Abstract
Intramembrane proteolysis is a core regulatory mechanism of cells that raises a biochemical paradox of how hydrolysis of peptide bonds is accomplished within the normally hydrophobic environment of the membrane. Recent high-resolution crystal structures have revealed that rhomboid proteases contain a catalytic serine recessed into the plane of the membrane, within a hydrophilic cavity that opens to the extracellular face, but protected laterally from membrane lipids by a ring of transmembrane segments. This architecture poses questions about how substrates enter the internal active site laterally from membrane lipid. Because structures are static glimpses of a dynamic enzyme, we have taken a structure–function approach analyzing >40 engineered variants to identify the gating mechanism used by rhomboid proteases. Importantly, our analyses were conducted with a substrate that we show is cleaved at two intramembrane sites within the previously defined Spitz substrate motif. Engineered mutants in the L1 loop and active-site region of the GlpG rhomboid protease suggest an important structural, rather than dynamic, gating function for the L1 loop that was first proposed to be the substrate gate. Conversely, three classes of mutations that promote transmembrane helix 5 displacement away from the protease core dramatically enhanced enzyme activity 4- to 10-fold. Our functional analyses have identified transmembrane helix 5 movement to gate lateral substrate entry as a rate-limiting step in intramembrane proteolysis. Moreover, our mutagenesis also underscores the importance of other residue interactions within the enzyme that warrant further scrutiny.
Keywords: cell signaling, presenilin, signal peptide peptidase, site-2 protease
Scission of peptide bonds within the membrane bilayer is an unexpected biochemical reaction that serves as a regulatory point of multiple cellular processes (1–3). The membrane-embedded proteases that catalyze these reactions are conserved in all branches of life, implying that they are ancient enzymes that assumed key roles early in the evolution of modern cells. Yet the function of these enzymes continues to pose fundamental questions about how hydrolysis is catalyzed and regulated within membrane environments.
Biochemical study of intramembrane proteases has been challenging, and our understanding of these enzymes remains rudimentary. Three superfamilies of intramembrane proteases have been discovered (1–3). The first in vitro assay was developed for γ-secretase, an aspartyl protease that catalyzes the final cleavage to generate Aβ, the component of senile plaques in Alzheimer's disease (4). The assay used detergent-extracted crude membranes, but γ-secretase is a complex of at least four components, with presenilin as the catalytic component (5, 6), and activity is difficult to reconstitute from a recombinant source (7). The related aspartyl protease signal peptide peptidase is thought to function as a single component, but its purification in active form has not been reported (8, 9). The membrane-embedded metalloprotease site-2 protease was discovered as the enzyme required for the release of membrane-tethered transcription factors required for fatty acid and sterol biosynthesis in humans (10). Recently, intramembrane proteolysis was reconstituted in vitro with purified RseP from Escherichia coli (11). This achievement demonstrated that zinc is the only cofactor required for proteolysis, and further work using this system is likely to reveal how intramembrane proteolysis is catalyzed by this class of enzymes.
Rhomboid proteins are intramembrane serine proteases that are among the most widely conserved membrane proteins known (12, 13). Despite significant sequence divergence, many rhomboid enzymes from both prokaryotes and eukaryotes share biochemical characteristics (14, 15). The first rhomboid enzyme was discovered in Drosophila and functions in cleaving transmembrane epidermal growth factor ligands including Spitz to initiate signaling, but bacterial forms also can cleave Spitz (15, 16). Mechanistic analyses revealed that Spitz is recognized by diverse rhomboid enzymes through the upper seven residues of its transmembrane domain, termed the Spitz substrate motif (14). These common properties facilitated development of a pure substrate–enzyme reconstitution system from recombinant source, revealing that rhomboid proteins function directly as proteases and do not require cofactors to catalyze intramembrane proteolysis (17, 18).
Recently structural analyses have been applied to model rhomboid enzymes from bacteria and provided a first glimpse of how nature has resolved the biochemical conundrum for at least one family of enzymes (19–22). The four structures revealed that the serine is sunk ≈10 Å into the plane of the membrane in a hydrophilic cavity that opens to the extracellular side, protected from membrane lipids by a ring of transmembrane protein segments and a partially submerged loop. This has provided the final proof that rhomboid proteins are intramembrane serine proteases, but it now raises questions about how the substrate enters the interior cavity of the active site laterally from the membrane.
Based on structural information, two general models have been proposed. The serine is surrounded on all sides by a ring of transmembrane domains, but helices 1 and 3 form a V-shape with a gap between them that is plugged by the half-submerged L1 loop (for example, see Fig. 2A). Based on this unusual structure, it was proposed that movement of the L1 loop would open a path between helices 1 and 3 to allow substrate access to the interior of the active-site cavity for catalysis (19). In support of the L1 loop gate hypothesis, comparing the rhomboid active site to that of the well studied serine protease chymotrypsin suggested that the orientation of the substrate would be similar if it enters the active site from the L1 loop direction (22).
