Abstract
The location of leukocytes in different microenvironments is intimately connected to their function and, in the case of leukocyte precursors, to the executed differentiation and maturation program. Leukocyte migration within lymphoid organs has been shown to be mediated by constitutively expressed chemokines, but how the bioavailability of these homeostatic chemokines is regulated remains unknown. Here, we report in vivo evidence for the role of a nonsignaling chemokine receptor in the migration of leukocytes under physiological, i.e., noninflammatory, conditions. We have studied the in vivo role of the silent chemokine receptor CCX-CKR1 by both loss- and gain-of-function approaches. CCX-CKR1 binds the constitutively expressed chemokines CC chemokine ligand (CCL)19, CCL21, and CCL25. We find that CCX-CKR1 is involved in the steady-state homing of CD11c+MHCIIhigh dendritic cells to skin-draining lymph nodes, and it affects the homing of embryonic thymic precursors to the thymic anlage. These observations indicate that the silent chemokine receptor CCX-CKR1, which is exclusively expressed by stroma cells, but not hematopoietic cells themselves, regulates homeostatic leukocyte migration by controlling the availability of chemokines in the extracellular space. This finding adds another level of complexity to our understanding of leukocyte homeostatic migration.
Keywords: cell trafficking, homeostasis, lymphopoiesis, thymus
The field of chemokines and their receptors has attracted much attention because the location of a hematopoietic cell is intimately linked to its function, in the case of mature cells, and the realization of its differentiation potential, in the case of hematopoietic precursors (1). Upon ligand binding of the classical chemokine receptors, a signaling cascade is initiated that causes a rapid increase of intracellular calcium resulting in directed migration, degranulation, and angiogenesis. Chemokines and their receptors can be categorized into so-called “inflammatory” chemokines that regulate the migration of leukocytes in response to an inflammatory challenge and “homeostatic” chemokines that are constitutively expressed to regulate the physiological migration of leukocytes and their precursors under steady-state conditions (2–4). Chemokines expressed in a given tissue are thought to confer directional cues by forming concentration gradients. These gradients are thought to positively induce the migration of immunocompetent cells that express the appropriate receptors into the microenvironment in which they are needed. Negative regulators of this process are poorly understood. In the inflammatory situation, proteases such as CD26/DPP IV have been proposed to modulate chemokine activity by either inactivating extracellular chemokines or by affecting their binding affinities (5). Furthermore, the silent chemokine receptor D6, which binds several inflammatory chemokines but fails to initiate the classical signaling cascade, has been shown to be involved in the resolution of postinflammatory skin lesions in vivo (6, 7). By contrast, an in vivo role for negative regulators of the physiological migration of cells within lymphoid organs has not been described.
Here, we investigated the in vivo role of the silent chemokine receptor CCX-CKR1. Silent chemokine receptors bind chemokines but are structurally incapable of signaling for migration (8, 9). Apart from CCX-CKR1, two silent chemokine receptors have been described, namely DARC (10) and D6 (11). In contrast to CCX-CKR1, DARC and D6 bind chemokines that are expressed only in response to inflammatory stimuli, indicating a distinct functional role of these receptors. By using gain- and loss-of-function approaches, we show that CCX-CKR1 modulates the physiological homing of dendritic cells (DCs) to skin-draining lymph nodes and the immigration of thymic precursors to the embryonic thymic anlage.
Results
Murine CCX-CKR1 Binds the Homeostatic Chemokines CC Chemokine Ligand (CCL)19, CCL21, and CCL25.
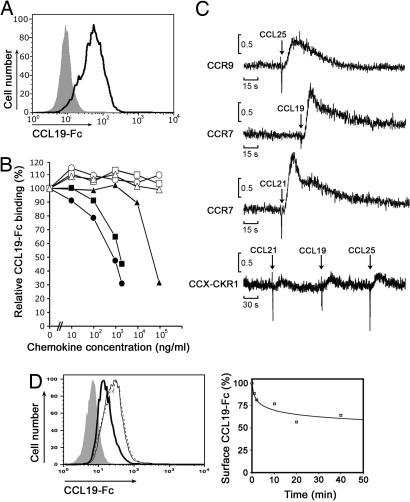
To identify molecules that mediate the directed migration of leukocytes, we screened EST databases for chemokine receptor related sequences. We found a murine EST (AI322657) with high homology to known chemokine receptors and determined the full-length cDNA by RACE. The resulting cDNA is most closely related to the bovine receptor PPR1 (12), with 81% identity on the nucleotide level, and encodes a predicted protein of 350 aa containing the DRY-motif characteristic for chemokine receptors. Consistent with previous reports, which have termed the encoded protein CCX-CKR1 (13, 14), we find that murine CCX-CKR1 binds the murine chemokines CCL19, CCL21, and CCL25, but not CXCL12, CCL17, or CCL22 (Fig. 1 A and B). Intriguingly, none of the CCX-CKR1 binding chemokines induced the typical rapid increase in intracellular calcium that is characteristic for chemokine receptor signaling (Fig. 1C). Thus, CCX-CKR1 is a surface receptor that binds the homeostatic chemokines CCL19, CCL21, and CCL25 but does not induce the classical calcium response. Nonsignaling chemokine receptors, also termed “silent” chemokine receptors, have been proposed to remove chemokines from the extracellular space by internalizing receptor-bound chemokines (8, 9). Consistent with this notion and a recent report on the human receptor (15), HEK293 cells transfected with murine CCX-CKR1 removed bound CCL19-Fc from the surface in a time-dependent manner when warmed to 37°C (Fig. 1D). The presented data therefore suggestd that CCX-CKR1 may control the availability of homeostatic chemokines in vivo.
