Abstract
Many enveloped viruses employ low-pH-triggered membrane fusion during cell penetration. Solution-based in vitro assays in which viruses fuse with liposomes have provided much of our current biochemical understanding of low-pH-triggered viral membrane fusion. Here, we extend this in vitro approach by introducing a fluorescence assay using single particle tracking to observe lipid mixing between individual virus particles (influenza or Sindbis) and supported lipid bilayers. Our single-particle experiments reproduce many of the observations of the solution assays. The single-particle approach naturally separates the processes of membrane binding and membrane fusion and therefore allows measurement of details that are not available in the bulk assays. We find that the dynamics of lipid mixing during individual Sindbis fusion events is faster than 30 ms. Although neither virus binds membranes at neutral pH, under acidic conditions, the delay between membrane binding and lipid mixing is less than half a second for nearly all virus-membrane combinations. The delay between binding and lipid mixing lengthened only for Sindbis virus at the lowest pH in a cholesterol-dependent manner, highlighting the complex interaction between lipids, virus proteins, and buffer conditions in membrane fusion.
INTRODUCTION
The boundaries of all living cells and their compartments are defined by lipid bilayers. Intracellular transport, cell entry, and secretion require vesicular lipid structures to fuse with target membranes. Membrane fusion proteins are involved in catalyzing almost every situation of biological membrane fusion. Despite decades of intense study, the precise molecular mechanism by which fusion proteins mediate membrane fusion remains a subject of much debate (1–11).
Among the best-studied membrane fusion protein machines are those present in enveloped viruses. Enveloped viruses have evolved highly efficient fusion proteins that allow the viral genome to penetrate targeted cells (12–18). For most enveloped viruses, environmental signals trigger these proteins to catalyze fusion of the viral membrane with the cell membrane. The decreased pH encountered along the endocytotic pathway is a common trigger for the fusion proteins of many enveloped viruses, most notably influenza (19,20). In a few cases, atomic resolution structures are available for viral fusion proteins under both neutral pH and low pH conditions (12) that have led to formulation of models of their molecular action (12,13).
Many enveloped viruses will fuse to protein free lipid bilayers (20–22). In vitro liposome fusion experiments have been extremely useful in characterizing the biochemical properties of viral fusion, including the pH and lipid species dependences (23,24). Bulk liposome measurements have also confirmed that viral fusion is often nonleaky and can fully mix both the lipids and the contents of fusing structures (25,26).
As valuable as these bulk assays have been in advancing our understanding of membrane fusion, they have limitations. Low-pH triggered viral membrane fusion occurs rapidly after acidification, with timescales measured in tens of seconds or minutes in the bulk liposome experiments(16,21). The stochastic nature of each occurrence of fusion prevents the precise synchronization of all of the individual virus-liposome fusion events within a cuvette-based bulk solution assay. These experimental challenges mask transient, intermediate states along the fusion pathway and obscure detailed analysis of the trajectory of the nonequilibrium processes driving the dynamics. Difficulty separating the linked processes of binding and fusion in bulk assays also complicates the interpretation of experimental results.
We have developed a fluorescence assay to make detailed measurements of the early stages of individual Sindbis and influenza virus particles fusing to supported lipid bilayers under acidic conditions. Our assay detects lipid mixing of octadecyl rhodamine (R18) between virus particles and the supported lipid bilayer. As R18 is known to “flip-flop” between the inner and outer leaflets of a labeled bilayer (27,28), this signal does not differentiate between hemifusion and full fusion. Throughout this article we will use hemifusion/fusion to indicate this ambiguity.
The supported lipid bilayer geometry is desirable for its compatibility with high-resolution optical measurements as well as its ease of integration with biotechnological instrumentation and sensors. Supported bilayers have been successfully applied in membrane fusion studies of SNARE proteins (29–31) and influenza virus (32–36). Illumination of the supported lipid bilayer by total internal refection yields a signal/noise ratio sufficient for precise measurements of individual virus particles during fusion. Continued development of this single-particle approach will allow detection of the transient, stochastic intermediate states that are commonly averaged in liposome fusion assays.
We have verified that our supported bilayer assay reproduces the general trends seen in the bulk liposome experiments for these viruses (16,21,37). Although preexposure to low pH for several minutes inactivates the virus for fusion (21) and infection (38), our results show that virus exposed to low pH immediately before encountering the target membrane maintains fusion capacity. Observation of individual virus particles indicates that lipid mixing follows binding rapidly, in under a half second for most conditions, and that the dynamics of lipid mixing in individual fusion events is faster than our instrumental resolution limit of 30 ms.
Cholesterol in the target membrane enhances the final extent of fusion for Sindbis virus. Cholesterol forms microdomains, or lipid rafts, when combined with sphingomyelin in lipid bilayers (39–42). These microdomains are not required for Sindbis virus fusion and may actually inhibit fusion (41,43) by reducing lipid mobility (44–49). Here, we find that inclusion of cholesterol in the target membrane increases the interval between binding and lipid mixing at pH values <5 for Sindbis but not for influenza; a result that emphasizes the complex interactions between lipids and proteins in membrane fusion and may point to subtle differences between the mechanisms of fusion for type 1 and type 2 viral fusion proteins.
MATERIALS AND METHODS
Virus and dye labeling
Influenza (A, X:31, A Aichi/68, H3N2) grown in fertilized chicken eggs and purified by density-gradient ultracentrifugation was purchased from Charles River Laboratories (Wilmington, MA) and used as provided (2 mg/ml viral protein).
Sindbis was grown in baby hamster kidney cells (BHK-21) cultured by standard methods in minimal essential media with Earl's salts containing 10% fetal bovine serum, 5% tryptose phosphate broth, and 2 mM glutamine. Cells were inoculated with Sindbis virus and incubated for 12 h at 37°C. Supernatant was then collected and clarified by low-speed centrifugation.
Sindbis virus was purified from the clarified supernatant by ultracentrifugation on a step density gradient followed by a continuous density gradient in phosphate-buffered saline (PBS, 10 mM phosphate, 140 mM sodium chloride, pH 7.4), containing variable amounts (15–35%) of potassium tartrate to adjust the density. Purified Sindbis solutions were adjusted to a concentration of 2 × 1012 particles/ml as calibrated by BCA Assay (Pierce Biotechnology, Rockford, IL). Sindbis was used immediately or stored at −80°C without noticeable loss of fusion capacity (50).
Both Sindbis and influenza were dye-labeled with the hydrophobic fluorescent dye R18 (octadecyl rhodamine B chloride) from Molecular Probes (Invitrogen, Carlsbad, CA). Aliquots of virus (100 μl each, Sindbis at 2 × 1012 particles/ml, influenza at 0.2 or 2 mg/ml viral protein) were rapidly mixed at room temperature with 3 μl of dye dissolved in ethanol at 1.4 mM. The dye-virus mix was incubated on ice for 2 h. Virus was purified from unincorporated dye by gel filtration (NAP 5, G.E. Biosciences, Piscataway, NJ) at room temperature in PBS (for Sindbis) or Hepes-buffered saline (HBS, 45 mM Hepes, pH 7.4, 100 mM NaCl) (for influenza). We estimate that gel filtration in NAP 5 columns diluted the samples to ∼⅓ the starting concentration (0.7 × 1012 particles/ml (Sindbis) or 0.07 or 0.7 mg/ml viral protein (influenza)).
Lipids, flow cells, bilayers, and buffer exchanger
All lipids (including total liver extract, brain phosphatidylethanolamine (PE), egg phosphatidylcholine (PC), brain sphingomyelin, and cholesterol) were purchased from Avanti Polar Lipids (Alabaster, AL). Chloroform was removed from solutions of lipids and cholesterol (mixed at ratios indicated in the text) under flowing argon leaving a film on the surface of a glass tube. The lipid films were placed in vacuum for at least 2 h and then hydrated with Tris-buffered saline (TBS, 25 mM Tris-HCL, pH 7.3, 150 mM NaCl) (51). Small unilamellar vesicles were prepared by extrusion through 100-nm-pore filters (miniextruder, Avanti Polar Lipids) at 5 mg/ml (liver extract), 4 mg/ml (mixes), or 20 mg/ml (PC).
Quartz microscope slides were cleaned with a sequence of bath sonication steps (soapy water, acetone, ethanol, 1 M potassium hydroxide, and water), and dried in a propane flame immediately before use. Flow-cell chambers were built by attaching glass coverslips to the quartz microscope slides, with channels defined by double-sided tape. The ends of the channels were sealed with 5-min epoxy. Fluid was introduced into the channel through holes drilled in the quartz slide at either end of the channel. To form supported bilayers on the walls of the channel, liposomes were incubated in the chamber for 5 min and then rinsed away. A second incubation with 20 mg/ml of PC liposomes for 1 h improved overall experimental reproducibility.
