Abstract
Glycoprotein VI (GPVI) is an essential platelet receptor for collagens that is exclusively expressed in the megakaryocytic lineage. Transcription of the human gene GP6 is driven largely by GATA-binding protein 1 (GATA-1), specificity protein 1 (Sp1), and Friend leukemia integration 1 (Fli-1). In this report, we show that GPVI expression during megakaryocytic differentiation is dependent on cytosine-phosphate-guanosine (CpG) demethylation that can be initiated by thrombopoietin (TPO). Sodium bisulfite genomic sequencing established that a CpG-rich island within the GP6 promoter region is fully methylated at 10 CpG sites in GPVI-nonexpressive cell lines, such as UT-7/EPO and C8161, but completely unmethylated in GPVI-expressive cell lines, including UT-7/TPO and CHRF288-11. To further confirm the relationship between CpG demethylation and expression of GPVI in primary cells, we treated human cord blood cells with TPO. The GP6 promoter is highly methylated in cord blood mononuclear cells (progenitors) but not in CD41+-enriched cells obtained after TPO differentiation. Furthermore, when UT-7/EPO-Mpl cells, which stably express human C-myeloproliferative leukemia virus ligand (c-Mpl), were treated with TPO, demethylation of the GP6 promoter was induced. In every case, demethylation of the GP6 promoter correlated with an increase in mRNA level. Thus, megakaryocyte-specific expression of the GP6 gene is regulated, in part, by CpG demethylation, which can be directly initiated by TPO.
Introduction
GP6 is uniquely expressed by cells of the megakaryocyte lineage and encodes platelet glycoprotein VI (GPVI), which figures prominently in collagen-dependent activation and signal transduction.1 GP6 transcription requires a number of factors, such as GATA-binding protein 1 (GATA-1), Friend leukemia integration 1 (Fli-1), and specificity protein 1 (Sp1).2,3 However, the mere presence of these is not sufficient to initiate transcription during megakaryocyte differentiation, since all of these transcription factors are already present in megakaryocyte-erythrocyte progenitors that do not express GPVI. UT-7/TPO is a thrombopoietin (TPO)-dependent human erythro-megakaryocytic human cell line that has an absolute requirement for TPO to induce and maintain proliferation. GPVI is expressed in UT-7/TPO cells cultured in the presence of TPO, but expression declines rapidly (within hours) after TPO starvation. Since the expression of these essential transcription factors is not changed before or after TPO starvation, other mechanisms must be involved in the regulation of megakaryocyte-specific expression of GPVI, and TPO likely initiates these mechanisms.
Methylation of cytosines within the dinucleotide sequence cytosine-phosphate-guanosine (CpG) is probably the most common epigenetic mechanism of transcriptional suppression in vertebrates,4 and it is widely involved in the establishment and maintenance of cell type-specific gene expression.5,6 In this study, we show that TPO can initiate the reversal of CpG methylation during human megakaryocyte differentiation leading to the expression of GPVI.
Materials and methods
Sequence analysis
DNA sequences were obtained using an Applied Biosystems ABI Prism Model 377 DNA Sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) by personnel in the DNA Core Laboratory of the Department of Molecular and Experimental Medicine, The Scripps Research Institute.
Cultured human cell lines
The megakaryocyte cell line CHRF-288-117 was a gift from Dr M. A. Lieberman (Cincinnati, OH). The human cell line C8161 was purchased from The American Type Tissue Culture (Manassas, VA). For treatment with 5-aza-2′-deoxycytidine (5-aza-dC; Sigma, St Louis, MO), 6 × 105 cells were cultured in the presence of 10 μM 5-aza-dC in dimethyl sulfoxide (DMSO) for 48 hours.
The erythropoietin (EPO)-dependent cell line UT-7/EPO and the thrombopoietin-dependent cell line UT-7/TPO were established from bone marrow cells obtained from a patient with acute megakaryocytic leukemia.8,9 UT-7/TPO cells were maintained in liquid culture with Iscoves modified Dulbecco medium (IMDM; Invitrogen, San Diego, CA) containing 10% fetal calf serum (FCS) and 10 ng/mL recombinant human (rh) TPO. UT-7/EPO cells were maintained in the same culture conditions as UT-7/TPO cells, except for the addition of 1 IU/mL rhEPO; both UT-7/EPO and UT-7/TPO were kindly provided by the Kirin Brewery (Gunma, Japan).