Fig. 1.
CCX-CKR1 binds the chemokines CCL19, CCL21, and CCL25. (A) CCX-CKR1-transfected HEK293 cells (dark line) bind a CCL19-Fc fusion protein. Mock transfected HEK293 cells (filled gray population) were used as control. (B) CCL19 (filled circles), CCL25 (filled squares), and CCL21 (filled triangles), but not CXCL12 (open circles), CCL17 (open squares), or CCL22 (open triangles) compete with CCL19-Fc for binding on CCX-CKR1 transfectants. (C) CCX-CKR1 does not induce an increase in intracellular calcium upon the addition of the indicated chemokines. Chemokine receptor transfectants loaded with the calcium-sensitive dye fura2-AM were exposed to the indicated chemokines, and the ratio of relative fluorescence intensities was measured over time. (D) (Left) CCX-CKR1 transfectants internalize CCL19-Fc in a time dependent manner. CCX-CKR1-HEK293 cells incubated at 4°C with CCL19-Fc were either warmed to 37°C (thick line) or not warmed (thin line) before cooling the cells to 4°C and adding an anti-Fc secondary reagent. To rule out the possibility that incubation at 37°C by itself reduced receptor levels, CCX-CKR1-HEK293 cells were warmed to 37°C in the absence of CCL19-Fc, cooled to 4°C, and stained with CCL19-Fc (dashed line). (Right) The reduction of surface CCL19-Fc caused by warming the cells (Left, thick line) relative to the amount of surface staining without prior addition of ligand (Left, dotted line) is plotted over time.
The Silent Chemokine Receptor CCX-CKR1 Is Not Expressed by Hematopoietic Cells.
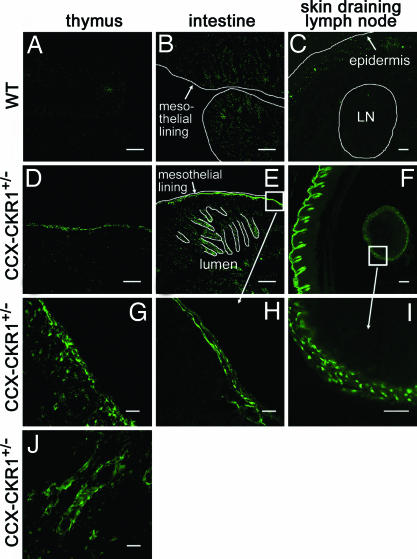
To investigate the in vivo role of CCX-CKR1 we generated CCX-CKR1-EGFP knockin mice [supporting information (SI) Fig. 5]. To this end, we replaced the N-terminal half of the single coding exon (exon 4) of the CCX-CKR1 gene (14) in frame by an EGFP cassette. To investigate the in vivo function of CCX-CKR1, it was crucial to unequivocally determine the cell type(s) that express this silent receptor because previous reports had produced contradictory results (13, 14, 16). We therefore analyzed heterozygous CCX-CKR1-EGFP knockin mice, which are phenotypically indistinguishable from wild-type mice (see below), for CCX-CKR1-driven EGFP (EGFPCCX-CKR1) expression. We found that CCX-CKR1 is not expressed by hematopoietic cells. No EGFPCCX-CKR1 expression was detectable in hematopoietic cells isolated from bone marrow, spleen, and lymph nodes by FACS. We did, however, by confocal microscopy, find EGFPCCX-CKR1 expression in nonhematopoietic cells in thymus, intestine, lymph node, and epidermis (Fig. 2), but not in heart, kidney, liver, spleen, and brain. In the embryonic thymus, subcapsular EGFPCCX-CKR1 expression is detectable starting at embryonic day 13.5 (E13.5) (SI Fig. 6). In the adult thymus, EGFPCCX-CKR1 is expressed exclusively in perivascular thymic epithelial cells (TECs) of the corticomedullary junction and the medulla and in the subcapsular epithelial layers (Fig. 2 and SI Fig. 7). This expression pattern marks the thymic niches that are thought to contain the most immature precursor cells in the adult thymus. In the intestine, large tubular structures in the submucosa, presumably lymph vessels, are marked by EGFPCCX-CKR1 expression. In lymph nodes draining the skin, EGFPCCX-CKR1 is found exclusively in stroma cells lining the marginal sinus and in the skin, EGFPCCX-CKR1 expression is restricted to the epidermis (Fig. 2 and SI Fig. 8).
Fig. 2.