A home-built buffer exchanger pumped solutions through the flow cells during observations with the microscope. Virus and acidic buffers were contained in separate syringes and mixed in a tee immediately before injection into the flow cell. A variable-speed motor actuated the syringes. Enough solution was perfused to ensure that ∼2.5 channel volumes were injected through the flow cell within ∼5 s (∼20 μl/s) during each experiment.
Sindbis virus will stick to many surfaces. We minimized surface adsorption by constructing the buffer exchange apparatus using PEEK (polyetheretherketone) tubing and fittings from Upchurch Scientific (Oak Harbor, WA) and disposable syringes (Norm-Ject, Henke Sass Wolf, Tuttlingen, Germany). Clear Tygon tubing was used immediately before the flow-cell entrance to diagnose the presence of air bubbles.
Lipid mixing hemifusion/fusion assays
For bulk measurements, equal volumes of liposome solution (concentrations as extruded, except for PC which was used at 4 mg/ml) and dye-labeled virus solution (∼0.7 × 1012 particles/ml for Sindbis; 0.07 or 0.7 mg/ml viral protein for influenza) were mixed in a fluorescence cuvette. A fluorimeter (Shimadzu, Kyoto, Japan) detected fluorescence emission from the cuvette using excitation at 510 nm and emission at 570 nm. After the initial emission intensity (Io) was recorded, the mixture was acidified by addition of an equal volume of an acidic buffer pretitrated to yield the desired final pH. The emission intensity (I) was recorded for 3–5 min, and at the end of the experiment, ∼1% vol/vol of 100 mM dodecyl-maltoside was added to dissolve the samples and yield the unquenched intensity (If)(16,21,22). Dequenching was calculated as (I − (Io/2))/If and converted into a percentage for plotting.
Single virus particles were observed with a prism-type total internal reflection florescence microscope. Samples were illuminated with 8–10 mW laser light (532 nm). Fluorescence emission was collected by a water immersion objective lens (PlanApo 60×, NA 1.2, Olympus, Melville, NY), filtered by long-pass filter (HQ545lp, Chroma Technologies, Rockingham, VT), and detected with a CCD camera (Cascade 512B, Photometrics, Roper Scientific, Tucson, AZ) at 10 or 67 frames/s. Custom programs written in MATLAB (The MathWorks, Natik, MA) identified individual spots within movies representing individual virus particles. Intensity time traces were extracted from the movies at those locations by integrating the background-subtracted signal from a 7-pixel-diameter circle (1.8 μm in the sample plane) centered on the identified spots and averaging by the integrated number of pixels. Virus solutions with low pH were injected into the flow cell while movies were recorded. For injection into flow cells, virus solution at neutral pH (∼0.7 × 1012 particles/ml for Sindbis; 0.07 or 0.7 mg/ml viral protein for influenza) was contained in one syringe and acidic buffer was held in a second syringe. When injection was triggered, a motor activated both syringes. The two solutions mixed in a tee (mixing time <1 s) and then traveled to the entrance of the flow cell. We measured a 1-s delay between solution leaving the tee and appearing in the field of view in a flow cell mounted on the microscope. The virus was thus acidified 1–2 s before it was exposed to the lipid bilayer. Particles then encountered the bilayer at random times after their free diffusion in the flow cell.
Different acidification buffers were used depending on the virus and the desired final pH. For Sindbis experiments, 0.1 M Mes/0.2 M Acetic acid/145 mM NaCl was added to achieve the lowest final pH of 4.1. For all other final pH values, 100 mM Mes/150 mM NaCl buffer with pH adjusted with NaOH was diluted with variable amounts of TBS to yield a final pH as indicated in the text when mixed with virus samples in PBS. Control experiments at neutral pH used TBS (pH 7.3). For influenza experiments, the second syringe contained 500 mM Mes/145 mM NaCl adjusted to various pH to yield the indicated final pH when mixed with virus in HBS. HBS (pH 7.3) was used for the influenza control experiments. Both bulk and single-virus-particle experiments were performed at room temperature.
The precise values of our ensemble measurements of single-particle behavior (see Table 1, and Figs. 1, 2, and 6) were variable over the months of experimentation of this project. Different purifications of Sindbis virus or different manufacturer lots of influenza virus exhibit different infectivity and fusion capacity. Different substrates and different manufacturer lots of lipids generate heterogeneity in the properties and defects in the supported bilayers. To minimize the effects of the variability of our samples on the ensemble measurements presented here, all data for any given figure were acquired from the same sample, on the same day, on substrates prepared in batch with identical cleaning treatment and using the same supply of lipids.
TABLE 1.
Hemifusion/fusion of Sindbis virus with bilayers formed from mixtures of purified lipids
| Membrane composition | Molar ratio | Final % dequenching (bulk solution) pH 4.6 | Final % dequenching (bulk solution) pH 5.5 | Residence time single virus: bilayer pH 4.6 |
|---|---|---|---|---|
| PC/PE | 1:1 | 0 | 0 | 0.2 s |
| PC/PE/SM | 1:1:1 | 1 | 0 | 0.26 s |
| PC/PE/CH | 1:1:1.5 | 17 | 2 | 0.75 s |
| PC/PE/SM/CH | 1:1:1:1.5 | 20 | 8 | 0.5 s |
Liposomes of the indicated mixtures of lipids (CH, cholesterol; SM, sphingomyelin) were used in bulk solution assays or to form supported lipid bilayers for single particle studies. The final extent of dequenching from bulk fusion experiments illustrates the strict cholesterol requirement for fusion with Sindbis virus. The essential nature of sphingomyelin is evident at pH 5.5 but is reduced at pH 4.6. The column of residence times is from single-particle bilayer experiments at pH 4.6. In the absence of cholesterol, there were fewer events in agreement with the lower final dequenching extent in the bulk studies, but residence times could be determined from the few events that did occur. The residence time increased in the presence of cholesterol only for experiments with pH <5.
FIGURE 1.
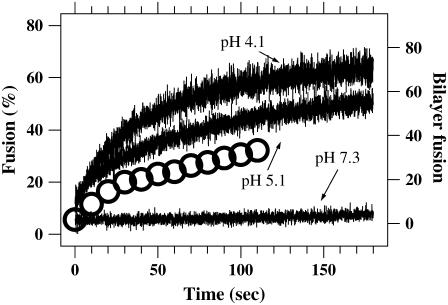
Bulk solution hemifusion/fusion of Sindbis virus with liposomes formed from total liver extract triggered by low-pH conditions. The continuous lines show how fluorescence emission from self-quenched dye in the virus membrane increases after hemifusion/fusion to liposomes dilutes the dye. Buffer is added at time zero to change the pH to the indicated final value. Lipid mixing does not occur at neutral pH. The large circles are the accumulated number of dequenching events as a function of time during the single-particle bilayer experiment from Fig. 5 B. Similar ensemble kinetics are observed in the bulk experiments and the single-particle bilayer experiments.
FIGURE 2.
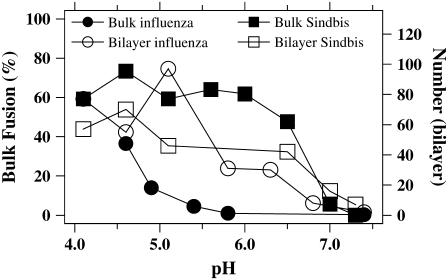
Comparison of bulk solution to supported lipid bilayer measurements of the pH response of membrane hemifusion/fusion for Sindbis and influenza viruses. The final extents of dye dequenching from bulk solution measurements with liposomes as in Fig. 1 (liver extract) are plotted against the left axis. Plotted on the right axis is the total number of individual dequenching events on a 100 μm × 100 μm area within 12 min after injection of virus and acidic buffer in supported lipid bilayer experiments (liver extract).
FIGURE 6.
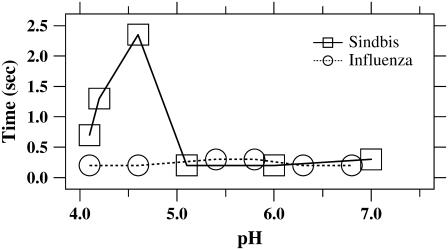
Residence times for Sindbis and influenza virus as a function of pH. The median residence time (as defined in Fig. 3 B and the text) accumulated from many experiments for Sindbis virus increases sharply by a factor of 10 at low pH compared to the near-constant residence time for influenza. Note that most events occur quickly after virus binds to the bilayer. No pH dependence was observed for influenza, but at the lowest pH values, the residence time for Sindbis virus increased by more than a factor of three.
