Abstract
Proteolytic processing of the amyloid precursor protein by β-secretase yields A4CT (C99), which is cleaved further by the as yet unknown γ-secretase, yielding the β-amyloid (Aβ) peptide with 40 (Aβ40) or 42 residues (Aβ42). Because the position of γ-secretase cleavage is crucial for the pathogenesis of Alzheimer’s disease, we individually replaced all membrane-domain residues of A4CT outside the Aβ domain with phenylalanine, stably transfected the constructs in COS7 cells, and determined the effect of these mutations on the cleavage specificity of γ-secretase (Aβ42/Aβ40 ratio). Compared with wild-type A4CT, mutations at Val-44, Ile-47, and Val-50 led to decreased Aβ42/Aβ40 ratios, whereas mutations at Thr-43, Ile-45, Val-46, Leu-49, and Met-51 led to increased Aβ42/Aβ40 ratios. A massive effect was observed for I45F (34-fold increase) making this construct important for the generation of animal models for Alzheimer’s disease. Unlike the other mutations, A4CT-V44F was processed mainly to Aβ38, as determined by mass spectrometry. Our data provide a detailed model for the active site of γ-secretase: γ-secretase interacts with A4CT by binding to one side of the α-helical transmembrane domain of A4CT. Mutations in the transmembrane domain of A4CT interfere with the interaction between γ-secretase and A4CT and, thus, alter the cleavage specificity of γ-secretase.
The main proteinaceous component of the amyloid plaques found in the brains of patients with Alzheimer’s disease (AD) is β-amyloid (Aβ; refs. 1 and 2), an ≈4-kDa peptide that is derived from the larger amyloid precursor protein (APP; ref. 3). APP processing by the as yet unidentified protease activities, termed α-, β-, and γ-secretases, leads to a variety of different soluble and membrane-bound proteins (for reviews, see refs. 4 and 5). The α-secretase activity cleaves APP within the Aβ domain and thus precludes the generation of Aβ. This cleavage yields secretory α-APPs, comprising most of the N-terminal ectodomain of APP, and the remaining membrane-bound C-terminal fragment p3CT. Alternatively, APP can be cleaved by the β-secretase activity at the N terminus of Aβ, generating a truncated, soluble β-APPs and a C-terminal fragment of 99 residues (A4CT, C99). The β-secretase product A4CT contains the entire Aβ domain, the transmembrane domain, and the cytoplasmic tail of APP and represents the direct precursor for Aβ (6, 7).
Both membrane-bound C-terminal fragments of APP, A4CT and p3CT, are cleaved by the γ-secretase activity within their transmembrane domains at the C terminus of Aβ or p3, thus releasing the 40- and 42-residue Aβ peptides (Aβ40 and Aβ42) and the 24–26 residue p3 peptides (p340 and p342) (8–11). Most cells secrete both peptides Aβ and p3 into the conditioned medium. In neuronal cells, as in primary hippocampal neurons and in kidney 293 cells, Aβ, but not p3, also can be found intracellularly and does not seem to be secreted (12–16).
The major Aβ species secreted by cultured cells expressing wild-type (wt) APP is Aβ40; the minor species is Aβ42 (17). Mutations in the APP close to the γ-cleavage site have been shown to alter the cleavage specificity of the γ-secretase activity (Aβ42/Aβ40 ratio; refs. 14 and 18–20). However, the factors that determine this cleavage specificity are unknown. Experiments with inhibitors of γ-secretase activity suggest that distinct proteases generate the Aβ40 and Aβ42 peptides (11, 21–23), but it is not known whether these enzymes are related or not.
Furthermore, although γ-cleavage occurs in the transmembrane domain of A4CT, it is not clear whether the cleavage occurs while A4CT is inserted into the membrane or after release of A4CT from the membrane. Understanding the substrate specificity of the γ-secretase activity is of great importance, because the cleavage at residue 42 of Aβ is strongly linked to the disease. Therefore, the γ-cleavage constitutes an obvious target for disease prevention and for understanding the basic molecular mechanisms underlying AD (for a review, see ref. 5).
To dissect the substrate specificity of the γ-secretase activity in more detail, we replaced all residues in the transmembrane domain of A4CT that are outside the Aβ domain with phenylalanine, and measured the influence of these mutations on the ratio of Aβ42/Aβ40 in the conditioned medium of A4CT-expressing COS7 cells.
MATERIALS AND METHODS
Cell Culture and Transfections.
COS7 cells were cultured according to published protocol (20), except that DMEM was used instead of a 1:1 mixture of MEM and F12. Cell culture media were obtained from Sigma. The pCEP4 vector (Invitrogen) with the signal peptide SPA4CT cDNA inserts was transfected into COS7 cells by using Lipofectin (GIBCO/BRL) as described (24). For each construct, two or more independent transfections were analyzed.
Antibodies.
The monoclonal antibodies W02 (for the precipitation of all Aβ peptides, regardless of the specific C terminus), G2–10 (specific for Aβ ending at residue 40), and G2–11 (specific for Aβ ending at residue 42) were raised against synthetic peptides (25). The polyclonal antibody CT13 was raised against a synthetic peptide comprising the C-terminal 13 residues of A4CT (Eurogentech, Brussels, Belgium).
Metabolic Labeling and Immunoprecipitation.