CCX-CKR1 is expressed in stromal but not hematopoietic cells in thymus, intestine, lymph node and epidermis. (A–C) Sections from wild-type mice were negative. (A–F) Overview images. (G–J) Higher magnifications of images. For the thymus, EGFPCCX-CKR1 expression is shown for the subcapsular zone (G) and the cortico-medullary junction (J). (Scale bars: in A–F, 200 μm; in G–J, 40 μm.)
CCX-CKR1 Regulates Steady-State Homing of Dendritic Cells to Skin-Draining Lymph Nodes.
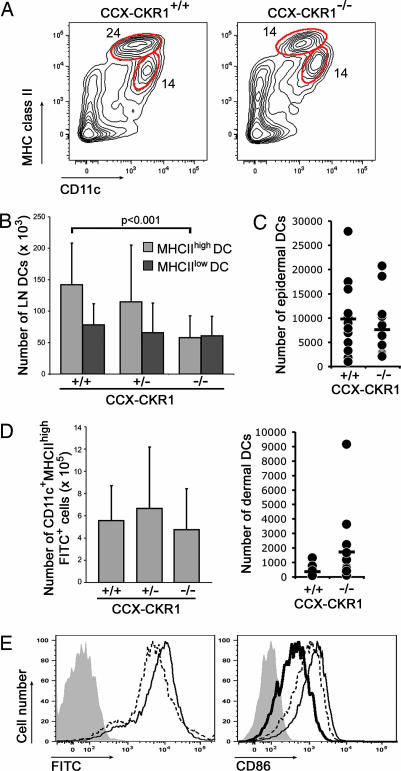
The lymph node marginal sinus collects fluid and antigen-bearing cells from afferent lymphatics. CD11c+MHCIIhigh DCs in skin-draining lymph nodes are known to enter lymph nodes by way of afferent lymph under steady-state conditions through a largely CC chemokine receptor (CCR)7-dependent process, whereas CD11c+MHCIIlow DCs enter lymph nodes by way of the blood (17–20). The expression of CCX-CKR1 in lymph node marginal sinus and epidermis suggested that this silent chemokine receptor may be involved in the homing of leukocytes that enter lymph nodes by way of the afferent lymph. The analysis of DCs in skin-draining lymph nodes showed that this is indeed the case. The microanatomy of spleen and lymph nodes was found to be normal by hematoxylin/eosin and immunohistochemical stains, and there was a trend to lower total numbers of splenocytes and lymph node cells in CCX-CKR1-deficient mice (SI Fig. 9A and data not shown). Importantly, there was a significant reduction in the absolute number of CD11c+MHCIIhigh DCs as well as in their relative proportion among non-B and non-T cells isolated from skin-draining lymph nodes (Fig. 3 A and B and SI Fig. 9B). This reduction affected both dermal and epidermal DCs in skin-draining lymph nodes (SI Fig. 9C). In contrast, the number of blood-derived CD11c+MHCIIlow DCs was normal in CCX-CKR1-deficient mice (Fig. 3B). This reduction in steady-state CD11c+MHCIIhigh DC levels could have been caused by the disturbed production and homing of DC precursors to the skin or a general block of DC migration. But no evidence for these possibilities was found. The number of CD11c+MHCIIhigh DCs that could be isolated from epidermal sheets prepared from the ears of individual mice by proteolytic digestion was normal when comparing CCX-CKR1-deficient mice with their wild-type littermates (Fig. 3C). For dermal DCs, a nonsignificant increase (P = 0.08, two-sided t test) was found in CCX-CKR1-deficient mice, which may suggest that the steady-state migration of these cells to the draining lymph nodes is disturbed, leading to accumulation in the dermis (Fig. 3C). Consistently, there was no obvious difference in the number and distribution of MHCII-positive DCs in epidermal sheets of wild-type and CCX-CKR1-deficient mice (SI Fig. 9D). Furthermore, a homing defect of CD11c+MHCIIhigh DCs 48 h after the epicutaneous application of the skin sensitizer FITC that causes the activation and mobilization of skin-resident DCs (18–21) was not observed (Fig. 3 D and E). Wild-type and CCX-CKR1-deficient CD11c+MHCIIhigh DCs accumulated in skin-draining lymph nodes to the same level, acquired the same amount of engulfed FITC, and up-regulated the costimulatory molecule CD86 to similar levels. These observations indicated that CCX-CKR1-deficient CD11c+MHCIIhigh DCs are present in the epidermis at normal numbers and respond to activating stimuli like wild-type cells. Determining CD11c+MHCIIhigh DC numbers after FITC treatment over time we found that FITC+ wild-type and CCX-CKR1-deficient DCs disappeared from skin-draining lymph nodes with the same kinetics (226 ± 4 × 103 versus 223 ± 9 × 103 DCs after 96 h). This finding ruled out the possibility that CD11c+MHCIIhigh DCs have a shorter half-life in CCX-CKR1-deficient mice than in wild-type mice, which could explain the reduction in the steady-state situation. We conclude that the function of CCX-CKR1 is required to maintain normal numbers of CD11c+MHCIIhigh DCs that enter lymph nodes by way of afferent lymphatics under steady-state conditions. In contrast, CCX-CKR1 function is dispensable for the homing of DCs that enter peripheral lymph nodes by way of the blood. No effect was seen on DC homing after FITC treatment, suggesting that DC activation initiated additional homing mechanisms that compensated for the lack of CCX-CKR1.
