Abstract
tmRNA and small protein B (SmpB) are essential trans-translation system components. In the present study, we determined the crystal structure of SmpB in complex with the entire tRNA domain of the tmRNA from Thermus thermophilus. Overall, the ribonucleoprotein complex (tRNP) mimics a long-variable-arm tRNA (class II tRNA) in the canonical L-shaped tertiary structure. The tmRNA terminus corresponds to the acceptor and T arms, or the upper part, of tRNA. On the other hand, the SmpB protein simulates the lower part, the anticodon and D stems, of tRNA. Intriguingly, several amino acid residues collaborate with tmRNA bases to reproduce the canonical tRNA core layers. The linker helix of tmRNA had been considered to correspond to the anticodon stem, but the complex structure unambiguously shows that it corresponds to the tRNA variable arm. The tmRNA linker helix, as well as the long variable arm of class II tRNA, may occupy the gap between the large and small ribosomal subunits. This suggested how the tRNA domain is connected to the mRNA domain entering the mRNA channel. A loop of SmpB in the tRNP is likely to participate in the interaction with alanyl-tRNA synthetase, which may be the mechanism for the promotion of tmRNA alanylation by the SmpB protein. Therefore, the tRNP may simulate a tRNA, both structurally and functionally, with respect to aminoacylation and ribosome entry.
Keywords: crystal structure, small protein B, tmRNA, trans-translation
Trans-translation is an important quality control process in bacterial cells that recycles ribosomes accidentally stalled by defective mRNAs (1, 2). This system is ubiquitous in Bacteria, and is facilitated by tmRNA. Alanyl-tmRNA is delivered to the empty A site of the ribosome. Translation then resumes, using the mRNA portion of tmRNA, which encodes a tag targeted by a specific protease. Small protein B (SmpB), another key molecule for trans-translation (3), is highly conserved among all bacteria and some organelle genomes [supporting information (SI) Fig. 6]. The multifunctional roles of SmpB include alanylation enhancement of tmRNA and association with tmRNA entering the empty A site of the ribosome (4–6). The β-barrel structure of SmpB, revealed from two bacterial species, seems to have adapted to interact with the tmRNA to facilitate their association with translational components (7, 8). The structure of the T arm and a portion of the D loop domain of tmRNA in complex with SmpB was reported, using the Aquifex aeolicus sequence (9), which revealed that the surface of the SmpB β-barrel structure strongly bound to the single-stranded D loop. To clarify how the tmRNA interacts with SmpB and to determine the functional mechanism on the ribosome, we solved the crystal structure of the entire tRNA domain with SmpB from Thermus thermophilus HB8.
Results and Discussion
Structure Determination.
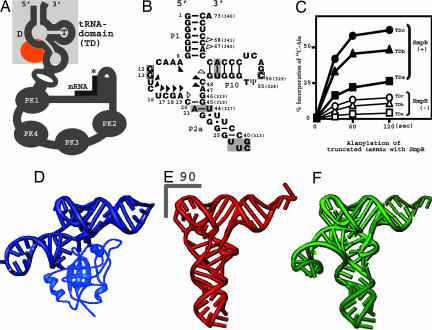
To create a stable, but still functional, tRNA domain of tmRNA (Fig. 1 A and B), several stem mutants, for slipless folding in vitro, were tested for the activation of alanylation in the presence and absence of SmpB (Fig. 1C). The candidate named tmRNA-TDc, which enhanced alanylation more than the wild-type tmRNA, was used in crystallization trials (Fig. 1B). The 3′ CCA terminus of tmRNA-TDc and the reportedly unfolded C-terminal tail of SmpB were truncated for crystallization. The successfully determined crystal structure of tmRNA-TDc with SmpB clearly showed that SmpB corresponded to the anticodon and D stem of the L-shaped tRNA (Fig. 1 D and E). This structure agrees with a biochemical report of SmpB acting as an anticodon arm of tRNA for GTP hydrolysis of EF-Tu on the ribosome (6). We also found that the linker helix (P2a in Fig. 1B) between the tRNA and mRNA domains was located behind the L-shaped region of the tRNA (Fig. 1D). This structure most closely mimics those of the class II tRNAs, which have longer variable arms (Fig. 1F). To verify this idea, we superimposed the structure of tmRNA-TDc with SmpB onto all three (serine-, tyrosine-, leucine-tRNA) kinds of bacterial class II tRNAs (SI Fig. 7). The data showed good consistency between them, especially in terms of the orientation of the variable arms against the tertiary structure of the L-shaped region. The overall structure of SmpB-bound tmRNA-TDc is most consistent with that of tRNASer, which has the longest variable arm among the class II tRNAs (10).
