Abstract
CD4+ T cells selected by the type 1 diabetes associated class II MHC I-Ag7 molecules play a critical role in the disease process. Multivalent MHC/peptide tetramers have been used to directly detect antigen-specific T cells. Detection of autoantigen-activated CD4+ T cells with tetramers should be very helpful in the study of the roles of these cells in diabetes. We report here the generation of tetramers of I-Ag7 covalently linked to two glutamic acid decarboxylase (GAD) peptides and the detection of GAD peptide-activated T cells from nonobese diabetic (NOD) mice. The I-Ag7 heterodimers can form stable complexes with a covalently bound GAD peptide and can stimulate antigen specific T cells. Furthermore, I-Ag7/GAD peptide tetramer can detect most if not all of the antigen-specific CD4+ T cells from immunized NOD mice. Antigen-specific T cells detected by the tetramers can up-regulate their CD4 expression on the cell surface after being restimulated with the GAD peptides in vitro. In contrast, the tetramers can detect a percentage of T cells in lymph nodes and spleens and T cells infiltrating islets from nonimmunized mice that is not significantly above the background. Therefore, T cells specific for the GAD peptides are present in NOD mice at a frequency too low to be detected, but immunization of NOD mice can facilitate their detection by tetramers.
Type 1 diabetes is a T cell-mediated autoimmune disease characterized by the destruction of insulin-producing cells in the islets of the pancreas (1–10). In NOD mice, an animal model for human type 1 diabetes, the class II MHC I-Ag7 complex must be present in both alleles for diabetes to develop (11–15). Expression of non-I-Ag7 class II can protect NOD mice from diabetes, which may be caused by the selection of different T cell antigen receptor (TCR) specificities, deletion of diabetogenic T cells, or immune suppression of lymphocytes (16–18). Previous biochemical studies of the I-Ag7 complex have suggested that purified I-Ag7 complex is intrinsically unstable and is a poor peptide binder in vitro (19, 20). These properties have made it difficult to determine how I-Ag7 selects for autoantigen-specific autoreactive T cells in NOD mice.
To study how autoantigen-specific T cells arise and how they can lead to diabetes, it is necessary to identify the autoantigens, to characterize the functional response of autoantigen-specific T cells, and to determine their roles in the disease progress. It has recently been shown that the detection of antigen-specific T cells could be achieved by using multivalent MHC/peptide tetrameric complexes as the staining reagents (21–23). Among the I-Ag7 complex presented autoantigens that may participate in the disease, the GAD protein represents a major type of autoantigen involved in the initial stages of diabetes development in both human and mouse (2, 24–32). In an initial effort to study the roles of different GAD peptides, to identify and characterize I-Ag7-selected GAD peptide-reactive T cells, and to understand the mechanisms of the selection of GAD peptide-reactive T cells, we have generated soluble recombinant I-Ag7 complexes covalently linked to two GAD peptides, p206 and p524. It has been shown previously that these two immunodominant peptides may participate in the selection of autoreactive T cells during the early development of type 1 diabetes (24, 33). The p206 peptide represents a major immunodominant epitope recognized by hybridoma cells from NOD mice immunized with GAD 65 protein (33). The p524 peptide has been identified as a dominant epitope in the early phases of the disease, and transfer of a p524-reactive T cell line can transfer the disease to recipient mice (31). The current results show that I-Ag7 heterodimer can form functional I-Ag7/GAD peptide tetramers, which can detect antigen-specific T cells derived from immunized mice, but not from lymph nodes, spleens, and islets of nonimmunized NOD mice. In addition, the results have shown that most if not all antigen-specific T cells from immunized mice could be detected by the tetramers, and they expressed higher levels of CD4 after being restimulated in vitro.
Materials and Methods
Mice.
The NOD and BALB/c mice were purchased from the Jackson Laboratory and housed in a specific pathogen free environment in the animal facility at the Beckman Research Institute, City of Hope.
Production of T Cell Hybridomas.
T cell hybridomas were derived from NOD mice immunized with 100 μg of synthetic peptides (HPLC purified with at least 90% purity) plus complete Freund's adjuvant (CFA) as previously described (34). For stimulating cells with the recombinant I-Ag7/GAD peptide proteins, the recombinant proteins were coated at the bottom of Immulon 4 96-well flat-bottomed plates (Dynatech Laboratories, Chantilly, VA).
Isolation and Growing of Tetramer-Positive T Cells.
T cells from immunized NOD female mice were cultured with the antigenic peptide and then in IL2 medium before analysis. Tetramer-positive and tetramer-negative CD4+ cells were sorted by using fluorescence-activated cell sorting (FACS). The purity of cells after sorting was greater than 98%.
Production of Soluble I-Ag7 Molecules with Covalently Linked Peptides and I-Ag7 Tetramers.