Reverse transcriptase-polymerase chain reaction (RT-PCR)
RNAwas extracted using ISOGEN (Nippongene, Tokyo, Japan) according to the manufacturer's instructions. Total RNA was reverse transcribed using the GeneAmp RNA PCR kit (Applied Biosystems Japan, Tokyo, Japan). The PCR primers used were as follows: GPVI forward, 5′-CTCAGGACAGGGCTGAGGAA-3′; GPVI reverse, 5′-GGATGAAGAGGACTGCCTGA-3′; glyceraldehyde phosphate dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGT-3′; and GAPDH reverse, 5′-CTTCTACCACTACCCTAAAG-3′.
Bisulfite treatment of DNA samples
Genomic DNA (1 μg) was denatured in 50 μL 0.2-M NaOH at 37°C for 20 minutes and then mixed with 30 μL 10-mM hydroquinone (Sigma) and 520 μL 3-M sodium bisulfite (pH 5.0; Sigma). Reactants were incubated at 55°C for 16 hours. Bisulfite-modified DNA was purified using the Wizard purification resin and a Vacuum Manifold (Promega, Madison, WI) and eluted into 50 μL water. After addition of 5.5 μL 3-M NaOH (final 0.3 M), samples were let to stand at 37°C for 20 minutes, then precipitated in ethanol and dissolved in 20 μL water. A fragment of genomic DNA encompassing the CpG island of GP6 (-264 to +35) was amplified using the following primer pair: forward, 5′-GTGATATTAGGGAGTTTATGGGAGT-3′; and reverse, 5′-AAAACATAATTCCTCAACCCTATCC-3′.
For methylation-specific PCR, dmsp-R1 (5′-TTTCCTAATTAAAACTCATCAAACCA-3′) or msp-R2 (5′-CGACCTTTCCTAATTAAAACTCATCG-3′) was used in combination with the same forward primer described in the previous paragraph.
In the case of established cell lines, PCR products were directly sequenced; when primary mononuclear or CD41-enriched cells, or UT-7/EPO Mpl cells, were studied, the PCR products were gel purified using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) and cloned into the pGEM-T easy vector (Promega).
Serum-free liquid culture system for in vitro analysis of megakaryocytopoiesis
Culture was carried out as previously described, with modifications.10 Briefly, venous cord blood from the umbilical vessels of healthy, full-term infants was collected immediately after delivery and mixed with one quarter volume of 6% hydroxyethyl starch. Erythrocytes were allowed to settle at room temperature for 30 to 45 minutes. Mononuclear cells (MNCs) in the supernatant were then isolated using a Ficoll-Hypaque density gradient, washed, and resuspended for culture, as previously described.11 The culture system consisted of 1 × 106 MNCs/mL in Iscoves modified Dulbecco medium (Irvine Scientific, Irvine, CA), supplemented with 1% bovine serum albumin (BSA); 75 μM α-thioglycerol; 40 μg/mL each of linoleic acid, lecithin, and cholesterol; 1% Nutridoma Hu (Roche Boehringer Mannheim, Indianapolis, IN); and 30 ng/mL recombinant human thrombopoietin (rhTPO, 288-TPN-005; R&D Systems, Minneapolis, MN). The total number of suspension cells in culture was determined using a CELL-DYN 1600 multiparameter hematology analyzer (Abbott Laboratories, Abbott Park, IL). The day-8 cells were used for the CD41+ cell purification.
Isolation of CD41+ cells
CD41+ cells were isolated using Dynabeads Pan Mouse immunoglobulin G (IgG; Dynal Biotech, Lake Success, NY) and murine anti-CD41 monoclonal antibody (mAb, clone 5B12; DAKO, Carpinteria, CA), according to the manufacturer's instruction (Dynal Biotech). Briefly, Dynabeads Pan Mouse IgG and murine anti-CD41 mAb (0.5-l μg anti-CD41 mAb/107 beads) were incubated at 4°C for 30 minutes and washed. Cultured cells (4 × 107) were then incubated for 30 minutes at 4°C with the anti-CD41-coated beads, and those cells adherent to the beads were separated magnetically.