Fig. 1.
Structural mimicry of a long-variable-arm tRNA by tmRNA with SmpB. (A) Schematic diagram showing the secondary structure of tmRNA. The tRNA domain is highlighted in a shaded square. The SmpB-binding site is colored orange. PK means pseudoknot structure of RNA. The asterisk shows the stop codon of the short mRNA. (B) The secondary structure of the tRNA domain of T. thermophilus tmRNA. The substituted base-pairs in the P2a and P10 stems, and the connecting UUCG tetra-loop are shaded. The actual nucleotide numbers for the 3′-terminus of the tmRNA are in parentheses. The squares indicate the nucleotides involved in the interaction between the T and D loops. Triangles show nucleotides that interact with SmpB (SI Fig. 6B and SI Table 2). (C) Alanylation of truncated tmRNAs by alanyl-tRNA synthetase, in the presence (closed symbols) and the absence (open symbols) of SmpB. (D) Overall structure of the entire tRNA domain of tmRNA complexed with SmpB, determined in this study. (E and F) Yeast tRNAPhe (33) and T. thermophilus tRNASer (10) represent short (class I) and long (class II) variable-arm tRNAs, respectively. The disordered acceptor stem, anticodon and variable-loop structures of tRNASer were completed by using yeast tRNAPhe and a typical 6nt loop (34).
Interaction Between the T Loop and the D Loop of tmRNA.
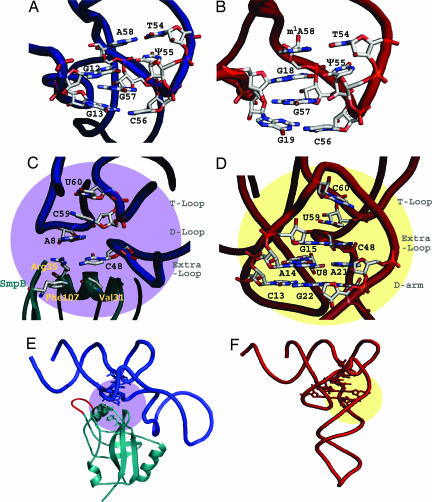
Despite the shortness of the D arm of tmRNA, the interaction between the T and D loops of the tmRNA was almost the same as that in a normal tRNA (Fig. 2 A and B). Because tmRNA has the same modified tRNA bases in the T loop (11), we prepared tmRNA-TDc with modified bases to stabilize the RNA structure for crystallization, by connecting the 5′-half of the RNA transcript with synthesized RNA containing T54 (327) and Ψ55 (328) (Fig. 1B). The T54 (327) base made a reverse-Hoogsteen base pair with A58 (331) inside the T loop (12). The Ψ55 (328) and C56 (329) dinucleotide of the T loop formed base pairs with G12 and G13 of the shortened D loop, consistent with 2D NMR analyses for the minimal tRNA-like domain of A. aeolicus tmRNA (13). The G12 and G13 dinucleotide in the D loop is conserved among almost all tmRNAs, as is the G18 and G19 dinucleotide in the D loop of tRNAs. The base stacking of A58 (331, T loop), G12 (D loop), G57 (330, T loop), and G13 (D loop) in tmRNA also contributes toward stabilizing the interaction between the T and D loops, in the same manner as in the tRNA (m1A58, G18, G57, and G19) (Fig. 2 A and B).
Fig. 2.
