Abstract
Meiosis is the developmental program by which diploid organisms produce haploid gametes capable of sexual reproduction. Here we describe the yeast gene AMA1, a new member of the Cdc20 protein family that regulates the multisubunit ubiquitin ligase termed the anaphase promoting complex/cyclosome (APC/C). AMA1 is developmentally regulated in that its transcription and splicing occur only in meiotic cells. The meiosis-specific processing of AMA1 mRNA depends on the previously described MER1 splicing factor. Several results indicate that Ama1p is required for APC/C function during meiosis. First, coimmunoprecipitation assays indicate that Ama1p associates with the APC/C in vivo. Second, Ama1p is required for the degradation of the B-type cyclin Clb1p, an APC/C substrate in both meiotic and mitotic cells. Third, ectopic overexpression of AMA1 is able to stimulate ubiquitination of Clb1p in vitro and degradation of Clb1p in vivo. Mutants lacking AMA1 revealed that it is required for the first meiotic division but not the mitotic-like meiosis II. In addition, ama1 mutants are defective for both spore wall assembly and the expression of late meiotic genes. In conclusion, this study indicates that Ama1p directs a meiotic APC/C that functions solely outside mitotic cell division. The requirement of Ama1p only for meiosis I and spore morphogenesis suggests a function for APC/CAma1 specifically adapted to germ cell development.
Gametogenesis requires the execution of several interrelated events including genetic exchange, haploidization, and cellular differentiation. Haploidization is achieved through two consecutive nuclear divisions, meiosis I (reductional) and meiosis II (equational). During the reductional division, replicated sister chromatids stay attached and segregate as a single unit to the same pole. The second meiotic division resembles mitosis in that the centromeres of replicated sisters bind to spindles emanating from opposite poles and separate at anaphase II. Finally, during gametogenesis, differentiation programs instruct the formation of specialized cells that are capable of sexual reproduction. In yeast, the haploid products are encapsulated in spores, which have the capacity to mate after they germinate and reenter the mitotic cell cycle (1).
Several studies have indicated that the basic mitotic cell cycle machinery is required for many aspects of meiosis (reviewed in ref. 2). For example, the budding yeast mitotic cell cycle is driven by the cyclin-dependent protein kinase Cdc28p (3). Cdc28p is activated by a conserved family of proteins termed cyclins (4) with the four B-type cyclins (Clb1-4p) regulating the G2/M transition. Similarly, the normal execution of meiosis I and II also requires the Cdc28p-Clb1p and Cdc28p-Clb4p kinases (5–7). However, the production of haploid products during meiosis requires two events that are strictly prohibited by mitotic checkpoint pathways (8). First, replicated sister chromatids stay paired during meiosis I rather than segregate to the opposite poles as they do in mitosis. Second, haploidization requires the execution of two chromosome divisions without an intervening S phase. These differences suggest the existence of meiosis-specific regulators able to use the basic cell cycle machinery to permit haploidization.
The mitotic metaphase/anaphase transition and exit from mitosis are triggered by the sequential destruction of the anaphase inhibitor Pds1p (9, 10) and the B-type cyclins (11, 12), respectively. This temporal proteolytic control is directed by the multisubunit ubiquitin ligase termed the anaphase promoting complex/cyclosome (APC/C) (13). In mitotic yeast cells, APC/C specificity is provided by two Cdc20 family members (Cdc20p and Hct1p/Cdh1p; reviewed in ref. 14). The Cdc20p-APC/C complex (APC/CCdc20) directs the degradation of Pds1p (9, 15) whereas APC/CHct1 selectively targets Clb2p (16).
The present study provides evidence for a meiosis-specific APC/C in yeast. We identified AMA1, a new member of the Cdc20 family of proteins that is transcribed and spliced only in meiotic cells. Ama1p associates with the APC/C in vivo and is required for the destruction of the B-type cyclin Clb1p but not the anaphase inhibitor Pds1p. Mutational studies revealed that Ama1p is required for the meiosis I reductional division and spore formation but not for meiosis II. A meiosis-specific regulator of the core APC/C complex may be an example of how the mitotic cell machinery can be modified to direct the unique nuclear divisions associated with meiosis.
Experimental Procedures
Strains/Plasmid Constructions.
The strains used in this study are derivatives of homozygous diploid RSY335 (MATa/MATα cyh2R-z ho∷LYS2 leu2∷hisG lys2 trp1∷hisG ura3-1). All disruption alleles or epitope tags were constructed by using PCR-based gene replacement (17) unless otherwise specified. The GAL1-AMA1 genomic DNA (pGAL1-AMA1g) and cDNA (pGAL1-AMA1c) expression constructs were tagged with the T7/6HIS epitope using pGALSET351 (18). The expression from these constructs was induced by adding 2% galactose to raffinose grown cultures. The high-copy AMA1 plasmid (pAMA1-HC) was constructed by inserting the NsiI AMA1 genomic fragment into the PstI site of pRS424 (19). The ENO1-AMA1 fusion plasmid (pENO1-AMA1) was constructed by inserting the ENO1 cDNA and the CYC1 terminator into the high-copy plasmid pJS21-C (20). The CLB1 destruction box (21) was mutated (RxxL to AxxA) by using oligonucleotide-directed mutagenesis (22). All introduced mutations were verified by DNA sequence, and their functionality was determined by complementation assays. The myc-tagged Ume3p expression construct (pKC337) was described previously (23). O. Cohen-Fix (National Institutes of Health) provided the epitope-tagged Pds1p-HA expression construct (pOC40).