Isolation of DNA from mononuclear cells and enriched CD41+ cells
DNA from cord blood MNCs or CD41+-enriched cells was purified using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN), according to the manufacturer's instruction.
Measurement of surface expression of GPVI
Surface expression of GPVI was measured by flow cytometry using mouse anti-GPVI monoclonal antibody (204-11).12 Cells were incubated for 20 minutes at 4°C with the appropriately diluted antibody. After a first washing, the cells were incubated with a fluorescein-labeled second antibody for 20 minutes at 4°C. After a second washing, bound antibody was detected using a Facscalibur flow cytometer (Becton Dickinson, San Jose, CA), and data were reported as the geometric mean fluorescence intensity (GMFI).
Results and discussion
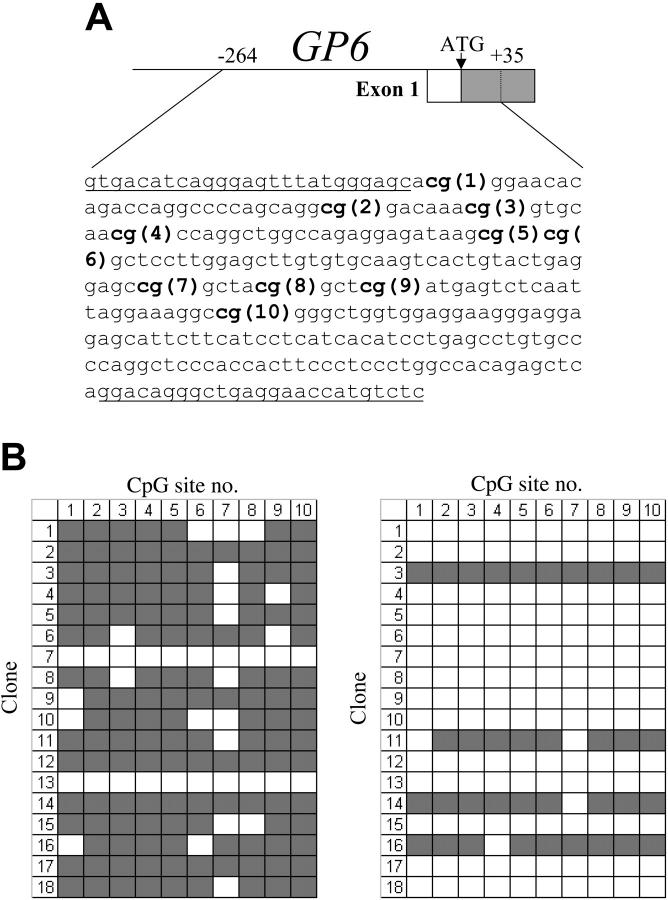
As shown in Figure 1A, the proximal 5′-regulatory region of GP6 contains 10 CpG sites within the segment from -238 to -90, which overlaps the core promoter. We used bisulfite sequencing to analyze the methylation status of these CpG sites in the GP6 promoter of DNA isolated from various established cell lines. Primers were designed that are complementary to sequences flanking this region and lacking CpG dinucleotides, in order to permit amplification of all genomic sequences, regardless of the status of methylation.
Figure 1.
(A) Schematic diagram showing the sequence encompassing the CpG islands within the proximal 5′-regulatory region of human GP6. The CpG sites are indicated in bold and numbered consecutively in the 5′ to 3′ direction. The sequences of the flanking primer pairs used to amplify each CpG-rich segment are underlined. (B) Methylation status of the GP6 CpG island in human cord blood mononuclear cells (left) or enriched CD41+ cells (right). In each grid, each CpG site is represented by a column and indicated at the top, while individual cloned DNA sequences are represented by rows and numbered to the left.  indicates methylated CpG sites; □, unmethylated sites. Numbers at the top correspond to the same CpG sites shown in panel A. In MNC precursors, 16 (89%) of 18 clones showed methylation at 7 or more of the 10 sites; and in the CD41+-enriched population, only 4 (22%) of 18 clones showed methylation at 8 or more sites.
indicates methylated CpG sites; □, unmethylated sites. Numbers at the top correspond to the same CpG sites shown in panel A. In MNC precursors, 16 (89%) of 18 clones showed methylation at 7 or more of the 10 sites; and in the CD41+-enriched population, only 4 (22%) of 18 clones showed methylation at 8 or more sites.