Detailed structures of tmRNA and SmpB, compared with yeast tRNAPhe. The tmRNA and SmpB are colored blue and blue-green, respectively (A, C, and E), and the tRNAPhe is red (B, D, and F). (A and B) The T loop and D loop connection for tmRNA and yeast tRNAPhe are viewed from an appropriate angle to show the base-stacking. The acceptor and T stems are in the upper areas of both figures. (C and D) The base stacking of the central core is shown for tmRNA and yeast tRNAPhe, respectively. The amino acid residues of SmpB, which participate in the base stacking, are labeled in light-yellow in C. (E and F) The regions of the central cores are shown on the overall structure of tmRNA (purple) and tRNAPhe (yellow). Both figures (E and F) are horizontally rotated by 30° from C and D, respectively. The central loop of SmpB is colored red (E). The structure of yeast tRNAPhe is the most suitable to compare the detailed structure with tmRNA, among all of the known tRNA structures to date. The long-variable-arm (class II) tRNAs, despite their lower resolution, have almost the same structure as that of yeast tRNAPhe in both the T loop to D loop connection and the central core region, besides a base (position 46) in the long-variable arm (10, 35, 36).
The Central Core of the tmRNA·SmpB Complex.
The base arrangement in the central core region is dissimilar between the tRNA domain of tmRNA and a normal tRNA (Fig. 2 C–F). However, despite the deficient D stem, we found that tmRNA adopted a similar base-stacking organization as in tRNA. In the normal tRNA, the G15-C48 Levitt pair, which connects the D loop and the variable loop, stacks with U59 of the T loop (Fig. 2D). The U8 base, between the acceptor stem and the D stem in the cloverleaf, participates in the A14-A21 base pair of the D arm, and also forms base-stacking interactions below the G15-C48 Levitt pair. On the other hand, the A8 of tmRNA directly stacks with C59 (332) of the T loop alone, because tmRNA has no bases corresponding to C13, A14, and G15 of the normal D arm of tRNA (Fig. 2C). Instead, surprisingly, the side chains of three amino acids (Arg-35, Phe-107, and Val-31) of the SmpB protein play the role of the D-arm bases in the canonical tRNA. The side chain of Arg-35, which is conserved among all bacteria, stacks with the A8 of tmRNA, and approaches C48 (321) to mimic base pairing (Fig. 2C and SI Table 2). Furthermore, Phe-107 and Val-31 of SmpB, instead of the C13-G22 pair in the D stem of tRNA, form a consecutive stacking structure by interacting with the side chain of Arg-35 and C48 (321) of tmRNA, respectively. The function of these two residues are conserved with branched chain and/or aromatic amino acids in the sequence alignment (SI Fig. 6A), as they are generally used as stacking structures with nucleic acids. These findings suggest that the tmRNA terminus and the SmpB mimic tRNA by their collaboration, not only in the overall structure but also at the level of the detailed structure.
The Central Loop of SmpB.
