Abstract
African sleeping sickness is a fatal disease that is caused by the protozoan parasite Trypanosoma brucei. Polyamine biosynthesis is an essential pathway in the parasite and is a validated drug target for treatment of the disease. S-adenosylmethionine decarboxylase (AdoMetDC) catalyzes a key step in polyamine biosynthesis. Here, we show that trypanosomatids uniquely contain both a functional AdoMetDC and a paralog designated prozyme that has lost catalytic activity. The T. brucei prozyme forms a high-affinity heterodimer with AdoMetDC that stimulates its activity by 1,200-fold. Both genes are expressed in T. brucei, and analysis of AdoMetDC activity in T. brucei extracts supports the finding that the heterodimer is the functional enzyme in vivo. Thus, prozyme has evolved to be a catalytically dead but allosterically active subunit of AdoMetDC, providing an example of how regulators of multimeric enzymes can evolve through gene duplication and mutational drift. These data identify a distinct mechanism for regulating AdoMetDC in the parasite that suggests new strategies for the development of parasite-specific inhibitors of the polyamine biosynthetic pathway.
Keywords: gene duplication, S-adenyosylmethionine decarboxylase, prozyme, Trypanosoma brucei
African sleeping sickness is a neglected disease that is caused by the protozoan parasite Trypanosoma brucei. It is epidemic in subSaharan Africa, where 60 million people are at risk, and tens of thousands die annually of this disease (1). Without treatment, the disease is always fatal. Current drug therapies are limited by toxicity and the requirement for complex treatment regimes (2). Polyamines are essential for cell growth in all organisms. They influence chromatin structure, modify translation initiation factor eIF5A, and regulate gene expression through a number of mechanisms (3–6). In trypanosome parasites, the polyamine spermidine is conjugated with glutathione to form a unique cofactor termed trypanothione that functions in cellular redox reactions (7). Excess polyamines lead to cancer in mammalian cells (4), and inhibitors of polyamine biosynthesis have been widely studied for their potential as antiproliferative agents (6, 8). The most successful clinical application of these inhibitors is the treatment of African sleeping sickness with the ornithine decarboxylase suicide inhibitor α-difluoromethylornithine (eflornithine) (9). Eflornithine is the only therapy for sleeping sickness with a known mechanism of action, and its effectiveness demonstrates the importance of the polyamine pathway to the parasite.
S-adenosylmethionine decarboxylase (AdoMetDC) is required for the formation of the precursor used for the synthesis of spermidine and spermine from putrescine, and in mammalian cells it is a rate-limiting step in polyamine formation (10). Knockout of the AdoMetDC gene in the trypanosomatid Leishmania donovani led to spermidine auxotropy (11). The suicide inhibitor of AdoMetDC, MDL73811, has demonstrated efficacy in animal models of both T. brucei (12) and a related parasite Trypanosoma cruzi (13), which is the causative agent of Chagas disease. Thus, AdoMetDC is considered a promising, but as yet unexploited target for the development of new anti-trypanosomal drugs.
AdoMetDC is a pyruvoyl-dependent enzyme and uses this cofactor to stabilize the carbanion intermediate formed during the decarboxylation reaction. The pyruvoyl-moiety derives from an autocatalytic cleavage reaction that generates the active enzyme consisting of two chains (βα), with the pyruvoyl group formed at the N terminus of the α-chain (14, 15). Human AdoMetDC is a homodimer, and both the processing reaction and decarboxylation of AdoMet are stimulated by putrescine (10, 16). The x-ray structure shows that the active sites sit in a large cleft between β-sheets distal from the dimer interface and that the putrescine-binding sites are formed by a group of acidic residues in the β-sandwich core ≈15 Å from the active sites (14, 17). This site is eliminated in the structure of the monomeric plant enzyme, which is fully active without putrescine (18). The putrescine-binding site is partially conserved in the trypanosomatid enzymes. Putrescine stimulates the activity of the recombinant T. cruzi enzyme, but it is not required for processing (19–23). Perplexingly, the putrescine-activated T. cruzi enzyme has significantly lower catalytic efficiency than the enzyme from mammals and plants, thus suggesting the possibility that other regulatory factors are necessary for enzyme function.
The polyamine biosynthetic and catabolic enzymes are tightly regulated in animals, plants, and yeast (3, 4, 24). Unusually, in the trypanosomatid parasites, analogous regulatory mechanisms for the control of polyamine biosynthesis have not been identified. Here, we show that T. brucei AdoMetDC is activated by formation of a heterodimer with a catalytically inactive regulatory subunit termed prozyme that arose in the trypanosomatids as a gene duplication of the ancestral enzyme. The regulation of AdoMetDC by an inactive homolog is unique to the trypanosomatid parasites. The finding has implications for both the regulation of polyamines in the parasite and for the development of enzyme inhibitors that will block this essential pathway.
Results
Genomic Analysis of the Trypanosomatid AdoMetDC Family.