In each experiment, similar numbers of cells were used. After 45 min of preincubation in methionine-free MEM, stably transfected COS7 cells were incubated for 48 h in methionine-free MEM containing 10% fetal bovine serum and 133 μCi/ml [35S]methionine (Amersham Pharmacia). The conditioned medium was centrifuged at 4°C for 1 min at 4,000 × g and divided into three 1-ml samples. Aβ and p3 were immunoprecipitated over 6 h with 100 μg of Protein G Agarose (Boehringer Mannheim) and antibody G2–10 (12.5 μg/ml), G2–11 (17.3 μg/ml), or W02 (5 μg/ml; W02 binds Aβ, but not p3). The immunoprecipitated proteins were separated on 10% Tris/Tricine {N-[tris(hydroxymethyl)methyl]glycine} gels (26). The intensity of the bands was quantified with a Fuji phosphorimager (BAS 1000).
To determine whether the same amount of Aβ was generated from the wt and mutated A4CT proteins, the Aβ immunoprecipitated from the conditioned medium and the A4CT immunoprecipitated from the cell lysate (24) were quantified as described above. For the immunoprecipitation of Aβ and A4CT, antibodies W02 and CT13 were used, respectively. The ratios of Aβ/A4CT were determined in two independent experiments.
Plasmid Construction.
Plasmids pBS/SPA4CT with the mutations V44F, I45F, I45V, I47F, T48F, T48A, L49F, V50F, M51F, and L52F were generated with the Quik Change Site Directed Mutagenesis Kit (Stratagene), suitable oligonucleotides, and pBS/SPA4CTrev. (20) as the DNA template. The KpnI/SpeI fragments of these constructs were cloned into the pCEP4 vector that was digested with KpnI/NheI. The generation of plasmids pCEP/SPA4CT-wt, SPA4CT-T43F, SPA4CT-V46F, APP695-wt, APP695-V46F, and pBS/APP695 has been described (20). pBS/APP695-I45F and V50F were generated by cloning the EcoRI/ClaI fragments of the pBS/SPA4CT plasmids into pBS/APP695-wt that was digested with EcoRI/ClaI. The obtained plasmids were digested with SmaI/SpeI, and the fragments cloned into pCEP4 (digested with NheI/PvuII) to obtain pCEP/APP695-I45F and V50F. The identity of the constructs obtained by PCR was confirmed by DNA sequencing.
Mass Spectrometric Analysis of Immunoprecipitated Aβ.
COS7 cells were incubated overnight in DMEM (Sigma). Aβ peptides were immunoprecipitated from the conditioned medium by using monoclonal antibody 4G8 (Senetek, Maryland Heights, MO) and Protein G Plus/Protein A agarose beads (Oncogene Science). Immunoprecipitated Aβ peptides were analyzed by matrix-assisted laser desorption/ionization mass spectrometry by using a time-of-flight mass spectrometer (Voyager-DE STR BioSpectrometry Workstation, PerSeptive Biosystems, Framingham, MA) as described (17).
RESULTS
Phenylalanine-Scanning Mutagenesis of A4CT and Generation of Aβ.
Point mutations within the transmembrane domain of A4CT at amino acid residues Thr-43 and Val-46 have been shown to alter the cleavage specificity of the γ-secretase activity (18–20) by leading to an altered product ratio of Aβ42/Aβ40. To analyze whether other residues, besides Thr-43 and Val-46, in the transmembrane domain of A4CT are also able to influence the cleavage specificity of γ-secretase, we introduced three kinds of mutations into the A4CT sequence and measured their effects on the ratio of Aβ42/Aβ40 in the conditioned medium of COS7 cells.
First, residues 43–52 of A4CT, which directly follow the γ-cleavage site in the transmembrane domain, were replaced individually with phenylalanine: A4CT-T43F, V44F, I45F, V46F, I47F, T48F, L49F, V50F, M51F, and L52F (Fig. 1). Second, amino acid Ile-45 was mutated to valine (A4CT-I45V), because this mutation has been found recently in the APP gene of a patient with familial Alzheimer’s disease (27). Third, Thr-48 was replaced by alanine (A4CT-T48A, Fig. 1). All A4CT constructs were transfected stably as SPA4CT (APP SP-Leu-Glu-A4CT; ref. 24) in COS7 cells (Fig. 1).
Figure 1.
Schematic representation of SPA4CT. SPA4CT consists of the SP of APP followed by leucine and glutamic acid and the C-terminal 99 aa (A4CT, C99) of APP. Amino acids are shown in the one-letter code and are numbered according to the A4CT-sequence (1–99). The vertical bars within the A4CT-sequence indicate the cleavage sites of γ-secretase after residues 40 and 42. The shaded area represents the transmembrane domain. The horizontal black bars indicate the synthetic peptides that were used for the generation of the three monoclonal antibodies W02 (which binds all Aβ species), G2–10 (which is specific for Aβ40 and p340), and G2–11 (which is specific for Aβ42 and p342).