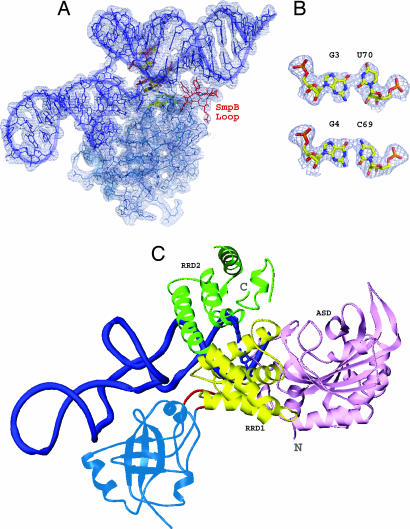
The central loop of SmpB is trypsin-sensitive in the absence of tmRNA (Y.B., R.S., S.-I.S., H. Sakai, M. Kawazoe, C.H.-T., M.S., S.K., S.Y., unpublished data). The two NMR structures of SmpB alone also revealed that the central loop is dynamically flexible, and a particular structure could not be determined (7, 8). In the 1.7 Å resolution structure of T. thermophilus SmpB [Protein Data Bank (PDB) ID code 1WJX], the central loop is disordered, showing that it must be flexible when the protein is alone. The cocrystal of the partial tRNA domain and SmpB from A. aeolicus also lacked structural assignments for the loop of SmpB (9). Here, we could spatially assign the loop, because the structure of the entire tRNA domain became obvious. The main and/or side chains of Gly-66 and Ser-67 contact the backbone of A67 (340) and C68 (341) in the acceptor stem (Fig. 1B and SI Table 2). These contacts are stabilized by the interaction between the side chain of Tyr-63 (supported by Pro-62) and G49 (322) of the T stem, near the acceptor stem. Despite the low sequence homology in this SmpB loop, the Gly-66 (Ala) and Ser-67 (Thr or Asn) residues are weakly conserved among bacteria, considering the structural information. The Pro-62 and Tyr-63 residues are well conserved among bacteria. Because the alanylation of tmRNA is activated by SmpB, the SmpB loop would contribute toward stabilizing the tertiary structure of tmRNA, especially the coaxial structure of the T stem and the acceptor stem. The base pair U50 (323)-A64 (337), which was replaced by a GC base pair to avoid slippage in the T stem, could be involved in the alanylation of tmRNA, in a structural manner (Fig. 1 B and C). The bases of tmRNA that contact the loop of SmpB are adjacent to the G3-U70 (343) wobble pair (Fig. 3 A and B), which is an important tRNA identity determinant of alanyl-tRNA synthetase (AlaRS). We found that the catalytic domain of AlaRS docked with tRNA is superimposable with the complex of tmRNA and SmpB, without steric hindrance (Fig. 3C). In the model, the loop of SmpB is close to the conserved helix of the AlaRS RRD1 domain (14), suggesting a collaboration between the proteins for the alanylation of tmRNA. This agrees with the results of a biochemical assay, which revealed that the C-terminal tail of SmpB is not required for the enhancement of aminoacylation (15).
Fig. 3.
Alanyl-activation model of tmRNA caused by SmpB. (A) Stick model of tmRNA-TDc in complex with SmpB, shown on the electron density map (contour level 1.0 σ). The nucleotides and amino acid residues of the central core region are depicted by the yellowish elemental models. The residues (from Tyr-63 to Asn-70) in the central loop of SmpB are colored red near the lower acceptor stem of tmRNA. (B) The G3-U70 (343) wobble base pair, which is the most important aminoacyl-discriminator by AlaRS, is compared with the G4-C69 (342) Watson–Crick base pair. (C) Docking model of the tmRNA-TDc with SmpB and the N-domain of A. aeolicus AlaRS (14). The tmRNA-TDc and SmpB are colored blue and blue-green, respectively. The central loop of SmpB is red. For AlaRS, three domains (ASD, active site domain; RRD1, RNA recognition domain 1; RRD2, RNA recognition domain 2) are colored pink, yellow, and green, respectively. This model is an imitation of the original superimposed method for AlaRS and tRNA (14); that is, it represents individual superpositions of AlaRS (PDB ID code 1RIQ) and tmRNA-TDc, on the structure of the complex of AspRS and tRNAAsp (PDB ID code 1C0A) (37).
The SmpB·tmRNA Complex on the Ribosome.