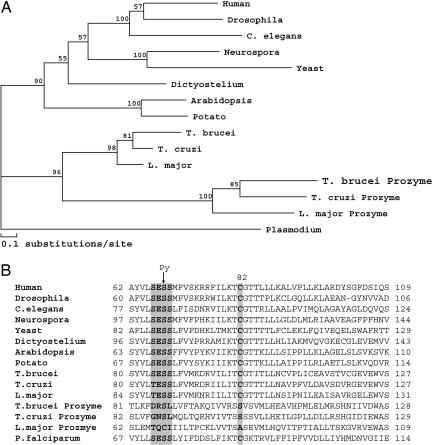
Evolutionary analysis of the AdoMetDC family indicates that the trypanosomatids, T. brucei, T. cruzi, and Leishmania major, encode two types of AdoMetDC genes: an ortholog to the functional enzyme in other species, AdoMetDC, and a paralog defined herein as prozyme [Fig. 1 and see supporting information (SI) Fig. 4], named in analogy to the ornithine decarboxylase inhibitory protein, antizyme (25). AdoMetDC and prozyme are found in close proximity in the genome; however, they have diverged significantly; prozyme shares only ≈30% amino acid sequence identity with the AdoMetDC from the same trypanosomatid species. Sequence analysis suggests that prozyme is not present outside of the trypanosomatid lineage. Thus, it appears to have arisen by gene duplication of the ancestral enzyme after the divergence of the trypanosomatids from other eukaryotes. Northern blot analysis demonstrates that both AdoMetDC and prozyme are expressed in blood form and procyclic T. brucei parasites, suggesting that they both will have a functional role in the parasites (Fig. 2A). However, the prozyme is missing several critical residues required for processing and catalytic activity (10, 14, 15, 17, 20). Therefore the prozyme is unlikely to display AdoMetDC activity and must have a unique function.
Fig. 1.
Evolutionary analysis of the AdoMetDC family. (A) Phlyogenetic tree analysis of representative eukaryotic AdoMetDCs. (B) Partial sequence alignment of AdoMetDCs. The processing site (highlighted in gray) demarcates the serine residue (S68 in human AdoMetDC numbering; S86 in T. brucei AdoMetDC) converted to pyruvate (Py) during autocatalytic cleavage to generate the functional βα subunit. The catalytic base (C82) is also highlighted. In T. brucei AdoMetDC, the α-chain contains residues 86–370, and the smaller β-chain residues 1–85. The full sequence alignment and accession numbers are presented in SI Fig. 4.
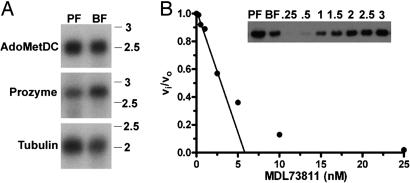
Fig. 2.
Analysis of AdoMetDC activity and expression in T. brucei lysates. (A) Northern blot analysis of mRNA (1 μg per lane) isolated from T. brucei cells for both AdoMetDC (Top) and prozyme (Middle) in procyclic form (PF), and blood form (BF) trypanosomes; tubulin (Bottom) is shown as a loading control. Molecular weight markers are given in kilobases. (B) Active-site titration of AdoMetDC in trypanosome BF cell lysates. Lysate (0.04 mg total protein; 0.01 ml at 4 mg/ml) was incubated with MDL 73811 (0–25 nM) for 1 h at 37°C. Remaining enzyme activity was determined (vi) and compared with the uninhibited activity (vo) in the lysate by quantitation of the amount of 14CO2 liberated from 1-14CO2-AdoMet. The concentration of AdoMetDC in lysates was estimated from the linear portion of the graph by the x-intercept (AdoMetDC = 6 nM). (Inset) Western blot of T. brucei AdoMetDC in PF and BF trypanosome cell lysates (0.04 mg of total protein per lane). Intensity of the 34-kDa band was compared with that measured for a titration of recombinant AdoMetDC (0.25–3 ng of protein) providing an estimate of the AdoMetDC protein concentration in the BF lysates (AdoMetDC = 5 nM). Based on the MDL 73811 titration and the Western blot analysis, the specific activity of AdoMetDC in the BF lysate can be estimated to be 0.8 s−1 (1.1 μmol/min/mg) at 0.04 mM AdoMet, which would correspond to ≈3 s−1 at saturating substrate (assuming the Km reported in Table 1).
Recombinant T. brucei AdoMetDC Has Low Catalytic Efficiency.
T. brucei AdoMetDC was expressed and purified from Escherichia coli to evaluate its kinetic properties. Recombinant T. brucei AdoMetDC is processed to generate the pyruvoyl cofactor (Fig. 3A), and it catalyzes the decarboxylation of AdoMetDC (Table 1 and SI Fig. 5). Like the T. cruzi enzyme (19–23), T. brucei AdoMetDC is stimulated by putrescine but not sufficiently to generate a catalytically efficient enzyme. The kcat of the putrescine-stimulated enzyme remains 320-fold lower than for human AdoMetDC. To determine whether the low activity could be caused by a change in activator specificity, several other polyamines were tested as activators. Norspermidine, spermidine, and cadaverine also activate T. brucei AdoMetDC; however, the activation was similar to what was observed for putrescine, suggesting that activator-specificity differences do not account for the low activity of the trypanosome enzymes (SI Fig. 5).