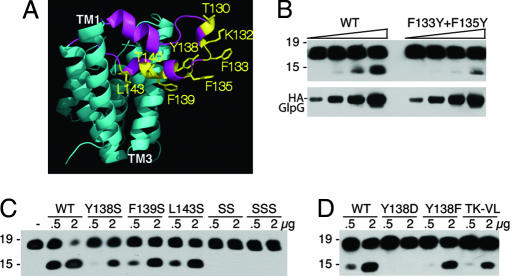
Fig. 2.
L1 loop residues that face toward membrane lipids are required for protease activity. (A) Lateral view of GlpG (2NRF) with L1 loop in magenta and its residue side chains that are expected to contact membrane lipid highlighted in yellow. (B) Effect of changing F133 and F135 to tyrosine on protease activity was compared with wild-type GlpG in a limiting enzyme dilution series (amounts used for each were 100 ng, 200 ng, 400 ng, and 800 ng). Anti-Flag Western blot analyses are shown, with mass standards in kilodaltons depicted to the left of each panel. GlpG levels in the reactions were compared directly by anti-HA Western blot analysis. (C) Y138, F139, and L143 were mutated to serine, and the effect on protease activity against C100Spitz-Flag was assessed. SS is a double Y138S+F139S mutant, whereas SSS carries all three residues mutated to serine. Two different amounts of enzyme (in micrograms) were assayed for each GlpG variant for 1 h, with the highest level of GlpG being approximately equimolar to substrate levels, and resulted in almost complete substrate cleavage by wild type (to assess whether mutants abolished activity). (D) Mutation of Y138 to aspartate or phenylalanine, as well as a double mutant T130V+K132L (TK-LV), reduced activity.
Conversely, analysis of different crystal forms revealed that, although the L1 loop remains constant in all structures, helix 5 on the opposite side of the molecule adopts a different orientation, tilting its top ≈35° laterally from the enzyme core, exposing a path to the catalytic serine (20) (for an example see Fig. 6C). Although one caveat is that helix movement could result from a lack of lateral pressure that is normally exerted by membrane lipids, this observation led us to propose that helix 5 is the substrate gate. A further attractive point of this model is that the catalytic serine lies on the face of helix 4 closest to helix 5 but opposite to the L1 loop. Moreover, one structure revealed a lipid bound between helices 5 and 2, with the phosphate group adopting what could be the position of the substrate tetrahedral intermediate of the cleavage reaction (21).
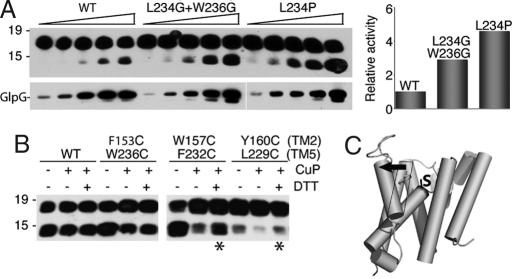
Fig. 6.
Effect of increasing or decreasing transmembrane helix 5 flexibility. (A) Mutating L234, which is on the opposite face of helix 5 and is not involved in forming the helix 5:helix 2 interaction (see Fig. 5A), to proline increased enzymatic activity 5-fold, whereas changing both L234 and W236 to glycine increased activity ≈3-fold, compared with wild-type GlpG. GlpG levels were varied from 50 to 800 ng in 2-fold increments, and resulting enzyme amounts in the reaction were verified by anti-HA Western blot analysis. Activity stimulation was quantified by densitometry and is depicted graphically (Right). (B) Corresponding pairs of residues on helices 2 and 5 were changed to cysteine, and activity of the untreated, oxidized (CuP), and oxidized and subsequently reduced (DTT) enzymes were assessed. Asterisks denote enzyme activity that could be restored by DTT after oxidation. Note that the double mutant containing mutated Y160 had a lower enzymatic activity, as observed for this mutation in conjunction with other helix 5 mutants. (C) Model of intramembrane substrate gating by rhomboid enzymes: lateral tilting (black arrow) of the top of transmembrane helix 5 opens a path for substrate entry to the catalytic serine (labeled S).
Because these structures are static glimpses of an otherwise dynamic enzyme, we have taken a structure-based protein engineering approach to differentiate between substrate gating models; both models make a number of predictions that can be tested by analyzing the effect of engineered mutants on enzyme activity. This approach revealed that the L1 loop plays an important structural role in the protease, during either folding or function. Conversely, mutation of transmembrane helix 5 to allow it to move away more readily resulted in a protease with a 4- to 10-fold increase in activity, suggesting that helix 5 movement to accommodate substrate entry is rate-limiting. Our functional studies therefore implicate helix 5 as the substrate gate for rhomboid proteases.