Soluble recombinant MHC class II molecules were prepared by infecting insect cells (SF9 and Hi5 cells; PharMingen, San Diego) with baculoviruses according to the manufacturer's instructions. DNA constructs were generated by cloning a DNA fragment encoding a GAD peptide to the 5′ end of the I-Ag7 β chain, which lacked the transmembrane and intracellular region, using a dual-promoter vector pAcUW51 as described previously (35). The 3′ end of the I-Ag7 β chain also contained sequences encoding a factor Xa recognition site, a stretch of histidine residues, and a peptide that can be biotinylated by the Escherichia coli enzyme BirA (21, 36). The baculovirus expression vector was a generous gift from J. Kappler (National Jewish Medical and Research Center, Denver; ref. 35). The cDNAs encoding the I-Ag7 α and β chains were a generous gift from H. McDevitt (Stanford University Medical Center; ref. 37). Soluble I-Ag7/peptide complexes were purified with an affinity column containing Ni beads purchased from Qiagen (Valencia, CA) and a Superdex 200 h 10/30 column (Amersham Pharmacia). The sequences of the two GAD65 peptides, p206 and p524, used in making T cell hybridomas and I-Ag7/peptide tetramers were TYEIAPVFVLLEYVT (p206) and SRLSKVAPVIKARMMEYGTT (p524). Procedures for preparing tetramers have been described previously (21–23). Briefly, purified I-Ag7/peptide complexes were biotinylated using a kit containing the enzyme BirA according to the manufacturer's instructions (Avidity, Denver, CO). Biotinylated proteins were incubated with streptavidin-phycoerythrin (SAPE) (PharMingen), and multivalent complexes were purified using a Superdex-200 column.
Flow Cytometry Analysis.
Surface staining of cells with tetramers was done by incubating cells at 37°C for 2–3 h with the tetramers in RPMI medium containing 10% FCS in the presence of blocking reagents, including normal mouse serum and cell culture supernatant containing the anti-Fc receptor antibody 2.4G2 (38). The final concentration of the tetramers used in the staining reactions was 30 μg/ml. Unconjugated H57 antibody was also used at 1 μg/106 cells together with the tetramers to enhance the staining of tetramers to TCRs.
Isolation of Pancreatic Islets and Lymphocytes.
Pancreatic islets were isolated as described previously (39). In brief, the islets were isolated by stationary digestion of pancreatic tissues with collagenase (Boehringer Mannheim) and Ficoll density-gradient purification (Sigma). Lymph nodes were removed from the islet preparations by hand picking of the isolated islets. Islets were then made into single cells by trypsin/EDTA treatment and washed with Hanks' balanced salt solution.
Results
Generation of Recombinant I-Ag7/Peptide Tetramers.
We have generated soluble recombinant I-Ag7 heterodimer covalently linked to one of two GAD peptides, p206 or p524, by using a method described previously (23, 35). Purified recombinant proteins showed a peak for a dimeric form of I-Ag7/peptide complex at around 65 kDa on a gel exclusion column (data not shown). The recombinant complexes were dissociated in the presence of SDS, consistent with previous reports that I-Ag7 heterodimer is unstable in the presence of SDS (19, 20, 40). However, despite this potential instability, the recombinant proteins were able to form stable I-Ag7/p206 and I-Ag7/p524 tetramers (tet-I-Ag7/p206 and tet-I-Ag7/p524). Functional assays showed that these two recombinant complexes could stimulate antigen-specific NOD mouse hybridoma cells (NOD206 and NOD524 cells as described below) (Fig. 1A). These results indicate that these recombinant I-Ag7/GAD peptide heterodimers were stable and functional under the in vitro conditions used.
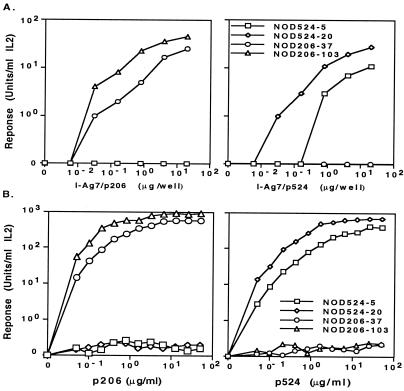
Figure 1.
Analysis of IL2 production by antigen-specific T cell hybridomas. (A) Stimulation of NOD mouse T cell hybridomas with various concentrations of plate-bound I-Ag7/p206 and I-Ag7/p524 proteins. The proteins were diluted with a 5-fold serial dilution starting with 20 μg/ml. The responses of two representative hybridoma cells for each recombinant protein, I-Ag7/p206 (NOD206–37 and NOD206–103 cells) and I-Ag7/p524 (NOD524–5 and NOD524–20 cells), at different concentrations, are shown. The results are an average of two independent experiments. (B) The IL-2 production responses of T cell hybridomas for p206 or p524. The results shown are typical of two independent experiments.
To test the function and antigen specificity of the soluble recombinant I-Ag7/GAD peptide complexes, we prepared T cell hybridomas specific for these peptides. The results showed that the hybridoma cells reacted only to their specific immunizing peptide and not to the other peptide. The hybridoma cells reactive to peptide p206 were named “NOD206,” and those reactive to peptide p524 were named “NOD524.” Therefore, hybridomas NOD206–37 and NOD106–103 responded only to p206, and, conversely, NOD524–5 and NOD524–20 responded only to p524 (Fig. 1B). It is interesting to note that it was relatively easy to raise T cell hybridomas responding to these two self-antigens from NOD mice. This result is consistent with previous studies that a high frequency of autoreactive T cells is present in NOD mice (41). This result indicated that autoreactive T cells reactive to these two self-antigenic GAD peptides were present in NOD mice.