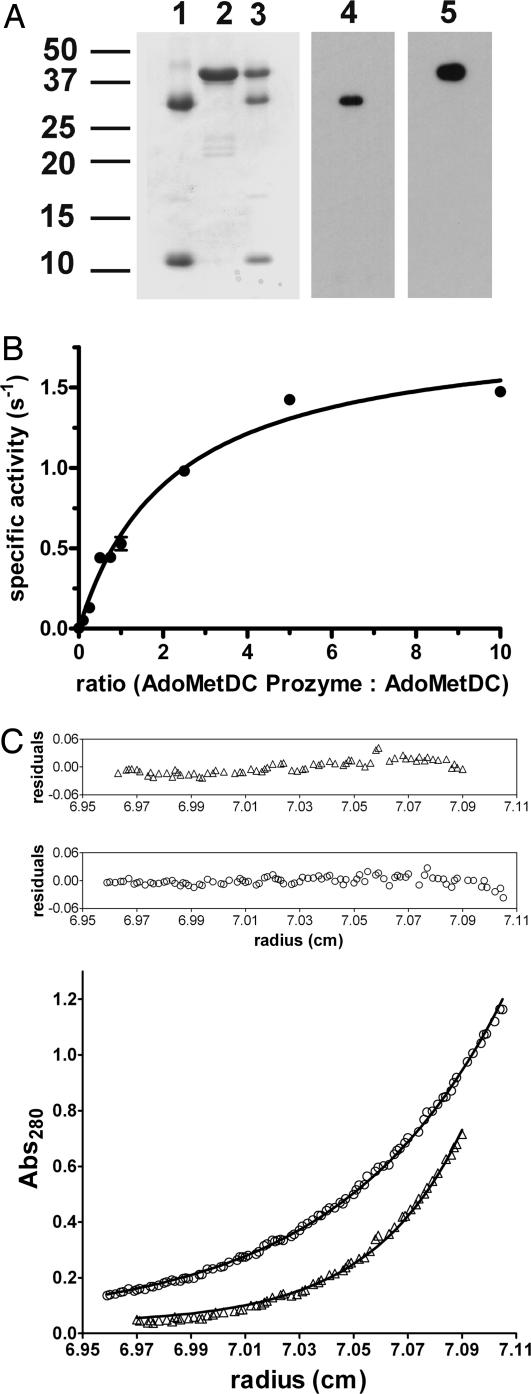
Fig. 3.
T. brucei AdoMetDC forms a heterodimer with prozyme leading to activation of the enzyme. (A) SDS/PAGE and Western blot analysis of AdoMetDC, prozyme, and AdoMetDC/prozyme complex. Coomassie-stained gel of purified AdoMetDC (lane 1), prozyme (lane 2), and the copurified complex (lane 3). Western blot of copurified complex showing His-tagged AdoMetDC probed with α-T. brucei AdoMetDC (lane 4) and Flag-tagged prozyme probed with α-Flag antibody (lane 5). For AdoMetDC, both the α subunit of 34 kDa and 11.9 kDa β-subunit are observed on the Coomassie-stained gel, but only the α-subunit is recognized by the antibody. (B). Activation of catalysis by titration of purified recombinant prozyme (0–2 μM) into purified AdoMetDC (0.2 μM) at saturating (1 mM) AdoMet. (C) Sedimentation equilibrium by analytical ultracentrifugation of the copurified AdoMetDC/prozyme complex. Data were collected at 15,000 (○) and 20,000 (Δ) rpm at 6 μM complex. The data were globally fitted to a single ideal-species model, and the molecular mass of the species was calculated to be 80,101 Da (95% confidence interval 79,212–80,997), which corresponds well to the predicted heterodimer weight of 81,400. There was no evidence for dissociation under the experimental conditions, thus the Kd for dimer dissociation is below the level of detection of the method (Kd < 0.5 μM).
Table 1.
Steady-state kinetic analysis of purified recombinant T. brucei AdoMetDC and the copurified AdoMetDC/prozyme complex
| No putrescine |
5 mM putrescine |
|||||
|---|---|---|---|---|---|---|
| kcat, s−1 | Km, mM | kcat/Km, M−1 s−1 | kcat, s−1 | Km, mM | kcat/Km, M−1 s−1 | |
| AdoMetDC | 0.0013 ± 0.0004 | 0.38 ± 0.15 | 3.4 ± 0.5 | 0.0082 ± 0.0015 | 0.24 ± 0.06 | 35 ± 3 |
| AdoMetDC/prozyme | 1.4 ± 0.1 | 0.11 ± 0.02 | 1.2 ± 0.1 × 104 | 1.7 ± 0.1 | 0.17 ± 0.02 | 1.1 ± 0.05 × 104 |
Data were collected in triplicate, and errors are the standard error of the mean. For human AdoMetDC, the steady-state kinetic parameters in the presence of putrescine were previously reported to be kcat = 2.6 s−1; kcat/Km = 4.4 × 104 M−1s−1 (23).
Characterization of AdoMetDC Activity in Blood Stream Form T. brucei Parasites.