To estimate the expected variability in the quantitative values from our experimental protocol, we simulated multi-experiment runs all in one day, but instead of scanning parameter space, as was done to generate the data in the figures of this article, we repeated one condition in parameter space many times. From these repeated measurements of a single parameter space point, we estimate the standard deviation for number of fusions at 44% (Sindbis) and 36% (influenza). Standard deviations for the median residence times are 25% (Sindbis) and 21% (influenza). The fact that both Sindbis virus and influenza have similar quantitative variability suggests that these effects are random. The spread in our parameter-space data generally agrees with this variability in regions of parameter space where previous bulk solution experiments suggest there should be no systematic variation. The main ensemble effects that we claim in this article (increased fusion at low pH and increased residence time for Sindbis at pH below 5 for cholesterol containing target membranes) are effects substantially larger than these random run-run variations. The standard deviation of the final dequenching percentages, upon multiple attempts at identical bulk solution virus-liposome fusion experiments, was ∼10% for both viruses in our lab.
RESULTS
Bulk solution experiments
Fusion of virus with liposomes in bulk solution has been studied with fluorescence dequenching for many years. Before developing a single-particle fusion assay, we confirmed that our Sindbis and influenza viruses exhibit fusion behavior consistent with this large body of past work. In bulk assays, samples of virus labeled with self-quenching concentrations of lipophilic, fluorescent dyes are mixed in a fluorescence cuvette with liposomes. Upon acidification of the solution in the cuvette, virus particles fuse with liposomes, resulting in dilution of the fluorescent dye and release of the self-quenching of the fluorescent dye. The intensity increase due to the dequenching of most of the lipophilic dyes used in these bulk fusion experiments (as well as our single-particle assay described below) only reports lipid mixing, but this signal has correlated with content mixing in some cases (21,25,37).
The release of the self-quenching of membrane dye during bulk fusion experiments leads to an increase in the fluorescence emission intensity that reaches a steady value typically within a few minutes. The emission is scaled to the completely de-quenched value measured upon addition of detergent to completely dissolve the vesicular structures in the cuvette. Time courses of typical bulk fusion experiments using our Sindbis virus are displayed as the continuous traces in Fig. 1. Similar results are found when using influenza. No de-quenching from membrane fusion is detected in control experiments where the pH is maintained near neutral (Fig. 1, lowest curve).
Fig. 2 (solid symbols) displays the pH dependence of the final extent of the dequenching signal resulting from fusion for Sindbis and influenza viruses, as deduced from bulk measurements with liposomes. In agreement with previous published work, we find that fusion with a physiological blend of lipids in liposomes (total liver extract) is rapid and nearly complete for Sindbis virus for pH <∼6.5, whereas influenza requires a lower pH of <6.0 to reach highly efficient fusion with the same type of liposomes (16,20,21,52,53). Table 1, columns 3 and 4, contains results of bulk fusion experiments where the composition of the target liposome is varied. Again consistent with previous work (21), we find an enhancement of fusion efficiency for Sindbis virus when the target membrane contains both cholesterol and sphingomyelin. The data in Table 1 only include a small region of the parameter space relevant to fusion. For example, it is clear that the composition of the target membrane determines the optimal pH for fusion, as demonstrated by the observation that influenza can fuse to liposomes at neutral pH when specific blends of lipids are used to form target membranes (54). The agreement between these bulk studies using virus in our lab and the published work of other labs validates our starting point to design a single-particle assay for viral membrane fusion.
Single-virus assay
To exploit the advantages afforded by the single-particle experimental approach (55,56), we developed an assay to observe individual Sindbis and influenza virus particles fusing to a supported lipid bilayer. For this assay, we formed a bilayer on a quartz surface within a liquid flow cell. The bilayer was illuminated with total internal reflection of laser light, which results in fluorescence emission only from dye-labeled virus particles that are less than a few hundred nanometers from the bilayer. We observed the illuminated region of the bilayer with a sensitive fluorescence microscope capable of single-molecule detection.
Membrane fusion is triggered by low pH for both Sindbis and influenza viruses. The priming of the particles for fusion upon low-pH exposure is transient and within a few minutes of initial acidification they are rendered incompetent for fusion. We therefore built a two-chamber buffer exchange system to change the pH of the virus solution immediately before it was pumped into the flow cell. Virus was contained at neutral pH in one chamber, separate from the low-pH solution in the other chamber. Upon activation, the buffer exchanger mixed the solutions in a tee and pumped the mixture into the flow cell containing the supported lipid bilayer while the flow cell was being observed with the microscope. The pumping was slow to allow mixing of the solutions in the tee. The delay between solutions leaving the mixing tee and arriving in the flow chamber was ∼1 s (∼30 μl in the connecting lines and tee).
Virus samples were labeled with self-quenching concentrations of fluorescent membrane dyes in the same manner as in the bulk fusion studies. After injection of the virus sample into the flow cell, individual particles were detected around the bilayer as their Brownian motion took them into the evanescent light field that extends 100–200 nm beyond the supported bilayer. Individual particles diffused into the illuminated region and then back into bulk solution in control experiments using neutral pH (pH 7.3). Sindbis and influenza virus did not bind to the protein free lipid bilayers at neutral pH. For those few particles that did bind the bilayer, hemifusion/fusion was very rare at neutral pH for either virus. Of the few bound particles, only 3% of Sindbis and 7% of influenza eventually hemifused/fused when the pH was >7.0.
When the final pH was in the range 4.1–6.5, both viruses quickly bound to and efficiently hemifused/fused with the supported bilayer. We found that 60% of bound Sindbis hemifused/fused, whereas 30% of bound influenza hemifused/fused in the low-pH experiments. (These values are determined by averaging the ratio of hemifused/fused particles to total bound particles for experiments in Fig. 2 that lie between pH 4.1 and 6.5. We find (hemifused/fused)/(total bound) = 0.6 (SD 0.05) for Sindbis and (hemifused/fused)/(total bound) = 0.30 (SD 0.12) for influenza.) This value for influenza efficiency agrees with published reports using the same influenza product from the identical supplier that found “two-thirds of the viruses were intrinsically fusion defective” (19).
Background-subtracted time courses of emission intensity integrated over a 1.7-μm-diameter spot centered on two Sindbis virus particles that bind to the bilayer at low pH are displayed in Fig. 3, B and C. Binding of a virus particle from solution leads to a small, localized emission emerging from the background. In the absence of lipid mixing, these particles appear as diffraction-limited spots with very low mobility. The intensity of a bound and unfused virus particle remains localized and stable for minutes (Fig. 3 C), unbinding only in rare cases.
FIGURE 3.
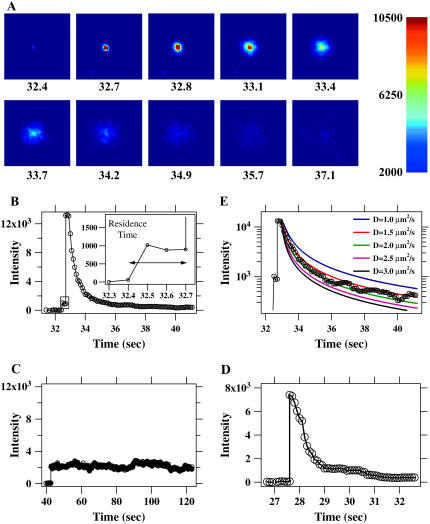
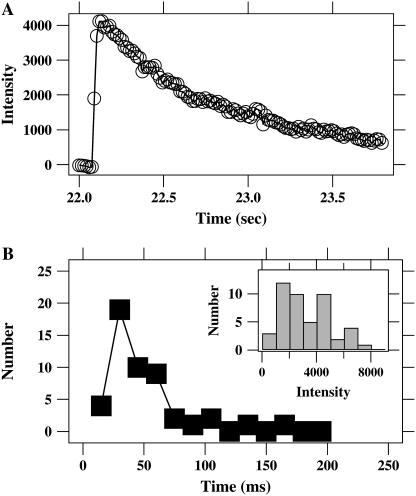
Single-particle hemifusion/fusion events. (A) Images extracted from a 10 frames/s movie (Movie S1) of a single Sindbis virus particle labeled with R18 dye hemifusing/fusing with a supported lipid bilayer (liver extract). The pH is 5.5 and virus has been in solution in the flow cell for ∼30 s. Time in seconds is indicated under each frame. Individual dye molecules are visible in the later images as they diffuse away from the initial fusion site. The edges of the squares are 13 μm long. The image is uncorrected camera output values that have been false-colored using the jet color table (MATLAB) with the indicated pixel value mapping. (B) Background-subtracted intensity time course for the particle in A. The particle binds the supported bilayer at ∼32.5 s. The particle remains attached but unfused for less than a third of a second. The interval between binding and hemifusion/fusion is indicated by a rectangle, zoomed in the inset, and denoted as the residence time. The sudden increase in intensity arises from the dequenching of the membrane dye label as it mixes with the supported bilayer after hemifusion/fusion. The dye disperses by diffusion away from the hemifusion/fusion site, leading to the slower intensity decrease. The smooth line connects the data points to guide the eye. (C) Intensity time course for a Sindbis virus particle on a supported lipid bilayer (liver extract) at pH 5.0 that binds but does not hemifuse/fuse. The intensity from virus particles that do not fuse is stable for much longer than is required for the dye to diffuse away after fusion events. (D) Intensity time course for a hemifusing/fusing influenza virus particle on a supported lipid bilayer (liver extract) at pH 4.6. (E) Logarithmic plot of the event from B overlaid with theoretical diffusion curves (see text) for varying diffusion constants, D. D ≈ 1.5–2.0 μm2/s fits the data.