Peptide titration showed that NOD206–103 cells reacted to a smaller amount of p206 than the NOD206–37 cells, and the NOD524–20 cells reacted to a smaller amount of the p524 than the NOD524–5 cells (Fig. 1B). The difference in the antigen response sensitivity was not attributable to a difference in TCR level (data not shown). The NOD206–103 TCR probably has a greater affinity for the I-Ag7/p206 ligand than does the NOD206–37 TCR. Similarly, the TCR expressed on the NOD524–20 cells probably has a greater affinity for the I-Ag7/p524 ligand than does the TCR expressed on the NOD524–5 cells.
We then wanted to know whether we could detect the TCRs expressed on I-Ag7-selected hybridoma cells by using the tetramers. In the following experiments, we used the NOD206–103 and NOD524–20 cells. For comparison, we used NOD206–37 and NOD524–5 cells, which might express relatively lower affinity TCRs than the other two hybridoma cells. The results showed that the tet-I-Ag7/p206 tetramer detected the TCRs expressed on NOD206 but not NOD524 hybridoma cells (Fig. 2A). The tetramer could also detect other p206-specific hybridomas tested (data not shown). Therefore, the tet-I-Ag7/p206 tetramer could stain antigen-specific T cells. When staining NOD524 cells by using the tetramers, we found that tet-I-Ag7/p524 tetramer weakly detected the TCRs expressed on the NOD524–20 cells and barely detected the TCR expressed on the NOD524–5 cells (Fig. 2C). Staining of cells using the tet-I-Ag7/p524 tetramer was also antigen specific, as it did not stain NOD206 hybridoma cells.
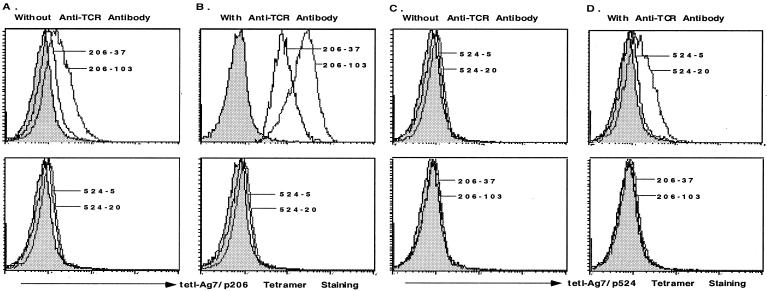
Figure 2.
Staining of T cell hybridomas from NOD mice. Antigen-specific T cell hybridomas derived from NOD mice could be detected by the tet-I-Ag7/GAD peptide tetramers. The staining intensity could be enhanced by the addition of the H57 antibody. (A and B) Hybridoma cells were stained with the tet-I-Ag7/p206 tetramer in the absence (A) or presence (B) of the H57 antibody. (C and D) Hybridoma cells were stained with the tet-I-Ag7/p524 tetramer in the absence (C) or the presence (D) of the H57 antibody. The results are representative of at least three experiments.
In an effort to increase the staining intensity of T cells by tetramers, we added the anti-TCR H57 antibody to the cells (Fig. 2 B and D), as has been shown before (42). The results showed that H57 antibody improved the staining of cells by the tetramers. The addition of the H57 antibody has a better enhancement effect on the staining of NOD206 cells than with NOD524 cells with their antigen-specific tetramers. These results suggest that the binding of the tet-I-Ag7/p206 tetramer to p206-specific TCRs is generally better than the binding of the tet-I-Ag7/p524 tetramer to p524-specific TCRs. For the following experiments we added the H57 antibody to the staining reactions to increase the staining intensity and percentage of cells detected by the tetramers.
Detection of Activated NOD Mouse T Cells with Tetramers.
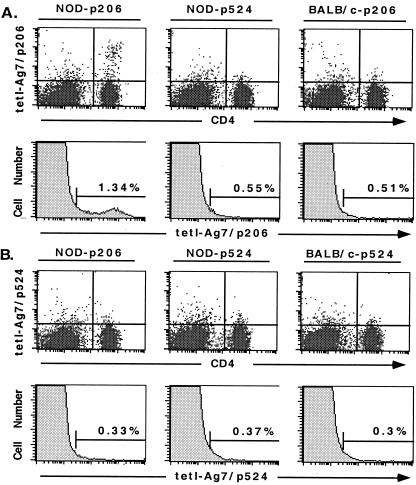
The tetramers were then used to detect GAD peptide-activated NOD mice T cells. Staining of T cells from lymph nodes and spleens of immunized 12-week-old NOD mice showed that the tet-I-Ag7/p206 tetramer detected a population of T cells representing 0.6–1% of CD4+ T cells from p206 immunized mice, in comparison with control BALB/c mice cells (Fig. 3A). CD4− and CD8+ cells did not bind the tetramer (Fig. 3A and data not shown). Similar results were obtained from mice of different ages (data not shown).
Figure 3.
Staining of T cells from immunized NOD mice. (A) The tet-I-Ag7/p206 tetramer stained a small population of CD4+ T cells (0.6–1% as compared with the negative controls) derived from NOD mice immunized with the p206 peptide. (B) The tet-I-Ag7/p524 tetramer could barely stain T cells derived from NOD mice immunized with the p524 peptide. NOD mice were immunized with p206 (NOD-p206) or p524 (NOD-p524). BALB/c mice were immunized with p206 (BALB/c-p206) or p524 (BALB/c-p524). The results are representative of at least three experiments.