Results
Intramembrane Cleavage of C100Spitz by GlpG.
We sought to study the mechanism of intramembrane substrate gating by rhomboid proteases by testing the effect of engineered mutants on substrate cleavage. As such, the choice of substrate for these experiments was a critical consideration, because purified GlpG has been shown to have the capacity to cleave soluble proteins in in vitro assays (11, 19). Because this probably involves insertion of an extended loop into the active-site cavity from the extracellular face of the enzyme, such artificial substrates could lead to erroneous conclusions regarding lateral gating of transmembrane substrates.
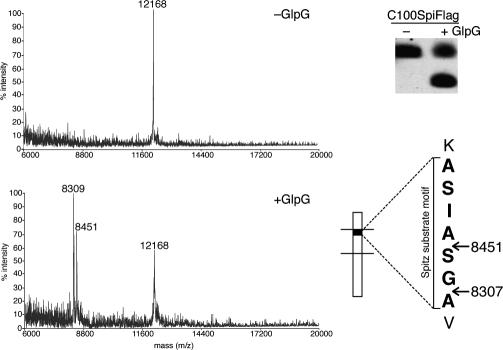
No pure substrate for any rhomboid protease is currently known to be cleaved in its transmembrane domain in vitro. We therefore examined the cleavage of C100Spitz-Flag, a substrate that we previously adapted for the study of diverse rhomboid proteases in vitro (17). Rhomboid proteases from a variety of organisms display strong substrate specificity, and GlpG was able to cleave C100Spitz-Flag, but not C100-Flag, the fragment of Alzheimer's β-amyloid precursor protein lacking Spitz transmembrane residues (4). We thus examined the cleavage site of C100Spitz-Flag by mass spectrometry and discovered two cleavage sites within the Spitz substrate motif, four and six residues into the transmembrane domain (Fig. 1). Cleavage was between the alanine–serine and glycine–alanine residues. This is a direct determination of a cleavage site for a Spitz-type substrate by a rhomboid protease, and it further indicates that proteolysis can occur at multiple positions, which is analogous to multiple cleavage sites observed for both γ-secretase and signal peptide peptidase (9, 23). Importantly, the cleavage sites generated in our in vitro assay are intramembrane, suggesting that this substrate is well suited for studying intramembrane protease gating.
Fig. 1.
C100Spitz-Flag substrate is cleaved at two intramembrane sites. Pure C100Spitz-Flag was incubated for 2 h at 37°C with GlpG or buffer alone, and the C-terminal cleavage products were captured and analyzed in parallel by MALDI-TOF mass spectrometry (Left) and Western blot (Upper Right). The predicted mass of intact C100Spitz-Flag is 12,166 Da. The Spitz substrate motif (bracketed) is shown, with an external lysine N-terminal to the transmembrane domain above. Predicted masses of cleavage products designated by arrows are shown, and they correspond well to the peaks that appear in the mass spectrum after incubation with GlpG but not buffer alone.
Engineered Variants Suggest a Structural Rather than a Gating Role for the L1 Loop.
The L1 loop gating hypothesis proposes that displacement of the L1 loop opens a gate to allow substrate entry into the active site (19, 22). One prediction of this model is that loosening some of the contacts made by the L1 loop might allow it to move away more readily, resulting in mutants with enhanced substrate proteolysis. To test this hypothesis, we engineered three categories of variants: mutants that destabilize L1 loop:lipid interactions, interactions within the L1 loop itself, and L1 loop interactions with other parts of the enzyme. All GlpG enzymes were expressed in bacteria as full-length proteins and purified (17).
Residues F133, F135, Y138, F139, and L143 in the L1 loop face outwards and directly contact lipid, thus helping to stabilize the L1 loop in its membrane-submerged conformation (19) (Fig. 2A). We reasoned that reducing the hydrophobicity of these residues might make the membrane-submerged conformation less stable, facilitating displacement of the L1 loop and thus enhanced substrate access and increased protease activity. Contrary to this prediction, we found that mutation of F133 and F135 both to tyrosine (Fig. 2B), or any of the other residues singly to serine, resulted in a strong decrease in enzyme activity, while double and triple serine mutants abolished activity (Fig. 2C). Moreover, even subtle mutations such as Y138F had inhibitory effects on enzymatic activity, while mutating more distal L1 loop residues T130 and K132 also failed to increase enzyme activity (Fig. 2D).