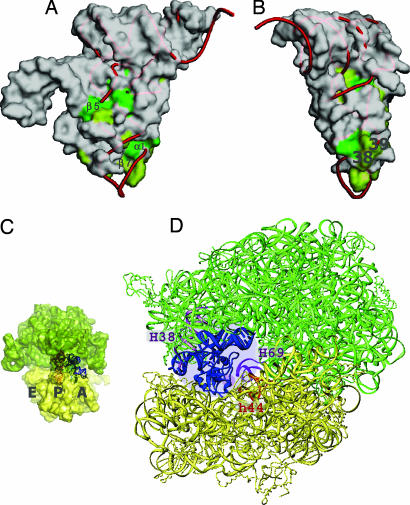
The conserved residues on the surface of SmpB are concentrated in the region near the variable arm of tmRNA (β5) and in the corresponding region of the anticodon loop (α1 and β7), especially position 38 of the tRNA (Fig. 4 A and B). The superimposed model of tmRNA-TDc on the A-site tRNA in the 70S ribosome revealed that the C-terminal region of SmpB was close to the decoding region of the 30S ribosomal subunit (Fig. 4 C and D). The region of SmpB corresponding to the 3′-part of the anticodon loop might be important, in the same way that tRNA eventually contacts the helices of rRNA or ribosomal proteins (16, 17). On the β5 strand of SmpB, the conserved residues orient the linker helix (P2a) in the proper direction (Fig. 4A). Therefore, the linker helix of tmRNA-TDc can fit well into the space between the 50S and 30S subunits (Fig. 4D). Especially, the side chain of Leu-80 functions as a wedge, by mimicking the base of a nucleotide again and stacking with the A19 and U16 bases (SI Fig. 7D). The tandem GU wobble pairs (G22-U43 (316), G23-U42 (315) in Fig. 1B) bend the linker region to the bottom side of the molecule. This could cause the mRNA region of the tmRNA to be directed to the decoding region of the empty A site during trans-translation. From the view looking down from the acceptor stem, the P2a linker helix of tmRNA-TDc bends to the left side, as in other long-variable-armed tRNAs (SI Fig. 7C). This helix might need to enter the limited space of the A site of the ribosome, because the mRNA domain of tmRNA is located on the entry site of the ribosome in a cryo-EM structure (18). Because the linker helix is spatially equivalent to the long-variable arm of a class II tRNA, the tmRNA could be moved from the A site to the P site (19) by the linker region passing beneath the bridge of the A-site finger (helix 38 of 23S rRNA), as in a class II tRNA. This suggests that the tRNA domain could be in the P site, without necessarily dissociating from SmpB. This idea is supported by the recent report describing the footprinting of SmpB with the ribosome, which showed that SmpB could bind the rRNA around the P (E) site in addition to the A site (20).
Fig. 4.
Functional mimicry of a canonical tRNA by tmRNA and SmpB. (A and B) Molecular surface of SmpB, represented as with tmRNA. The conserved residues on the surface of SmpB are colored green (complete) or light green (partial). The tRNAPhe was superimposed on the tRNA domain of the tmRNA by using the nucleotides of the acceptor stems and the T arms, and the central cores with the residues of SmpB corresponding to the nucleotides (Fig. 2C). Positions 38 and 39 of the tRNA are labeled in B. (C and D) tmRNA with SmpB on the ribosome. The 50S and 30S subunits are colored green and yellow, respectively. tmRNA and SmpB are superimposed on the A-site tRNA of the 70S ribosome (16). The yeast tRNAPhe is shown in the P-site of the ribosome in C. The corresponding region of SmpB for nucleotide positions 38 and 39, shown in A and B, is indicated by a small purple circle around H69 and h44 of the ribosome in D.
Interaction of tmRNA with tRNA.
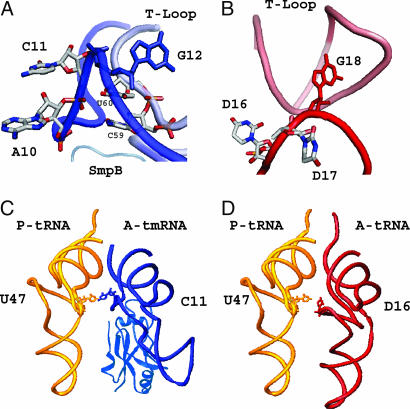
Although the spatial arrangement of the D loop of tmRNA was completely different from that of a canonical tRNA, it still shared some structural conservation with the canonical tRNA (Fig. 5 A and B). In the D-loop region of a canonical tRNA, the G18 base-paired with Ψ55 makes an acute turn from the previous nucleotide (D17), which causes the D16 and D17 bases to point toward the outside of the tRNA (Fig. 5B). This orientation of D16 would be important to stabilize the aminoacyl-tRNA in the A site, because the D16 base of the aminoacyl-tRNA and the U47 base of the peptidyl-tRNA are within hydrogen-bonding distance in the crystal structure of the 70S ribosome (Fig. 5D) (16). The G12 in tmRNA, which is base-paired with Ψ55 (328), also makes an acute turn from the previous C11, as in normal tRNA (Fig. 5A). Because of the lack of a D stem in the tmRNA, the A10 and C11 in the D loop are directly followed by the acceptor stem, and both bases face the outside of the tRNA domain, because the C59 (332) and U60 (333) bases of the T loop stabilize the backbones of A10 and C11 (Figs. 2C and 5A). Despite the different trace from that of the tRNA, the A10 and C11 bases of the aminoacyl-tmRNA could point toward the peptidyl-tRNA and be stabilized in the A site of the ribosome, as in normal translation (Fig. 5C).