Hemifusion/fusion of a bound virus particle leads to lipid mixing between the viral membrane and the bilayer. The dilution of the membrane-incorporated dye as it diffuses into the supported bilayer is reported by sudden dequenching of the dye emission by a factor of ∼10. In Fig. 3, A and B, the small intensity increase due to binding of a Sindbis particle is followed by a larger intensity increase 0.2 s later that is due to membrane hemifusion/fusion (see Fig. 3 B, inset). Influenza hemifusion/fusion was similar to Sindbis (Fig. 3 D). Hemifusion/fusion is confirmed by the observation in the two-dimensional image data of free diffusion of the lipid dye isotropically into the bilayer away from the site of binding after the large dequenching (Fig. 3 A and Supplementary Material, Movie 1). The motion of the dye in the bilayer was generally consistent with diffusion using a diffusivity of ∼1–2 μm2/s for all events measured. (Diffusion after the sudden dequenching is discussed in detail below.) In all measurements described below we confirmed hemifusion/fusion events by verifying both the sudden dequenching of the integrated intensity and visualization of the subsequent radial spreading of the membrane dye as it diffused into the supported bilayer.
Dynamics of sudden R18 dequenching due to lipid mixing during individual events
Lipids mixed quickly during individual hemifusion/fusion events. Using our 10-Hz imaging mode, the sudden dequenching of dye emission due to lipid mixing during fusion occurred within a single 100-ms time bin for all Sindbis fusion events. Seventy-five percent of influenza events were consistent with dequenching within a single 100-ms bin, whereas the remaining 25% extended up to three time bins (300 ms). As no lipid dye remained at the docking site after hemifusion/fusion (with the exception of a few very rare cases described below and in Movie S2), the sudden dequenching indicates that lipid mixing of the membranes occurs in <100 ms.
To further constrain the dynamics of lipid mixing during individual hemifusion/fusion events we increased the data rate to 67 frames/s (our current instrumental limit). Laser illumination intensity was increased by a factor of 3 above the value used in the 10-Hz movies to increase dye emission levels and maintain an acceptable signal/noise ratio. The brighter illumination increased the photobleaching rate. For these measurements, focusing on the short interval between binding and hemifusion/fusion, photobleaching was not a limiting factor. An example of a fusion event recorded at the 67 frames/s rate is shown in Fig. 4 A. Even when using 15-ms time steps, dequenching for this Sindbis virus event occurred in two frames (Fig. 4 B). The dequenching intervals for many events are plotted as a histogram in Fig. 4 B. All of these events dequenched to approximately the same final intensity as illustrated in the inset of Fig. 4 B. The events in this graph were derived from 10 movies using five independently prepared experiments. Only 2 out of the 51 total events exceeded the 0.2-s interval plotted here.
FIGURE 4.
Measurements of the dynamics of lipid mixing between individual virus particles and a supported bilayer, at 15 ms/frame. (A) The intensity versus time for a single R18-labeled Sindbis virus particle on a supported lipid bilayer (liver extract). The particle binds the supported bilayer, then hemifuses/fuses in less than two 15-ms time steps. (B) A histogram of dequenching times (10–90%) for 49 of 51 fusion events taken over 10 movies from five slides (two were >200 ms). Most events occur in two time steps. Some events that are fast enough to be completed within one time bin will be counted in the group requiring two time bins, because those events may begin toward the end of one time bin and finish during the next bin. An exponential fit omitting the first time bin yields a time constant of 28 ms. The histogram in the inset displays the maximum intensity after dequenching for the fusion events analyzed in B. The distribution in the inset shows that the events used to measure the initial dequenching rate did not vary by more than a factor of 3–4 in final intensity, although there is some heterogeneity in the efficiency of particle labeling.
Following Liu et al. (29), we note that for fast kinetics the lack of synchronization of fusion events with the edge of a time bin in the detector reduces the number of events observed to transition within one time bin. Dequenching transitions due to fusion that start close to the end of one detector integration interval may finish during the following integration interval. Such events will thus span two time bins even if the inherent dynamics are faster than one time bin. Such a suppression of the first 15 ms bin in the distribution of dequenching transition times is clear in Fig. 4 B. The remaining bins of the histogram match a simple exponential decay with a time constant of 28 ms (or ∼2 time bins). Due to the similarity of the observed dynamics to our experimental resolution, we conclude that our observation of the lifetime of states leading to lipid mixing during membrane fusion in these viral systems remains instrument-limited and that 30 ms is an upper bound.
Virus particles appeared to undergo multiple fusions in a few rare cases. These viruses would hemifuse/fuse to the bilayer, leaving behind a smaller, but still bright, center after the initial burst of diffusing dye in the bilayer dispersed and faded. Another sudden dequenching event followed by diffusion of membrane dye into the bilayer occurred a few seconds later from that bright center. In a few cases, a third hemifusion/fusion event occurred from the same spot (Movie S2). We interpret these multiple hemifusion/fusion events as being caused by aggregates of virus in which different virus particles in the aggregate hemifused/fused at different moments. It has also been suggested that aggregation may explain the observation of multiple fusions at single sites for SNARE-driven fusion of liposomes to supported bilayers (29).
Diffusion of R18 in the supported bilayer after sudden dequenching
After sudden dequenching of the dye within the virus particles, we observed that the R18 dye spread in the supported bilayer uniformly away from the fusion site. The concentration (number of dyes per area), C(r,t), of R18 in a two-dimensional bilayer that spreads from a sudden point source of No molecules of dye deposited at r = 0 and t = 0 is described by 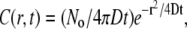 where D is the diffusivity of R18 in the bilayer (57). Integration of this concentration over a circle of radius R around the initial fusion site yields a function proportional to the intensity we observe in the time traces (Fig. 3, B and D) in our experiment,
where D is the diffusivity of R18 in the bilayer (57). Integration of this concentration over a circle of radius R around the initial fusion site yields a function proportional to the intensity we observe in the time traces (Fig. 3, B and D) in our experiment, 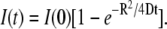 This function is plotted in Fig. 3 E for R =1.2 μm (our experimental parameter), along with the data from Fig. 3 B, as a log-linear plot for values of D ranging from 1 to 3 μm2/s. D ≈ 1.5–2.0 μm2/s fits the data well, capturing the nonexponential behavior. This diffusivity agrees with published reports of R18 mobility in bilayers (33), with our measurements of R18 diffusivity in bilayers using single-particle tracking and fluorescence recovery after photobleaching (data not shown), and also with the mobility of many different fluorescent lipid analogs in purified lipid bilayers (58,59).
This function is plotted in Fig. 3 E for R =1.2 μm (our experimental parameter), along with the data from Fig. 3 B, as a log-linear plot for values of D ranging from 1 to 3 μm2/s. D ≈ 1.5–2.0 μm2/s fits the data well, capturing the nonexponential behavior. This diffusivity agrees with published reports of R18 mobility in bilayers (33), with our measurements of R18 diffusivity in bilayers using single-particle tracking and fluorescence recovery after photobleaching (data not shown), and also with the mobility of many different fluorescent lipid analogs in purified lipid bilayers (58,59).
We often observed a delay between the sudden dequenching due to initial lipid mixing and the eventual spreading of lipid dye by free diffusion. Such a delay is evident in Fig. 3 B, where a nearly constant intensity is observed for 0.3 s (three time bins) between the sudden dequenching rise and the eventual spread of dye into the bilayer by diffusion. Not all fusion events showed this sort of delay; for example, Fig. 3 D does not have a measurable delay between dequenching and free diffusion. Several fusion events (55% of Sindbis events and 44% of influenza events) did not have the delay at the top, whereas the remaining events showed a subdiffusive pause after the sudden dequenching. These pauses were of variable duration, with the most common delays ∼0.2–0.5 s (2–5 time bins), but a few pauses extended up to ∼1 s. This delay may indicate that the viral fusion proteins at a site of hemifusion/fusion alter the free flow of lipids between the viral particle and the target bilayer (60–63).