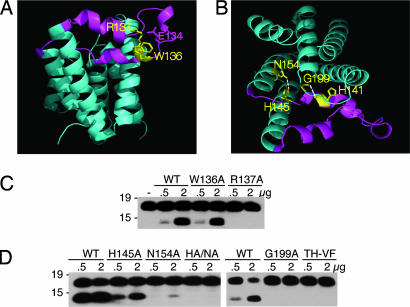
Second, a tryptophan–arginine pair in the L1 loop is conserved in rhomboid proteases across evolution, and their mutation in Drosophila Rhomboid-1 results in activity loss when assayed in living cells (12). Structural analysis revealed that these residues contribute to interactions within the L1 loop itself, with the arginine donating five hydrogen bonds (19, 20) (Fig. 3 A and B). Although weakening this interaction might increase L1 loop mobility, mutation of the arginine abolished activity whereas mutation of the tryptophan had a mild effect (Fig. 3C).
Fig. 3.
Disrupting L1 loop:core interactions reduces protease activity. (A) Side view of GlpG (2NRF) with the L1 loop highlighted in magenta and the conserved WR motif within the L1 loop shown in yellow. R137 makes a series of hydrogen bonds to upper regions of the loop and to E134. (B) Top view of GlpG (2NRF) with the L1 loop:core hydrogen bonds between N154 of transmembrane domain 2 and H145 of the L1 loop, and the backbone of H141 of the loop and G199 above helix 4. (C) Mutagenesis of W136 had a mild effect on protease activity of GlpG, whereas changing R137 to alanine abolished protease activity. Western blot analysis of C100Spitz-Flag cleavage is shown. (D) The effect of disrupting L1 loop:core contacts by mutagenesis of H145 and N154 individually to alanine, or their double mutant (HA/NA), as well as G199 to alanine and H141 to phenylalanine with the neighboring T140 to valine (TH-VF), tested for proteolytic activity.
The L1 loop also makes a series of internal interactions within the protease (Fig. 3B). The conserved histidine-145 near the end of the loop is hydrogen-bonded to the conserved asparagine-154 on transmembrane helix 2 (19). This interaction might help to keep the L1 loop folded over by tethering it to the second transmembrane domain. Similarly, glycine-199, which lies two residues above the active site serine, also contacts the L1 loop through hydrogen bonding with its backbone to the backbone of H141 in the loop. Although mutation of these residues would thus be expected to allow greater freedom for L1 loop displacement and substrate access, mutation of the three residues singly to alanine or H141 to phenylalanine (bulkier side chain to change backbone position) strongly reduced cleavage (Fig. 3D).
Therefore, despite targeting three classes of L1 loop interactions with 16 engineered GlpG mutants in all, we have not been able to generate a single enzyme with enhanced intramembrane proteolysis activity. In fact, almost all mutations strongly decreased enzymatic activity, suggesting an important structural role for the L1 loop.
Local Perturbations Around the Active Site Hinder Proteolytic Activity.
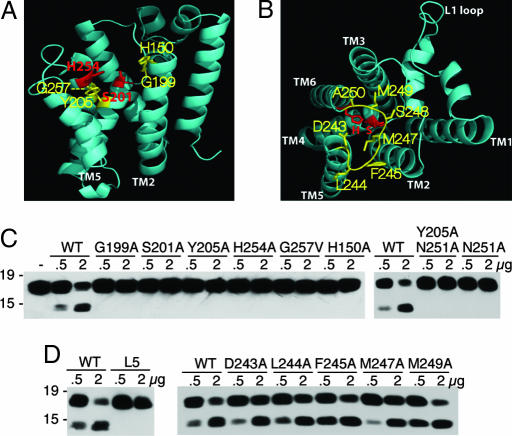
The catalytic serine lies on the face of helix 4 that is opposite to the L1 loop. As such, it has been proposed that some local rearrangement could be required for proteolysis if substrate entered from the L1 loop direction (19, 20, 22). To test this possibility, we disrupted some of the interactions surrounding the active site (Fig. 4). First, the active-site histidine is stabilized by base stacking onto tyrosine-205 one helical turn below the catalytic serine, and this tyrosine was also observed to form a hydrogen bond with asparagine-251 through a water molecule in one structure (19). To simulate a local perturbation of helix 4, we mutated Y205 and/or N251 to alanine to disrupt this base-stacking interaction. This resulted in a strong decrease in substrate cleavage for Y205A and a modest decrease in activity for N251A (Fig. 4C). Similarly, mutating G199, which lies two residues above the serine, to an alanine also greatly reduced enzyme activity. Helix 6, which contributes the active-site histidine, makes a close association with helix 4, which contributes the catalytic serine, through a universally conserved GxxxG transmembrane dimerization motif below the histidine (12, 19, 20). To simulate a local movement of helix 4, we mutated the first glycine of the GxxxG motif to valine (G257V). Again, the resulting enzyme lost all detectable proteolytic activity (Fig. 4C). Collectively, these results argue that local perturbations surrounding the active serine, as might be expected to occur if substrate enters from the L1 direction, strongly disrupt proteolytic activity (although these mutants could also affect folding). This is contrary to the predictions made by the L1 loop gating model.