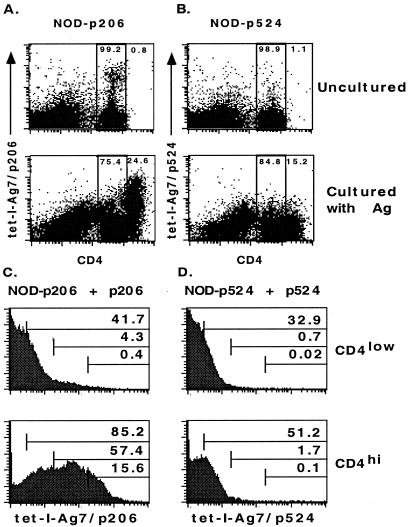
In an attempt to improve the detection of antigen-specific T cells, the T cells from immunized mice were restimulated with the peptide in vitro. After culturing with p206, around 25% of CD4+ cells up-regulated their CD4 expression and became CD4hi cells (Fig. 4A), consistent with the idea that CD4 expression can be up-regulated on activated T cells (43). Moreover, when compared with CD4low cells, more than half of the CD4hi cells (57%) were detected by the tet-I-Ag7/p206 tetramer with intermediate to high staining intensity (Fig. 4C). Surprisingly, a significant portion of CD4hi T cells were still weakly detected (28%) or were not detected (15%) by the tetramer (Fig. 4C). It is likely that these antigen-activated CD4hi T cells may express TCRs whose affinities for the ligands were too low to be detected by the tetramer or that they were not specific for the antigen.
Figure 4.
Staining of T cells restimulated by antigens in vitro. T cells were derived from NOD mice immunized with (A) p206 (NOD-p206) and (B) p524 (NOD-p524). These T cells were either stained with the tetramers directly without further treatment (Upper) or incubated with the antigen for 4 days before being stained with the tetramers (Lower). For cells cultured with antigens, as shown in A, around 24.6% of CD4+ T cells up-regulated their CD4 expression after they were restimulated with p206. Among these CD4hi cells, more than 50% of them (see C) were detected by the tetramer with intermediate to high staining intensities. As shown in B, around 15% of CD4+ T cells up-regulated their CD4 expression after they were restimulated with p524. The average staining intensity of CD4hi cells, although it was still weak, was 2- to 3-fold higher than that of CD4low cells (see D). CD4hi and CD4low T cells were further gated and analyzed for the staining intensity by the tetramer. In each histogram, the cells were arbitrarily divided into four different populations based on their relative tetramer staining intensities (negative, low, intermediate, and high). The numbers shown in each histogram represent the percentage of cells above the indicated minimum staining intensity for each subpopulation. The average staining intensity and percentage of CD4hi cells in C is significantly higher than those of CD4low cells. In D, the average tetramer staining intensity of CD4hi cells (the mean channel value was 5–6) was 2- to 3-fold higher than that of CD4low cells (the mean channel value was 2–3). The total percentage of CD4hi cells detected with weak to high intensity (≈50%) was also higher than that of CD4low cells (≈30%). In addition, the percentage of CD4hi cells detected with intermediate to high intensities was 1% higher than that of CD4low cells.
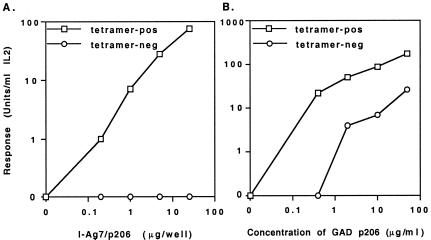
We then performed experiments to determine whether tetramer-positive T cells were indeed specific for the p206 peptide, and whether all of the p206-reactive T cells were detected by the tetramer. Tetramer-positive and tetramer-negative CD4+ T cells from immunized mice were isolated by sorting using FACS. The reactivity of these cells to synthetic p206 peptide and to plate-bound recombinant I-Ag7/p206 protein at different concentrations was then determined. The results in Fig. 5A showed that only tetramer-positive but not tetramer-negative CD4+ T cells were reactive with the recombinant I-Ag7/p206 protein. This selective reactivity suggested that the tetramer-positive phenotype of antigen-specific T cells correlated with the function of these cells and that the tetramers could detect most if not all of the antigen-specific T cells from immunized mice. On the other hand, both tetramer-positive and tetramer-negative cells responded to the synthetic peptide, and the response of tetramer-negative T cells was weaker than that of tetramer-positive cells (Fig. 5B). Why tetramer-negative cells responded differently to synthetic peptide and recombinant protein was unclear. The difference in response may be because of contaminants present in the peptide preparation, and T cells reactive to such contaminants were not specific for the p206 peptide because they did not respond to the recombinant I-Ag7/p206 protein. Alternatively, there was no contaminant in the peptide preparation, and the response was indeed peptide specific because the tetramers simply failed to detect/stimulate all T cells bearing TCRs of various affinities for the selecting MHC/peptide combination.
Figure 5.