The ensemble of particles in solution
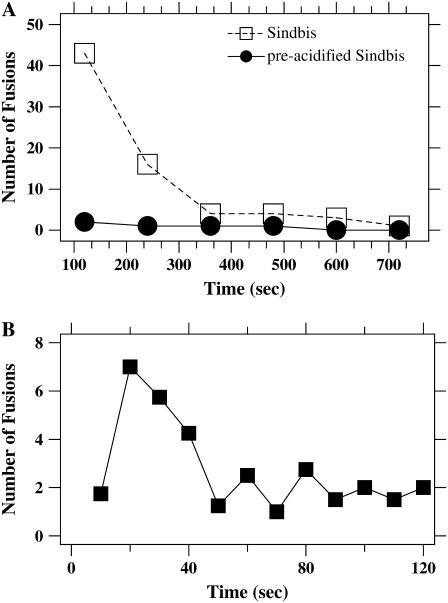
Immediately after acidification and injection into the flow cell, we observed a burst of activity, with virus particles binding and hemifusing/fusing with the supported bilayers. After continued exposure to acidic conditions for only 1–2 min, the frequency of events leading to lipid mixing declined. Sporadic events were detected out to 12 min. By recording the time of each fusion event relative to the initial introduction of virus into the flow cell, we were able to compare the time course and parameter dependence of fusion in our bilayer assay with bulk fusion assays. As shown in Fig. 5 A, almost all of the fusion activity of Sindbis virus was completed within 4 min, in agreement with bulk fusion studies (21). Fig. 5 B displays the rate of fusion for earlier times, demonstrating that there is a burst of activity within the first half-minute of exposure to acidic conditions. In Fig. 1, the accumulation of dequenching events from the data in Fig. 5 B is plotted as large circles along with the bulk solution experiments. Similar overall kinetics is observed in the bulk solution and bilayer experiments, with most fusion being completed in the first minute or two although there is a small amount of fusion continuing for several more minutes.
FIGURE 5.
Time course of single particle-bilayer experiments. (A) Each of the open symbols (dashed line) corresponds to the total number of hemifusions/fusions observed in the preceding 2-min interval for Sindbis virus interacting with supported lipid bilayers (liver extract) at pH 4.6. Virus was introduced into the flow cell at time zero. The solid line (solid symbols) reports an identical measurement of virus that was exposed to pH 4.6 for 6 min before the injection into the flow cell. Sindbis loses its fusion capacity quickly after exposure to acidic conditions. (B) Results from a different experiment at pH 5.5 (without acid exposure to the virus before injection to the flow cell) binned into events within the preceding 10 s illustrates that the majority of lipid mixing events takes place within the first minute of the experiment. The large circles in Fig. 1 plot this same data as the cumulative number of hemifusion/fusions as a function of time.
The total number of individual hemifusion/fusion events occurring on supported bilayers within 12 min (analogous to the final extent of dequenching in bulk solution experiments) for experiments at varying pH is reported in Fig. 2 (open symbols). The integrated activity in bilayer experiments matches the trends observed in the final extent of dequenching for bulk fusion experiments despite the substantial difference in curvature of vesicles and supported bilayers.
The decline of the hemifusion/fusion rate is due to both the overall depletion of virus from solution due to hemifusion/fusion, as well as unfused virus in solution losing its fusion competence after incubation in acidic conditions. In control experiments where virus was mixed with low pH for 6 min before being introduced onto the bilayer, fusion was almost completely eliminated (Fig. 5 A, solid symbols). In control experiments at neutral pH, the rate of binding and lipid-mixing events was much lower, although the few individual events that did occur could be analyzed for their detailed behavior as described in Figs. 2 and 6.
The residence time: the interval between membrane binding and membrane fusion for individual virus particles
The single-particle approach allowed us to measure the time interval between binding of an individual virus particle to the bilayer and the hemifusion/fusion of that particle to the bilayer (Fig. 3 B, inset, arrow). Measurements of the median of this time interval, which we call the residence time, for ensembles of fusing influenza and Sindbis particles are plotted in Fig. 6 for varying pH. Note that in our assay, viral priming and activation processes due to acidic exposure are completed before the membrane is engaged by the particles. Therefore, our residence time is not equivalent to the delay time reported in experiments where influenza fusion proteins (hemagglutinin (HA)) are prebound to a target membrane by receptor interactions at neutral pH. Those delay times, which are longer, include low-pH priming and activation steps (see Discussion). At pH near 6.9, fusion events were rare, but we were able to calculate median residence times from the few events that were observed. In most cases, hemifusion/fusion follows quickly after each virus particle binds the bilayer. Particles that hemifuse/fuse spend a median time of only ∼0.2 s on the bilayer. A few virions remain stably docked on the bilayer for half a minute or longer before eventually hemifusing/fusing.
The surprising exception to the short residence time is evident in Fig. 6 for Sindbis virus fusing with total liver extract bilayers below pH 5. In this particular system, the residence time was longer by more than a factor of 10. The only other condition in which longer residence times were observed was in the hemifusion/fusion to blends of purified lipids (Table 1, last column). The inclusion of cholesterol and sphingomyelin into mixtures of PC and PE lipids increase the residence time of Sindbis virus by a factor of 2–3 for pH 4.9.
DISCUSSION
Membrane fusion is a rapid, nonequilibrium process, and high-resolution observations of its dynamics will greatly advance our understanding of the mechanics underlying this important biological phenomenon. Membrane fusion is often described as triggerable by environmental conditions; for instance, neurotransmitter release is triggered by calcium influx during action potentials, or many viruses fuse membranes upon acidification of endosomes. In actuality, triggering is only a change of the probability (sometimes dramatically) that membranes will fuse. Even under highly promoting conditions, the occurrence of any single fusion event is still stochastically distributed. The single-particle approach is well suited to reveal the mechanistic details about protein-mediated membrane fusion because single-particle analysis avoids averaging the behavior of ensembles of particles and is therefore sensitive to the transient dynamics during membrane fusion.
Here, we describe an in vitro assay for high-precision measurements of lipid mixing during early stages of individual viruses fusing with supported lipid bilayers in response to low pH. In our assay, an ensemble of virus labeled with quenched fluorescent dye is acidified and immediately introduced into a flow cell with a protein-free, supported lipid bilayer on its surface. Particles encountered the bilayer by diffusion, so we used a fluorescence microscope to observe the bilayer for up to 12 min (Fig. 5). When particles encountered the bilayer, many bound and hemifused/fused. Lipid mixing commenced around a quarter-second after binding (Fig. 6). The sudden dequenching transitions during individual lipid mixing events were resolution-limited at 30 ms (Figs. 3 and 4). As the ensemble was monitored for the 12-min interval, the rate of individual hemifusion/fusion events decreased, largely due to the loss of fusion capacity after acidification of the virus (Fig. 5). Our assay has reproduced many results previously found in bulk solution experiments, including the pH dependence of fusion for Sindbis and influenza, the inactivation of fusion capacity by preexposure to acidic conditions, and the requirement for cholesterol and sphingomyelin in Sindbis fusion (37,39–43,45).
Our supported lipid bilayer assay differs from the bulk-solution-based fusion assays in several important aspects. In the bilayer experiments, it is clear that the virus is exposed to low pH before encountering the target membrane. In bulk fusion experiments, this distinction is difficult to verify. The successful crystallization of low pH conformations of trimers of type 2 fusion proteins, as are present in Sindbis, required the presence of liposomes and lipidlike detergents(64,65). This biochemical result suggests that the virus might require exposure to lipids before the acidification. Our single-particle approach allows the sequence of acidification, binding, and hemifusion/fusion to be observed, and demonstrates that acidification of virus particles before lipid exposure maintains hemifusion/fusion capacity.
Criticisms of spontaneous dye transfer between virus and target membranes independent of membrane fusion (66) are avoided in our assay. Such spontaneous transfer is easily rejected in our data analysis, as it is much slower than the dequenching due to fusion (67) and also involves a much smaller percentage of dye than our complete particle fusion signals (25,26).
In bulk experiments, fusion typically occurs between virus particles and highly curved, small liposomes. Supported lipid bilayers have zero curvature and more closely approximate the geometry of physiological situations where viruses fuse to larger structures. Different energetic concerns arise related to supported lipid bilayers. Fusion of a virus particle to a supported lipid bilayer requires compression of the bilayer to accommodate the extra lipids. Also, interactions with the supporting surface will alter the energy balance between the virus particle and the target membrane. It is not clear how relevant any of these considerations are to the in vivo situation of viral entry in living cells, but the rapid, efficient hemifusion/fusion observed in our experiments suggests that these effects are not substantial barriers.