Fig. 4.
Importance of residue interactions neighboring the active site on protease activity. (A) Lateral view of GlpG (2NRF) with catalytic residues in red and neighboring transmembrane residues chosen for mutagenesis in yellow. (B) Top view of GlpG (2IC8) with catalytic residues in red and Loop5 (L5) residues chosen for mutagenesis in yellow. (C) Mutagenesis of residues G199 and Y205 to alanine and G257 to valine that are near the active site, and the catalytic S201 and H254 to alanine, abolished protease activity. Note that, under longer incubation, Y205A displayed residual proteolytic activity. Mutating N251, which interacts with Y205 in the closed form, to alanine reduced but did not abolish activity under prolonged incubation (data not shown) unless it was combined with Y205. (D) Mutating residues 243–250 of the L5 loop all to glycine (L5) resulted in a strong decrease in activity, whereas mutating L5 residues to alanine individually had mild effects.
Opening the Gate: Mutations Within Helix 5 Enhance Proteolytic Activity.
The same logic could be applied to test predictions made by the helix 5 gating model: loosening the interactions between helix 5 and the rest of the molecule should enhance gate opening and result in stimulation of proteolytic activity. Consistent with this prediction, we were able to engineer three independent classes of helix 5 mutants that enhanced substrate cleavage.
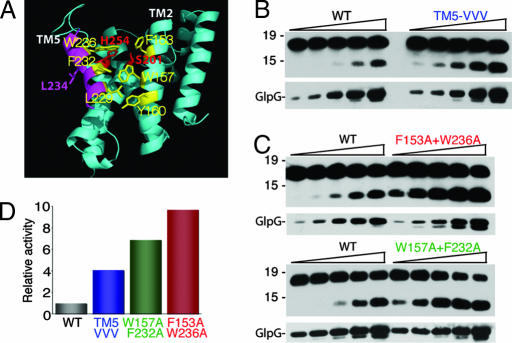
First, helix 5 interacts with its neighboring helix 2 via a series of hydrophobic interactions mediated by large hydrophobic or aromatic residues that zipper together (Fig. 5A). We mutated three of these residues on the helical face of helix 5 that interacts with helix 2, namely W236, F232, and L229, each to valine, thus retaining significant hydrophobic nature for these transmembrane residues. Strikingly, this triple mutant displayed a 400% increase in activity compared with wild-type GlpG in an enzyme dilution series (Fig. 5B), consistent with the helix 5 gating model.
Fig. 5.
Mutations in transmembrane helix 5 dramatically stimulate protease activity. (A) Lateral view of the open conformation of GlpG (2NRF) with catalytic serine and histidine in red, helix 5 in magenta, and helix 5 and helix 2 residues that form the interface highlighted in yellow. The L234 side chain that lies on the opposite face of helix 5 is shown in magenta. (B) Mutation of helix 5 residues L229, F232, and W236 to valine in a triple mutant increase protease activity ≈4-fold in an enzyme dilution assay (titrated 50–800 ng in 2-fold increments, also in C). GlpG levels in the reactions were assessed by anti-HA Western blot analysis (Lower). Note that fold stimulation was evident by both quantifying cleavage products for the same concentration of GlpG and comparing which dilution of the mutant yielded activity similar to wild type. (C) Mutation of helix 5 residue W236 and helix 2 residue F153 both to alanine increased enzyme activity ≈10-fold compared with wild-type GlpG, whereas alanine substitutions at 232 and 157 increased activity ≈7-fold. (D) Enzyme activity of wild type and GlpG mutants was quantified by densitometry for several different enzyme concentrations, and average fold increase was plotted.
We next examined whether mutation of residues on neighboring helix 2 that contact helix 5 also increased activity. We therefore targeted contact points mediated by pairs of residues on helix 5 and helix 2. Mutating the top residue pairs (236 on helix 5 and 153 on helix 2) to alanine, a very small residue, dramatically stimulated GlpG activity ≈10-fold (Fig. 5 C and D). Movement of helix 5 would also result in concomitant displacement of the L5 Cap loop that is connected to the top of helix 5. Interestingly, the side chain of the L5 Cap residue F245 wedges between helices 2 and 5, and its mutation displayed a weak enhancement of proteolysis (Fig. 4D), providing further evidence for the helix 5 gating model. Notably, mutation of residues F232 and W157, which are deep within the transmembrane segment of these helices, also stimulated activity by ≈7-fold (Fig. 5 C and D). These observations suggest that interactions between helices 2 and 5 along their length are important for helix 5 gating.