The kinetic data on the recombinant trypanosomatid AdoMetDCs suggested that these enzymes either have intrinsically lower activity or they are activated by a previously uncharacterized mechanism. To address this question, the specific activity of AdoMetDC in the blood form T. brucei parasites was determined. Decarboxylation was followed by the standard assay using 14CO2-AdoMet as the substrate, and the concentration of AdoMetDC protein in the cell extract was estimated first by titrating the number of active sites with the AdoMetDC suicide inhibitor MDL73811 (26, 27) and second by Western blot (Fig. 2B). Both methods of determining AdoMetDC concentration in the extracts yielded similar results and demonstrated that the specific activity of AdoMetDC in blood stream form T. brucei parasites is ≈3 s−1 (4 μmol/min/mg), a value that is ≈ 400-fold higher than the kcat measured for the putrescine-stimulated recombinant enzyme (Table 1). These data provide evidence that the trypanosomatid enzymes in vivo have activities similar to those reported for AdoMetDC from other species [e.g., human AdoMetDC (23)].
Prozyme Is a Regulatory Subunit of T. brucei AdoMetDC.
The finding that AdoMetDC in T. brucei cell extracts has higher activity than the recombinant enzyme suggested that an unaccounted for factor is present in the parasites that regulates the activity of the trypanosomatid AdoMetDCs. These results led us back to question the role of the prozyme. To determine whether prozyme might regulate the activity of AdoMetDC, recombinant T. brucei prozyme was expressed and purified from E. coli. The recombinant protein is a single polypeptide chain of 38 kDa that is not processed by the self-cleavage reaction (Fig. 3A). Consistent with the predictions from the sequence analysis, the purified recombinant prozyme is inactive and unable to decarboxylate AdoMet. However, when purified recombinant AdoMetDC is mixed with prozyme, the AdoMetDC activity is stimulated 1,200-fold, with maximum activity observed at a ratio of 1:4 AdoMetDC/prozyme (Fig. 3B). The requirement for excess prozyme to fully activate AdoMetDC can be attributed to the observation that prozyme is partially aggregated when purified in the absence of AdoMetDC (as demonstrated by gel-filtration analysis; SI Fig. 6).
The AdoMetDC/Prozyme Complex Is the Catalytically Functional Enzyme.
The stability and function of the AdoMetDC/prozyme complex was assessed by copurification of the recombinant His-tagged AdoMetDC with Flag-tagged prozyme. The proteins were expressed separately and then copurified by Ni+2-agarose and anion-exchange column chromatography. The two subunits in the purified complex elute as a single peak on gel filtration (SI Fig. 6), demonstrating that the complex remains associated through three successive column chromatography steps (e.g., Ni+2-agarose, anion exchange, and gel filtration). SDS/Page analysis suggests that prozyme and AdoMetDC are present in the purified complex at a 1:1 molar ratio and Western blot analysis using either antibody to AdoMetDC or the Flag-tag confirms the presence of both subunits (Fig. 3A). Sedimentation equilibrium analysis by analytical ultracentrifugation demonstrates that the complex is a heterodimer of AdoMetDC and prozyme, which is formed at high affinity (Kd < 0.5 μM; Fig. 3C). In contrast, the AdoMetDC and prozyme homodimers form with weaker affinity (Kd = 50 ± 6 and 4 ± 0.5 μM, respectively; SI Fig. 7).
The rate constant for the decarboxylation of AdoMet by the copurified heterodimer was determined by steady-state kinetic analysis, and the kcat of 1.4 s−1 (Table 1) is in good agreement with the activity estimated in the T. brucei extracts. Unlike for the T. brucei AdoMetDC homodimer, putrescine does not affect the activity of the heterodimeric enzyme (Table 1 and SI Fig. 5). In summary, AdoMetDC is activated 1,200-fold by formation of a functional heterodimer with prozyme. The AdoMetDC/prozyme complex has equivalent activity to orthologs from other species, and, like the monomeric plant enzyme, heterodimeric T. brucei AdoMetDC/prozyme is not activated by polyamines.
Discussion
The polyamine pathway is tightly regulated in many species, including mammals, plants, and yeast, where the biosynthetic enzymes are controlled by transcriptional and translational regulation, and by several posttranslational mechanisms, including regulation of protein stability (4). AdoMetDC from the trypanosomatid parasites has evolved to be regulated by a mechanism not found in any other species. Our data demonstrate that the functional AdoMetDC in the trypanosomatid parasites is a heterodimer between AdoMetDC and prozyme. The prozyme arose through gene duplication of the ancestral AdoMetDC gene after the trypanosomatids diverged from other eukaryotes. AdoMetDC and prozyme then apparently coevolved such that one subunit was subject to selective pressure to remain catalytically active, and the other was selected for its regulatory function while losing catalytic activity.
Our data suggest that regulators of enzyme function can evolve through gene duplication, followed by mutational drift that results in loss of catalytic function. In addition to catalyzing chemistry, many enzymes also form functional interactions with macromolecules. The interaction binding surfaces may be maintained in these “pseudoenzymes,” providing a perfect scaffold for the evolution of novel regulatory functions. A reported analysis of the genomes of metozoan species found that inactive homologs are common and that they are present in a large variety of enzyme families (28). Few biochemical studies demonstrating the function of pseudoenzymes have been published, however several examples suggest that they function as regulators of the active enzymes. Our discovery of a pseudoenzyme in a protozoan parasite suggests that the hijacking of inactive homologs to regulate function is a general mechanism that will be found throughout evolution.