Fig. 3.
The absence of CCX-CKR1 impairs the steady-state homing of DCs entering lymph nodes by way of afferent lymphatics but not the homing of DCs entering by way of the blood. (A) Flow cytometric analysis of DCs in skin-draining lymph nodes of wild-type and CCX-CKR1-deficient mice. Contour plots are gated on B220-negative Thy1-negative cells. Gates are set to distinguish CD11c+MHCIIhigh DCs that enter lymph nodes by way of afferent lymphatics from CD11c+MHCIIlow DCs that enter by way of the blood. (B) The steady-state number of CD11c+MHCIIhigh, but not CD11c+MHCIIlow DCs, in skin-draining lymph nodes is reduced in CCX-CKR1-deficient mice. At least 10 7-week-old mice were analyzed per genotype. (C) Enumeration of CD11c+MHCIIhigh DCs isolated from dermal and epidermal sheets of individual CCX-CKR1-deficient mice and wild-type littermates subjected to proteolytic digestion. Each dot represents one mouse, and the bars indicate the mean. (D) Epicutaneous application of FITC normalizes the numbers of CD11c+MHCIIhigh DCs in CCX-CKR1-deficient mice. The number of CD11c+MHCIIhighFITC+ DCs 48 h after FITC treatment in six skin-draining lymph nodes per mouse was determined. At least seven 7-week-old mice were analyzed per genotype. (E) FACS analysis of wild-type (dashed line) and CCX-CKR1-deficient (thin line) CD11c+MHCIIhigh DCs 48 h after FITC treatment. Both histograms are gated on CD11c+MHCIIhigh DCs as shown in A. Filled gray populations are CD11c+MHCIIhigh DCs from a mouse 48 h after treatment with carrier alone (Left) and after FITC treatment without the CD86 stain (Right). The thick line indicates the levels of CD86 staining on CD11c+MHCIIhigh DCs without FITC treatment.
CCX-CKR1 Regulates the Migration of Thymic Precursor Cells in Vivo.
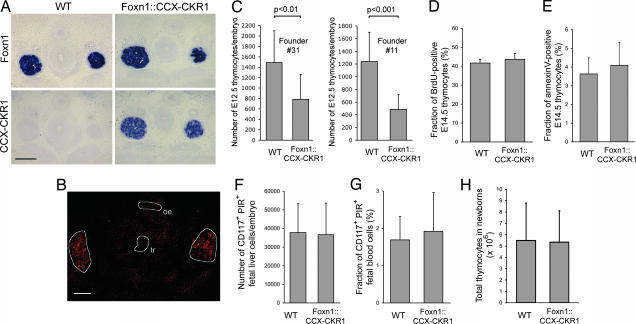
The CCX-CKR1 expression pattern in the thymus suggested that CCX-CKR1 may play a role in the migration of thymocyte precursors or developing thymocytes. But, T cell development was found to be unaffected in CCX-CKR1-deficient mice (SI Fig. 10). To reveal the function of CCX-CKR1 in the thymus we next opted for a gain-of-function approach. To this end we generated mice that overexpress CCX-CKR1 specifically in TECs. This was achieved by placing the full-length CCX-CKR1 cDNA under the control of a 27.9-kb Foxn1 promoter fragment for which we have shown that it leads to faithful expression of a given cDNA in the Foxn1 expression domain (22). In situ hybridization of embryonic thymic anlagen showed that this was also true for CCX-CKR1 overexpressed in Foxn1::CCX-CKR1 transgenic embryos (Fig. 4A). CCX-CKR1 expression can be detected in wild-type embryos at this stage by RT-PCR (SI Fig. 11), but the expression levels are apparently below the sensitivity of in situ hybridization. Overexpression of CCX-CKR1 had no effect on the expression levels of thymic chemokines (SI Fig. 11) or on the expression pattern of CCL25 protein that is restricted to the developing thymic anlagen (Fig. 4B).
Fig. 4.
Overexpression of CCX-CKR1 impairs homing of hematopoietic thymic precursors to the embryonic thymic anlage. (A) In situ hybridization of E13.5 thymic anlagen of wild-type and Foxn1::CCX-CKR1 transgenic littermates. Sections stained with sense probes remained negative. (Scale bar: 200 μm.) (B) Immunohistochemical staining of E13.5 thymic anlagen of a Foxn1::CCX-CKR1 transgenic mouse for CCL25 expression. Staining is restricted to the thymic anlagen (circled in white) as in wild-type mice (our unpublished data). tr, trachea; oe, oesophagus. (Scale bar: 200 μm.) (C) The number of CD45-positive hematopoietic precursors that home to the E12.5 thymic anlage is reduced in Foxn1::CCX-CKR1 transgenic mice in comparison with wild-type littermates. (D) The fraction of BrdU-positive thymocytes was determined in E14.5 wild-type and Foxn1::CCX-CKR1 transgenic littermates. (E) The fraction of annexin V-positive thymocytes was determined in E14.5 littermates of the indicated genotypes. (F) The number of lineage marker-negative CD117-positive PIR-positive prethymic T lineage precursors in the fetal liver was determined for E14.5 littermates by FACS. (G) The fraction of lineage marker-negative CD117-positive PIR-positive thymic precursors was determined in the blood of E14.5 littermates by FACS. (H) Thymocyte numbers are normal in newborn Foxn1::CCX-CKR1 transgenic mice. (D–H) At least eight offspring of Foxn1::CCX-CKR1 founder no. 11 were analyzed per genotype.