In GPVI-expressive cell lines, such as CHRF-288-11 and UT-7/TPO, these CpG sites are unmethylated, while the corresponding tract in genomic DNA of the GPVI-nonexpressive cell lines C8161 and UT-7/EPO is fully methylated (data not shown). These results establish that the expression of GPVI correlates with the CpG methylation status of the proximal 5′-regulatory region.
To obtain a more accurate evaluation of a role for CpG methylation in expression of these genes during megakaryocytic differentiation, we turned to the study of primary human megakaryocytes. We isolated mononuclear cells (MNCs) from human cord blood, cultured these in the presence of rhTPO for 8 days, and then selected the CD41+ cells. The methylation status of the GP6 promoter region of CpG islands was then compared between rhTPO-stimulated CD41+-enriched (megakaryocyte enriched) cells and the MNCs from which they were derived.
In the initial MNC population, megakaryocytes and progenitors may represent no more than 5% to 10% of the total cell population. The allelic methylation status is shown in Figure 1B. In MNCs, 16 of 18 clones showed substantial methylation (at least 7 of the 10 possible CpG sites), whereas the majority of DNA clones obtained from the CD41+-enriched population now contained unmethylated CpG sites (14 of 18). These results suggest that megakaryocyte differentiation is characterized by a decrease in CpG methylation of the GP6 proximal 5′-regulatory region. Because of the difficulty in obtaining adequate numbers of primary megakaryocytes in sufficient purity, the CD41+-enriched population we used contains a significant proportion of CD41- or CD41-low cells. Thus, it is highly likely that contamination by these cells contributes to the proportion of unmethylated CpG detected in the CD41+-enriched preparations.
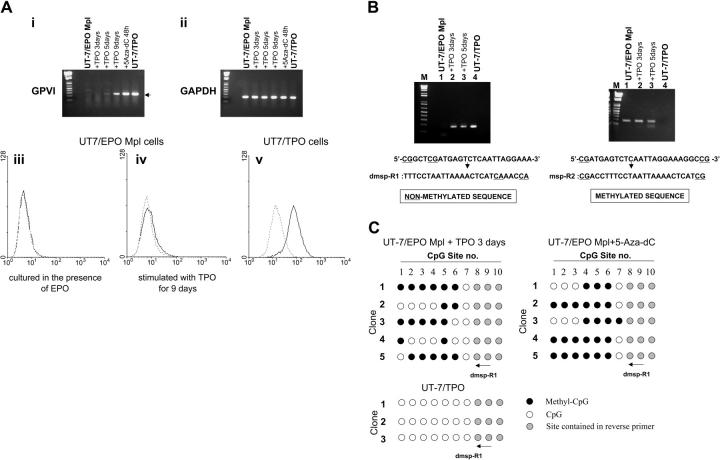
To directly show that CpG methylation plays an important role in the regulation of GPVI expression during megakaryocyte differentiation, we elected to differentiate a homogeneous non-megakaryocytic cell line with TPO. UT-7/EPO is an erythropoietin (EPO)-dependent human leukemic cell line that does not express endogenous c-Mpl. The related cell line UT-7/EPO-Mpl stably expresses exogenously introduced c-mpl cDNA and can be differentiated by TPO.13 We treated UT-7/EPO-Mpl with TPO and examined the expression of GP6 by RT-PCR (Figure 2A, upper panel). UT-7/EPO-Mpl cells do not express GPVI when they are grown in the presence of EPO. Expression of GPVI was detected when cells were treated with 5-aza-dC for 48 hours, which suggests that inactivation of GP6 in this cell line is caused by methylation. De novo expression of GPVI was also detected when the cells were cultured in the presence of TPO but not EPO for 5 days. We also analyzed surface expression of GPVI by flow cytometry and detected very low but reproducible expression of GPVI after TPO differentiation (Figure 2A, lower panel).