Stimulation of tet-I-Ag7/p206 tetramer-positive and tetramer-negative CD4+ T cells with various concentrations of recombinant I-Ag7/p206 protein (A) or synthetic p206 peptide (B). Cells isolated from immunized mice were restimulated in vitro with the peptide and cultured in the presence of IL-2 before analysis. Sorted tetramer-positive and tetramer-negative CD4+ T cells were incubated with plate-bound recombinant protein or with the synthetic peptide that was diluted with a 5-fold serial dilution starting with 25 μg/well and 50 μg/ml, respectively. Tetramer-pos, tetramer positive cells; tetramer-neg, tetramer-negative cells.
Our results also showed that the tet-I-Ag7/p524 tetramer could barely detect T cells from p524 immunized NOD mice. The very small percentage (0.03–0.05%) of CD4+ cells detected may not be regarded as significantly above the background (Fig. 3B). Incubation of T cells with p524 in vitro resulted in a population of T cells that became T cell blasts (data not shown), and a significant portion of CD4+ T cells (15%) also increased their CD4 expression and became CD4hi T cells (Fig. 4B). These CD4hi T cells were still weakly stained with the tetramer (Fig. 4D). However, the average staining intensity of total CD4hi T cells (with a mean channel value of 5–6) was 2–3 times higher than that of CD4low T cells (with a mean channel value of 2–3). In addition, more CD4hi T cells (1.7%) than CD4low T cells (0.7%) showed intermediate to high staining intensities (Fig. 4D). These results suggest that further activation of p524-reactive CD4+ T cells in vitro can also up-regulate their CD4 expression, and the tetramer can bind weakly to TCRs of almost all p524-reactive T cells.
To find out whether p206- and p524-reactive T cells were detectable in untreated NOD mice, we stained lymph node and splenic T cells from 4-, 6-, 8-, and 12-week-old NOD mice and 18-week-old diabetic NOD mice. The T cells were barely detectable with the tetramers, and the very small percentage (0.03–0.05%) of CD4+ cells detected weakly by the tetramers may not be regarded as significantly above the background, i.e., the staining of T cells from BALB/c mice (data not shown). This lack of detection was consistent with a previously published study that showed that a class I MHC Kd/insulin peptide tetramer also did not detect a significant population of antigen-specific T cells in the lymph nodes or spleens (44). To test whether CD4+ T cells specific for the two GAD peptides were present in the islets at a higher frequency, we purified islets from 4-, 6-, 8-, and 12-week-old NOD mice. The results showed that two to three times more CD4+ T cells than CD8+ T cells were present in the islets of NOD mice, consistent with the results of previous reports that more CD4+ T cells infiltrated the islets (45, 46). However, the tetramer staining results showed that the tetramers detected T cells infiltrating the islets of NOD mice with a percentage that is not significantly above the background (data not shown).
These results suggest that a spontaneous population of p206- or p524-reactive T cells were present in NOD mice at a very low frequency, or their TCR affinity for the tetramer was too low to be detected. Immunization of NOD mice, similar to immunization of normal mice (47, 48), may result in the selection/expansion of antigen-reactive T cells and may facilitate their detection by the tetramers more easily.
Discussion
We report here the generation of I-Ag7 tetramers specific for two different GAD peptides and the use of these tetramers to stain T cells derived from NOD mice. The two soluble recombinant I-Ag7/GAD peptide complexes were stable and functional in the in vitro conditions used, except when SDS was present, consistent with the results of other reports (40, 49, 50). The results suggest that I-Ag7/GAD peptide tetramers were able to detect most if not all of the antigen-specific T cells from immunized NOD mice because tetramer-positive but not tetramer-negative CD4+ T cells responded to the recombinant I-Ag7/p206 complex. Interestingly, our results also showed that synthetic peptides could stimulate tetramer-negative T cells. It is not known how the differences in the response may occur. It is likely that the tetramer-negative T cells were not specific for p206 but for some contaminants in the peptide preparation. If this conclusion is true, then removal of tetramer-negative T cells may be necessary before it is possible to study tetramer-positive T cell functions with synthetic peptides. It is also likely that there is no contaminant in the peptide preparation, that the response was indeed peptide specific, and that the tetramers did not detect/stimulate T cells bearing TCRs of various affinities for the selecting MHC/peptide complex. Although the two tetramers could detect T cells from immunized NOD mice with differential staining intensities, neither tetramer detected significant numbers of antigen-specific T cells from nonimmunized mice. The reason for this difference in detection is unclear. Several possibilities may explain the results. Is tetramer analysis inherently unsuited for the direct visualization of endogenously primed GAD-reactive CD4+ cells in NOD mice using FACS? Is it because of the result of the low-affinity T cell repertoire in these mice, or because of the low frequency of such GAD peptide-reactive T cells? Alternatively, are the endogenously primed T cells not detected because of the different types of GAD peptide configurations displayed by synthetic peptides vs. naturally processed peptides (51)?