Meiotic Timecourse Experiments.
The meiotic timecourse experiments were conducted as described (23) except for cdc16-1 cultures. They were maintained at 23°C for 3 h after transfer to sporulation medium to allow the cells to exit mitosis and enter the meiotic program before shifting the culture to the nonpermissive temperature (34°C). In this strain background, meiosis I is completed by 9 h and meiosis II by 12–15 h. Total RNA was prepared as described (24) from samples taken at times after the transfer to sporulation medium as indicated. Reverse transcription (RT)-PCR was performed on the RNA samples using the Titan One Tube System (Roche) as suggested by the manufacturer. Ume3p-myc was detected in immunoprecipitates as previously described (23). Pds1p-hemagglutinin (HA) and Clb1p-3HA were visualized by Western analysis of 50 μg soluble protein. For Ama1p and Cdc16p coimmunoprecipitation studies, 1 mg of soluble extract was used for each sample. Western blot signals were detected using goat anti-mouse secondary antibody conjugated to alkaline phosphatase (Sigma) and the CDP-Star chemiluminescence kit (Tropix, Bedford, MA).
In Vitro Ubiquitination Assays.
The in vitro ubiquitin assays were performed essentially as described (25) with the following modifications. The extracts were prepared from hydroxyurea (HU)-arrested cultures (3 h at 23°) harboring either pENO1-AMA1 or the vector alone. The cells were lysed using zymolyase 100T (167 μg/ml, Seikagaku America, Rockville, MD). The Clb1p substrate was added (2 μl of 30 mg/ml) from extracts prepared from galactose grown cultures overexpressing GAL1-CLB1-T7/His6. Additional ubiquitin activating (E1) and conjugating (E2) enzymes were supplied from concentrated extracts prepared from a cdc16 culture harvested at the restrictive temperature to inactivate the endogenous APC/C (13).
Cell Imaging Protocols.
Spindle morphology was determined by indirect immunofluorescence as described (26). Chromosome behavior was analyzed in the spo13-1 ama1Δ double mutant using the insertion of tandem Tet operators and the Tet repressor-green fluorescent protein (GFP) fusion protein as described (27). Quantitation of meiosis I and II completion was obtained by direct counting of at least 200 4′,6-diamidino-2-phenylindole (DAPI)-stained cells.
Results
AMA1 Encodes a Developmentally Regulated Cdc20 Family Member.
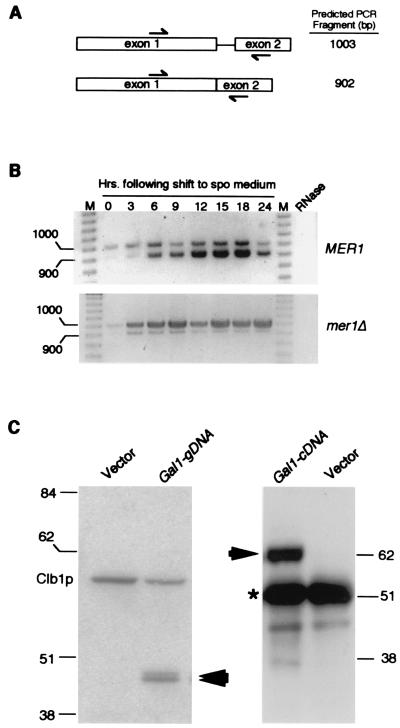
The AMA1 gene was identified as a low-copy suppressor of the rut2 mutation (for regulator of ume3p turnover) identified in this laboratory. Ume3p (Srb11p/Ssn8p) is the yeast C-type cyclin that, along with its cyclin-dependent kinase partner Ume5p, negatively regulates diverse gene sets required for meiosis [e.g., SPO13 (24, 28)] and the stress response [e.g., SSA1 (23)]. To relieve Ume3p-Ume5p-dependent repression, the cyclin is destroyed early in meiosis or in response to stress (23, 29). The rut2 mutant fails to destroy Ume3p in response to heat shock and displays a meiotic defect (unpublished results). Sequence analysis revealed that AMA1 contains SPO70, a previously reported meiotic gene (30). However, the SPO70 ORF did not contain suppressor activity (data not shown). Further inspection of the genomic sequence revealed consensus splice signals, suggesting that SPO70 was actually exon 1 of a spliced gene. This possibility was confirmed by RT-PCR analysis that was able to distinguish spliced from nonspliced transcripts (Fig. 1 A and B). Moreover, this processing requires the meiosis-specific splicing protein Mer1p (31) (Fig. 1B).