Fig. 5.
Structural function of the shortened D arm of tmRNA. (A) The A10 and C11 bases in the deficient D loop of tmRNA are shown by stick models. G12, a key base connecting the D loop with the T loop, is shown in solid blue. C59 (332) and U60 (333) of the T loop are shown to indicate that their bases stabilize the backbone of A10 and C11. (B) D16, D17, and G18 of yeast tRNAPhe are colored in the same manner as the corresponding bases of tmRNA (A). G18, which interacts with Ψ55, is colored solid red, instead of solid blue for the G12 of tmRNA. (C and D) The SmpB-bound tmRNA-TDc is superimposed with the A-site tRNA of the 70S ribosome, in the same manner as in Fig. 4 C and D. In the A site of the ribosome, the tmRNA with SmpB (C) is colored blue, and the tRNAPhe (D) is red. The P-site tRNAPhe is colored yellow in both C and D. C11 of the A-site tmRNA, which spatially corresponds to D16 of the A-site tRNAPhe, is shown as a stick model.
Functional Mimicry of tRNA by the SmpB·tmRNA Complex.
These findings all indicate that the SmpB-bound tmRNA functionally mimics a canonical tRNA as a ribonucleoprotein complex (tRNP), during aminoacylation and entry into the ribosome. Because the acceptor stem of the tmRNA was refined in this study, we could accurately specify the location of the SmpB on the ribosome by superimposition. Although the structure of SmpB is truncated in the crystal structure, it is obvious that the C-terminal tail of SmpB should play an important role in the trans-translation activity of the ribosome (6, 21), because the β7 strand of SmpB structurally corresponds to the anticodon loop (Fig. 4 A and B). On the superimposed model, the tip of the anticodon loop is empty in the tmRNA and SmpB, which reminds us that the C-terminal tail could occupy this area on the ribosome. The trans-translation system would be maintained by coevolution between the tmRNA and its supporting cofactor SmpB, with adaptation to multiple counterparts in the series of reactions in bacteria.
Materials and Methods
Protein Expression and Purification.
The C-terminally truncated SmpB (123 of 144 amino acids) from T. thermophilus HB8 was expressed from pET-11b (Novagen, Madison, WI) in E.coli strain BL21-CodonPlus(DE3)-RP (Stratagene, La Jolla, CA). The harvested cells were resuspended in 50 mM Hepes buffer (pH 7.5) containing 1 M NaCl, 1 mM EDTA, 5% glycerol, 6 mM 2-mercaptoethanol, and 0.5 mM PMSF. After sonication and centrifugation at 15,000 × g for 30 min, the supernatant was heat-treated at 70°C for 30 min and purified by a series of Butyl-Toyopearl, HiTrap-butyl, heparin-Toyopearl, and source-15S column chromatography steps (Tosoh, Montgomeryville, PA). The buffer of the purified protein sample was finally exchanged by dialysis to 50 mM Hepes buffer (pH 7.0) containing 100 mM KCl. The yield was ≈10 mg per 1 g wet cells.
RNA Preparation.