The structural basis for the regulation of AdoMetDC by prozyme is likely to arise from the induction of an allosteric transition. Previous data on the T. cruzi enzyme suggested that putrescine regulates the activity by an allosteric mechanism (23). The current observations support a model whereby putrescine induces a partial conformational change from the inactive structure toward the active one; however, the fully active conformation is realized only upon binding to the prozyme. The unusual requirement for two gene products to generate AdoMetDC activity provides a means to regulate the production of polyamines in the cell by regulating the expression level of prozyme. It remains to be determined whether the parasite utilizes this regulation in a dynamic way to change the flux through the polyamine pathway under different environmental challenges.
AdoMetDC provides an alternative and likely very effective target for the development of new antitrypanosomal drugs within a proven pathway. Our discovery of the mechanism by which AdoMetDC is activated is a key finding that will aid in the identification of potent inhibitors of this enzyme and, thus, in the generation of lead compounds that can exploit this target. Inhibitors developed against the heterodimeric enzyme may be more fully complimentary to the active site than inhibitors of the homodimer alone. In addition, our data suggest new approaches for inhibiting AdoMetDC in trypanosomes, either by blocking formation of the AdoMetDC–prozyme complex and/or by stabilizing the inactive conformation of AdoMetDC. Successful strategies for these approaches have recently been described for other proteins (29–31).
Although the evolution of the prozyme occurred by a unique gene duplication event in the trypanosomatids, gene duplication has played a key role in the evolution of the polyamine pathway and its regulation. Structural data suggests that the eukaryotic AdoMetDCs themselves arose from a gene duplication and gene fusion of the bacterial enzymes (32). Substrate-specificity changes have evolved by gene duplication in both the spermidine/spermine synthase family (33) and in the group IV decarboxylase family (34). Ornithine decarboxylase in mammalian and yeast cells is regulated by a protein inhibitor termed antizyme, which appears to be a duplication of a catabolic enzyme in the pathway, spermine–spermidine N1-acetyltransferase (35). Antizyme is, in turn, regulated by antizyme inhibitor, which is itself a pseudoenzyme, having arisen by gene duplication of ornithine decarboxylase, followed by loss of catalytic activity (36). The polyamine pathway thus provides a paradigm for the evolution of metabolic pathways and their regulation by gene duplication and divergence.
Materials and Methods
Multiple Sequence Alignment and Phylogenic Tree Generation.
AdoMetDC sequences were compiled by using the National Center for Biotechnology Information program Blast with the T. brucei AdoMetDC protein sequence as the search query. The prozyme sequence was found in the genome databases of the trypanosomatids (www.genedb.org/genedb/tryp), where it is annotated as putative AdoMetDC-like. Blast analysis with the T. brucei AdoMetDC prozyme as the query identified the prozyme sequence in the trypanosomatids but not in any other organisms. AdoMetDC sequences were aligned by using the CLUSTAL W program (version 1.83, www.ebi.ac.uk/clustalw/index.html). The phylogenetic tree was generated by using ProtML and Njdist of the MOLPHY package (http://bioweb.pasteur.fr/seqanal/interfaces/prot_nucml.html) with default settings (37–39). The reliability of the tree was determined by the RELL method of MOLPHY (40).
T. brucei Cell Growth and Preparation of Lysates.
Trypanosome parasites were cultured in HMI-9 media (90-13 bloodstream form) or SDM-79 media (29-13 procyclic form) supplemented with 10% FBS and the appropriate antibiotics as described (41–43). Cells were pelleted by centrifugation, washed with cold PBS (pH = 7.4), resuspended in lysis buffer [50 mM Hepes (pH 8)/100 mM NaCl/5 mM 2-mercaptoethanol/2 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin/2 μg/ml antipain/10 μg/ml benzamidine/1 μg/ml pepstatin/1 μg/ml chymostatin], lysed by freeze/thaw cycles, and the lysate clarified by centrifugation.
Expression and Purification of AdoMetDC and Prozyme.
The T. brucei AdoMetDC and prozyme genes were amplified by PCR from genomic DNA and were cloned into the pET 15b vector (Novagen, San Diego, CA) to generate N-terminal His6-tag fusions. Additionally, prozyme was cloned into the pT7-Flag1 (Sigma, St. Louis, MO) vector to generate an N-terminal Flag tag. Cloning primers are provided in SI Table 2. AdoMetDC or prozyme were expressed individually in E. coli BL21/DE3 cells containing the respective construct, and purified in two steps by Ni2+-agarose (Qiagen, Valencia, CA) and anion-exchange column chromatography (Mono Q 5/50 GL column; Amersham, Piscataway, NJ) as described (19), except that the buffer was exchanged after elution from Ni+2 agarose and Mono Q by a HiPrep 26/10 desalting column (Amersham) equilibrated in storage buffer [50 mM Hepes (pH 8.0)/50 mM NaCl/1 mM DTT]. Flag-tagged prozyme was copurified with His6-tagged AdoMetDC from BL21 cells that contained the individual constructs. Cell pellets were resuspended in lysis buffer, the lysates were mixed together, and the proteins were copurified as above. Purified proteins were quantified by using their respective extinction coefficients: AdoMetDC 65.3 mM−1 cm−1; prozyme, 35.9 mM−1cm−1; and AdoMetDC/prozyme complex, 102.7 mM−1 cm−1. SDS/PAGE and analytical gel filtration were used to assess purity.
Western Blot Analysis.