Figure 1.
AMA1 exhibits meiosis-specific transcription and splicing. (A) The AMA1 genomic locus. The arrows indicate the primer locations and predicted sizes of RT-PCR reactions with spliced and unspliced transcripts. Exon 1 contains the ORF described for SPO70 (29). (B) AMA1 is transcribed and spliced during meiosis. The RT-PCR products generated from total RNA samples taken from the wild-type and (MER1) and mutant (mer1Δ) culture during vegetative growth (0 h) and at the times indicated after transfer to sporulation medium. The molecular weight standards (M) in base pairs are indicated. The signals observed are specific for AMA1 mRNA as they are absent when the template RNA is treated with RNase A. (C) Ama1p is not synthesized in vegetative cells. Western blot analysis of soluble extracts (25 μg) or immunoprecipitates (250 μg) prepared from a wild-type strain harboring either the pGAL1-AMA1g (genomic DNA) or pGAL-AMA1c (cDNA), respectively. Ama1p-specific bands are indicated by the arrows. A 6HIS/T7 epitope-tagged Clb1p derivative served as an internal size standard and loading control. The vector lanes control for nonspecific cross hybridization of the antibody. Molecular weight standards (kDa) are given on the sides of the gels. The asterisk denotes the heavy chain present in the immunoprecipitates required for visualizing full-length Ama1p.
To determine whether the splicing reaction is restricted to meiotic cells, the genomic sequence (gDNA) and the cDNA were placed under the control of the GAL1 promoter to permit regulated expression in vegetative cells. Ectopic expression of the gDNA produced a protein the size predicted for SPO70 [43 kDa (30)], whereas the cDNA produced a 63-kDa protein indicating that the spliced mRNA is fully translated (Fig. 1C). Sequence analysis of the AMA1 cDNA revealed a gene encoding a 63.7-kDa protein that is 34% identical to Cdc20 (Fig. 2). Based on these findings and the results described below, this spliced gene has been named AMA1, for activator of meiotic APC/C.
Figure 2.
Ama1p is a Cdc20p family member. The predicted protein sequences for the AMA1 cDNA and other Cdc20 family members are presented. The arrows indicate the exon 1-exon 2 junction. Positions of predicted WD repeats are indicated by the solid triangles above the sequence. The black and gray shaded regions signify identical residues and conservative substitutions, respectively. The underlined sequence indicates additional amino acids identified by our sequence analysis that differ from the yeast genome project. Cdc20 family members representing human [55CDC (54)], Drosophila, [fzy2 (55)], and two from budding yeast [Hct1p/Chd1p (15, 16) and Cdc20p (56)] are presented.
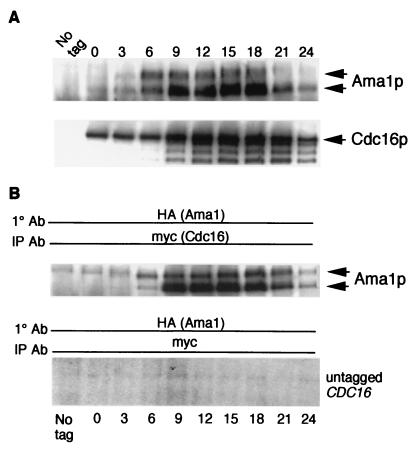
Ama1p Associates with the APC/C in Vivo.
APC/C activation requires the direct association of Hct1p or Cdc20p (32). To determine whether Ama1p binds the APC/C, coimmunoprecipitation studies were conducted. A strain containing chromosomally tagged versions of Ama1p (AMA1-HA) and Cdc16p (CDC16-myc), a core APC/C component (33), was induced to enter meiosis, and samples were taken at various timepoints. Western blots of immunoprecipitates prepared from each timepoint confirmed the synthesis of each tagged protein and revealed that the levels of both proteins peak at approximately the time of the meiotic divisions (12–15 h) (Fig. 3A). The Cdc16p-myc immunoprecipitation blot was stripped and reprobed for the presence of Ama1p-HA. Ama1p-HA was detected in the Cdc16-myc immunoprecipitates, indicating that these two proteins interact in vivo (Fig. 3B). The presence of Ama1p-HA in the Cdc16p-myc immunoprecipitates depended on the tagged CDC16 allele. Furthermore, similar to Cdc20p during mitotic cell division (15, 34), Ama1p binds the APC/C continuously throughout meiotic development.
Figure 3.
Ama1p associates with the APC/C in vivo. (A) Extracts were prepared from a meiotic timecourse of a wild-type diploid harboring chromosomally tagged AMA1-HA and CDC16-myc. Ama1p-HA (Upper) and Cdc16-myc (Lower) accumulation in these extracts was determined by immunoprecipitation and Western blot analysis. Meiosis I and II were completed by 9 and 12 h, respectively. (B) Ama1p associates with Cdc16p in vivo. The Cdc16p-myc blot shown in A was stripped and probed for Ama1p-HA. Ama1p-specific bands are indicated by the arrows (Upper). No Ama1p-HA was detected in myc immunoprecipitates from a strain lacking the CDC16-myc allele (Lower).
