Abstract
Nitrogen regulatory protein C (NtrC) of enteric bacteria activates transcription of genes/operons whose products minimize the slowing of growth under nitrogen-limiting conditions. To reveal the NtrC regulon of Escherichia coli we compared mRNA levels in a mutant strain that overexpresses NtrC-activated genes [glnL(Up)] to those in a strain with an ntrC (glnG) null allele by using DNA microarrays. Both strains could be grown under conditions of nitrogen excess. Thus, we could avoid differences in gene expression caused by slow growth or nitrogen limitation per se. Rearranging the spot images from microarrays in genome order allowed us to detect all of the operons known to be under NtrC control and facilitated detection of a number of new ones. Many of these operons encode transport systems for nitrogen-containing compounds, including compounds recycled during cell-wall synthesis, and hence scavenging appears to be a primary response to nitrogen limitation. In all, ≈2% of the E. coli genome appears to be under NtrC control, although transcription of some operons depends on the nitrogen assimilation control protein, which serves as an adapter between NtrC and σ70-dependent promoters.
Enteric bacteria initially perceive external nitrogen limitation as a decrease in the internal concentration of glutamine (1). One consequence is activation of transcription of genes under control of nitrogen regulatory protein C (NtrC), the products of which ameliorate slowing of growth. The products of NtrC-activated genes have three roles: they mediate assimilation of ammonium into the two central intermediates glutamate and glutamine, yield these intermediates catabolically, or spare the requirement for them. Products of NtrC-activated genes include an ammonia transporter (AmtB), glutamine synthetase (encoded by glnA), and amino acid permeases and catabolic enzymes (see references in Table 1). NtrC also activates transcription of genes encoding regulatory proteins: ntrB and ntrC (glnL and glnG, respectively), glnK, and nac (2). The nitrogen assimilation control (Nac) protein activates transcription of σ70-dependent genes, whereas NtrC activates transcription of σ54-dependent genes. Thus, Nac, which has no known coregulator other than its DNA-binding sites (3), serves as an adapter between NtrC and σ70-dependent operons.
Table 1.
NtrC/Nac-activated operons
| b no. | Coding strand | Name | Description | Control* | References
for control
|
|
|---|---|---|---|---|---|---|
| Nitrogen | Other | |||||
| b0336-7 | + | codBA | Cytosine transport, deaminase | Nac | (2) | (19)† |
| b0450-1 | + | glnK-amtB | N-regulation and NH3transport | NtrC | (13) | |
| b0652-5 | − | gltIJKL | Glutamate transport | NtrC?‡ | (11) | (20) |
| b0809-11 | − | glnHPQ | Glutamine transport | NtrC | (21, 22) | (21–23) |
| b0854-7 | + | potFGHI | Putrescine transport | NtrC | (24) | |
| b0929 | − | ompF | Outer membrane protein F | Nac?§ | (25)† | |
| b1006-12 | − | ycdGHIJKLM | Hypothetical proteins | NtrC | ¶ | |
| b1217-8 | + | chaBC | Cation transport regulator | NtrC?‡ | ||
| b1243-7 | + | oppABCDF | Oligopeptide transport | Nac?§ | (20, 26–29)† | |
| b1440-4 | + | ydcSTUV | Putrescine/spermidine transport (30) | Nac | (23) | |
| b1483-8 | − | ddpXABCDE | d-ala-d-ala dipeptide transport and dipeptidase | NtrC | ¶ | (10, 31) |
| b1744-8 | − | astCADBE | Arginine catabolism | NtrC | (32–34) | (32) |
| b1783-4 | + | yeaGH | Hypothetical proteins | NtrC? | ¶ | † |
| b1932 | + | yedL | Hypothetical protein | Nac | ||
| b1987 | − | cbl | Regulator for sulfur metabolism | NtrC‖ | (35) | |
| b1988 | − | nac | N-regulation | NtrC‖ | (2, 36) | |
| b2306-9 | − | hisJQMP | Histidine transport | NtrC** | (37–39) | (38, 39)† |
| b2310 | − | argT | Basic amino acid transport | NtrC | (37–39) | (38, 39)† |
| b2393 | + | nupC | Nucleoside transport | Nac | (40) | |
| b2661-4 | + | gabDTPC | γ-aminobutyric acid transport and catabolism | Nac | (41–43) | (23, 42, 43) |
| b3073 | + | ygjG | Probable ornithine aminotransferase | NtrC | † | |
| b3268-71 | + | yhdWXYZ | Polar amino acid transport (30) | NtrC | ||
| b3540-4 | − | dppABCDF | Dipeptide transport | Nac?§ | (20, 29, 44)† | |
| b3868-70 | − | glnALG | NH3 assimilation and N-regulation | NtrC | (45–47) | (47)† |
| b4207-8 | + | fklB-cycA | Peptidyl-prolyl isomerase; d-ala,d-ser,gly transport | Nac | ||
| Total: 25 operons; 75 genes | ||||||
Derived from Table 3. To avoid making the summary list artificially long, six operons (11 genes) in this table were excluded because it was difficult to assign their control and little is known about them. These are b1034-35, b1296, b1384 (feaR), b2875-76, b2882-85, and b3512. See Xi et al. (48) for recent information on b2875-76 and b2882-85. In addition, annotations (6) for the following operons have been changed: gltIJKL, oppABCDF, yeaGH, hisJQMP, gabDTPC, yhdWXYZ, and fklB-cycA.