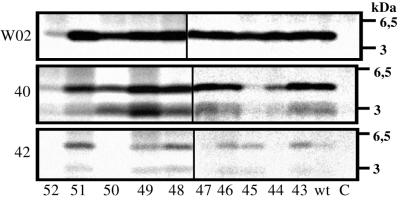
Metabolically labeled COS7 cells stably transfected with an SPA4CT construct secrete Aβ as seen by immunoprecipitation with the monoclonal antibody W02 (Fig. 2), which precipitates all Aβ species regardless of their individual C-terminal residue (Aβ42 or Aβ40; the gels for T48A and I45V are not shown). Thus, the γ-secretase activity seems to have a broad sequence specificity, as none of the mutations abolished γ-secretase cleavage. We cannot exclude that the mutations might affect slightly the overall amount of secreted Aβ (Fig. 2). But when normalizing the amount of Aβ generated to the expression level of A4CT in each particular transfection, no significant differences were observed between wt and mutant A4CT proteins (data not shown). Control cells stably transfected with the expression vector alone did not secrete detectable amounts of Aβ. Interestingly, COS7 cells expressing SPA4CT-L52F secreted only low amounts of Aβ (Fig. 2). To find out whether A4CT-L52F (derived from SPA4CT-L52F by signal peptidase) was a poor substrate for the γ-secretase activity, we immunoprecipitated A4CT from the cell lysate. In contrast to A4CT-wt and the other A4CT mutants, A4CT-L52F was barely detectable in the cell lysate (results not shown). The reason for this low amount of A4CT-L52F is not known. However, residue 52 is the last amino acid in the transmembrane domain of A4CT and is located directly in front of the three lysine residues (Lys-53–Lys-54–Lys-55) that function as a stop-transfer sequence for A4CT (Fig. 1). Thus, the large aromatic phenylalanine at position 52 might interfere with the correct membrane insertion of A4CT-L52F. However, A4CT-L52F was not detected in the conditioned medium (data not shown). Therefore, it seems possible that the phenylalanine at position 52 at least leads to a change in the protein structure that could be followed by a rapid degradation of A4CT-L52F. Because of the low amounts of A4CT-L52F and Aβ, it was not possible to determine whether this mutation L52F has a direct influence on the γ-secretase cleavage.
Figure 2.
Processing of the mutant A4CT proteins to Aβ. COS7 cells were transfected stably with the indicated SPA4CT constructs (the numbers indicate the residue in the A4CT-sequence that was replaced by phenylalanine) and metabolically labeled with [35S]methionine. Aβ was immunoprecipitated from the conditioned medium by using the antibodies shown on the left side of the gels. W02, an antibody that binds all Aβ species; 40, antibody G2–10 that binds Aβ40 and p340; 42, antibody G2–11 that binds Aβ42 and p342; C, vector control. Vertical black bars distinguish the two separate electrophoresis gels used.
Generation of Aβ42 and Aβ40.
To determine whether the mutated A4CT constructs were processed in the same manner as A4CT-wt, we used the Aβ42- and Aβ40-specific antibodies G2–11 and G2–10, respectively, to immunoprecipitate Aβ from the conditioned medium (Fig. 2). Besides the 4.5-kDa peptide band (Aβ), two additional peptide bands were immunoprecipitated, with apparent molecular masses of 3 kDa (p3) and 3.5 kDa (p3.5). The N terminus of both p3.5 and p3 could be generated by the same α-secretase activity (28), and therefore, we use the term p3 for both peptides, p3.5 and p3.
Fig. 2 shows that A4CT-wt and all A4CT-mutants, including A4CT-L52F, were processed by the γ-secretase activity to the peptides Aβ and p3 ending with residue Val-40 (Aβ40 and p340). The lowest amount of both peptides was found for A4CT-I45F. The peptides ending in residue Ala-42 (Aβ42 and p342) were detected for all A4CT proteins except A4CT-V44F, I47F, V50F, and L52F (Fig. 2). As the overall amount of Aβ was very low for the L52F mutant (Fig. 2), it is possible that this mutant was still processed to Aβ42 and p342 but that the amounts of these peptides were below the detection limit.
Ratios of Aβ42/Aβ40 and p342/p340.
To study the effects of the different A4CT mutations on the cleavage specificity of γ-secretase (ratios Aβ42/Aβ40 and p342/p340), the amounts of the four peptides (Aβ40, Aβ42, p340, and p342) were quantified by phosphorimaging. The ratios of Aβ42/Aβ40 and p342/p340, as determined here, are expressed as percentages and represent relative but not absolute ratios, because the antibodies have different affinities for the corresponding peptides.
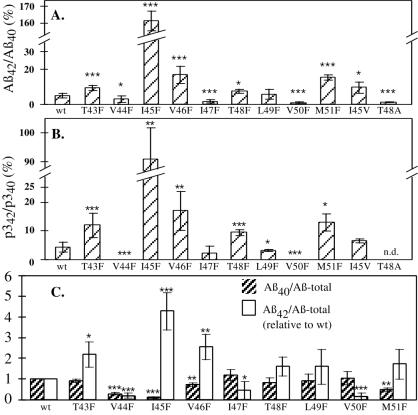
In comparison to A4CT-wt (relative ratio of Aβ42/Aβ40 = 4.7 ± 1.3%; ref. 20), the phenylalanine mutations at Val-44, Ile-47, and Val-50 led to decreased peptide ratios of Aβ42/Aβ40 (Fig. 3A); thus, γ-cleavage occurred more efficiently after Aβ40 and less efficiently after Aβ42. Compared with A4CT-wt, the ratio of Aβ42/Aβ40 was reduced to 60% for the mutation V44F (Aβ42/Aβ40 = 2.9 ± 1.9%; P < 0.05) and to 30% for the mutation I47F (Aβ42/Aβ40 = 1.4 ± 0.9%; P < 0.001). The largest decrease was found for the mutation V50F (Aβ42/Aβ40 = 0.8 ± 0.6%; P < 0.001), corresponding to 20% of the ratio measured for A4CT-wt.
Figure 3.