Total protein from lysates (40 μg per lane) and purified recombinant T. brucei AdoMetDC, prozyme or copurified AdoMetDC/prozyme complex (10 ng per lane) were separated by SDS/PAGE and transferred to a PVDF membrane (Hybond-P; Amersham). Membranes were blocked by standard methods and incubated with either rabbit polyclonal antibody to T. brucei AdoMetDC at 1:2,500 or with monoclonal anti Flag-M2 (Sigma) at 1:1,000. Horseradish peroxidase-linked donkey anti-rabbit or anti-mouse IgG secondary antibodies (Amersham Biosciences) were used at 1:10,000. Antigen was visualized by using the ECL Western Blotting Analysis system (Amersham). Rabbit antibody to T. brucei AdoMetDC was raised (by Berkeley Antibody, Richmond, CA) to His6-tagged protein purified from the insoluble fraction of lysed E. coli cells expressing T. brucei AdoMetDC. Protein was purified by Ni+2-agarose under denaturing conditions (8 M urea), renatured on the column as recommended by the manufacture (Qiagen), and eluted with a gradient of imidazole (0–250 mM). As a final step, protein was applied to a Superdex 200 HR 10/30 (Amersham) size-exclusion column as described (SI Fig. 6).
Preparation of mRNA and cDNA from T. brucei Cells.
mRNA was isolated from at least 3 × 108 trypanosome cells by using the micro polyA purist kit (Ambion, Austin, TX). cDNA to prozyme and AdoMetDC were cloned and sequenced (SI Fig. 4B). The splice leader for prozyme and AdoMetDC is inserted 70 and 161 base pairs, respectively, upstream of the ATG start sites. These data confirm that the full-length ORFs were correctly predicted by the annotation of the genes in GeneDB.org.
Northern Blot Analysis.
mRNA (1 μg) was separated by denaturing 1% agarose gel electrophoresis, transferred to a positively charged nylon membrane (BrightStar-Plus, Ambion) and cross-linked. Probe templates were prepared from plasmid DNA and radiolabeled ([32P]dATP; MP Biomedicals, Irvine, CA) probes were prepared by using the Strip-EZ PCR kit (Ambion).
Steady-State Kinetic Analysis.
Steady-state kinetics were performed by trapping liberated 14CO2 on a filter paper soaked in saturated barium hydroxide as described (19, 23). For homodimeric AdoMetDC, reactions were performed over a range of enzyme (1–4 μM) and 1-14C-CO2-AdoMet (Amersham) concentrations (10–160 μM monomer) with or without saturating putrescine (5 mM) or higher order polyamines (0.05 to 5 mM) at 37°C in buffer [100 mM Hepes (pH 8.0)/50 mM NaCl/1 mM DTT]. Reactions were allowed to proceed for various times (5–40 min) before quenching with 6 M HCl. For assay of the heterodimer, the AdoMetDC/prozyme copurified complex (25–400 nM, based on monomer concentration) was incubated with 14C-AdoMet (25 μM) and unlabeled AdoMet (0–975 μM) for various time points (2.5–10 min) before quenching as above. Data were fitted to the Michaelis–Menten equation to determine the steady-state kinetic parameters by using Prism (GraphPad, San Diego, CA).
AdoMetDC Active-Site Titration with MDL 73811 in T. brucei Cell Lysates.
MDL 73811 (0–100 nM) was incubated with 0.01 ml (40 μg total protein) T. brucei blood form cell lysate at 37°C for 30 min to 2 h in assay buffer, which allowed the reaction to proceed to completion. After incubation, the enzymatic reaction was initiated by the addition of 40 μM 14C-AdoMet substrate, followed by incubation for 30 min before quenching with 6 M HCl. The remaining enzyme activity was determined (vi) and compared with the uninhibited activity (vo) in the lysate. The concentration of AdoMetDC in lysates was estimated from these data (Fig. 2) by using standard protocols for the estimation of active-site concentration by titration with tight binding or irreversible inhibitors (44).
Determination of Molecular Weight of the AdoMetDC/Prozyme Complex.
The molecular weights of the complex, and the individual subunits were determined by equilibrium sedimentation analysis using a XLI analytical ultracentrifuge (Beckman, Fullerton, CA) equipped with an AN60 Ti rotor. Samples in buffer [50 mM Hepes (pH 8.0)/50 mM NaCl/1 mM 2-mercaptoethanol) were loaded into a six-sector equilibrium centerpiece and equilibrated for data collection at 15,000 and/or 20,000 rpm. After equilibrium was reached (≈24 h), absorption data were collected at 280 nm through sapphire windows by using a radial step size of 0.001 cm. Baseline absorbance readings for each cell were acquired by over speed at 42,000 rpm. Data sets were analyzed by using equations (SI Fig. 7) describing a single ideal species model or a monomer–homodimer model as described (45) or, for the AdoMetDC/prozyme complex, globally fitted to a single ideal species model by using the Beckman XL-A/XL-I Data Analysis Software version 6.0. Both analyses gave similar results.
Synthesis of 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine (MDL 73811).
MDL 73811 was synthesized by a previously undescribed method (SI Scheme 1) that improved yield over the published method (26, 27).