Assessed as described in Materials and Methods.
RNA levels in an ntrC strain were ≥2-fold different for cells grown on glutamine versus ammonia (experiment i).
Some data from the second set of PCR products of poor quality; hard to assess NtrC versus Nac control.
At least one gene in these operons also appears to be NtrC-controlled, which is unlikely biologically; Nac control is in accord with known forms of control for these operons (see references), which involve sigma factors other than σ54.
The nac promoter is NtrC-activated (2). The cbl gene is probably expressed from the nac promoter under nitrogen-limiting conditions.
The NtrC-controlled promoter may be upstream of argT (38).
Studies with DNA microarrays (4, 5), together with use of new computer tools, allowed us to detect all genes/operons known to be under NtrC control and a number of new ones. These studies also allowed us to assess which of the new operons depended on the adapter Nac. Some of the largest and most interesting responses on microarrays were confirmed by other means, including direct detection of proteins.
Materials and Methods
Strains, Growth Conditions, and Assays.
Parental strain MG1655 (CGSC6300) was obtained from the Escherichia coli Genetic Stock Center. Unexpectedly, it differs in some regards from the strain that was sequenced (ref. 6; E.S., unpublished work). The glnA1859 allele was introduced by P1vir-mediated transduction based on linkage to zih-102∷Tn10 (7). The glnL2302 allele [glnL(Up)] (8) and the glnG10∷Tn5 allele (9) then were introduced by transduction to glnA+, yielding NCM3292 and NCM3285, respectively. Transductants that had lost zih-102∷Tn10 were chosen. The nac-28 disruption (2), which also deletes the nac promoter, was introduced directly into NCM3292 to yield the kanamycin-resistant strain NCM3596. The glnL(Up) and glnG alleles also were introduced into strain IADL310 (10) to yield strains NCM3700 and NCM3701, respectively. IADL310 carries at the λ attachment site a fusion of the ddp promoter-regulatory region (12–287 bp upstream of the translational start for ddpX) to lacZ.
Strains were grown with aeration at 37°C to midexponential phase on N−C− minimal medium (11) with 0.4% glucose as carbon source and 10 mM NH4Cl or 5 mM glutamine as nitrogen source (http://coli.berkeley.edu/∼zimmer/PNAS_NtrC_2000). On NH4Cl, glnL(Up) strains grew more slowly than glnG strains (representative doubling times of 90 and 75 min, respectively). The nac disruption suppressed the growth defect of glnL(Up) strains. The basis for slow growth and its suppression are not understood. On glutamine, glnL(Up) strains grew faster than glnG strains (representative doubling times of 110 and 180 min, respectively, after midexponential phase). Slow growth of glnG strains on glutamine is due, at least in part, to poor expression of the high-affinity glutamine transport system (glnHPQ).
Differential rates of synthesis of β-galactosidase (units/ml per OD600) were determined as described (12, 13). Levels of full-length mRNAs in glnL(Up) and glnG strains were assessed by priming first-strand cDNA synthesis in reverse transcriptase PCRs from the 3′ ends of genes (Sigma Genosys E. coli ORFmer primer pairs). Periplasmic fractions were obtained by treating whole cells with chloroform essentially as described (11, 14).
Preparation of Microarrays and Data Analysis.
Two sets of PCR products for E. coli ORFs were synthesized by using different sets of primers, and each was printed several times on glass slides as described (ref. 4 and http://coli.berkeley.edu/∼zimmer/PNAS_NtrC_2000). Growth of cultures was quenched on ice that had been cooled to −80°C (15) or with 1/10 volume of 5% phenol in ethanol (16) and RNA was extracted with hot phenol/SDS (15). Synthesis of cDNAs containing fluorescent nucleotide analogues, hybridization to microarrays, and scanning and normalization were as described (4).