Relative amounts of the Aβ and p3 species in the conditioned medium of COS7-cells expressing SPA4CT. The amounts of p340, p342, Aβ40, Aβ42, and Aβ-total (immunoprecipitation with antibody W02) in Fig. 2 were quantified by phosphorimaging in 4–17 (A) or 3 (B) independent experiments. (A) Relative ratios of Aβ42/Aβ40. (B) Relative ratios of p342/p340. (C) Relative ratios of Aβ42/Aβ-total and Aβ40/Aβ-total. The ratios Aβ40/Aβ-total and Aβ42/Aβ-total were divided by the corresponding ratios obtained for A4CT-wt in the same experiment [(Aβ40/Aβ-total)mutant/(Aβ40/Aβ-total)wt]. Thus, the divided ratios for A4CT-wt are 1.0. Columns represent the mean values. Black error bars give the standard deviation. The asterisks indicate the significance (Student’s t test) relative to A4CT-wt (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001). n.d., not determined.
The mutations T43F, I45F, V46F, T48F, L49F, and M51F produced increased ratios of Aβ42/Aβ40 compared with A4CT-wt (Fig. 3A); thus, γ-cleavage occurred more efficiently after Aβ42 and less efficiently after Aβ40. Replacement of Thr-43 by phenylalanine led to an increase of Aβ42/Aβ40 by a factor of 2.0 (Aβ42/Aβ40 = 9.5 ± 1.5%; P < 0.001; ref. 20). Substitution of Ile-45 by phenylalanine produced a dramatic increase (34-fold) in the Aβ42/Aβ40 ratio (Aβ42/Aβ40 = 161.9 ± 5.7%; P < 0.001). For comparison, the mutation V46F that was found originally in a familial case of Alzheimer’s disease, increased the ratio of Aβ42/Aβ40 by a factor of 3.6 (Aβ42/Aβ40 = 17.1 ± 4.8%; P < 0.001; ref. 20). The mutation T48F showed an increase by a factor of 1.7 (Aβ42/Aβ40 = 7.7 ± 1.1%; P < 0.05). However, when Thr-48 was replaced by the small, aliphatic amino acid alanine, the Aβ42/Aβ40 ratio (1.3 ± 0.2%; P < 0.001) was decreased to 30% of the ratio found for A4CT-wt. A slight increase (1.3-fold), which was not statistically significant, was measured for the mutation L49F (Aβ42/Aβ40 = 5.8 ± 2.8%). Substitution of Met-51 by phenylalanine led to an increase of the ratio Aβ42/Aβ40 by a factor of 3.4 (Aβ42/Aβ40 = 15.4 ± 1.6%; P < 0.001).
The recently discovered mutation I45V, which causes familial Alzheimer’s disease (27), increased the ratio of Aβ42/Aβ40 by a factor of 2.1 (Aβ42/Aβ40 = 9.7 ± 3.2%; P < 0.05) compared with A4CT-wt. Our results, which were obtained with COS7 cells, are thus in agreement with the recent data of Eckman et al. (27), who found an increase by a factor of 1.3 (with Chinese hamster ovary cells) and by a factor of 1.6 (with 293 cells).
The mutations analyzed did not alter only the ratios of Aβ42/Aβ40 but also the ratios of p342/p340 (Fig. 3B). For most of the mutations, the extent of the alteration in the ratio of p342/p340 was similar to the corresponding alterations that were found for the Aβ42/Aβ40 ratios. Differences in both ratios likely are caused by the fact that the p3 concentration in the conditioned medium of SPA4CT-expressing COS7 cells is lower than the concentration of Aβ (Fig. 2); thus, the determination of the 42/40 ratios is less precise for p3 than for Aβ (Fig. 3B).
The mutations I45F, V46F, and V50F were also introduced into APP-695. The mutations did not affect the maturation of these proteins compared with wt APP-695 (data not shown). Moreover, the 42/40 ratio of Aβ derived from APP was within the range of standard deviation identical to the ratio found for Aβ derived from the corresponding A4CT protein (data not shown).
Ratios of Aβ42/Aβ-Total and Aβ40/Aβ-Total.
Compared with A4CT-wt, the A4CT mutants analyzed in our study altered the peptide ratio of Aβ42/Aβ40. If no Aβ peptides were generated in addition to Aβ42 and Aβ40, then an altered ratio of Aβ42/Aβ40 implies that the relative amount of one of the two peptides (Aβ42/Aβ-total; Aβ40/Aβ-total) must be increased, whereas the relative amount of the other peptide must be decreased at the same time.
To address this issue, Aβ was immunoprecipitated from three equal volumes of conditioned medium by using antibodies G2–11 (specific for Aβ42), G2–10 (specific for Aβ40), or W02 (which binds all Aβ species, corresponding to Aβ-total). As the antibodies have different affinities to the peptides, the ratios of Aβ42/Aβ-total and Aβ40/Aβ-total are relative ratios. With A4CT-wt as standard, the ratios Aβ42/Aβ-total and Aβ40/Aβ-total were divided by the corresponding ratios found for A4CT-wt determined in the same set of experiments. This division shows whether the amounts of Aβ42 and Aβ40 among Aβ-total were altered because of the phenylalanine mutations. Fig. 3C indicates that the alterations of these ratios were as expected. For those mutations that caused an increased ratio of Aβ42/Aβ40 (i.e., T43F, I45F, V46F, T48F, L49F, and M51F; Fig. 3A), the ratio of Aβ42/Aβ-total also was increased, whereas the ratio of Aβ40/Aβ-total was decreased compared with A4CT-wt (Fig. 3C). For the mutations that caused a decreased ratio of Aβ42/Aβ40 (i.e., I47F and V50F; Fig. 3A), the ratio of Aβ42/Aβ-total also was found to be decreased, whereas the ratio of Aβ40/Aβ-total was increased relative to A4CT-wt (Fig. 3C). Because of the restricted number of measurements, not all the changes in the ratios of Aβ42/Aβ-total and Aβ40/Aβ-total were statistically significant.