(Z)-1-BOCamino-4-chloro-2-butene: (Z)-1-amino-4-chloro-2-butene (10.0 g, 70.4 mmol) was dissolved in tetrahydrofuran (50 ml), di-tert-butyldicarbonate (16.13 g, 73.9 mmol) was added as a solid, and the reaction was stirred overnight at room temperature. THF was removed, and the solid was dissolved in CH2Cl2 and extracted three times with deionized water. The CH2Cl2 layer was dried with anhydrous MgSO4 and filtered. CH2Cl2 was removed, yielding a white solid (12.2 g). 1H NMR (Varian AS400) spectra (d6-DMSO) δ 1.3 (s), 3.6 (m), 4.2 (d), 5.5 (m), 5.6 (m), and 7.0 (s).
The 5′-deoxy-5′-methylaminoadenosine: 5′-deoxy-5′-chloroadenosine (0.5 g) was added to a microwave vial with 20 ml of the methylamine/methanol solution (2 M). The reaction vial was sealed and heated in a Biotage Initiator 60 microwave to 120 C for 2 h. After cooling, the methanol and methylamine were removed. The resulting yellow syrup was purified by column chromatography on silica gel. First a mobile phase of CH2Cl2/MeOH/Et3N (66/30/4) was used to remove starting material and impurities, then MeOH/Et3N (96/4) was used to elute the product. Fractions were collected and the solvent removed to yield a tan solid (0.34 g). LC/MS [an 1100 Series Chromatograph (Agilent Technologies, Palo Alto, CA) equipped with a PE API2000 mass spectrometer (Sciex, South San Francisco, CA)] confirmed purity and identity. 1H NMR spectra (d6-DMSO) δ 2.2 (s), 2.6–2.8 (m), 3.9 (bs), 4.1 (bs), 4.6 (bs), 5.2 (bs), 5.4 (bs), 5.8 (d), 7.3 (s), 8.1 (s), and 8.3 (s).
MDL 73811: 5′-deoxy-5′-methylaminoadenosine (1.0 g, 3.57 mmol) was dissolved in methanol. K2CO3 (0.617 g, 4.46 mmol) and (Z)-1-tert-butoxycarbonylamino-4-chloro-2-butene (0.661 g, 3.21 mmol) were added as solids to the reaction, which was stirred at room temperature for 72 h and then filtered through paper and concentrated to an oil. The oil was purified by column chromatography on silica gel. The isolated fractions were dried and dissolved in HCl (1 M) for 48 h to remove the BOC group. Water was removed, and the solid was purified by column chromatography with CH2Cl2/MeOH/Et3N (60/35/5). Fractions were combined and dried to yield a white solid (0.42 g). LC/MS confirmed purity and identity. The 1H NMR spectra were (d6-DMSO) δ 2.2 (s), 2.6–2.7 (m), 2.9 (d), 3.1 (d), 3.9 (m), 4.1 (m), 4.6 (bs), 5.3–5.4 (m), 5.4–5.6 (m), 5.7 (s), 5.8 (d), 7.3 (s), 8.1 (s), and 8.3 (s).
Supplementary Material
Acknowledgments
We thank Lisa Kinch for advice on phylogenetic tree analysis, Nicholas A. Malmquist for help with analytical ultracentrifugation, Lizbeth Hedstrom for critical reading of the manuscript, and Anthony J. Michael for suggesting the nomenclature of prozyme and for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant R01 AI34432 (to M.A.P.) and Predoctoral Fellowship GM007062 (to E.K.W.) and Welch Foundation Grant I-1257 (to M.A.P.).
Abbreviations
- AdoMetDC
S-adenosylmethionine decarboxylase
- BF
blood stream form trypanosomes
- PF
procyclic form trypanosomes
- MDL 73811
5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine
- AdoMetDC/prozyme refers to the purified complex between AdoMetDC and prozyme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.E.P. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EF545594 (Prozyme) and EF545595 (AdoMetDC)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0701111104/DC1.