Our assay only diagnoses hemifusion between the supported lipid bilayer and the virus particle. R18 rapidly flips between the inner and outer leaflet of a labeled bilayer membrane, and therefore the complete lipid mixing between individual virus particles and the supported bilayer that we observe is expected for either arrested hemifusion or full fusion. The geometry of the supported bilayer clearly prevents the late stages of fusion from progressing naturally, as there is not room to accommodate the viral capsid between the bilayer and the solid support. It is possible that this restriction will prevent formation of the final fusion pore and limit this assay to studies of the initial lipid mixing aspects of protein-mediated membrane fusion. Further experiments will attempt to confirm full membrane continuity using an aqueous content dye or a lipid dye that asymmetrically differentiates between the inner and outer leaflets. Adaptation of our single-virus-particle assay to fusion with immobilized liposomes (our unpublished results and Yoon et al. (68)) will also address the issue of hemifusion versus full fusion in this assay.
Imaging individual events allows us to measure kinetic features of the rapid transitions during the hemifusion/fusion process. By recording time trajectories for each particle we can synchronize fusion dynamics for many particles after the data have been recorded. In this way, we directly confirm that lipid mixing occurs during individual Sindbis virus fusion events faster than 30 ms and that hemifusion/fusion rapidly follows membrane binding. Influenza hemifuses/fuses with similarly fast kinetics. Lipid mixing is complete, with no dye remaining at the fusion site after the dispersal by diffusion into the bilayer after a few seconds. Niles and Cohen (33) imaged fusion of individual influenza particles with planar lipid bilayers with results that are consistent with our measurements. They observed the initial sudden dequenching to be complete on the order of 0.1 s and the dye to radially disperse in a few seconds. Niles and Cohen also observed that the overall rate of fusion declined to zero over 3–5 min after injection into the low-pH solution around the bilayer. Our higher resolution allowed us to further constrain the upper limit of the dequenching event to 30 ms and to determine the intensity trajectory of fusion events for quantitative comparison to diffusion theory.
Diffusion theory accurately described the radial transport of the dye in the bilayer away from the initial particle attachment site after the sudden dequenching due to a lipid mixing event (Fig. 3 E). Careful examination of the data for the few tenths of a second between the sudden dequenching and the eventual diffusive flow revealed subdiffusive lipid spreading (Fig. 3 B). Other published work examining lipid mixing during membrane fusion of HA cells to red blood cells (RBC) or fusion of influenza virus to RBC (32,60,63) have also found subdiffusive lipid dye mixing. Those experiments using RBC as the target membrane saw dequenching and lipid mixing that continued for minutes. Studies of the fusion of influenza virus with protein-free bilayers found less restriction of lipid transfer than the RBC experiments, with the lipid dye dispersing at a speed consistent with free diffusion in the bilayer in a matter of seconds (33). These results led to the suggestion that the viral proteins could restrict lipid flow at a fusion site (61–63).
Our observation of a brief, subdiffusive pause between initial lipid dequenching and subsequent diffusive lipid spread in the bilayer using influenza hemifusing/fusing with protein free bilayers supports the idea that HA restricts lipid flow at a fusion site. That we observed roughly half of hemifusion/fusion events for both Sindbis virus and influenza virus with similar subdiffusive, subsecond pauses suggests that the restriction of lipid mixing by fusion proteins occurs for both type1 fusion proteins and type 2 fusion proteins, and thus supports the idea that the mechanisms of action for type 1 and type 2 fusion proteins are similar (64,65).
As a result of many different types of experimentation, a unified model of the sequence of events underlying influenza-HA-mediated membrane fusion is emerging (13,14,69,70). At neutral pH, HA on the influenza particle bind their receptor, sialic acid, which is present on glangliosides and proteins in a targeted cell membrane. After acidification of the medium around this receptor-HA complex, HA molecules undergo conformational rearrangement into a primed, or activated, state. This activated state is metastable, and after tens of seconds to a few minutes, in the absence of membrane fusion, HA decays to a dead-end state incapable of catalyzing fusion. It is expected that activated HA trimers must aggregate until a threshold density (estimated to be ∼3–8 molecules per fusion site (71,72)) is achieved that is capable of catalyzing fusion through further conformational changes. In the event that the rapid fusion transition is triggered, a number of transient intermediate structures form, including small pores capable of conducting ionic currents, hemifused stalks permitting lipid mixing between the membranes, and full fusion pore formation capable of transferring larger molecules between the two fusing compartments (10,73).
Different assays applied to influenza-HA-mediated membrane fusion are sensitive to different aspects of this overall progression. The most common assays detect fluorescence dequenching or ionic current when whole influenza virus (18,33,60,74), mammalian cells engineered to express HA on their surface (HA cells) (32,63,74–76), or purified HA reconstituted into liposomes (77) fuse with target membranes in liposomes, planar lipid bilayers, or, commonly, RBCs. Experimental configurations detect fusion of either in bulk solution or spatially resolved individual viruses or individual cells.
In assays where virus is not prebound to a target membrane, the diffusional search for a virus to encounter a target membrane typically limits fusion rates. Many assays that use neutral-pH prebinding of virus or HA-expressing cells to a target membrane via receptor interactions have found a delay between acidification and fusion, often called the lag time. The lag time is found to be ∼1–5 s by stopped-flow measurements (74) or single fusion imaging of whole influenza virus fusing with RBCs (60) but lengthens to the range of 20 s to several minutes for HA cells fusing with RBCs (18,60,63,74,76). A series of experiments varying the levels of HA expression in transfected cell lines found shorter lag times as the concentration of functional HA in the membrane increased. These results led to the conclusion that at least some part of the lag time is due to diffusion and oligomerization of primed fusion proteins to a sufficient density to catalyze the fusion transition. The lag time depends upon the composition of the target membrane (52,74), the temperature (18,63), and pH (74), suggesting that the lag time may be a convolution of several different kinetic processes.
In our assay, virus is not prebound to the target bilayer. For influenza, the supported bilayer assay is easily extendable to allow prebinding by incorporating its receptor molecule, sialic acid on glycolipids, into the bilayer (18,33,34,36,75). In contrast, Sindbis cannot be prebound because the relevant receptor molecule is unconfirmed. (In the event that the receptor molecule is identified, it could likely be incorporated into a single virus bilayer fusion assay.) To allow direct comparison to Sindbis virus, we did not include influenza receptor in the bilayers. Thus, unlike the prebound experiments, virus in our assay is exposed to low pH before encountering the membrane.
We measured the time interval between bilayer binding and hemifusion/fusion for many individual virus particles. In Fig. 3 B, we denote this interval as the residence time. Our residence time differs from the previously described delay time for prebound experiments, because in our assay we expect that low-pH priming reactions occur while the virus is in solution. Under most conditions, for both viruses, we find that the residence time is around a quarter-second. Measurement of the residence time for Sindbis virus interacting with liposomes has previously been attempted using coflotation during density-gradient centrifugation (21). Our results qualitatively agree with these earlier experiments that found that not all virus binds to liposome, and additionally not all of the bound virus will fuse with the bilayer. However, our assay measured a much shorter delay between the binding and lipid mixing than was determined with the bulk assay. The inherent delay and insensitivity to weak binding of the centrifugation measurements may account for these differences.
The delay time observed in prebound experiments is longer than our residence time. The prebound virus experiments measure the additional delays of protonation and priming after acidification that in our assay occur before particles encounter the targeted membrane. In all cases, the dynamic transition from a primed and membrane-engaged fusion complex to lipid mixing is rapid and remains temporally unresolved by any experiment. We emphasize that our assay primes virus particles before they are bound to the membrane, the opposite of the situation in assays where virus is prebound to the target membrane at neutral pH by receptor interactions.
The residence time for Sindbis virus increased in the presence of cholesterol at the lowest pH values tested. The role of cholesterol and sphingomyelin in virus entry is complex (41). The combination of cholesterol and sphingomyelin in sufficient concentrations in membranes is known to cluster into microdomains, or lipid rafts (78,79), that some viruses including Ebola and human immunodeficiency virus (HIV) require in the target membrane for infection (80,81). Membrane fusion by influenza does not require cholesterol in the target membrane (20), although cholesterol in the viral membrane has recently been implicated to be important for HA trimerization (82). We did not observe any cholesterol dependent change in the residence time of influenza comparable to what we observed with Sindbis.
Fusion by Sindbis and other alphaviruses strictly requires cholesterol and sphingomyelin in the target membrane (21,83–85), but lipid raft formation is not relevant to this property (43). Rather, the 3β-OH group on cholesterol has been shown to be essential for low-pH binding to membranes (85), possibly by interacting with the alphavirus fusion protein E1 to stabilize an uncharacterized intermediate conformation that is essential for membrane binding (42). Any sterol with the 3β-OH group substitutes for cholesterol, and furthermore, a point mutation in E1 can eliminate this requirement, presumably by stabilizing the necessary intermediates without the sterol involvement (86,87). Although the 3β-OH group plays a specific role in binding, a fusogenic role for cholesterol cannot be ruled out (44,88).