Figure 2.
(A, i-ii) TPO induced expression of GPVI on UT-7/EPO Mpl cells. Total RNA was extracted from cells treated with 10 ng/mL TPO for 3, 5, or 9 days or with 10 μM 5-aza-dC for 48 hours. GPVI (i) and GAPDH (ii) mRNA were then amplified by RT-PCR. By this method, GPVI mRNA was detected in cells treated with TPO for 5 or 9 days or in cells treated with 5-aza-dC. (iii-v) Surface expression of GPVI by UT-7/EPO Mpl cells treated with TPO, as determined by flow cytometry. — represents binding of the murine monoclonal antihuman GPVI antibody 204-11; --- corresponds to staining with fluorescein isothiocyanate-labeled secondary antibody alone. (iii) No binding was observed when UT-7/EPO Mpl are cultured in the presence of EPO. (iv) Weak, but reproducible, binding was observed following culture of UT-7/EPO Mpl in the presence of TPO for 9 days. (v) Intense binding is observed when UT-7/TPO are cultured in the presence of TPO. These results are representatives of identical findings made in 3 independent experiments. (B) Methylation-specific PCR of the GP6 promoter CpG island. Reverse primers were designed to distinguish the unmethylated (left) or methylated (right) DNA. The methylation-negative primer dmsp-R1 includes the eighth and ninth CpG sites converted to TG; in the methylation-positive primer msp-R2, the ninth CpG site is conserved as CG. The GP6 product (170 bp) is obtained using dmsp-R1 only when the cells are treated with TPO. (C) Methylation status of GP6 CpG sites in the product amplified with dmsp-R1 and shown in panel B. DNA was isolated from UT-7/EPO Mpl cells stimulated with TPO for 3 days or 5-aza-dC for 48 hours and treated with sodium bisulfite. PCR was performed with the same forward primer used in Figure 1B and dmsp-R1 used as the reverse primer. Amplified DNA was cloned into pGEM-T easy vector, and the methylation status was determined by DNA sequencing. • indicates methylated CpG sites; ○, unmethylated sites; and, sites contained within the reverse primers. Numbers at the top correspond to the same CpG sites shown in Figure 1A.
To confirm that TPO-induced expression of GPVI occurs via demethylation of the GP6 promoter, we next assessed the methylation status of the GP6 promoter region (Figure 2B). In this case, sodium bisulfite-treated genomic DNA was amplified with 2 kinds of reverse primers, which anneal specifically to either unmethylated (dmsp-R1) or methylated (msp-R2) sequences. We used these primers in order to more efficiently detect the small population of demethylated alleles that would otherwise have been obtainable by sequencing a large number of alleles amplified with the primers used in the experiments described in Figure 1B. By this approach, product is obtained with the dmsp-R1 primer (specific for the unmethylated sequence) only when UT-7/EPO-Mpl cells are cultured in the presence of TPO (Figure 2Bi). Nothing is detected when they are cultured in the presence of EPO. In contrast, a detectable product is obtained from UT-7/EPO-Mpl cells with the msp-R2 primer (specific for the methylated sequence) when these are cultured either in the presence of rhEPO or rhTPO (Figure 2Bii). At the same time, no product is obtained from UT-7/TPO cells.
Thus, we detected PCR product amplified with dmsp-R1 when UT-7/EPO Mpl cells were maintained in the presence of TPO. This product was cloned into the pGEM-T easy vector, and individual DNA clones were characterized by direct sequencing. As shown in Figure 2C, 43% of CpG sites in the 5′-regulatory region of GP6 were unmethylated in the alleles amplified with dmsp-R1. These results confirmed that TPO induced partial demethylation and expression of the GP6 gene in UT-7/EPO-Mpl cells. Although GPVI mRNA is maximally up-regulated when these cells are differentiated with TPO for 9 days, the methylation status is not significantly changed between 3 and 9 days of TPO differentiation. Additional factors, not expressed by this cell line, may be required for complete demethylation and full expression of GPVI.
