protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela, C., Velasco, C., Ptushkina, M., McCarthy, J.E. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 2000;19:4372–4382. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Kiledjian, M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., Brown, P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002a;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., Kiledjian, M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 2002b;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wilson, T., Treisman, R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Wilusz, J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., Peltz, S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wyers, F., Minet, M., Dufour, M.E., Vo, L.T., Lacroute, F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 2000;20:3538–3549. doi: 10.1128/mcb.20.10.3538-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., Darnell J.E., Jr Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Warner, J.R. Expression of a micro-protein. J. Biol. Chem. 2001;276:33821–33825. doi: 10.1074/jbc.M103772200. [DOI] [PubMed] [Google Scholar]
- Yu, J.H., Yang, W.H., Gulick, T., Bloch, K.D., Bloch, D.B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaric, B., Chami, M., Remigy, H., Engel, A., Ballmer-Hofer, K., Winkler, F.K., Kambach, C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J. Biol. Chem. 2005;280:16066–16075. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R., Storz, G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Wassarman, K.M., Ortega, J., Steven, A.C., Storz, G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]