Studies around pH 5.5 have found that small amounts (a few percent) of sphingomyelin are required in the target membrane for fusion, but not for binding of alphaviruses (89). It is also postulated that sphingomyelin interacts with E1 to facilitate conformational changes leading to membrane fusion (90). We observe that Sindbis virus fusion increases in the absence of sphingomyelin for pH well below the optimal acidic levels (Table 1). We have found that pH below the optimal partially overcomes the strict requirement for sphingomyelin to mediate fusion, but under these conditions the delay between binding and fusion increases. The specific mechanisms responsible for the delicate interplay between the fusion proteins, the lipids, and the buffer remain unknown, but the pH dependence supports the model that certain lipid species have specific interactions with the fusion proteins.
In this work, we have compared the fusion behavior of influenza and Sindbis viruses, prototypical examples of the type 1 and type 2 viral fusion protein families. In their neutral pH conformations, type 1 and type 2 proteins show little organizational similarity, but recent structural studies at low pH have led to the suggestion that these two families of proteins achieve membrane fusion by a unified mechanism (12). In our experiments, we find that the rapid lipid mixing kinetics, the residence time and a subdiffusive lipid mixing pause during hemifusion/fusion of Sindbis and influenza are similar for most conditions, supporting a common mechanism for the action of type 1 and type 2 fusion proteins. We have also identified a region of parameter space with the lowest pH and cholesterol where the delay between binding and fusion is lengthened for Sindbis but not for influenza.
To conclude, the single-particle approach is well suited for studies of membrane fusion because it overcomes difficulties associated with high-precision synchronization of independent fusion events. Continued improvements of single-particle membrane fusion assays will allow experimental access to the dynamics of transient, intermediate states that occur during any individual event. Our results on the fusion of influenza and Sindbis virus with supported bilayers emphasize the importance of quantitative, high-resolution data, as from single-particle experiments, in helping to unravel the molecular mechanisms of membrane fusion.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Dennis Brown, Raquel Hernandez, and Gongbo Wang for assistance in producing Sindbis virus and for useful discussions.
Keith Weninger is partially supported by a Ralph E. Powe Award from Oak Ridge Associated Universities, and a Career Award at the Scientific Interface from the Burroughs Wellcome fund.
Editor: Lukas K. Tamm.
References
- 1.Chen, E. H., and E. N. Olson. 2005. Unveiling the mechanisms of cell-cell fusion. Science. 308:369–373. [DOI] [PubMed] [Google Scholar]
- 2.Chernomordik, L. V., and M. M. Kozlov. 2005. Membrane hemifusion: crossing a chasm in two leaps. Cell. 123:375–382. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, F. S., and G. B. Melikyan. 2004. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 199:1–14. [DOI] [PubMed] [Google Scholar]
- 4.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell. 112:519–533. [DOI] [PubMed] [Google Scholar]
- 5.Kozlovsky, Y., and M. M. Kozlov. 2002. Stalk model of membrane fusion: solution of energy crisis. Biophys. J. 82:882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lentz, B. R., and J. K. Lee. 1999. Poly(ethylene glycol) (PEG)-mediated fusion between pure lipid bilayers: a mechanism in common with viral fusion and secretory vesicle release? Mol. Membr. Biol. 16:279–296. [DOI] [PubMed] [Google Scholar]
- 7.Markin, V. S., and J. P. Albanesi. 2002. Membrane fusion: stalk model revisited. Biophys. J. 82:693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller, M., K. Katsov, and M. Schick. 2003. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys. J. 85:1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel, D. P. 1993. Energetics of intermediates in membrane fusion: comparison of stalk and inverted micellar intermediate mechanisms. Biophys. J. 65:2124–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sollner, T. H. 2004. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16:429–435. [DOI] [PubMed] [Google Scholar]
- 11.Yang, L., and H. W. Huang. 2002. Observation of a membrane fusion intermediate structure. Science. 297:1877–1879. [DOI] [PubMed] [Google Scholar]
- 12.Schibli, D. J., and W. Weissenhorn. 2004. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 21:361–371. [DOI] [PubMed] [Google Scholar]
- 13.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kielian, M., and F. A. Rey. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sieczkarski, S. B., and G. R. Whittaker. 2005. Viral entry. Curr. Top. Microbiol. Immunol. 285:1–23. [DOI] [PubMed] [Google Scholar]
- 16.Stegmann, T., D. Hoekstra, G. Scherphof, and J. Wilschut. 1986. Fusion activity of influenza virus. A comparison between biological and artificial target membrane vesicles. J. Biol. Chem. 261:10966–10969. [PubMed] [Google Scholar]
- 17.Stegmann, T., I. Bartoldus, and J. Zumbrunn. 1995. Influenza hemagglutinin-mediated membrane fusion: influence of receptor binding on the lag phase preceding fusion. Biochemistry. 34:1825–1832. [DOI] [PubMed] [Google Scholar]
- 18.Stegmann, T., J. M. White, and A. Helenius. 1990. Intermediates in influenza induced membrane fusion. EMBO J. 9:4231–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA. 100:9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White, J., J. Kartenbeck, and A. Helenius. 1982. Membrane fusion activity of influenza virus. EMBO J. 1:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smit, J. M., R. Bittman, and J. Wilschut. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J. Virol. 73:8476–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haywood, A. M., and B. P. Boyer. 1982. Sendai virus membrane fusion: time course and effect of temperature, pH, calcium, and receptor concentration. Biochemistry. 21:6041–6046. [DOI] [PubMed] [Google Scholar]
- 23.White, J., and A. Helenius. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc. Natl. Acad. Sci. USA. 77:3273–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, T., K. Kawasaki, and S. Ohnishi. 1981. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA. 78:4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit, J. M., G. Li, P. Schoen, J. Corver, R. Bittman, K. C. Lin, and J. Wilschut. 2002. Fusion of alphaviruses with liposomes is a non-leaky process. FEBS Lett. 521:62–66. [DOI] [PubMed] [Google Scholar]
- 26.Hoekstra, D., T. de Boer, K. Klappe, and J. Wilschut. 1984. Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry. 23:5675–5681. [DOI] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., B. N. Deriy, D. C. Ok, and F. S. Cohen. 1996. Voltage-dependent translocation of R18 and DiI across lipid bilayers leads to fluorescence changes. Biophys. J. 71:2680–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenhouts, J. M., and B. De Kruijff. 1995. Membrane potential-driven translocation of a lipid-conjugated rhodamine. Biochim. Biophys. Acta. 1237:121–126. [DOI] [PubMed] [Google Scholar]
- 29.Liu, T., W. C. Tucker, A. Bhalla, E. R. Chapman, and J. C. Weisshaar. 2005. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys. J. 89:2458–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fix, M., T. J. Melia, J. K. Jaiswal, J. Z. Rappoport, D. You, T. H. Sollner, J. E. Rothman, and S. M. Simon. 2004. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. USA. 101:7311–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen, M. E., K. Weninger, A. T. Brunger, and S. Chu. 2004. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J. 87:3569–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niles, W. D., and F. S. Cohen. 1991. Fusion of influenza virions with a planar lipid membrane detected by video fluorescence microscopy. J. Gen. Physiol. 97:1101–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niles, W. D., and F. S. Cohen. 1991. The role of N-acetylneuraminic (sialic) acid in the pH dependence of influenza virion fusion with planar phospholipid membranes. J. Gen. Physiol. 97:1121–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinterdorfer, P., G. Baber, and L. K. Tamm. 1994. Reconstitution of membrane fusion sites. A total internal reflection fluorescence microscopy study of influenza hemagglutinin-mediated membrane fusion. J. Biol. Chem. 269:20360–20368. [PubMed] [Google Scholar]
- 37.Smit, J. M., B. L. Waarts, R. Bittman, and J. Wilschut. 2003. Liposomes as target membranes in the study of virus receptor interaction and membrane fusion. Methods Enzymol. 372:374–392. [DOI] [PubMed] [Google Scholar]
- 38.Edwards, J., E. Mann, and D. T. Brown. 1983. Conformational changes in Sindbis virus envelope proteins accompanying exposure to low pH. J. Virol. 45:1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeagle, P. L. 1985. Cholesterol and the cell membrane. Biochim. Biophys. Acta. 822:267–287. [DOI] [PubMed] [Google Scholar]