One exception was the mutation V44F. This mutation caused a decreased ratio of Aβ42/Aβ40 (Fig. 3A; decrease to 60% compared with A4CT-wt). Fig. 3C shows that the ratio of Aβ40/Aβ-total and the ratio of Aβ42/Aβ-total were also decreased. The decrease of Aβ42/Aβ-total was more pronounced than the decrease of Aβ40/Aβ-total, reflecting the decrease in the ratio of Aβ42/Aβ40 (Fig. 3A). However, the relative amounts of both peptides (Aβ42 and Aβ40) to Aβ-total were decreased compared with A4CT-wt, indicating that a further Aβ species must have been generated in addition to Aβ40 and Aβ42. This new Aβ species must have a different C terminus than Aβ40 and Aβ42.
Processing of A4CT-V44F to Aβ38.
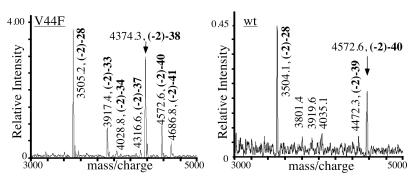
To determine the C terminus of this new Aβ species, Aβ in the conditioned medium of COS7 cells expressing A4CT-V44F was first immunoprecipitated and then analyzed by mass spectrometry. As a control, Aβ derived from A4CT-wt was analyzed. The spectrum obtained for A4CT-wt showed a strong signal at a molecular mass of 4572.6 Da corresponding to Aβ40 (Fig. 4) and confirming data obtained earlier (17) for Aβ derived from full-length APP. Aβ42 was not detected. Because Aβ42 easily forms aggregates (29), these aggregates might have escaped the detection in the mass spectrometer. The only difference between Aβ from A4CT-wt and Aβ from APP (17) is that the former shows slightly less N- and C-terminal heterogeneity. For A4CT-V44F, a prominent Aβ signal was found with a molecular mass of 4374.3 Da, corresponding to Aβ38; low signal intensities were found for Aβ37, Aβ40, and Aβ41 (Fig. 4). Thus, the mutation V44F shifted the main cleavage site of γ-secretase by two residues in the N-terminal direction.
Figure 4.
Mass spectrum of Aβ derived from A4CT-V44F and A4CT-wt. By using antibody 4G8, Aβ was first immunoprecipitated from the conditioned medium of COS7 cells expressing SPA4CT-wt or V44F and then analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. In addition to the measured molecular masses, peaks in the spectrum were labeled with Aβ peptide sequence numbers. The main peaks are labeled with arrows. The peptides start at the N terminus with the two additional amino acids leucine and glutamic acid (Fig. 1).
For A4CT-I45F, no Aβ signals were found, although Aβ was detected in the conditioned medium by western blot analysis (data not shown). Because the Aβ derived from A4CT-I45F is mainly Aβ42, which aggregates rapidly, it might escape the detection in the mass spectrometer as discussed above.
DISCUSSION
Analyzing the Cleavage Specificity of γ-Secretase.
Proteolytic processing of APP by β-secretase yields A4CT (C99), which is cleaved further by the as yet unknown protease called γ-secretase activity (Fig. 1). γ-secretase cleaves mainly after residue 40 of A4CT and partly after residue 42, thus generating the peptides Aβ40 and, to a lesser extent, Aβ42. Because γ-secretase cleaves at different peptide bonds, it is reasonable to divide the term substrate specificity of γ-secretase into two subfactors: the sequence specificity and the cleavage specificity. The term sequence specificity indicates whether the activity of γ-secretase, and hence the amount of generated Aβ, depends on the amino acid sequence of A4CT. In contrast, the term cleavage specificity refers to whether the cleavage site of γ-secretase (Aβ42/Aβ40 ratio) depends on the amino acid sequence of A4CT. Of these two factors, the cleavage specificity of γ-secretase is particularly crucial for the pathogenesis of Alzheimer’s disease, because Aβ42 is strongly linked to the disease (for a review, see ref. 5). Additional examples of enzymes with a cleavage specificity distinct from the sequence specificity are restriction enzymes that cleave outside of their recognition sequence. These enzymes bind to a clearly defined recognition sequence and, thus, have a high sequence specificity. However, the distant cleavage site does not depend on the particular nucleotide sequence around the cleavage site. Thus, these enzymes have a broad cleavage specificity.
An appropriate way to study the cleavage specificity of the γ-secretase activity is to express mutant precursors for Aβ in eukaryotic cells and analyze the effect of these mutations on the generation of both Aβ40 and Aβ42. In the case of full-length APP, the mutations might not affect only γ-secretase cleavage but also β-secretase cleavage. Using A4CT, which is the direct precursor for Aβ (6), offers the advantage of obviating the β-secretase cleavage. A4CT is processed to Aβ42 and Aβ40 in the same way as full-length APP (20). To ensure correct insertion of A4CT into the membrane of the endoplasmic reticulum, an N-terminal SP was added to A4CT (SPA4CT; Fig. 1; ref. 24), which is cleaved during membrane insertion by signal peptidase.
Phenylalanine-Scanning Mutagenesis.