Third, we also examined the nature of the mutations that enhanced proteolytic activity. If helix 5 tilting away from the core is required to open the substrate gate, then introducing helix-breaking residues into helix 5 should have a stimulatory effect on proteolysis. Consistent with this prediction, mutagenesis of leucine-234, which is near the top of helix 5 but faces away from the helix 2–helix 5 interface (Fig. 5A), to proline, a helix-breaking residue, dramatically enhanced proteolytic activity ≈5-fold compared with the wild-type enzyme (Fig. 6A). Similarly, mutagenesis of tryptophan-236 and leucine-234 both to glycine enhanced proteolytic activity compared with wild type. These observations suggest that helical flexibility within helix 5, as well as decreasing the helix 5–helix 2 interaction, increases helix 5 lateral opening, thus enhancing proteolytic activity.
Closing the Gate: Tethering Helix 5 to Helix 2 in the “Closed” Conformation Abrogates Activity.
In addition to predicting mutants that enhance activity, the helix 5 gating model also predicts mutants that should abrogate activity: if helix 5 is the substrate gate, then tethering helix 5 onto helix 2 and thus decreasing its ability to move out laterally should strongly decrease proteolytic activity. To test this prediction, we incorporated cysteine residues in pairs along the length of helices 2 and 5. Inducing chemical cross-linking through oxidation to form a disulfide bridge resulted in a strong decrease in activity for both the 157/232 and 160/229 mutant pairs, although not for 153/236 (Fig. 6B), presumably because these side chains are too far apart to be cross-linked efficiently. The activity could be restored by addition of reducing agent, confirming that the lack of activity of these mutants was due to immobilization of helix 5. Moreover, treating wild-type enzyme with oxidant or reductant had no effect on proteolytic activity. Thus, immobilizing helix 5 by cross-linking it to helix 2 at either of two transmembrane points strongly reduced proteolytic activity, further arguing that lateral movement of helix 5 is responsible for gating and substrate access.
Discussion
We have taken a structure-based enzymology approach to test models for substrate gating by rhomboid intramembrane proteases. Despite making in total >40 engineered GlpG variants in transmembrane domains 2, 4, 5, and 6 and Loops 1 and 5, only mutants that are predicted to increase helix 5 flexibility in different ways resulted in enhanced enzyme activity from 4- to 10-fold. Conversely, tethering helix 5 to helix 2, as occurs in the “closed” conformation, abrogated enzyme activity. Mutations affecting L1 loop interactions or interactions near the active site strongly decrease enzyme activity. Collectively, these structure–function analyses contradict the possibility that the L1 loop performs a gating function and indicate that helix 5 plays a rate-limiting dynamic function as the lateral substrate gate for intramembrane proteolysis (see model in Fig. 6C).
Our initial caveat to the functional significance of observing lateral helix displacement in GlpG crystallized in detergent is that it reflects lack of lateral pressure that is normally applied to proteins by the membrane itself. Our mutagenesis experiments provide functional evidence against this potential artifact: introduction of transmembrane mutations that decrease helix 5 interactions with the rest of the molecule dramatically increase proteolytic activity instead of denaturing the enzyme. In fact, cleavage of C100Spitz-Flag can occur as far as six residues into the substrate transmembrane domain, which would require significant intramembrane helix displacement for gating, not just at its very top. Consistent with this notion, mutating contacts between helices 2 and 5 in the middle region of the membrane resulted in a dramatic 7-fold stimulation of activity. However, the bottom of helix 5 is required to remain stationary; mutating lower contacts between helices 2 and 5 hindered activity. It should be noted that, although rare, analogous lateral gating mechanisms by transmembrane helices are not unprecedented: membrane protein insertion by the protein-conducting translocon heterotrimer is also thought to open laterally to allow transmembrane segment release between helices TM2b and TM7/TM8 (24). Rhomboid intramembrane proteases may therefore perform an analogous function in reverse: disengagement of contacts between helix 5 from its neighboring helix 2 would allow lateral movement of helix 5 to open a path for substrate to enter the internal active site of the protease. This step is likely rate-limiting, and thus engineering mutants that facilitate helix 5 opening dramatically increase enzyme activity. Another attractive point of this model is that movement of helix 5 to accommodate substrate entry would also drag the L5 Cap along as they are connected, which is also thought to be required for catalysis (25).
What is not clear is the sequence of events in gate opening and substrate docking, but once the helix 5 gate opens, the top of the substrate transmembrane domain likely unwinds into the active site for cleavage. This is consistent with the notion that rhomboid substrates depend on helix-disrupting residues within the top of their transmembrane domain for cleavage (14, 17). In fact, this cleavage occurs between the glycine–alanine pair, which has been defined to be the main helix-breaking sequence (14).