Conceivably, autoreactive T cells have previously been activated by autoantigens, and the number or frequency of these activated T cells in the total T cell population may increase in NOD mice. Despite these possibilities, our results suggest that T cells specific for these two GAD peptides are present in lymph nodes, spleens, and pancreatic islets of NOD mice at a very low frequency, probably less than one per 1000 CD4+ T cells. This frequency is too low to be detected by the I-Ag7 tetramers, which can reliably detect antigen-specific T cells of more than 0.1% in a T cell population from NOD mice. GAD peptide-specific T cells can be detected by the tetramers once they are expanded in the mice after immunization with the peptide. Another possibility is that CD4+ T cells specific for the two GAD peptides bear low-affinity TCRs for their ligands and cannot be detected by the tetramers. In addition, it is possible that the covalently bound GAD peptide might not be flexible enough in the I-Ag7 groove to bind various TCRs with a broad range of affinities. It is also possible that the T cells from immunized NOD mice may be functionally different from the spontaneous population of GAD peptide-reactive T cells arising during the development of type 1 diabetes (26, 52, 53). It has been shown before that there may be two different types of hybridoma cells derived from mice immunized with synthetic peptides (51). Most hybridomas may recognize the peptides, but not the peptide/MHC complexes generated via natural processing of the protein antigen (51). The difference in antigenicity between synthetic peptides and naturally processed peptides may also explain why T cells from peptide-immunized NOD mice stained with tetramers, but the endogenously primed peptide reactive T cells did not. In addition, unlike MHC class I tetramers, which included synthetic peptides, the covalently linked peptides presented by the recombinant I-Ag7 heterodimers were generated as part of the I-Ag7 β chain inside the cells. Therefore, these covalently linked peptides may not behave in exactly the same way as synthetic peptides, as shown in our current studies.
Immunization may expand antigen-specific T cells or select for cells expressing relatively higher affinity TCRs, allowing them to be detected more easily by tetramers (47, 48). Restimulation of antigen-specific T cells with the antigenic peptides in vitro can further expand the antigen-specific T cells, and these activated cells can up-regulate their CD4 expression on the cell surface. Almost all of the cells detected by the tet-I-Ag7/p206 tetramer have up-regulated their CD4 expression and become CD4hi T cells. In addition, the percentage of CD4+ cells detected by the tetramer also increases. Although many CD4hi cells are brightly stained by the tetramer, a large portion of the CD4hi cells are stained very weakly or are not detected by the tetramer. These activated tetramer-negative CD4hi T cells may not be specific for p206 because they do not react with recombinant I-Ag7/p206. It is possible that these cells are stimulated by contaminants in synthetic peptides and were not truly GAD peptide-specific cells. Therefore, the use of tetramer can help to remove such nonspecific T cells. Alternatively, they may bear TCRs of lower affinities than can be detected by the tetramers. Further expansion of p524-reactive T cells can also be achieved by restimulation of T cells with p524 in vitro. Analysis of these restimulated p524-reactive T cells shows that, similar to p206-reactive T cells, many T cells have increased their cell size and have become blast cells (data not shown). Furthermore, a significant portion of cultured T cells also increased their cell surface CD4 expression, suggesting that they have reacted to p524. Although stained weakly, these CD4hi cells are detected by the tetramer better than the CD4low cells, and the staining intensity of these CD4hi cells by the tetramer increases by 2- to 3-fold.
Our results showed that the two GAD peptide-specific tetramers stain antigen-specific T cells with differential staining intensities. There are several possible explanations for the results. First, the relatively higher staining intensity of tet-I-Ag7/p206 tetramer for p206 reactive cells results from the tet-I-Ag7/p206 tetramer being more stable than the tet-I-Ag7/p524 tetramer. This greater stability could be attributable to better binding affinity of the p206 for I-Ag7 than that of the p524 for I-Ag7. However, the protein yield of the I-Ag7/p524 complex (≈1–2 mg/liter) is better than that of the I-Ag7/p206 complex (≈0.3–0.5 mg/liter), suggesting that this is unlikely to be the reason. A second possibility is that the proportion of tetramers whose I-Ag7 bound with p524 in the best position for interacting with the TCRs is low, and some of them are essentially trimers or dimers rather than tetramers. This low proportion of binding in the best position may be attributable to the fact that p524 (20 aa) is longer than p206 (15 aa). If this is the case, then incubating cells with a higher concentration of tetramers can significantly improve the staining intensities. However, a further increase of the concentration or incubation time from the ones used in this study does not improve the staining significantly (data not shown). These results suggest that if inactive molecules are present, they are present at similar levels in the two tetramer preparations. The third possibility is that the affinity of I-Ag7-selected TCRs specific for p206 is generally greater than the affinity of TCRs specific for p524. It has been shown that the level of staining intensity of class II MHC tetramers for TCRs correlates with the affinity of the TCRs for the MHC/peptide ligands (23). Our results from staining T cell hybridomas and T cells from immunized mice with these two tetramers are consistent with such a correlation.
Although the expression of GAD may be critical to the development of diabetes in NOD mice (24, 25, 32), it is not clear which GAD peptide(s) can select for diabetic T cells and cause the disease. It is possible that T cells specific for different autoantigenic peptides play differential roles in the development of type 1 diabetes. Therefore, T cells specific for p206, p524, and other GAD peptides may contribute differently to the disease. Use of the tetramers made in this study and the generation of more tetramers specific for peptides derived from GAD and other potential autoantigens will allow us to determine in more detail the roles of different autoantigenic peptides and T cells specific for these various autoantigenic peptides in type 1 diabetes.