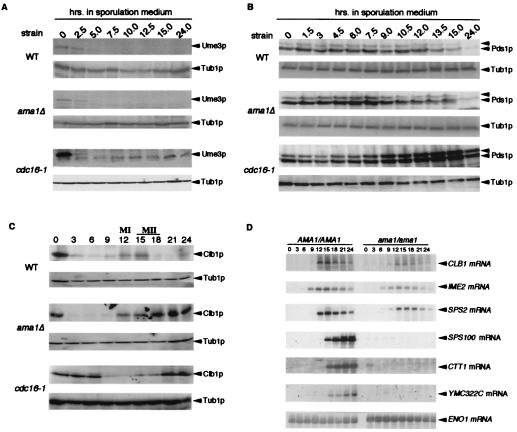
Ama1p Is Required for Destruction of Clb1p in Meiotic Cells.
If Ama1p activates the APC/C in meiosis, then it should be required for the degradation of meiotic regulatory proteins. Therefore, we examined the levels of Ume3p, Clb1p, and Pds1p in diploids deleted for the first exon of AMA1 (ama1Δ). As described previously (23), Western blot analysis revealed that Ume3p is destroyed as cells enter meiosis (Fig. 4A). However, no difference in the degradation kinetics was observed in the ama1Δ or cdc16-1 mutant extracts, indicating that Ume3p destruction is independent of both Ama1p and the APC/C. Next, the involvement of Ama1p in regulating Pds1p was tested. In the wild-type strain, Pds1p levels increased gradually until the completion of meiosis I (12 h) and then declined as the cells completed the second division (15 h; Fig. 4B). The Pds1p accumulation profile was similar in wild-type and ama1Δ mutant cells. However, Pds1p was stabilized in the cdc16 mutant, indicating that, similar to vegetative cells, the APC/C is required for Pds1p destruction during meiosis. These findings suggest that another Cdc20 family member is involved in targeting Pds1p for destruction in meiotic cells.
Figure 4.
Ama1p is required for Clb1p degradation during meiosis. Wild-type (WT), ama1Δ, or cdc16-1 mutants harboring plasmids expressing Ume3p-myc (pKC337), Pds1p-HA (pOC40), or chromosomally integrated CLB1-3HA allele were induced to enter meiosis and timepoints taken as indicated. Western blot analyses of protein extracts (see Experimental Procedures) were conducted to detect the proteins indicated (arrows, A–C). MI and MII indicate the approximate times of meiosis I (MI) and meiosis II (MII) completion in wild-type strains as determined by DAPI analysis. Tub1p serves as protein loading control. (D) Northern blot analysis from the timecourse described in C. Northern blots were probed for the genes indicated. IME2 represents the early expression class, CLB1 and SPS2 middle genes and SPS100, CTT1, YMC322C are late genes. ENO1 served as a loading control.
Finally, we assessed the requirement of Ama1p for Clb1p destruction. As observed previously (5, 6), Clb1p levels rapidly declined as the cells entered the meiotic program and then rebounded transiently during meiosis I and meiosis II (12–15 h; Fig. 4C). In the ama1Δ mutant, Clb1p levels declined normally as the culture entered meiosis but remained elevated after meiotic induction. Similar results were obtained with the cdc16-1 strain, although initial Clb1p degradation was somewhat delayed in this strain. This finding is probably due to the relatively slow entry of cdc16-1 mutants into the meiotic program as determined by mRNA expression profiles (data not shown). The increased level of Clb1p in the ama1Δ mutant was not due to enhanced mRNA levels as determined by Northern blot analysis (Fig. 4D). These findings indicate that changes in Clb1p levels in the ama1Δ mutant occurred posttranscriptionally, consistent with a role for Ama1p in protein turnover. We conclude that Clb1p is a substrate for the APC/C in meiotic cells and that its destruction requires Ama1p.
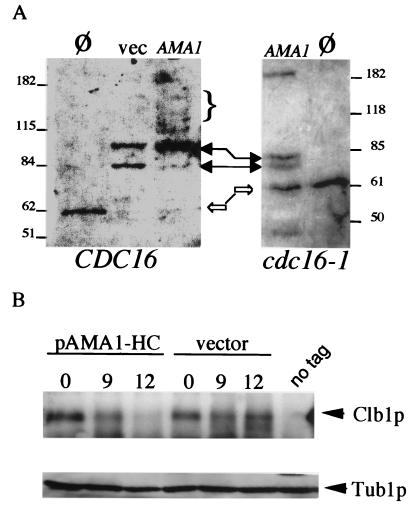
To further investigate the requirement of Ama1p for Clb1p destruction, two approaches were taken. We first examined the ability of ectopically expressed Ama1p to stimulate ubiquitination of Clb1p in vitro. A transformant harboring the ENO1-AMA1 cDNA expression construct was arrested in S phase by HU to inactivate the APC/C controlled by Cdc20p and Hct1p (16, 34). Extracts were prepared and incubated with Clb1p in the presence of free ubiquitin. A background level of ubiquitination was observed in the vector control extracts (Fig. 5A, solid arrows), which is probably due to incomplete HU inactivation of APC/C or residual activity from concentrated E1/E2 supplemental extracts (see Experimental Procedures). Extracts prepared from cultures expressing the ENO1-AMA1 cDNA produced two modest changes in Clb1p mobility. First, Clb1p was chased into the slower migrating background ubiquitination species. Second, a low, but detectable level of higher molecular weight forms of Clb1p was observed (Fig. 5A, bracket). These two effects were dependent on a functioning APC/C, because they were absent in extracts prepared from a cdc16 strain-expressing ENO1-AMA1 (Fig. 5A Right). These results suggest that Ama1p is able to direct the ubiquitination of Clb1p.