The thymic anlage at E12.5 is not vascularized and hematopoietic precursors colonize the anlage from the surrounding mesenchyme. We and others have found that hematopoietic precursors do not enter the thymic anlage of nude mice that lack the expression of CCL21 and CCL25 (23, 24). More recent data point at the combination of CCR7 and CCR9 and their ligands that regulate the immigration of hematopoietic precursors to the embryonic thymic anlage before vascularization (25). We therefore hypothesized that overexpression of CCX-CKR1, which binds the ligands of both CCR7 and CCR9, should impair the colonization of the embryonic thymic anlage by hematopoietic precursors if CCX-CKR1 had a role in homeostatic leukocyte migration. We attempted to quantify the amount of bioavailable CCL25 that is secreted from microdissected thymic anlagen by ELISA but found bioavailable CCL25 to be below the level of detection even when several anlagen were pooled. But significantly reduced numbers of hematopoietic precursors in the E12.5 thymic anlage were indeed observed in the two transgenic lines that were analyzed (Fig. 4C). This observation suggested that CCX-CKR1 regulates the homing of hematopoietic precursors to the avascular embryonic thymic anlage. To rule out the possibility that CCX-CKR1 overexpression inhibited the proliferation of thymic precursors or caused their premature death, we analyzed embryonic thymocytes of wild-type and Foxn1::CCX-CKR1 mice for BrdU incorporation and annexin V staining (Fig. 4 D and E). No significant difference was found. Reduced numbers of thymic precursors could also be explained if thymic precursors failed to be generated in the fetal liver. Staining for prethymic T lineage precursors in fetal liver and blood (26) revealed that these were present in Foxn1::CCX-CKR1 transgenic mice at wild-type levels showing that overexpression of CCX-CKR1 did not hamper their production (Fig. 4 F and G). Furthermore, the hematopoietic cells entering the E12.5 thymic anlage of Foxn1::CCX-CKR1 transgenic mice are T lineage precursors (SI Fig. 11) ruling out the possibility that CCX-CKR1 overexpression leads to the aberrant homing of a precursor lacking T cell lineage potential. In contrast to the embryonic stages (SI Fig. 11), thymocyte numbers in newborn transgenic mice reached normal levels (Fig. 4H), indicating that overexpression of CCX-CKR1 showed no effect in later stages of thymus development. Consistently, T cell development in adult Foxn1::CCX-CKR1 transgenic mice was found to be normal (our unpublished data), showing that thymic precursors develop normally in a thymus overexpressing CCX-CKR1. Thus, overexpression of CCX-CKR1 impairs the physiological immigration of embryonic precursors into the E12.5 thymic anlage.
Discussion
We report in vivo evidence for the role of a nonsignaling chemokine receptor in the migration of leukocytes under physiological, i.e., noninflammatory, conditions. In vitro analyses demonstrated that CCX-CKR1 binds the constitutively expressed chemokines CCL19, CCL21, and CCL25 without signs of chemokine receptor signaling. We find that the lack of CCX-CKR1 impairs the steady-state homing of DCs to lymph nodes by way of afferent lymphatics but not the homing of DCs that enter by way of the blood. Furthermore, overexpression of CCX-CKR1 affects the homing of embryonic thymic precursors to the thymic anlage. These observations demonstrate a role for the silent chemokine receptor CCX-CKR1 in leukocyte homing. CCX-CKR1 is exclusively expressed by stroma cells, but not hematopoietic cells themselves, and regulates homeostatic leukocyte migration presumably by controlling the availability of chemokines in the extracellular space. These findings add another level of complexity to our understanding of leukocyte homeostatic migration.