Acknowledgments
We thank Dr. J. Shively for help with protein purification and Drs. J. Kappler and H. McDevitt for generously providing us with DNA reagents. We are indebted to the K-M laboratory for invaluable discussions. This work was supported in part by National Institutes of Health Grants AI44429 and AI44143 (to C.-P.L.) and Cancer Center Core Grant CA33752, and by the Juvenile Diabetes Foundation International.
Abbreviations
- NOD
nonobese diabetic
- GAD
glutamic acid decarboxylase
- SAPE
streptavidin-phycoerythrin
- TCR
T cell antigen receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250390997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250390997
References
- 1.Castano L, Eisenbarth G S. Annu Rev Immunol. 1990;8:647–680. doi: 10.1146/annurev.iy.08.040190.003243. [DOI] [PubMed] [Google Scholar]
- 2.Nepom G T. Curr Opin Immunol. 1995;7:825–830. doi: 10.1016/0952-7915(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 3.Wicker L S, Todd J A, Peterson L B. Annu Rev Immunol. 1995;13:179–200. doi: 10.1146/annurev.iy.13.040195.001143. [DOI] [PubMed] [Google Scholar]
- 4.Tisch R, McDevitt H. Cell. 1996;85:291–297. doi: 10.1016/s0092-8674(00)81106-x. [DOI] [PubMed] [Google Scholar]
- 5.Gill R G, Coulombe M, Lafferty K J. Immunol Rev. 1996;149:75–96. doi: 10.1111/j.1600-065x.1996.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 6.Haskins K, Wegmann D. Diabetes. 1996;45:1299–1305. doi: 10.2337/diab.45.10.1299. [DOI] [PubMed] [Google Scholar]
- 7.Bach J F, Mathis D. Res Immunol. 1997;148:285–366. doi: 10.1016/s0923-2494(97)87235-5. [DOI] [PubMed] [Google Scholar]
- 8.Wong F S, Janeway C A J. Res Immunol. 1997;148:327–332. doi: 10.1016/s0923-2494(97)87242-2. [DOI] [PubMed] [Google Scholar]
- 9.Tian J, Olcott A P, Hanssen L R, Zekzer D, Middleton B, Kaufman D L. Immunol Rev. 1998;164:119–127. doi: 10.1111/j.1600-065x.1998.tb01214.x. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson M A, Leiter E H. Nat Med. 1999;5:601–604. doi: 10.1038/9442. [DOI] [PubMed] [Google Scholar]
- 11.Reich E P, Sherwin R S, Kanagawa O, Janeway C A., Jr Nature (London) 1989;341:326–328. doi: 10.1038/341326a0. [DOI] [PubMed] [Google Scholar]
- 12.Lund T, O'Reilly L, Hutchings P, Kanagawa O, Simpson E, Gravely R, Chandler P, Dyson J, Picard J K, Edwards A, et al. Nature (London) 1990;345:727–729. doi: 10.1038/345727a0. [DOI] [PubMed] [Google Scholar]
- 13.Slattery R M, Kjer-Nielsen L, Allison J, Charlton B, Mandel T E, Miller J F A P. Nature (London) 1990;345:724–726. doi: 10.1038/345724a0. [DOI] [PubMed] [Google Scholar]
- 14.Nepom G T, Erlich H. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 15.Quartey-Papafio R, Lund T, Chandler P, Picard J, Ozogbe P, Day S, Hutchings P R, O'Reilly L, Kioussis D, Simpson E, et al. J Immunol. 1995;154:5567–5575. [PubMed] [Google Scholar]
- 16.Schmidt D, Verdaguer J, Averill N, Santamaria P. J Exp Med. 1997;186:1059–1075. doi: 10.1084/jem.186.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luhder F, Katz J, Benoist C, Mathis D. J Exp Med. 1998;187:379–387. doi: 10.1084/jem.187.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdaguer J, Amrani A, Anderson B, Schmidt D, Santamaria P. J Immunol. 1999;162:4614–4626. [PubMed] [Google Scholar]
- 19.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue E R. J Immunol. 1996;156:450–458. [PubMed] [Google Scholar]
- 20.Carrasco-Marin E, Kanagawa O, Unanue E R. Res Immunol. 1997;148:291–301. doi: 10.1016/s0923-2494(97)87237-9. [DOI] [PubMed] [Google Scholar]
- 21.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 22.Gutgemann I, Fahrer A M, Altman J D, Davis M M, Chien Y H. Immunity. 1998;8:667–673. doi: 10.1016/s1074-7613(00)80571-3. [DOI] [PubMed] [Google Scholar]
- 23.Crawford F, Kozono H, White J, Marrack P, Kappler J. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman D L, Care-Salzler M, Tian J, Forsthuber T, Ting G S P, Robinson P, Atkinson M A, Sercarz E E, Tobin A J, Lehmann P V. Nature (London) 1993;366:69–72. doi: 10.1038/366069a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tisch R, Yang X-D, Singer S M, Libiau R S, Fugger L, McDevitt H O. Nature (London) 1993;366:72–75. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 26.Elliott J F, Qin H Y, Bhatti S, Smith D K, Singh R K, Dillon T, Lauzon J, Singh B. Diabetes. 1994;43:1494–1499. doi: 10.2337/diab.43.12.1494. [DOI] [PubMed] [Google Scholar]