It is tempting to speculate whether analogous lateral gating mechanisms might be used by other intramembrane proteases. Although these enzymes are not evolutionarily related to rhomboid, they likely faced similar biophysical constraints in developing intramembrane proteolysis mechanisms. Based on current data, it seems possible that both site-2 protease and signal peptide peptidase could use a similar lateral gating mechanism, because both require helix-breaking residues in substrates (26, 27), and both are also thought to function alone without the need for other proteinaceous cofactors (8, 11). The likely exception to this potentially common mechanism is γ-secretase. This enzyme is able to cleave virtually any transmembrane helix, acting as the proteasome of the membrane, and appears to prefer helical substrates (28–30). In fact, Spitz residues that convert C100 into a substrate for rhomboid proteases abolish its ability to be cleaved in vitro by γ-secretase (17). Moreover, γ-secretase is known to function as a multicomponent enzyme (5, 7, 31), with at least nicastrin acting as a “gate-keeper,” regulating substrate entry into the active site by binding the N-terminal ectodomain stub of substrates through its aminopeptidase-like sequence (32). The recent low-resolution structure by electron microscopy revealed a very large inner cavity, from which two visible channels emanate (33, 34). These early observations raise the possibility that γ-secretase uses a different mechanism for substrate access and gating than rhomboid enzymes.
Beyond substrate gating by helix 5, our structure–function analysis also suggests important functions for residues that form interactions within the molecule, including within the active site and the L5 Cap. Residues that stabilize the active-site histidine traditionally play important functions in serine proteases. Although the conserved asparagine in transmembrane domain 2 was initially proposed by analogy to soluble serine proteases to stabilize the histidine (12), structural analysis revealed that this role is likely provided by Y205 onto which the histidine ring stacks (19–22). Although the functional importance of this interaction was unclear, we found that substituting Y205 with alanine causes a dramatic decrease in proteolytic activity, validating the structural proposal. All rhomboid enzymes also contain a conserved GxxxG motif two residues below the active-site histidine (12), but this motif had not been studied directly. We have mutated the first glycine to valine (G257V), and we observed a complete loss of enzyme activity, again highlighting the importance of this absolutely conserved residue that mediates a close juxtaposition of helices 4 and 6 to bring the catalytic serine and histidine together. This effect was also observed with mutagenesis of the same residue in a metazoan rhomboid protease analyzed in living cells (R.P.B. and S.U., unpublished data). One unexpected observation is the importance of the Loop 5 Cap residues: Although individual residue mutations had subtle effects, mutation of all eight of these residues to glycine, such that the length of the loop was maintained but the side chains were removed, strongly reduced activity (Fig. 4D). Although in the closed conformation several Loop 5 residues insert into the active site from above, it is not obvious why this mutation might have such profound consequences. Finally, the L1 loop is an unusual structure, and enzymatic activity is very sensitive to its mutation; thus, it remains possible that the L1 loop functions also beyond stabilizing the molecule.
Also of note are several conserved transmembrane residues that are part of the active site but are not catalytic directly. Although structural analysis has not yet unambiguously identified the oxyanion-stabilizing pocket, it has been proposed that helix 2 residues may contribute to it (21, 22, 25). Accordingly, we found that mutating H150 completely abrogated activity (Fig. 4C), and mutation of N154 strongly hindered activity, whereas mutation of H145 had a more mild effect (Fig. 3D). Thus, it is possible that H150 and N154 constitute the oxyanion pocket. Although the glycine two residues upstream of the serine was initially proposed to function as the oxyanion-binding hole by analogy to chymotrypsin, its function is currently unclear. Our mutation of glycine-199 to alanine, however, underscores the importance of this residue, because this subtle mutation completely abolished enzyme activity.
In conclusion, our structure–function analysis of a rhomboid protease has implicated helix 5 as the lateral substrate gate. The strong effect of mutations within other regions also indicates that more remains to be learned about the detailed function of this complex enzyme.
Materials and Methods
Recombinant Rhomboid Expression and Purification.
Wild-type and mutant GlpG proteins were expressed as N-terminal GST-fusion proteins in E. coli C43(DE3), solubilized from isolated membranes with 1% dodecyl-β-d-maltoside, and purified by using glutathione-Sepharose affinity chromatography as described by Urban and Wolfe (17), with the exception of cysteine-substituted GlpG mutants (see Fig. 6B), which were expressed as His-tagged fusion proteins and purified as described by Wu et al. (20). GlpG yields were standardized by Coomassie staining after SDS/PAGE and quantified by using LiCor infrared scanning and by incorporation of a single HA tag at their extreme N terminus. Mutations encoding specific amino acid substitutions were introduced into GlpG by using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA), and all constructs were verified by sequencing the entire ORF.