Chemokines and their receptors can be categorized into so-called inflammatory chemokines that regulate the migration of leukocytes in response to an inflammatory challenge and homeostatic chemokines that are constitutively expressed to regulate the physiological migration of leukocytes and their precursors under steady-state conditions (2). This distinction also applies to silent chemokine receptors as the binding properties of the inflammatory silent receptors DARC and D6 do not overlap with that of CCX-CKR1, the only known silent chemokine receptor that binds constitutively expressed chemokines. Silent chemokine receptors have been proposed to internalize and degrade bioavailable chemokines from the extracellular space (8, 9, 15, 27). This notion is based on in vitro studies using receptor transfected cell lines and should therefore be considered with caution. Although there is no doubt that silent receptors bind the chemokines in question, the kinetics with which the receptor is internalized and bound chemokine is separated from the receptor and possibly degraded will likely depend on the transfected cell line. Internalization of seven-transmembrane receptors is regulated by a plethora of downstream components, and it remains an open question whether in vitro findings can be extrapolated to the behavior of DARC on vascular endothelial cells, D6 on endothelial cells of lymphatic vessels, and CCX-CKR1 on TECs and stroma cells lining the lymph node marginal sinus. Other possibilities have to be considered. There are data that implicate DARC in the transcytosis of chemokines through endothelial cells (9, 28), and instead of degrading chemokines, silent chemokine receptors may indeed concentrate and present bound chemokines in specific microenvironments. Immunohistochemical analyses of the CCL25 expression pattern in CCX-CKR1-deficient and wild-type mice has so far not revealed discernible differences (our unpublished data), and further work will be required to quantify bioavailable chemokine levels in distinct microenvironments to elucidate the biochemical basis for the observed in vivo effects. This project is complex in itself because it is still unclear how chemokine receptors on leukocytes follow directional cues within lymphoid organs in vivo. Although expression of chemokine mRNA is restricted to distinct thymic microenvironments (23), chemokine protein shows a much broader expression pattern (ref. 29 and our unpublished data), making it difficult to envisage how chemokine concentration gradients function as directional cues. Methodology will have to be developed that allows the quantification of the bioavailable amount of free chemokine in the extracellular space.
The expression pattern of CCX-CKR1 in the thymus is particularly striking. CCX-CKR1 expression is restricted to TECs ensheathing the blood vessels of the medulla, the cortico-medullary junction, and to subcapsular zone TECs. This expression pattern overlaps but is not identical with the location of type I epithelium that has been classified on the basis of ultrastructural analyses (30–32), because type I epithelial cells ensheathing cortical blood vessels are CCX-CKR1-negative. CCX-CKR1 marks the thymic niche to which hematopoietic precursors are recruited from the blood. The niche that contains the earliest hematopoietic precursors to the thymus remains morphologically poorly defined. The expression pattern revealed by the CCX-CKR1-EGFP knockin and the functional demonstration that CCX-CKR1 affects embryonic precursor homing suggests that adult thymic precursors home to and specify within a thymic microenvironment formed by CCX-CKR1-positive TECs.
Materials and Methods
Chemokine Receptor Transfectants.
Chemokine receptor transfectants were generated by cloning the coding sequences of CCR9, CCR7, and CCX-CKR1 into pcDNA3 and stably transfecting HEK293 cells by using standard methodology. For chemokine competition experiments, 5 × 104 CCX-CKR1-HEK293 cells were incubated with the indicated concentrations of chemokines (R & D Systems, Minneapolis, MN) at 4°C for 20 min followed by staining with CCL19-Fc and a phycoerythrin-labeled anti-human IgG secondary reagent (Jackson ImmunoResearch, West Grove, PA). For calcium measurements, chemokine receptor transfectants were loaded with fura2-AM (Molecular Probes, Eugene, OR) and exposed to the indicated chemokines in a LS 50B photometer equipped with a filter wheel (PerkinElmer, Boston, MA). Ligand internalization was measured by incubating CCX-CKR1-HEK293 cells in the presence of CCL19-Fc for 15 min at 4°C, warming the cells for 15 min to 37°C followed by cooling to 4°C and visualization of surface CCL19 by anti-human IgG staining. The amount of bound ligand in the absence of ongoing internalization was determined by staining CCX-CKR1-HEK293 cells without warming to 37°C. To rule out the possibility that warming the transfectants reduces the amount of surface binding, CCX-CKR1-HEK293 cells were incubated in the absence of CCL19-Fc at 37°C for 15 min, cooled to 4°C, and stained with CCL19-Fc and secondary reagent.
Mice.
A CCX-CKR1 genomic clone from a 129/SvJ BAC library (Genome Systems, St. Louis, MO) was used to determine the genomic structure of the mouse CCX-CKR1 gene. For the targeting vector, 578 bp of coding sequence corresponding to nucleotides 45–622 of the mRNA (AF306532) contained in the major CCX-CKR1 coding exon was replaced in frame by an EGFP together with a neomycin resistance cassette. The 5′ and 3′ flanking arms comprised 1.2 kb and 7 kb of genomic sequence, respectively. KpnI-linearized construct was electroporated into the CCR9-targeted R1 ES cell clone 20/3 (33). Specific integration was confirmed by Southern blotting. Multiple integrations were excluded by Southern blot analysis by using an internal probe. Targeted ES cell clones were injected into C57BL/6 blastocysts and chimeric mice crossed onto C57BL/6 background to separate CCR9-targeted and CCX-CKR1-targeted alleles. Mice were kept under specific pathogen-free conditions in the mouse facility of the Max Planck Institute for Immunobiology. Experimentation and animal care was in accordance with the guidelines of the Max Planck Institute for Immunobiology. Genotyping was done on genomic DNA from tail biopsies by PCR using the following primers: P1, 5′-TAGGATTTAGTGACTAAGAGC-3′; P2, 5′-CACACACAGCAACAGATGATCC-3′; and P3, 5′-TGAACTTGTGGCCGTTTACGTC-3′. For the generation of Foxn1::CCX-CKR1 transgenic mice, the CCX-CKR1 cDNA including positions 46–1,104 of the mRNA (AF306532) were cloned downstream of the Foxn1 promoter as described in ref. 22. Linearized construct was injected into FVB pronuclei according to standard protocols. Transgenic Foxn1::CCX-CKR1 founders were crossed with C57/Bl6 mice and genotyped by PCR using the following primers: P1, 5′- GGCAAACAACAGATGGCCTCG-3′; and P2, 5′-ATGGCCGCCATCTTGCTGAGC-3′. Unless stated otherwise, age-matched 3- to 6-week-old mice were used for experiments.