Point mutations within the transmembrane domain of A4CT at amino acids Thr-43 and Val-46 have been shown to alter the cleavage specificity of γ-secretase (Aβ42/Aβ40; refs. 18–20). Thr-43 is located directly at the γ-cleavage site, where the pathogenic long Aβ species Aβ42 is generated (Fig. 1). Val-46 is in close proximity to this γ-cleavage site (Fig. 1). To analyze whether other mutations in the transmembrane domain of A4CT also affect the position of the γ-secretase cleavage site, we performed a phenylalanine scan (substitution of residues by phenylalanine) of all membrane domain residues of A4CT outside the Aβ-domain (residues 43 to 52). In similar studies with different proteins, the residues of the transmembrane domain are often replaced by alanine or cysteine (30, 31). However, we chose phenylalanine for the substitution for three reasons. First, the mutation V46F occurs in some AD patients and strongly influences the cleavage specificity of γ-secretase (18). Second, phenylalanine is one of the largest amino acids, carries an aromatic side chain, and, thus, is clearly different from the other residues in the transmembrane domain of APP. Therefore, we expected the phenylalanine mutations to have a more pronounced effect on the cleavage specificity of γ-secretase than substitutions with isoleucine or valine. This assumption was validated by the finding that the mutation I45F led to a 34-fold increase in the ratio of Aβ42/Aβ40, compared with a 2.1-fold increase found for the mutation I45V. Third, the amino acid phenylalanine fits into α-helical protein structures and also is found in the transmembrane domains of other type I membrane proteins (32).
Our phenylalanine scan of residues 43 to 52 of A4CT showed that all A4CT mutants were still processed to Aβ and that the overall amount of secreted Aβ did not vary significantly between the mutants. Thus, the γ-secretase activity must have a broad sequence specificity, which is in agreement with recent studies (18–20, 33).
In contrast to the broad sequence specificity, the cleavage specificity of γ-secretase (Aβ42/Aβ40 ratio) strictly depends on the amino acid sequence of the transmembrane domain of A4CT as shown by the results of our phenylalanine scan; all mutations altered the cleavage specificity of γ-secretase, and, depending on the mutation, the Aβ42/Aβ40 ratio was between 0.2- and 34-fold greater than the ratio found for A4CT-wt. Moreover, the magnitude of the alteration of the Aβ42/Aβ40 ratio depended on the amino acid used. Mutation I45F increased the Aβ42/Aβ40 ratio 34-fold, whereas mutation I45V increased the ratio 2.1-fold. The finding that the cleavage specificity of γ-secretase is strictly sequence-dependent gains additional support by the two mutations at Thr-48, which led to either an increased (T48F) or a reduced (T48A) Aβ42/Aβ40 ratio. Further confirmations come from previous studies, where we and others have shown that substitutions at residues 43 and 46 also led to different Aβ42/Aβ40 ratios, depending on the residue that was used for the substitution (18–20).
Most interestingly, in the phenylalanine scan, not only mutations close to the cleavage site, but even mutations at residues 50 and 51, which are located at the end of the transmembrane domain of A4CT, influenced the cleavage specificity, indicating that γ-secretase could directly interact with the whole C-terminal half of the transmembrane domain of A4CT.
Taken together, our experimental data show that the amino acid composition of the C-terminal half of the transmembrane domain of A4CT is a major factor determining the cleavage specificity of γ-secretase. Whether the γ-secretase activity could also interact with the N-terminal half of the transmembrane domain of A4CT remains to be studied.
Similar to A4CT-wt, most A4CT mutants were cleaved mainly after residue 40, despite the fact that all A4CT mutants altered the cleavage specificity of γ-secretase (Aβ42/Aβ40 ratio). However, there were two exceptions: A4CT-V44F was processed mainly to Aβ38, and A4CT-I45F was processed mainly to Aβ42. Thus, of the residues in the transmembrane domain of A4CT, residues 44 and 45 have a particularly important influence on the cleavage specificity of γ-secretase. This influence might be explained by the fact that in the structure of the transmembrane domain of A4CT, which is assumed to be in an α-helical conformation, both residues 44 and 45 are in close proximity to the γ-cleavage sites after residues 40 and 42 (Fig. 5B).
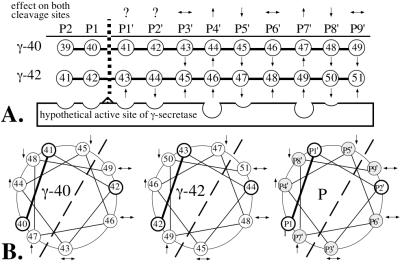
Figure 5.
Schematic representation of the amino acid positions (P) of A4CT relative to the cleavage site of γ-secretase. (A) Linear arrangement of the amino acids relative to the cleavage site after residue 40 (γ-40) or 42 (γ-42). The arrows (based on the data in Fig. 3) indicate whether the phenylalanine mutants of A4CT were cleaved at one site (γ-40 or γ-42) more (↑) or less (↓) efficiently than at the other site, compared with A4CT-wt. The scissile peptide bond, which is shown by a broken vertical line, is constituted by the residues in positions P1 and P1′. The arrows above the P positions show whether the mutations at the individual positions had the same effect (↑ or ↓) or the opposite effect (↔) on both cleavage sites. Question marks indicate that the effect of the mutation has been determined only for one cleavage site. (B) Helical wheel arrangement of amino acids 43 to 52 of A4CT with respect to the cleavage sites after residues 40 (γ-40) and 42 (γ-42). Helical arrangement P shows the amino acid positions in the transmembrane domain of A4CT in general. The scissile peptide bond (P1–P1′) is shown with a solid bold line. Those amino acid positions for which the effect of the phenylalanine mutations has been determined for both cleavage sites are shaded. The dashed line separates the helical wheel into two halves, one of which is supposed to interact directly with γ-secretase, whereas the other half is likely to face away from γ-secretase.
Helical Model for γ-Secretase Cleavage.