In Vitro Proteolysis Assays.
GlpG proteolytic activity was assayed by using the recombinant C100Spitz substrate with a C-terminal Flag tag, which was purified as described previously (17). Pure enzyme and ≈1 μg of substrate were combined in a final volume of 20 μl consisting of 50 mM Tris (pH 7.5), 150 mM NaCl, and 0.1% dodecyl-β-d-maltoside for 1 h at 37°C. Reactions were terminated by the addition of SDS/PAGE sample buffer, and 5 μl of the stopped reactions was detected by anti-Flag Western blot analysis.
Mass Spectrometry.
Substrate and C-terminal reaction products were captured by using anti-Flag immunoprecipitation after in vitro cleavage for 2 h at 37°C. A total of 0.5 μl of the resulting samples was spotted onto a stainless steel plate with sinapinic acid matrix (10 mg/ml in 50% acetonitrile and 0.05% trifluoroacetic acid) and analyzed on a Voyager DE-STR MALDI-TOF mass spectrometer operating in linear mode with 200 shots per spectrum and a low mass gate of 3,000. Before each session, the instrument was calibrated with bovine insulin, equine cytochrome c, and apomyoglobin as mass standards.
Rhomboid Intramolecular Disulfide Cross-Linking.
GlpG mutants were generated with cysteine substitutions for three different pairs of residues in helices 2 and 5 (F153C+W236C, W157C+F232C, and Y160C+L229C). Cysteine-substituted (and for comparison wild-type) GlpG were treated with 0.25 mM copper phenanthroline for 15 min at room temperature followed by quenching with 5 mM EDTA and subsequent purification by gel filtration. Oxidized proteins were assayed for C100Spi-Flag cleavage in parallel with nontreated controls in standard reaction buffer or supplemented with 10 mM DTT.
Structural Analysis and Imaging.
Atomic coordinates 2IC8 and 2NRF of published structures were obtained from the Protein Data Bank and analyzed and imaged by using Pymol.
Acknowledgments
We are grateful to the Institute for Basic Biomedical Sciences Proteomics Facility at Johns Hopkins for use of the Voyager instrument, Jeff Corden and Jessica Ames for use of the LiCor, Raquel Lieberman for demonstrating Pymol, and Chelsea Newhouse for administrative assistance. This work was supported by National Institutes of Health Grant R01AI066025 (to S.U.) and a career award from the Burroughs-Wellcome Fund (to S.U.). Work in the Y.S. laboratory is supported by Princeton University and grants from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8199.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe MS, Kopan R. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 3.Urban S. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 4.Li YM, Lai MT, Xu M, Huang Q, DiMuzio-Mower J, Sardana MK, Shi XP, Yin KC, Shafer JA, Gardell SJ. Proc Natl Acad Sci USA. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 6.Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Proc Natl Acad Sci USA. 2003;100:6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 8.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 9.Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, Wolfe MS. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 10.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama Y, Kanehara K, Ito K. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 13.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urban S, Freeman M. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 15.Urban S, Schlieper D, Freeman M. Curr Biol. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 16.Gallio M, Sturgill G, Rather P, Kylsten P. Proc Natl Acad Sci USA. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urban S, Wolfe MS. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maegawa S, Ito K, Akiyama Y. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Zhang Y, Ha Y. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Shem A, Fass D, Bibi E. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. Proc Natl Acad Sci USA. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraering PC, Ye W, Strub JM, Dolios G, LaVoie MJ, Ostaszewski BL, van Dorsselaer A, Wang R, Selkoe DJ, Wolfe MS. Biochemistry. 2004;43:9774–9789. doi: 10.1021/bi0494976. [DOI] [PubMed] [Google Scholar]
- 24.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Ha Y. Proc Natl Acad Sci USA. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemberg MK, Martoglio B. Mol Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 27.Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2000;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopan R, Ilagan MX. Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 29.Das C, Berezovska O, Diehl TS, Genet C, Buldyrev I, Tsai JY, Hyman BT, Wolfe MS. J Am Chem Soc. 2003;125:11794–11795. doi: 10.1021/ja037131v. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenthaler SF, Wang R, Grimm H, Uljon SN, Masters CL, Beyreuther K. Proc Natl Acad Sci USA. 1999;96:3053–3058. doi: 10.1073/pnas.96.6.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, et al. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 32.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, III, Sudhof T, Yu G. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Proc Natl Acad Sci USA. 2006;103:6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogura T, Mio K, Hayashi I, Miyashita H, Fukuda R, Kopan R, Kodama T, Hamakubo T, Iwatsubo T, Tomita T, Sato C. Biochem Biophys Res Commun. 2006;343:525–534. doi: 10.1016/j.bbrc.2006.02.158. [DOI] [PubMed] [Google Scholar]