Figure 5.
Ama1p regulates Clb1p in vitro and in vivo. (A) Ama1p stimulates Clb1p ubiquitination in vitro. Extracts prepared from HU-arrested transformants harboring either the AMA1 cDNA expression plasmid (pENO1-AMA1) or the vector control (vec) were incubated with immunoprecipitated Clb1p-HA. The reactions were subjected to Western blot analysis probing for HA. Clb1p mobility without added extract (Ø) is indicated by the open arrow. Solid arrows indicate changes in Clb1p mobility by APC/C complex activity presumably because of incomplete HU arrest (32). The bracket indicates slower migrating species of Clb1p in pENO1-AMA1 overexpression extracts. These experiments were repeated in extracts prepared from cdc16-1 mutants (Right). The high molecular weight band in the pENO1-AMA1 lane results from the occasional retention of Clb1p in the wells of the gel. The molecular weight markers (kDa) are indicated on the outside of each panel. (B) Overexpression of AMA1 down regulates Clb1p in vivo. A wild-type diploid harboring either AMA1 on a high-copy plasmid (pAMA1-HC) or the vector control was induced to enter meiosis. Timepoints were taken before (0 h) and at times representing peak Clb1p expression periods. Clb1p and the Tub1p (loading control) signals are indicated adjacent to the lanes.
A caveat to this conclusion is that, for reasons that are unclear, the in vitro ubiquitination assays were difficult to perform reliably. To further test our hypothesis that Ama1p directly influences Clb1p levels, we took advantage of previous studies that found that overexpression of CDC20 or HCT1 increased degradation of their respective substrates in vivo (15, 32). To overexpress Ama1p, the gDNA was placed on a high-copy plasmid (pAMA1-HC) and introduced into a wild-type diploid strain. This strain and the vector control were induced to enter meiosis, and timepoints were taken during peak Clb1p expression. In the vector control, Clb1p was detected by 9 h after the shift to sporulation medium, and high levels were maintained through the 12 h timepoint (Fig. 5B). However, Clb1p levels were slightly reduced in the 9 h timepoint in the presence of pAMA1-HC and barely detectable by 12 h. Identical results were obtained when these experiments were repeated using a complete timecourse (data not shown). Overexpression of AMA1 did not alter sporulation kinetics or efficiency, indicating that this reduction in Clb1p levels did not significantly impede normal development. These findings, together with the in vivo association of Ama1p and Cdc16p, the stabilization of Clb1p in ama1 mutants, and the in vitro ubiquitination studies, argue that Ama1p is able to specifically target the degradation of Clb1p through the APC/C.
Ama1p Is Required for the Execution of Meiosis I.
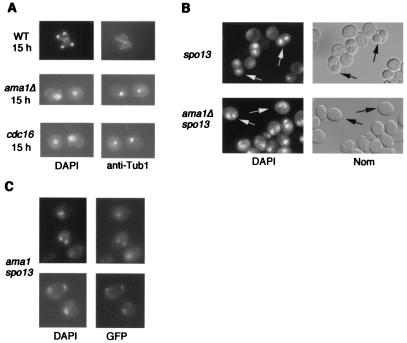
To investigate the requirement of AMA1 for meiosis, wild-type and isogenic ama1Δ cultures were induced to undergo synchronous meiotic divisions. FACS analysis demonstrated that both strains replicated their DNA with similar efficiency (data not shown). The analysis of nuclear divisions and spindle morphology indicated that, by 15 h, 75% of the wild-type cells contained four individually staining nuclei with elongated spindles diagnostic of the execution of meiosis I and meiosis II (Fig. 6A Top). However, by the same timepoint, 83% of the cells in the ama1Δ culture contained a single nucleus with a short spindle, indicating a failure to execute either nuclear division (Fig. 6A Middle). The mononucleated/short spindle phenotype persisted even after 24 h (data not shown), indicating that the arrest phenotype was terminal and not due to slowed meiotic kinetics. Cells displaying a single nucleus with an elongated spindle indicative of anaphase I were observed in only 4.5% of the mutant population. Of the ama1Δ mutants exhibiting normal appearing multinucleated cells (7.7%), no spore walls were detected (see below). A similar arrest phenotype was observed for cdc23-1 (data not shown) and cdc16-1 mutants (Fig. 6A Bottom). These results indicate that Ama1p and the APC/C are required for the execution of the first meiotic division. The similarity in phenotypes exhibited by ama1 and cdc16 strains is consistent with our model that Ama1p activates a meiotic APC/C required for the first meiotic division.