The demethylation of the GP6 promoter and surface expression of GPVI initially observed in primary megakaryocytes (Figure 1B) was mimicked by these established UT-7 cell lines, but the extent of methylation/demethylation was not as dramatic as in the primary cells. Since UT-7/EPO was established from UT-7 by prolonged selection in the presence of EPO, it may well be that this subline is so extensively differentiated into an erythroid lineage that it is not possible to completely dedifferentiate and redifferentiate the cell line into a full megakaryocyte lineage.
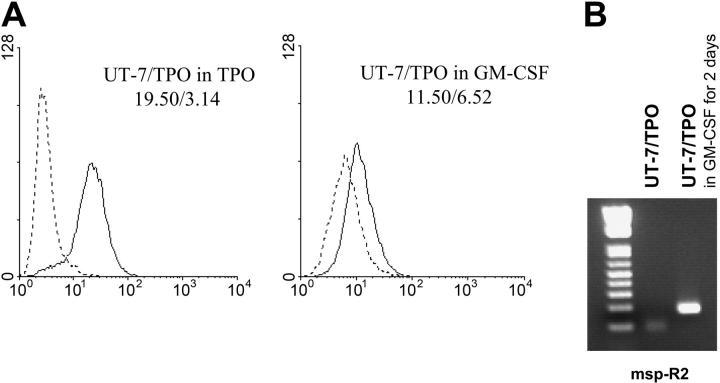
To distinguish whether demethylation of the GP6 promoter is directly linked to signal induction by TPO or merely a nonspecific result of the differentiation of these cells, we assayed the demethylation status of UT-7/TPO grown in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) for 2 days. As shown in Figure 3A, the surface expression of GPVI decreased following culture in the presence of GM-CSF. Using methylation-specific PCR, we also obtained a product with the msp-R2 primer only when UT-7/TPO cells were grown in the presence of GM-CSF (Figure 3B). This product was cloned into the pGEM-T easy vector for sequencing, and 17% of CpG sites were methylated in the alleles amplified with msp-R2 (data not shown). These results support the contention that demethylation and expression of GP6 are not an indirect effect of the differentiation state of the cells but a direct consequence of TPO signaling.
Figure 3.
Lack of effect of nonspecific differentiation upon demethylation. (A) Surface expression of GPVI by UT-7/TPO cells cultured in the presence of GM-CSF. UT-7/TPO cells were cultured in the presence of 1 ng/mL GM-CSF for 2 days, and surface expression of GPVI was analyzed by flow cytometry using antibody 204-11 (—). --- corresponds to staining with fluorescein isothiocyanate-labeled secondary antibody alone. The respective GMFI for each tracing is indicated in the respective panel, and these results are representative of 3 independent experiments. (B) Methylation-specific PCR of the GP6 promoter in UT-7/TPO cells cultured in the presence of GM-CSF. The reverse primer msp-R2 was used to specifically amplify methylated alleles. A product was obtained when UT-7/TPO were cultured in the presence of GM-CSF (right lane) but not when they were cultured in the presence of TPO (center lane).
Certain genes with restricted expression to hematopoietic cells, such as the genes for myeloperoxidase,14 globin,15 c-fms,16 and the granulocyte colony-stimulating factor (G-CSF) receptor,17 are regulated by methylation in a lineage- and differentiation-dependent manner. Megakaryocyte-specific gene induction, however, has not yet been associated with changes in promoter CpG methylation. As the CpG island is located in the core promoter of GP6 and some of CpG sites reside close to the binding sites for essential transcription factors, CpG methylation may directly interfere with the binding of transcriptional activators, or conversely, enhance the binding of transcriptional repressors.
Although TPO is the principal regulator of megakaryopoiesis and thrombocytopoiesis, it is not known whether it is directly involved in the mechanisms that regulate CpG methylation. Our results establish, for the first time, a role for TPO in dynamic changes in CpG methylation status that are involved in the epigenetic regulation of megakaryocyte-specific gene expression.
Acknowledgments
We thank Dr Jerry Ware (The Scripps Research Institute, La Jolla, CA) for his advice and support during these studies and Diana Rozenshteyn for her technical assistance. This is manuscript number 14045-MEM from The Scripps Research Institute.
Prepublished online as Blood First Edition Paper, February 8, 2005; DOI 10.1182/blood-2004-08-3109.