The tRNA domains of T. thermophilus tmRNA (tmRNA-TDa) and its mutants (tmRNA-TDb and tmRNA-TDc) were transcribed in vitro by using T7 RNA polymerase. The sequences used were 5′-GGGGGUGAAACGGUCUCGACGGGGGUC-GAGA-GACCUUCGGACGGGGGUUCGACUCCCCCCACCUCCACCA-3′ (tmRNA-TDa), 5′-GGGGGUGAAACGGUCUCGACAGGGGUC-GAGA-GACCUUUGGACGUGGGUUCGACUCCCACCACCUCCACCA-3′ (tmRNA-TDb), and 5′-GGGGGUGAAACGGUCUCGACAGGGG-UUCG-CCUUUGGACGUGGGUUCGACUCCCACCACCUCCACCA-3′ (tmRNA-TDc), respectively. The replaced base-pairs in the stems (italic) were selected by considering the preservation of conserved nucleotides among bacterial tmRNA and tRNAAla (22, 23).
Aminoacylation Assay.
We carried out the aminoacylation assay at 50°C, in a 50-μl reaction mixture containing 100 mM EPPS (pH 7.5), 50 mM KCl, 12 mM MgCl2, 5 mM ATP, 12 μM l-[U-14C]alanine (5.99 GBq/mmol), 100 nM T. thermophilus alanyl-tRNA synthetase (AlaRS), and 1.2 μM each examined tmRNA-transcript in complex with or without SmpB. At appropriate time points, aliquots of samples were mixed with a 10-fold volume of 5% trichloroacetic acid, and were incubated for 30 min on ice. The quenched samples were spotted on Whatman (Clifton, NJ) 3MM paper. The papers were washed at least twice with 5% trichloroacetic acid, and the radioactivity was measured with a liquid scintillation counter.
Crystallization and Data Collection.
For crystallization of the tmRNA-TDc complex with SmpB, the RNA was prepared by a T4 RNA ligase (24) reaction of a 5′-triphosphate transcript (41 nt) and a 22-nt synthetic RNA (Dharmacon, Lafayette, CO). The RNA has T54 (327) and Ψ55 (328) in the T loop and lacks C74 (347), A75 (348), and A76 (349) at the 3′-terminus, for stable folding and crystallization. The complex of T. thermophilus tmRNA-TDc and SmpB was crystallized by the hanging-drop vapor diffusion method, using a reservoir solution of 50 mM Hepes buffer (pH 7.0) containing 10 mM MgCl2, 0.3 mM hexaamine cobalt(III) chloride, 1.8 M ammonium sulfate, and 2% (vol/vol) glycerol. Rectangular-parallelepiped crystals grew to dimensions of 0.3 × 0.2 × 0.1 mm at 20°C in 7 days. For data collection, the crystals were frozen in liquid nitrogen with 16% glycerol as a cryoprotectant. All diffraction data sets were collected at the NW12A beamline at the Proton Factory Advanced Ring (Tsukuba, Japan), and were processed by the use of the HKL2000 software suite (25).
Structure Determination and Refinement.
We solved the structure by molecular replacement with the program MOLREP (26), using the 1.7-Å resolution structure of T. thermophilus SmpB (PDB ID code 1WJX) and the 3.2-Å resolution structure of A. aeolicus partial tRNA domain of tmRNA (9) as search models. The model (3.0 Å) was built and refined manually with the programs O (27) and CNS (28), using an operator (h, -k, -l) in the pseudomerohedral perfect twin (29). Data collection and refinement statistics for the structure are summarized in Table 1. The quality of the protein model was inspected by PROCHECK (30). Structure representations were prepared with PYMOL (www.pymol.org) and RIBBONS (31). Hydrophobic or hydrogen bonded contacts between the tmRNA-TDc and the SmpB protein were calculated by using LIGPLOT programs (32). Coordinates and structure factors have been deposited in the PDB (ID code 2CZJ).
Table 1.