In Situ Hybridization and Immunohistochemistry.
In situ hybridization was carried out as described in ref. 23. Immunohistochemistry was done as described in ref. 34. EGFP was detected on sections as described in ref. 35. Endothelial cells were stained by intravenously injecting 50 μg of biotin-labeled tomato lectin (Vector Laboratories, Burlingame, CA), euthanizing the mouse 10 min later and visualizing the label by using streptavidin-Cy3 (Jackson ImmunoResearch). Sections were visualized on a Leica (Heidelberg, Germany) TCS SP2 confocal microscope system or on a Zeiss (Jena, Germany) Axio Imager.Z1. Epidermal sheets were prepared as described in ref. 36. Lymph nodes and ear sheets were minced and stirred in 10 ml of RPMI medium 1640, 2% FCS, and 20 mM Hepes for 10 min. Tissue fragments were digested for 15 min at 30°C in RPMI medium 1640, 2% FCS, 20 mM Hepes, 0.2 mg/ml collagenase IV (Worthington Biochemicals, Lakewood, NJ), and 25 μg/ml DNase (MP Biomedicals, Aurora, OH) and subsequently for 25 min at 30°C in RPMI medium 1640, 2% FCS, 20 mM Hepes, 0.2 mg/ml collagenase IV, 25 μg/ml DNase, and 0.2 mg/ml dispase I (Roche, Mannheim, Germany) in a shaking water bath. Free cells were removed after each digestion step for counting by FACS.
Enumeration of Embryonic Thymic Precursors.
E12.5 thymic anlagen were microdissected, dissociated, stained, and the number of lineage-negative (B220, Ter119, Gr-1, CD11b, CD8, CD4) CD45-positive hematopoietic precursors per embryo was determined by FACS.
FACS Analysis.
FACS analyses were carried out as described in ref. 34. CCL19-Fc was a gift of S. Krautwald and U. Kunzendorf (both at University of Kiel, Kiel, Germany). The BrdU incorporation experiments and annexin V stainings were carried out as described in refs. 22 and 33.
FITC Sensitization.
FITC was applied to the shaved skin of mice in 0.4 ml of a 2% FITC (Sigma, St. Louis, MO) solution in a 1:1 (vol/vol) mixture of acetone/dibutyl phthalate. Control mice received carrier only. Lymphocytes and DCs were purified, counted, and stained for FACS analysis 48 h later from six skin-draining lymph nodes per mouse (two brachial, two axillary, and two inguinal) by pressing the lymph nodes through a 70-μm mesh cell strainer (BD Falcon, Bedford, MA). Contour plots are gated on Thy1-negative B220-negative cells.
Supplementary Material
Acknowledgments
We thank C. Sainz and M. Konrath for excellent technical support; A. Würch and J. Wersing for cell sorting; B. Kanzler for blastocyst and DNA injections; and T. Boehm for his support and discussions of the manuscript. This project was supported by Deutsche Forschungsgemeinschaft Grant SFB620/A7.
Abbreviations
- CCL
CC chemokine ligand
- CCR
CC chemokine receptor
- DC
dendritic cell
- En
embryonic day n
- TEC
thymic epithelial cell.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608274104/DC1.