- 40.Demel, R. A., and B. De Kruyff. 1976. The function of sterols in membranes. Biochim. Biophys. Acta. 457:109–132. [DOI] [PubMed] [Google Scholar]
- 41.Rawat, S. S., M. Viard, S. A. Gallo, A. Rein, R. Blumenthal, and A. Puri. 2003. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review). Mol. Membr. Biol. 20:243–254. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waarts, B. L., R. Bittman, and J. Wilschut. 2002. Sphingolipid and cholesterol dependence of alphavirus membrane fusion. Lack of correlation with lipid raft formation in target liposomes. J. Biol. Chem. 277:38141–38147. [DOI] [PubMed] [Google Scholar]
- 44.Tenchov, B. G., R. C. Macdonald, and D. P. Siegel. 2006. Cubic phases in phosphatidylcholine-cholesterol mixtures: cholesterol as membrane “fusogen”. Biophys. J. 91:2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Duyl, B. Y., D. Ganchev, V. Chupin, B. de Kruijff, and J. A. Killian. 2003. Sphingomyelin is much more effective than saturated phosphatidylcholine in excluding unsaturated phosphatidylcholine from domains formed with cholesterol. FEBS Lett. 547:101–106. [DOI] [PubMed] [Google Scholar]
- 46.Wassall, S. R., M. R. Brzustowicz, S. R. Shaikh, V. Cherezov, M. Caffrey, and W. Stillwell. 2004. Order from disorder, corralling cholesterol with chaotic lipids. The role of polyunsaturated lipids in membrane raft formation. Chem. Phys. Lipids. 132:79–88. [DOI] [PubMed] [Google Scholar]
- 47.Shaikh, S. R., A. C. Dumaual, A. Castillo, D. LoCascio, R. A. Siddiqui, W. Stillwell, and S. R. Wassall. 2004. Oleic and docosahexaenoic acid differentially phase separate from lipid raft molecules: a comparative NMR, DSC, AFM, and detergent extraction study. Biophys. J. 87:1752–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crane, J. M., and L. K. Tamm. 2004. Role of cholesterol in the formation and nature of lipid rafts in planar and spherical model membranes. Biophys. J. 86:2965–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veatch, S. L., I. V. Polozov, K. Gawrisch, and S. L. Keller. 2004. Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 86:2910–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paredes, A. M., D. Ferreira, M. Horton, A. Saad, H. Tsuruta, R. Johnston, W. Klimstra, K. Ryman, R. Hernandez, W. Chiu, and D. T. Brown. 2004. Conformational changes in Sindbis virions resulting from exposure to low pH and interactions with cells suggest that cell penetration may occur at the cell surface in the absence of membrane fusion. Virology. 324:373–386. [DOI] [PubMed] [Google Scholar]
- 51.Bangham, A. D., M. M. Standish, and J. C. Watkins. 1965. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 13:238–252. [DOI] [PubMed] [Google Scholar]
- 52.Bailey, A., M. Zhukovsky, A. Gliozzi, and L. V. Chernomordik. 2005. Liposome composition effects on lipid mixing between cells expressing influenza virus hemagglutinin and bound liposomes. Arch. Biochem. Biophys. 439:211–221. [DOI] [PubMed] [Google Scholar]
- 53.Stegmann, T., S. Nir, and J. Wilschut. 1989. Membrane fusion activity of influenza virus. Effects of gangliosides and negatively charged phospholipids in target liposomes. Biochemistry. 28:1698–1704. [DOI] [PubMed] [Google Scholar]
- 54.Haywood, A. M., and B. P. Boyer. 1985. Fusion of influenza virus membranes with liposomes at pH 7.5. Proc. Natl. Acad. Sci. USA. 82:4611–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu, S. 2003. Biology and polymer physics at the single-molecule level. Philos. Transact. A Math. Phys. Eng. Sci. 361:689–698. [DOI] [PubMed] [Google Scholar]
- 56.Weiss, S. 2000. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 7:724–729. [DOI] [PubMed] [Google Scholar]
- 57.Landau, L. D., and E. M. Lifshitz. 1987. Fluid Mechanics, 2nd ed. (Course of Theoretical Physics, Vol. 6). Pergamon Press, London.
- 58.Derzko, Z., and K. Jacobson. 1980. Comparative lateral diffusion of fluorescent lipid analogues in phospholipid multibilayers. Biochemistry. 19:6050–6057. [DOI] [PubMed] [Google Scholar]
- 59.Kusumi, A., C. Nakada, K. Ritchie, K. Murase, K. Suzuki, H. Murakoshi, R. S. Kasai, J. Kondo, and T. Fujiwara. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. [DOI] [PubMed] [Google Scholar]
- 60.Lowy, R. J., D. P. Sarkar, Y. Chen, and R. Blumenthal. 1990. Observation of single influenza virus-cell fusion and measurement by fluorescence video microscopy. Proc. Natl. Acad. Sci. USA. 87:1850–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 76:383–391. [DOI] [PubMed] [Google Scholar]
- 62.Zimmerberg, J., R. Blumenthal, D. P. Sarkar, M. Curran, and S. J. Morris. 1994. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J. Cell Biol. 127:1885–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibbons, D. L., M. C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 427:320–325. [DOI] [PubMed] [Google Scholar]
- 65.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature. 427:313–319. [DOI] [PubMed] [Google Scholar]
- 66.Ohki, S., T. D. Flanagan, and D. Hoekstra. 1998. Probe transfer with and without membrane fusion in a fluorescence fusion assay. Biochemistry. 37:7496–7503. [DOI] [PubMed] [Google Scholar]
- 67.Nunes-Correia, I., A. Eulalio, S. Nir, N. Duzgunes, J. Ramalho-Santos, and M. C. Pedroso de Lima. 2002. Fluorescent probes for monitoring virus fusion kinetics: comparative evaluation of reliability. Biochim. Biophys. Acta. 1561:65–75. [DOI] [PubMed] [Google Scholar]
- 68.Yoon, T. Y., B. Okumus, F. Zhang, Y. K. Shin, and T. Ha. 2006. Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl. Acad. Sci. USA. 103:19731–19736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569. [DOI] [PubMed] [Google Scholar]
- 70.Lakadamyali, M., M. J. Rust, and X. Zhuang. 2004. Endocytosis of influenza viruses. Microbes Infect. 6:929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, X., S. Kurteva, X. Ren, S. Lee, and J. Sodroski. 2006. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J. Virol. 80:4388–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bentz, J. 2000. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 78:227–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175–207. [DOI] [PubMed] [Google Scholar]
- 74.Clague, M. J., C. Schoch, and R. Blumenthal. 1991. Delay time for influenza virus hemagglutinin-induced membrane fusion depends on hemagglutinin surface density. J. Virol. 65:2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Razinkov, V. I., G. B. Melikyan, and F. S. Cohen. 1999. Hemifusion between cells expressing hemagglutinin of influenza virus and planar membranes can precede the formation of fusion pores that subsequently fully enlarge. Biophys. J. 77:3144–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mittal, A., E. Leikina, J. Bentz, and L. V. Chernomordik. 2002. Kinetics of influenza hemagglutinin-mediated membrane fusion as a function of technique. Anal. Biochem. 303:145–152. [DOI] [PubMed] [Google Scholar]
- 77.Imai, M., T. Mizuno, and K. Kawasaki. 2006. Membrane fusion by single influenza hemagglutinin trimers. Kinetic evidence from image analysis of hemagglutinin-reconstituted vesicles. J. Biol. Chem. 281:12729–12735. [DOI] [PubMed] [Google Scholar]
- 78.Kenworthy, A. K. 2005. Fleeting glimpses of lipid rafts: how biophysics is being used to track them. J. Investig. Med. 53:312–317. [DOI] [PubMed] [Google Scholar]
- 79.Rajendran, L., and K. Simons. 2005. Lipid rafts and membrane dynamics. J. Cell Sci. 118:1099–1102. [DOI] [PubMed] [Google Scholar]
- 80.Suzuki, T., and Y. Suzuki. 2006. Virus infection and lipid rafts. Biol. Pharm. Bull. 29:1538–1541. [DOI] [PubMed] [Google Scholar]
- 81.Nayak, D. P., and E. K. Hui. 2004. The role of lipid microdomains in virus biology. Subcell. Biochem. 37:443–491. [DOI] [PubMed] [Google Scholar]
- 82.Sun, X., and G. R. Whittaker. 2003. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 77:12543–12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mooney, J. J., J. M. Dalrymple, C. R. Alving, and P. K. Russell. 1975. Interaction of Sindbis virus with liposomal model membranes. J. Virol. 15:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scheule, R. K. 1987. Fusion of Sindbis virus with model membranes containing phosphatidylethanolamine: implications for protein-induced membrane fusion. Biochim. Biophys. Acta. 899:185–195. [DOI] [PubMed] [Google Scholar]
- 85.Kielian, M. C., and A. Helenius. 1984. Role of cholesterol in fusion of Semliki Forest virus with membranes. J. Virol. 52:281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu, Y. E., T. Cassese, and M. Kielian. 1999. The cholesterol requirement for sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 73:4272–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen, Z., and R. P. Rand. 1997. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys. J. 73:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nieva, J. L., R. Bron, J. Corver, and J. Wilschut. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 13:2797–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samsonov, A. V., P. K. Chatterjee, V. I. Razinkov, C. H. Eng, M. Kielian, and F. S. Cohen. 2002. Effects of membrane potential and sphingolipid structures on fusion of Semliki Forest virus. J. Virol. 76:12691–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.