Figure 6.
AMA1 is required for meiotic development. (A) Ama1p is required for meiosis I. Wild type (WT), ama1Δ, or cdc16-1 diploids were induced to enter meiosis and timepoints taken as indicated (h). The nuclei and spindles were visualized by DAPI staining or by indirect immunofluorescence using Tub1p antibodies, respectively (×1,000 final magnification). (B) Ama1p is not required for meiosis II. The spo13 single or ama1Δ spo13 double mutant was induced to enter meiosis then harvested after 24 h. Dyads (two spored asci) in the spo13 mutant (Upper) were visualized by Nomarski optics (Nom) or DAPI staining (arrows). Binucleated ama1 spo13 cells were identified by DAPI staining (arrows, Lower). (C) The ama1 spo13 mutant performs a single equational division. One homolog of chromosome V was marked with a tandem array of the Tet repressor. This chromosome was visualized by expressing the Tet repressor-GFP fusion protein and fluorescent microscopy.
Ama1p Is Not Required for the Meiosis II Chromosome Division.
To investigate whether Ama1p is also required for meiosis II, we took advantage of the spo13-1 mutation, which allows cells to bypass meiosis I but still execute meiosis II (35). This mutant forms two diploid spores (dyads) rather than the normal four haploid products (Fig. 6B Upper). An ama1Δ spo13-1 double mutant diploid was constructed and induced to enter meiosis. This strain produced binucleated cells with the same efficiency as the spo13 strain (54% and 50%, respectively) but still did not produce spores (Fig. 6B Lower). These results suggest that Ama1p is essential for the first meiotic division and spore wall assembly but not for meiosis II. To verify that the ama1Δ spo13-1 double mutant did indeed execute meiosis II, the Tet repressor-GFP system (27) was introduced into the ama1Δ spo13-1 strain to mark one copy of chromosome V. The analysis of binucleated meiotic cells revealed the presence of two GFP signals diagnostic for the meiosis II equational division (Fig. 6C). These findings confirm that Ama1p is dispensable for the second division.
Ama1p Is Required for Late Meiotic Gene Expression and Spore Formation.
The finding that the ama1Δ spo13-1 double mutant was able to undergo meiosis II but still displayed a defect in spore formation suggests that this defect is not simply a consequence of a meiosis I arrest. To further explore this issue, the mRNA levels of SPS100, a late expressing gene required for spore wall maturation (36), was examined in an ama1Δ mutant. Northern blot analysis revealed that SPS100 mRNA levels were below the limits of detection in the ama1Δ mutant (Fig. 4D). To determine whether this transcription defect was specific to SPS100 or affected other members of the late expression class, the mRNA levels of catalase (CTT1) and a predicted ORF (YMC322C) were examined. Both genes were identified in a microarray study (30) as late meiotic genes, which we confirmed (Fig. 4D). As observed for SPS100, both genes suffered a severe reduction in transcript accumulation, indicating that Ama1p is required for late meiotic gene expression. The expression of middle, but not late, genes suggested the possibility that the ama1 mutant is able to perform the initial steps in spore wall formation. This hypothesis is supported by ultrastructural studies that revealed prospore membrane deposition but not the mature spore (see supplemental Fig. 7, which is published as supplementary data on the PNAS web site, www.pnas.org).
Discussion
This report describes the isolation and analysis of Ama1p, a developmentally regulated member of the Cdc20 family of APC/C activators. Specifically, AMA1 transcription and splicing are restricted to meiotic cells. We also demonstrate that Ama1p is required for meiosis I, expression of the late class of meiotic genes, and spore morphogenesis. Finally, we present four pieces of data supporting a role for Ama1p in activating a meiotic APC/C. First, its amino acid sequence clearly places Ama1p into the Cdc20 protein family. Second, Ama1p is able to associate with a core component of the APC/C in vivo. Third, ama1Δ mutants arrest in meiosis with a phenotype similar to strains lacking the APC/C core component Cdc16p. Finally, Ama1p is required for the destruction of the B-type cyclin Clb1p, which we demonstrate is a target of a meiotic APC/C. Taken together, these results describe a specialized activator of the APC/C that controls the first meiotic division.
It is curious that the in vitro ubiquitination studies produce such variable results depending on the APC/C complex examined. APC/CHct1 readily ubiquitinates Clb2p in vitro as do several ligases derived from clam, Xenopus, and human extracts (32, 37, 38). However, APC/CCdc20 has not been shown to direct ubiquitination of Pds1p, although genetic studies clearly show its involvement in this process (15, 34). Similarly, we found APC/CAma1 ubiquitination activity toward Clb1p difficult to obtain. It is unclear why the in vitro APC/C activity varies depending on the associating Cdc20 family member. The finding that Cdc20p destruction is also dependent on the APC/C (39) may suggest that these complexes inactivate themselves under the proper in vitro conditions. We do not know whether Ama1p is a substrate for the APC/C, although it does contain destruction box motifs. A better understanding of both APC/C activation and inactivation may shed light on this issue.