- 27.Tian J, Atkinson M A, Clare-Salzler M, Herschenfeld A, Forsthuber T, Lehmann P V, Kaufman D L. J Exp Med. 1996;183:1561–1567. doi: 10.1084/jem.183.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian J, Clare-Salzler M, Herschenfeld A, Middleton B, Newman D, Mueller R, Arita S, Evans C, Atkinson M A, Mullen Y, et al. Nat Med. 1996;2:1348–1353. doi: 10.1038/nm1296-1348. [DOI] [PubMed] [Google Scholar]
- 29.Ma S W, Zhao D L, Yin Z Q, Mukherjee R, Singh B, Qin H Y, Stiller C R, Jevnikar A M. Nat Med. 1997;3:793–796. doi: 10.1038/nm0797-793. [DOI] [PubMed] [Google Scholar]
- 30.Wen L, Wong F S, Burkly L, Altieri M, Mamalaki C, Kioussis D, Flavell R A, Sherwin R S. J Clin Invest. 1998;102:947–957. doi: 10.1172/JCI2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zekzer D, Wong F S, Ayalon O, Millet I, Altieri M, Shintani S, Solimena M, Sherwin R S. J Clin Invest. 1998;101:68–73. doi: 10.1172/JCI119878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon J W, Yoon C S, Lim H W, Huang Q Q, Kang Y, Pyun K H, Hirasawa K, Sherwin R S, Jun H S. Science. 1999;284:1183–1187. doi: 10.1126/science.284.5417.1183. [DOI] [PubMed] [Google Scholar]
- 33.Chao C C, McDevitt H O. Immunogenetics. 1997;46:29–34. doi: 10.1007/s002510050238. [DOI] [PubMed] [Google Scholar]
- 34.Liu C-P, Parker D, Kappler J, Marrack P. J Exp Med. 1997;186:1441–1450. doi: 10.1084/jem.186.9.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozono H, White J, Clements J, Marrack P, Kappler J. Nature (London) 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 36.Schatz P J. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 37.Acha-Orbea H, McDevitt H O. Proc Natl Acad Sci USA. 1987;84:2435–2439. doi: 10.1073/pnas.84.8.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unkeless J C. J Exp Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gotoh M, Maki T, Kiyoizumi T, Satomi S, Monaco A P. Transplantation. 1985;40:437–438. doi: 10.1097/00007890-198510000-00018. [DOI] [PubMed] [Google Scholar]
- 40.Hausmann D H, Yu B, Hausmann S, Wucherpfennig K W. J Exp Med. 1999;189:1723–1734. doi: 10.1084/jem.189.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanagawa O, Martin S M, Vaupel B A, Carrasco-Marin E, Unanue E R. Proc Natl Acad Sci USA. 1998;95:1721–1724. doi: 10.1073/pnas.95.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotzin B, Falta M, Crawford F, Rosloniec E, Bill J, Marrack P, Kappler J. Proc Natl Acad Sci USA. 2000;97:291–296. doi: 10.1073/pnas.97.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridgway W, Fasso M, Fathman C G. J Immunol. 1998;161:714–720. [PubMed] [Google Scholar]
- 44.Wong F S, Karttunen J, Dumont C, Wen L, Visintin I, Pilip I M, Shastri N, Pamer E G, Janeway C A J. Nat Med. 1999;5:1026–3101. doi: 10.1038/12465. [DOI] [PubMed] [Google Scholar]
- 45.Signore A, Pozzilli P, Gale E A, Andreani D, Beverley P C. Diabetologia. 1989;32:282–289. doi: 10.1007/BF00265543. [DOI] [PubMed] [Google Scholar]
- 46.Faveeuw C, Gagnerault M C, Lepault F. J Immunol Methods. 1995;187:163–169. doi: 10.1016/0022-1759(95)00180-i. [DOI] [PubMed] [Google Scholar]
- 47.Savage P A, Boniface J J, Davis M M. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 48.Rees W, Bender J, Teague T K, Kedl R M, Crawford F, Marrack P, Kappler J. Proc Natl Acad Sci USA. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corper A L, Stratmann T, Apostolopoulos V, Scott C A, Garcia K C, Kang A S, Wilson I A, Teyton L. Science. 2000;288:505–511. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- 50.Latek R R, Suri A, Petzold S J, Nelson C A, Kanagawa O, Unanue E R, Fremont D H. Immunity. 2000;12:699–710. doi: 10.1016/s1074-7613(00)80220-4. [DOI] [PubMed] [Google Scholar]
- 51.Viner N J, Nelson C A, Deck B, Unanue E R. J Immunol. 1996;156:2365–2368. [PubMed] [Google Scholar]
- 52.Qin H Y, Elliott J F, Lakey J R T, Rajotte R V, Singh B. J Autoimmun. 1998;11:591–601. doi: 10.1006/jaut.1998.0243. [DOI] [PubMed] [Google Scholar]
- 53.Ridgway W M, Fasso M, Lanctot A, Garvey C, Fathman C G. J Exp Med. 1996;183:1657–1662. doi: 10.1084/jem.183.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]