References
- 1.Mackay CR. Nat Immunol. 2001;2:95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 2.Sallusto F, Lanzavecchia A, Mackay CR. Immunol Today. 1998;19:568–574. doi: 10.1016/s0167-5699(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 3.Petrie HT. Nat Rev Immunol. 2003;3:859–866. doi: 10.1038/nri1223. [DOI] [PubMed] [Google Scholar]
- 4.Cyster JG. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 5.Struyf S, Proost P, Van Damme J. Adv Immunol. 2003;81:1–44. doi: 10.1016/s0065-2776(03)81001-5. [DOI] [PubMed] [Google Scholar]
- 6.Jamieson T, Cook DN, Nibbs RJ, Rot A, Nixon C, McLean P, Alcami A, Lira SA, Wiekowski M, Graham GJ. Nat Immunol. 2005;6:403–411. doi: 10.1038/ni1182. [DOI] [PubMed] [Google Scholar]
- 7.Martinez de la Torre Y, Locati M, Buracchi C, Dupor J, Cook DN, Bonecchi R, Nebuloni M, Rukavina D, Vago L, Vecchi A, et al. Eur J Immunol. 2005;35:1342–1346. doi: 10.1002/eji.200526114. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A, Bonecchi R, Locati M. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 9.Nibbs R, Graham G, Rot A. Semin Immunol. 2003;15:287–294. doi: 10.1016/j.smim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Horuk R. Immunol Today. 1994;15:169–174. doi: 10.1016/0167-5699(94)90314-X. [DOI] [PubMed] [Google Scholar]
- 11.Nibbs RJ, Wylie SM, Yang J, Landau NR, Graham GJ. J Biol Chem. 1997;272:32078–32083. doi: 10.1074/jbc.272.51.32078. [DOI] [PubMed] [Google Scholar]
- 12.Matsuoka I, Mori T, Aoki J, Sato T, Kurihara K. Biochem Biophys Res Commun. 1993;194:504–511. doi: 10.1006/bbrc.1993.1848. [DOI] [PubMed] [Google Scholar]
- 13.Gosling J, Dairaghi DJ, Wang Y, Hanley M, Talbot D, Miao Z, Schall TJ. J Immunol. 2000;164:2851–2856. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- 14.Townson JR, Nibbs RJ. Eur J Immunol. 2002;32:1230–1241. doi: 10.1002/1521-4141(200205)32:5<1230::AID-IMMU1230>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Comerford I, Milasta S, Morrow V, Milligan G, Nibbs R. Eur J Immunol. 2006;36:1904–1916. doi: 10.1002/eji.200535716. [DOI] [PubMed] [Google Scholar]
- 16.Khoja H, Wang G, Ng CT, Tucker J, Brown T, Shyamala V. Gene. 2000;246:229–238. doi: 10.1016/s0378-1119(00)00076-7. [DOI] [PubMed] [Google Scholar]
- 17.Ruedl C, Koebel P, Bachmann M, Hess M, Karjalainen K. J Immunol. 2000;165:4910–4916. doi: 10.4049/jimmunol.165.9.4910. [DOI] [PubMed] [Google Scholar]
- 18.Henri S, Vremec D, Kamath A, Waithman J, Williams S, Benoist C, Burnham K, Saeland S, Handman E, Shortman K. J Immunol. 2001;167:741–748. doi: 10.4049/jimmunol.167.2.741. [DOI] [PubMed] [Google Scholar]
- 19.Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Forster R. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Villadangos JA, Heath WR. Semin Immunol. 2005;7:262–272. doi: 10.1016/j.smim.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Macatonia SE, Knight SC, Edwards AJ, Griffiths S, Fryer P. J Exp Med. 1987;166:1654–1667. doi: 10.1084/jem.166.6.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bleul CC, Boehm T. J Immunol. 2005;175:5213–5221. doi: 10.4049/jimmunol.175.8.5213. [DOI] [PubMed] [Google Scholar]
- 23.Bleul CC, Boehm T. Eur J Immunol. 2000;30:3371–3379. doi: 10.1002/1521-4141(2000012)30:12<3371::AID-IMMU3371>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Itoi M, Kawamoto H, Katsura Y, Amagai T. Int Immunol. 2001;13:1203–1211. doi: 10.1093/intimm/13.9.1203. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Saito F, Liu Z, Lei Y, Uehara S, Love P, Lipp M, Kondo S, Manley N, Takahama Y. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 26.Masuda K, Kubagawa H, Ikawa T, Chen CC, Kakugawa K, Hattori M, Kageyama R, Cooper MD, Minato N, Katsura Y, Kawamoto H. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonecchi R, Locati M, Galliera E, Vulcano M, Sironi M, Fra AM, Gobbi M, Vecchi A, Sozzani S, Haribabu B, et al. J Immunol. 2004;172:4972–4976. doi: 10.4049/jimmunol.172.8.4972. [DOI] [PubMed] [Google Scholar]
- 28.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, Auer M, Hub E, Rot A. Cell. 1997;91:385–395. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 29.Misslitz A, Pabst O, Hintzen G, Ohl L, Kremmer E, Petrie HT, Forster R. J Exp Med. 2004;200:481–491. doi: 10.1084/jem.20040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Wijngaert FP, Kendall MD, Schuurman HJ, Rademakers LH, Kater L. Cell Tissue Res. 1984;237:227–237. doi: 10.1007/BF00217140. [DOI] [PubMed] [Google Scholar]
- 31.Kendall MD. Thymus. 1989;13:157–164. [PubMed] [Google Scholar]
- 32.Kendall MD. J Anat. 1991;177:1–29. [PMC free article] [PubMed] [Google Scholar]
- 33.Benz C, Heinzel K, Bleul CC. Eur J Immunol. 2004;34:3652–3663. doi: 10.1002/eji.200425248. [DOI] [PubMed] [Google Scholar]
- 34.Boehm T, Scheu S, Pfeffer K, Bleul CC. J Exp Med. 2003;198:757–769. doi: 10.1084/jem.20030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shariatmadari R, Sipila PP, Huhtaniemi IT, Poutanen M. Biotechniques. 2001;30:1282–1285. doi: 10.2144/01306st06. [DOI] [PubMed] [Google Scholar]
- 36.Bergstresser PR, Juarez DV. Methods Enzymol. 1984;108:683–691. doi: 10.1016/s0076-6879(84)08127-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.