References
- 1.Word Health Organization. World Health Report. Geneva: WHO; 2004. [Google Scholar]
- 2.Fries D, Fairlamb AH. In: Burger's Medicinal Chemistry and Drug Discovery. Abraham D, editor. Vol 5. New York: Wiley; 2003. pp. 1033–1087. [Google Scholar]
- 3.Childs AC, Mehta DJ, Gerner EW. Cell Mol Life Sci. 2003;60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegg AE. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 5.Tabor C, Tabor H. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 6.Gerner EW, Meyskens FL., Jr Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 7.Fairlamb AH, Blackburn P, Chait BT, Cerami A. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 8.Marton LJ, Pegg AE. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 9.Bacchi CJ, Nathan HC, Hunter SN. Science. 1980;210:332–334. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- 10.Pegg AE, Xiong H, Feith DJ, Shantz LM. Biochem Soc Trans. 1998;26:580–586. doi: 10.1042/bst0260580. [DOI] [PubMed] [Google Scholar]
- 11.Roberts SC, Scott J, Gasteier JE, Jiang Y, Brooks B, Jardim A, Carter NS, Heby O, Ullman B. J Biol Chem. 2002;277:5902–5909. doi: 10.1074/jbc.M110118200. [DOI] [PubMed] [Google Scholar]
- 12.Bacchi CJ, Nathan HC, Yarlett N, Goldberg B, McCann PP, Bitonti A, Sjoerdsma A. Antimicrob Agents Chemother. 1992;36:2736–2740. doi: 10.1128/aac.36.12.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakubu MA, Majumder S, Kierszenbaum F. J Parasitol. 1993;79:525–532. [PubMed] [Google Scholar]
- 14.Ekstrom JL, Tolbert WD, Xiong H, Pegg AE, Ealick SE. Biochemistry. 2001;40:9495–9504. doi: 10.1021/bi010736o. [DOI] [PubMed] [Google Scholar]
- 15.Tolbert WD, Zhang Y, Cottet SE, Bennett EM, Ekstrom JL, Pegg AE, Ealick SE. Biochemistry. 2003;42:2386–2395. doi: 10.1021/bi0268854. [DOI] [PubMed] [Google Scholar]
- 16.Ekstrom JL, Mathews II, Stanley B, Pegg AE, Ealick SE. Structure (London) 1999;7:583–595. doi: 10.1016/s0969-2126(99)80074-4. [DOI] [PubMed] [Google Scholar]
- 17.Tolbert WD, Ekstrom JL, Mathews II, Secrist JA, III, Kapoor P, Pegg AE, Ealick SE. Biochemistry. 2001;40:9484–9494. doi: 10.1021/bi010735w. [DOI] [PubMed] [Google Scholar]
- 18.Bennett EM, Eckstrom JL, Pegg AE, Ealick SE. Biochemistry. 2002;41:14509–14517. doi: 10.1021/bi026710u. [DOI] [PubMed] [Google Scholar]
- 19.Kinch LN, Scott J, Ullman B, Phillips MA. Mol Biochem Parasitol. 1999;101:1–11. doi: 10.1016/s0166-6851(98)00181-9. [DOI] [PubMed] [Google Scholar]
- 20.Kinch LN, Phillips MA. Biochemistry. 2000;39:3336–3343. doi: 10.1021/bi991493r. [DOI] [PubMed] [Google Scholar]
- 21.Persson K, Aslund L, Grahn B, Hanke J, Heby O. Biochem J. 1998;333:527–537. doi: 10.1042/bj3330527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clyne T, Kinch LN, Phillips MA. Biochemistry. 2002;41:13207–13261. doi: 10.1021/bi026541d. [DOI] [PubMed] [Google Scholar]
- 23.Beswick TC, Willert EK, Phillips MA. Biochemistry. 2006;45:7797–7807. doi: 10.1021/bi0603975. [DOI] [PubMed] [Google Scholar]
- 24.Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. J Biol Chem. 2005;280:39229–39237. doi: 10.1074/jbc.M509340200. [DOI] [PubMed] [Google Scholar]
- 25.Heller JS, Fong WF, Canellakis ES. Proc Natl Acad Sci USA. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casara P, Marchal P, Wagner J, Danzin C. J Am Chem Soc. 1989;111:9111–9113. [Google Scholar]
- 27.Danzin C, Marchal P, Casara P. Biochem Pharmacol. 1990;40:1499–1503. doi: 10.1016/0006-2952(90)90446-r. [DOI] [PubMed] [Google Scholar]
- 28.Pils B, Schultz J. J Mol Biol. 2004;340:399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 29.Arkin MR, Wells JA. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 30.Noble ME, Endicott JA, Johnson LN. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 31.Scheer JM, Romanowski MJ, Wells JA. Proc Natl Acad Sci USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toms AV, Kinsland C, McCloskey DE, Pegg AE, Ealick SE. J Biol Chem. 2004;279:33837–33846. doi: 10.1074/jbc.M403369200. [DOI] [PubMed] [Google Scholar]
- 33.Korhonen VP, Halmekyto M, Kauppinen L, Myohanen S, Wahlfors J, Keinanen T, Hyvonen T, Alhonen L, Eloranta T, Janne J. DNA Cell Biol. 1995;14:841–847. doi: 10.1089/dna.1995.14.841. [DOI] [PubMed] [Google Scholar]
- 34.Shah R, Akella R, Goldsmith EJ, Phillips MA. Biochemistry. 2007;46:2831–2841. doi: 10.1021/bi6023447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman DW, Carroll D, Martinez N, Hackert ML. Biochemistry. 2005;44:11777–11785. doi: 10.1021/bi051081k. [DOI] [PubMed] [Google Scholar]
- 36.Mangold U. Cell Mol Life Sci. 2006;63:2095–2101. doi: 10.1007/s00018-005-5583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adachi J, Hasegawa M. Jpn J Genet. 1992;67:187–197. doi: 10.1266/jjg.67.187. [DOI] [PubMed] [Google Scholar]
- 38.Jones D, Taylor W, Thornton J. Comput Appl Biosci. 1992;8:275–283. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Kishino H, Hasegawa M. Methods Enzymol. 1990;183:550–570. doi: 10.1016/0076-6879(90)83036-9. [DOI] [PubMed] [Google Scholar]
- 41.Hirumi H, Hirumi K. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 42.Brun R, Schonenberger M. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 43.Wirtz E, Leal S, Ochatt C, Cross GA. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 44.Copeland R. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. New York: Wiley-VCH; 1996. [Google Scholar]
- 45.Lebowitz J, Lewis M, Schuck P. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.