Ama1p Is Required for Meiosis I.
Mutants lacking AMA1 complete DNA synthesis and recombination but arrest with a single nucleus and a short spindle. This phenotype is consistent with ama1 mutants arresting in late prophase or early metaphase. A similar phenotype is also observed when meiotic checkpoint pathways are activated (40–42). Two pieces of data argue that the ama1 arrest is not due to the activation of known checkpoint mechanisms. First, deleting RAD17 or MAD2, components of the DNA damage and spindle checkpoint pathways, respectively (40, 43), does not relieve the ama1 arrest (data not shown). In addition, it has been demonstrated that checkpoint activation eliminates transcription of the middle expression class of meiotic genes (44, 45). However, SPS2 and CLB1, two middle genes, are still transcribed with normal kinetics albeit at lower levels (Fig. 4D). Therefore, similar to mitotic APC/C mutants, the ama1 mutant arrest appears checkpoint independent. A more likely scenario based on studies in mitotically dividing cells is that APC/CAma1 is a target of checkpoint pathways. This possibility is currently being examined.
This study has demonstrated that the B-type cyclin Clb1p is destroyed through an Ama1p-dependent mechanism. Clb1p stabilization is not, however, the sole underlying reason for the meiotic defect associated with ama1Δ mutations. Neither expressing high levels of Clb1p throughout meiosis or introducing a nondegradable version of Clb1p in which the destruction box had been mutated caused a meiotic arrest (data not shown). This result was not too surprising given the finding in vegetative cells that continued expression of B-type cyclins in hct1 mutants does not confer mitotic arrest (16). Rather, the hct1 mutation, in combination with a mutation of the CDK inhibitor Sic1p, is required to induce cell cycle arrest because of elevated Cdc28 activity at the G2/M boundary (16, 46). Does this explanation hold true in meiosis as well? The analysis of both protein and transcript levels indicates that Sic1p is not present in meiotic cells undergoing either division (refs. 30 and 47, and our unpublished results). Although this possibility has not been directly tested, these expression studies argue against a role for Sic1p in down-regulating Cdc28-Clb activity during the meiotic divisions.
Activation of APC/CAma1.
In mitotic cells, Cdc20p expression is periodic, peaking in G2. Once synthesized, Cdc20p binds the APC/C core complex, but this event alone is insufficient for activation (39). In clam and Xenopus extracts, phosphorylation of APC/CCdc20 by cyclin B-Cdc2 is necessary for activation of the ubiquitin ligase (37, 38). In contrast, Hct1p is constitutively expressed, but its association with the APC/C is limited to late G2/M and G1 (32, 48). The regulation of Ama1p appears very similar to that observed for Cdc20p. AMA1 transcription and protein accumulation are restricted from premeiotic S through both meiotic divisions. Ama1p appears to associate with the APC/C on its synthesis, although this determination is somewhat difficult to make because of the relative asynchrony of meiotic cells. Therefore, similar to other APC/C complexes, we propose that APC/CAma1 activation also requires phosphorylation. Two candidate-activating protein kinases are Cdc28p and Ime2p. Genetic and biochemical studies indicate that both kinases are active when APC/CAma1 function is required (7, 47, 49–51).
If an activating kinase is required, one prediction would be that a mutation of the APC/CAma1 activating kinase should be consistent with the observed ama1 phenotypes. We show that Ama1p is required for late meiotic gene expression and spore formation but not meiosis II. However, temperature-shift experiments found that Cdc28p is required for meiosis II but not for spore formation (7, 49). These findings argue against Cdc28p activating APC/CAma1. Similar to Ama1p, Ime2p is essential for late meiotic gene expression and spore formation (52, 53). These observations are more consistent with Ime2p being the activating kinase.
Finally, Ama1p is required at two points during meiotic development, namely meiosis I and spore morphogenesis. These findings may suggest that APC/CAma1 is activated both early and late in development or remains active once stimulated. Cdc20p and Hct1p are inactivated in mitotic cells through degradation or disassociation from the APC/C, respectively (14). Because no changes were observed in Ama1p levels or with APC/C association between meiosis I and spore formation, we do not favor an activation-inactivation-reactivation model. Rather, the activation of APC/CAma1 only once early in meiosis seems more likely although subtle differences in Ama1p behavior may be masked by the asynchrony of the meiotic population.
This report establishes the role of a specialized APC/C in controlling reductional division and gametogenesis in yeast. The utilization of such an activity may allow the cells the flexibility necessary to perform cell divisions outside of the canonical cell cycle.