Data collection, phasing and refinement statistics
| Data set | Native |
|---|---|
| X-ray source | Photon Factory-NW12A |
| Wavelength, Å | 1.0000 |
| Resolution, Å | 50.0–3.0 |
| Space group | P21 |
| Cell parameters, Å | a = 84.776 b = 67.957 c = 178.662 |
| Cell parameters, deg. | α = 90.00 β = 90.07 γ = 90.00 |
| Unique reflections | 40370 |
| Redundancy | 6.3 |
| Completeness, % | 99.5 (98.6) |
| I/σ(I) | 23.7 (3.5) |
| Rsym* | 0.075 (0.366) |
| Refinement | |
| Resolution range, Å | 50.0–3.0 |
| Rwork†/Rfree‡, % | 25.5/32.0 |
| Number of protein atoms | 3932 |
| Number of nucleic acid atoms | 5292 |
| rmsd bond lengths, Å | 0.014 |
| rmsd bond angles, deg. | 2.20 |
| Average B-value, Å2 | 94.20 |
| Ramachandran plot | |
| Core, % | 71.5 |
| Allowed, % | 28.0 |
| Generously, % | 0.5 |
Values in parentheses are for the highest-resolution shells.
*Rsym = Σ | Iavg − Ii | / ΣIi.
†Rwork = Σ | Fo − Fc | / Σ | Fo |.
‡Rfree is the same as Rwork, but calculated by using a small fraction (7.4%) of randomly selected reflections with twinning rules.
Supplementary Material
Acknowledgments
We thank G. Kawai, N. Nameki, K. Hanawa, A. Muto, and H. Himeno for valuable discussions and T. Kaminishi and H. Sakai for technical advice. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, and the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviation
- SmpB
small protein B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: X-ray crystallographic structure factors and coordinates for the refined model have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2CZJ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700402104/DC1.
References
- 1.Keiler KC, Waller PR, Sauer RT. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 2.Karzai AW, Roche ED, Sauer RT. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 3.Karzai AW, Susskind MM, Sauer RT. EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu Y, Ueda T. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Ueda T. J Biol Chem. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- 7.Dong G, Nowakowski J, Hoffman DW. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, et al. FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- 9.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 10.Biou V, Yaremchuk A, Tukalo M, Cusack S. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 11.Felden B, Hanawa K, Atkins JF, Himeno H, Muto A, Gesteland RF, McCloskey JA, Crain PF. EMBO J. 1998;17:3188–3196. doi: 10.1093/emboj/17.11.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zagryadskaya EI, Doyon FR, Steinberg SV. Nucleic Acids Res. 2003;31:3946–3953. doi: 10.1093/nar/gkg448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudin C, Nonin-Lecomte S, Tisne C, Corvaisier S, Bordeau V, Dardel F, Felden B. J Mol Biol. 2003;331:457–471. doi: 10.1016/s0022-2836(03)00760-5. [DOI] [PubMed] [Google Scholar]
- 14.Swairjo MA, Otero FJ, Yang XL, Lovato MA, Skene RJ, McRee DE, Ribas de Pouplana L, Schimmel P. Mol Cell. 2004;13:829–841. doi: 10.1016/s1097-2765(04)00126-1. [DOI] [PubMed] [Google Scholar]
- 15.Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP. J Biol Chem. 2005;280:5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- 16.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 17.Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M. Nat Struct Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 18.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 19.Shpanchenko OV, Zvereva MI, Ivanov PV, Bugaeva EY, Rozov AS, Bogdanov AA, Kalkum M, Isaksson LA, Nierhaus KH, Dontsova OA. J Biol Chem. 2005;280:18368–18374. doi: 10.1074/jbc.M409094200. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH. Nucleic Acids Res. 2005;33:3529–3539. doi: 10.1093/nar/gki666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. Proc Natl Acad Sci USA. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gueneau de Novoa P, Williams KP. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C. Nucleic Acids Res. 2006;34:D163–168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QS, Unrau PJ. Biotechniques. 2002;33:1256–1260. doi: 10.2144/02336st03. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Yeates TO. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 31.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 32.Wallace AC, Laskowski RA, Thornton JM. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 33.Jovine L, Djordjevic S, Rhodes D. J Mol Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto T, Oguro A, Kawai G, Ohtsu T, Nakamura Y. Nucleic Acids Res. 2005;33:745–754. doi: 10.1093/nar/gki222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 37.Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D. EMBO J. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.