Supported by National Heart, Lung, and Blood Institute (NHLBI) grant HL46979 awarded to T.J.K.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Clemetson KJ, Clemetson JM. Platelet collagen receptors. Thromb Haemost. 2001;86: 189-197. [PubMed] [Google Scholar]
- 2.Furihata K, Kunicki TJ. Characterization of human glycoprotein VI gene 5′ regulatory and promoter regions. Arterioscler Thromb Vasc Biol. 2002;22: 1733-1739. [DOI] [PubMed] [Google Scholar]
- 3.Holmes ML, Bartle N, Eisbacher M, Chong BH. Cloning and analysis of the thrombopoietin-induced megakaryocyte-specific glycoprotein VI promoter and its regulation by GATA-1, Fli-1, and Sp1. J Biol Chem. 2002;277: 48333-48341. [DOI] [PubMed] [Google Scholar]
- 4.Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99: 4503-4508. [DOI] [PubMed] [Google Scholar]
- 5.Futscher BW, Oshiro MM, Wozniak RJ, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31: 175-179. [DOI] [PubMed] [Google Scholar]
- 6.Uhl J, Klan N, Rose M, et al. The 5-lipoxygenase promoter is regulated by DNA methylation. J Biol Chem. 2002;277: 4374-4379. [DOI] [PubMed] [Google Scholar]
- 7.Fugman DA, Witte DP, Jones CLA, Aronow BJ, Lieberman MA. In vitro establishment and characterization of a human megakaryoblastic cell line. Blood. 1990;75: 1252-1261. [PubMed] [Google Scholar]
- 8.Komatsu N, Kunitama M, Yamada M, et al. Establishment and characterization of the thrombopoietin-dependent megakaryocytic cell line, UT-7/TPO. Blood. 1996;87: 4552-4560. [PubMed] [Google Scholar]
- 9.Komatsu N, Yamamoto M, Fujita H, et al. Establishment and characterization of an erythropoietin-dependent subline, UT-7/Epo, derived from human leukemia cell line, UT-7. Blood. 1993;82: 456-464. [PubMed] [Google Scholar]
- 10.Chang M, Nakagawa PA, Williams SA, et al. Immune thrombocytopenic purpura (ITP) plasma and purified ITP monoclonal autoantibodies inhibit megakaryocytopoiesis in vitro. Blood. 2003;102: 887-895. [DOI] [PubMed] [Google Scholar]
- 11.Chang M, Suen Y, Lee SM, et al. Transforming growth factor-beta 1, macrophage inflammatory protein-1 alpha, and interleukin-8 gene expression is lower in stimulated human neonatal compared with adult mononuclear cells. Blood. 1994;84: 118-124. [PubMed] [Google Scholar]
- 12.Moroi M, Mizuguchi J, Kawashima S, et al. A new monoclonal antibody, mAb 204-11, that influences the binding of platelet GPVI to fibrous collagen. Thromb Haemost. 2003;89: 996-1003. [PubMed] [Google Scholar]
- 13.Takatoku M, Kametaka M, Shimizu R, Miura Y, Komatsu N. Identification of functional domains of the human thrombopoietin receptor required for growth and differentiation of megakaryocytic cells. J Biol Chem. 1997;272: 7259-7263. [DOI] [PubMed] [Google Scholar]
- 14.Lubbert M, Miller CW, Koeffler HP. Changes of DNA methylation and chromatin structure in the human myeloperoxidase gene during myeloid differentiation. Blood. 1991;78: 345-356. [PubMed] [Google Scholar]
- 15.Enver T, Zhang JW, Anagnou NP, Stamatoyannopoulos G, Papayannopoulou T. Developmental programs of human erythroleukemia cells: globin gene expression and methylation. Mol Cell Biol. 1988;8: 4917-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felgner J, Kreipe H, Heidorn K, et al. Lineage-specific methylation of the c-fms gene in blood cells and macrophages. Leukemia. 1992;6: 420-425. [PubMed] [Google Scholar]
- 17.Felgner J, Heidorn K, Korbacher D, Frahm SO, Parwaresch R. Cell lineage specificity in G-CSF receptor gene methylation. Leukemia. 1999;13: 530-534. [DOI] [PubMed] [Google Scholar]