Supplementary Material
Acknowledgments
We thank O. Cohen-Fix for the tagged PDS1 construct; V. Guacci for Tub1p antibodies, cdc16 strain, and help with the immunofluorescence studies; E. Winter for the ENO1 expression vector; W. Zachariae for the tagged CDC16 construct; and K. Nasmyth for the Tet repressor-GFP reporter system. We also thank V. Guacci, T. Yen, and P. Adams for helpful comments on this manuscript. This work was supported by grants from the National Science Foundation (MCB-9513479), the U.S. Army Breast Cancer Research Program (DAMD17-1-7296), and the National Institutes of Health (GM 57842) to R.S. and a postdoctoral award by the National Institutes of Health (CA-09035) to K.F.C.
Abbreviations
- APC/C
anaphase promoting complex/cyclosome
- RT-PCR
reverse transcription–PCR
- HU
hydroxyurea
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- AMA1
activator of meiotic APC/C
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250351297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250351297
References
- 1.Herskowitz I. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kupiec M, Byers B, Esposito R E, Mitchell A P. In: Meiosis and Sporulation in Saccharomyces cerevisiae. Pringle J R, Broach J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 889–1036. [Google Scholar]
- 3.Nasmyth K. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 4.Pines J. Trends Biochem Sci. 1993;18:195–197. doi: 10.1016/0968-0004(93)90185-p. [DOI] [PubMed] [Google Scholar]
- 5.Dahmann C, Futcher B. Genetics. 1995;140:957–963. doi: 10.1093/genetics/140.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandin N, Reed S I. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuster E O, Byers B. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray A W. Nature (London) 1992;359:599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- 9.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Genes Dev. 1996;10:3077–3080. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 10.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 11.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 12.Murray A W, Solomon M J, Kirschner M W. Nature (London) 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 13.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 14.Zachariae W, Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 15.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 16.Schwab M, Schulze Lutum A, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 17.Longtine M S, McKenzie A r, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Enomoto S, Chen G, Berman J. BioTechnology. 1998;24:782–786. doi: 10.2144/98245st01. [DOI] [PubMed] [Google Scholar]
- 19.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 20.Kaldis P, Pitluk Z W, Bany I A, Enke D A, Wagner M, Winter E, Solomon M J. J Cell Sci. 1998;111:3585–3596. doi: 10.1242/jcs.111.24.3585. [DOI] [PubMed] [Google Scholar]
- 21.Richardson H E, Lew D J, Henze M, Sugimoto K, Reed S I. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper K F, Mallory M J, Smith J S, Strich R. EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strich R, Slater M R, Esposito R E. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zachariae W, Nasmyth K. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pringle J R, Adams A E M, Drubin D G, Haarer B K. In: Immunofluoresence Methods for Yeast. Guthrie C, Fink G R, editors. Vol. 194. Boston: Academic; 1991. pp. 565–610. [DOI] [PubMed] [Google Scholar]
- 27.Michaelis C, Ciosk R, Nasmyth K. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 28.Surosky R T, Strich R, Esposito R E. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper K F, Mallory M J, Strich R. Mol Cell Biol. 1999;19:3338–3348. doi: 10.1128/mcb.19.5.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 31.Engebrecht J, Voelkel-Meiman K, Roeder G S. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 32.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 33.Lamb J R, Michaud W A, Sikorski R S, Hieter P A. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klapholz S, Esposito R E. Genetics. 1980;96:567–588. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law D T S, Segall J. Mol Cell Biol. 1988;8:912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer E R, Scheuringer N, Podtelejnikov A V, Mann M, Peters J M. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A. Biochem Biophys Res Commun. 1999;260:193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- 39.Prinz S, Hwang E S, Visintin R, Amon A. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- 40.Lydall D, Nikolsky Y, Bishop D K, Weinert T. Nature (London) 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 41.San-Segundo P A, Roeder G S. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 42.Leu J Y, Roeder G S. Mol Cell. 1999;4:805–814. doi: 10.1016/s1097-2765(00)80390-1. [DOI] [PubMed] [Google Scholar]
- 43.Shonn M A, McCarroll R, Murray A W. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- 44.Hepworth S R, Friesen H, Segall J. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu S, Herskowitz I. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 46.Knapp D, Bhoite L, Stillman D J, Nasmyth K. Mol Cell Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dirick L, Goetsch L, Ammerer G, Byers B. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- 48.Jaspersen S L, Charles J F, Morgan D O. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 49.Schild D, Byers B. Genetics. 1980;96:859–876. doi: 10.1093/genetics/96.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart D, Wittenberg C. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sia R A, Mitchell A P. Mol Cell Biol. 1995;15:5279–5287. doi: 10.1128/mcb.15.10.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida M, Kawaguchi H, Sakata Y, Kominami K, Hirano M, Harumasa S, Akada R, Yamashita I. Mol Gen Genet. 1990;221:176–186. doi: 10.1007/BF00261718. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein J, Jacobsen F W, Hsu-Chen J, Wu T, Baum L G. Mol Cell Biol. 1994;14:3350–3363. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawson I A, Roth S, Artavanis-Tsakonas S. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sethi N, Monteagudo M C, Koshland D, Hogan E, Burke D J. Mol Cell Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.