The proteolytic cleavages after residues 40 (γ-40 site) and 42 (γ-42 site) of A4CT are suggested to be carried out by distinct γ-secretases (11, 21, 22). Depending on whether the γ-40- or γ-42-secretase cleaves A4CT, different residues of A4CT are located at the same P position (for P nomenclature, see ref. 34). Fig. 5A shows whether a phenylalanine mutation at a certain residue—and, thus, at a specific P position—increased or decreased the cleavage of either the γ-40- or the γ-42-secretase. In terms of enzyme–substrate interaction, a decreased cleavage could be explained in such a way that the binding pocket at this position in the active site of γ-secretase is too small to accommodate the large phenylalanine residue (Fig. 5A; P5′ and P8′), resulting in a repulsion of the corresponding γ-secretase. Instead, the other γ-secretase would cleave more effectively.
A comparison of the γ-40- and γ-42-secretases in Fig. 5A shows that both proteases exhibit similar features. For both proteases, a phenylalanine mutation at P4′ and P7′ led to an increased cleavage, whereas the same mutation at P5′ and P8′ led to a reduced cleavage (Fig. 5A). The mutations at P3′, P6′, and P9′ had opposite effects on both cleavage sites (Fig. 5A).
The common features of both proteases are even more impressive if the transmembrane domain of A4CT is shown in its presumed α-helical conformation (Fig. 5B) and not in the linear arrangement of the residues (Fig. 5A). Those positions (P4′, P5′, P7′ and P8′) at which the phenylalanine mutations had the same effect for both γ-secretases (Fig. 5A) are located on the left side of the helix (Fig. 5B), where the cleavage site (P1–P1′) also is located and, thus, where the γ-secretases must bind the helix. Mutations on this side (P4′, P5′, P7′, and P8′) could affect the interaction between A4CT and the γ-secretases directly, resulting in an altered Aβ42/Aβ40 ratio as is found in the conditioned medium of cells expressing mutant A4CT or APP proteins (18–20). Those positions (P3′, P6′, and P9′) at which the phenylalanine mutations had opposite effects on both γ-secretase cleavage sites are located on the other side of the helix (Fig. 5B). These positions might exert their effect by a different mechanism, perhaps by interacting with a part of the γ-secretase that is different between the γ-40-secretase and the γ-42-secretase or by facing away from the γ-secretases. In the latter case, they could interact with the lipids of the membrane or with an additional protein, which might be part of a γ-secretase–A4CT complex. Such a protein could be presenilin 1, which is required for γ-secretase cleavage (35).
Our helical model has several possible implications. First, the γ-40- and the γ-42-secretases, although they are different enzymes, are likely to have a similar or the same active site. This idea is in good agreement with the findings that both proteases cleave preferentially within stretches of hydrophobic residues and might be inhibited by using the same peptide aldehydes and derivatives as inhibitors (11, 21–23). Interestingly, our helical model would be consistent even with the existence of only a single γ-secretase enzyme, cleaving either at one or the other side of the helix and thereby generating the Aβ40 and Aβ42 peptides. Second, the helical model is consistent with the γ-cleavage occurring within the membrane such that the γ-secretase activity itself could be a membrane protein. Similar to the γ-secretase cleavage of A4CT, two other proteases cleave their substrates within or close to their transmembrane domains: S2P (36) and m-AAA (37). Both proteases are believed to form channel-like structures in the membrane, providing the required space to accommodate the transmembrane domain of the substrate in an α-helical conformation. Similarly, the γ-secretase(s) might turn out to be new member(s) of a family of proteases involved in the degradation of transmembrane domains.
CONCLUSION
Our study could lead to the creation of new transgenic animal models of AD and to the design of efficient inhibitors for the γ-secretase activity. Among our mutations, A4CT-I45F was processed nearly exclusively to the pathogenic, rapidly aggregating Aβ42. Because the generation of large amounts of Aβ42 seems to be the most important requirement for the successful generation of transgenic animals that mimic the pathology of Alzheimer’s disease (38), A4CT-I45F might be highly valuable for generating such animals. To test this idea, transgenic mice should be evaluated by using the cDNA of A4CT-I45F.
The inhibitors of the γ-secretase activity that have been identified in published articles are derivatives of peptides that mimic the sequence around the γ-secretase cleavage site (7, 23, 39). However, such inhibitors are not very potent, because they are effective at concentrations in the 0.1–1 mM range. Our helical model for the interaction between A4CT and γ-secretase should enable the design of inhibitors that mimic the surface of the helical transmembrane domain of A4CT (Fig. 5B). Such peptides or peptidomimetics might turn out to be highly potent drugs for treatment or prevention of AD.
Acknowledgments
We thank Drs. Dirk Beher, Geneviève Evin, and Tobias Hartmann for helpful comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 317, a fellowship from the Fonds der Chemischen Industrie of Germany to S.F.L., and National Institutes of Health/National Institute on Aging Grant AG16065 to R.W.
ABBREVIATIONS
- Aβ
β-amyloid
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- Pn
amino acid position n
- wt
wild-type, SP, signal peptide
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 2574.
References
- 1.Glenner G G, Wong C W. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 2.Masters C L, Simms G, Weinman N A, Multhaup G, McDonald B L, Beyreuther K. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang J, Lemaire H G, Unterbeck A, Salbaum J M, Masters C L, Grzeschik K H, Multhaup G, Beyreuther K, Mueller-Hill B. Nature (London) 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Evin G, Beyreuther K, Masters C L. Amyloid. 1994;1:263–280. [Google Scholar]
- 5.Selkoe D J. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 6.Busciglio J, Gabuzda D H, Matsudaira P, Yankner B A. Proc Natl Acad Sci USA. 1993;90:2092–2096. doi: 10.1073/pnas.90.5.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higaki J, Quon D, Zhong Z, Cordell B. Neuron. 1995;14:651–659. doi: 10.1016/0896-6273(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 8.Haass C, Schlossmacher M G, Hung A Y, Vigo-Pelfrey C, Mellon A, Ostaszewski B L, Lieberburg I, Koo E H, Schenk D, Teplow D B, et al. Nature (London) 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 9.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C, et al. Nature (London) 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 10.Shoji M, Golde T E, Ghiso J, Cheung T T, Estus S, Shaffer L M, Cai X D, McKay D M, Tintner R, Frangione B, et al. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- 11.Citron M, Diehl T S, Gordon G, Biere A L, Seubert P, Selkoe D J. Proc Natl Acad Sci USA. 1996;93:13170–13175. doi: 10.1073/pnas.93.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook D G, Forman M S, Sung J C, Leight S, Kolson D L, Iwatsubo T, Lee V M, Doms R W. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann T, Bieger S C, Brühl B, Tienari P J, Ida N, Allsop D, Roberts G W, Masters C L, Dotti C G, Unsicker K, et al. Nat Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- 14.Tienari P J, Ida N, Ikonen E, Simons M, Weidemann A, Multhaup G, Masters C L, Dotti C G, Beyreuther K. Proc Natl Acad Sci USA. 1997;94:4125–4130. doi: 10.1073/pnas.94.8.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wild-Bode C, Yamazaki T, Capell A, Leimer U, Steiner H, Ihara Y, Haass C. J Biol Chem. 1997;272:16085–16088. doi: 10.1074/jbc.272.26.16085. [DOI] [PubMed] [Google Scholar]
- 16.Skovronsky D M, Doms R W, Lee V M. J Cell Biol. 1998;141:1031–1039. doi: 10.1083/jcb.141.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Sweeney D, Gandy S E, Sisodia S S. J Biol Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki N, Cheung T T, Cai X D, Odaka A, Otvos L, Jr, Eckman C, Golde T E, Younkin S G. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama K, Tomita T, Shinozaki K, Kume H, Asada H, Saido T C, Ishiura S, Iwatsubo T, Obata K. Biochem Biophys Res Commun. 1996;227:730–735. doi: 10.1006/bbrc.1996.1577. [DOI] [PubMed] [Google Scholar]
- 20.Lichtenthaler S F, Ida N, Multhaup G, Masters C L, Beyreuther K. Biochemistry. 1997;36:15396–15403. doi: 10.1021/bi971071m. [DOI] [PubMed] [Google Scholar]
- 21.Klafki H, Abramowski D, Swoboda R, Paganetti P A, Staufenbiel M. J Biol Chem. 1996;271:28655–28659. doi: 10.1074/jbc.271.45.28655. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki T, Haass C, Saido T C, Omura S, Ihara Y. Biochemistry. 1997;36:8377–8383. doi: 10.1021/bi970209y. [DOI] [PubMed] [Google Scholar]
- 23.Wolfe M S, Citron M, Diehl T S, Xia W, Donkor I O, Selkoe D J. J Med Chem. 1998;41:6–9. doi: 10.1021/jm970621b. [DOI] [PubMed] [Google Scholar]
- 24.Dyrks T, Dyrks E, Mönning U, Urmoneit B, Turner J, Beyreuther K. FEBS Lett. 1993;335:89–93. doi: 10.1016/0014-5793(93)80446-2. [DOI] [PubMed] [Google Scholar]
- 25.Ida N, Hartmann T, Pantel J, Schröder J, Zerfass R, Förstl H, Sandbrink R, Masters C L, Beyreuther K. J Biol Chem. 1996;271:22908–22914. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]
- 26.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 27.Eckman C B, Mehta N D, Crook R, Perez-tur J, Prihar G, Pfeiffer E, Graff-Radford N, Hinder P, Yager D, Zenk B, et al. Hum Mol Genet. 1997;6:2087–2089. doi: 10.1093/hmg/6.12.2087. [DOI] [PubMed] [Google Scholar]
- 28.Haass C, Selkoe D J. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 29.Jarrett J T, Berger E P, Lansbury P T., Jr Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 30.Ambesi A, Pan R L, Slayman C W. J Biol Chem. 1996;271:22999–23005. doi: 10.1074/jbc.271.38.22999. [DOI] [PubMed] [Google Scholar]
- 31.Lee G F, Dutton D P, Hazelbauer G L. Proc Natl Acad Sci USA. 1995;92:5416–5420. doi: 10.1073/pnas.92.12.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landolt-Marticorena C, Williams K A, Deber C M, Reithmeier R A. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 33.Tischer E, Cordell B. J Biol Chem. 1996;271:21914–21919. doi: 10.1074/jbc.271.36.21914. [DOI] [PubMed] [Google Scholar]
- 34.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 35.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 36.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 37.Leonhard K, Herrmann J M, Stuart R A, Mannhaupt G, Neupert W, Langer T. EMBO J. 1996;15:4218–4229. [PMC free article] [PubMed] [Google Scholar]
- 38.Duff K. Trends Neurosci. 1997;20:279–280. doi: 10.1016/s0166-2236(97)01093-x. [DOI] [PubMed] [Google Scholar]
- 39.Allsop D, Christie G, Gray C, Holmes S, Markwell R, Owen D, Smith L, Wadsworth H, Ward R V, Hartmann T, et al. In: Alzheimer’s Disease: Biology, Diagnosis and Therapeutics. Iqbal K, Winblad B, Nishimura T, Takeda M, Wisniewski H M, editors. New York: Wiley; 1997. pp. 717–727. [Google Scholar]