To visualize spots on microarrays in genome order, images representing the Cy3 and Cy5 fluorescence intensities of spots were cut from a portable network graphic (D.P.Z., unpublished work). The spots then were ordered from left to right and top to bottom according to their b numbers (6) to provide a new genome image. Genome images are 100 spot columns wide and 45 spot rows tall. Operons (6) under NtrC/Nac control were identified by the following criteria: (i) the mRNA level for at least one gene in the operon was ≥2.5-fold higher (see Results for rationale) in the glnL(Up) than the glnG strain on ammonia for arrays printed with at least one set of PCR products [red/green (R/G)median and (R/G)mean ≥ 2.5] (see Table 2, which is published as supplementary material on the PNAS web site, www.pnas.org; 99 genes in 43 operons); (ii) mRNA levels for genes that met the first criterion were also ≥2.5-fold higher in the glnL(Up) strain on glutamine for the same set of PCR products (see Table 3, which is published as supplementary material on the PNAS web site, www.pnas.org; 86 genes in 31 operons). With the exceptions indicated in the legend, operons containing genes that met both criteria are included in Table 1 (Results).
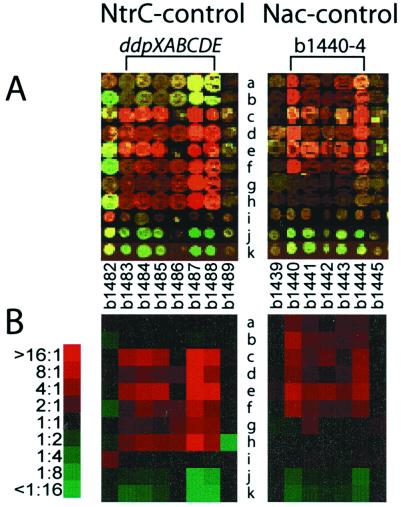
Eleven experiments (designated a–k) were performed with different strains under different growth conditions: a and b, glnL(Up) (Cy5) versus glnL(Up) nac (Cy3), NH3, and glutamine as nitrogen source, respectively (second set of PCR products); c and d, glnL(Up) (Cy5) versus glnG (Cy3), NH3, (first and second set of PCR products, respectively); e and f, same as c and d with glutamine; g and h, glnL(Up) nac (Cy5) versus glnG (Cy3), NH3, and glutamine, respectively (second PCR); i, glnG, glutamine (Cy5) versus NH3 (Cy3) (first PCR); j, MG1655, glutamine (Cy3) versus NH3 (Cy5) (first PCR) (note reversal of dyes); and k, same as j with 0.4% glycerol as carbon source. Higher mRNA levels in the first strain in experiments a–f are indicative of Nac activation, whereas higher levels in the first strain in experiments c–h are indicative of NtrC activation. This qualitative statement was quantified as follows: For each gene in an operon we determined ratios of RNA levels between the two strains [(R/G)median] in experiments a–h (see example in Fig. 2B). If all ratios in experiments a–f were >1, we assigned Nac control, whereas if all were >1 in experiments c–h, we assigned NtrC control. If the majority of genes in an operon were Nac-activated, we assigned Nac control to the operon and similarly for NtrC (Table 1, Results). Ambiguities are noted in footnotes to Table 1. Log ratios were used in hierarchical clustering analysis (17).
Figure 2.
Determination of NtrC versus Nac control. (A) Spot images for genes of the b1440–44 operon and the ddp (b1483–88) operon and adjacent genes in experiments a–k. (B) Profiles (log ratios) for these genes visualized as red and green intensities by using the program treeview (17) (http://rana.stanford.edu/software). The patterns expected for Nac or NtrC activation are red in experiments a–f or c–h, respectively. For either Nac or NtrC control green spots are expected in experiments j and k. If there is no control other than NtrC/Nac, yellow or black spots (A) or black squares (B) are expected in experiment i. Note that genes flanking each operon have expression patterns different from those of the operon and that the first PCR product for b1486 (experiments c, e, i, j, and k) was apparently bad. Red and green intensities representing induction and repression ratios, respectively, are indicated to the left of B.
Results
Initial Microarray Experiments.
To determine members of the NtrC regulon of E. coli we first compared mRNA levels in a glnL(Up) strain, which is called ntrB(Con) in other bacteria (Cy5; red) to those in a glnG strain, which is called ntrC in other bacteria (Cy3; green) for cells grown in minimal medium with ammonium chloride as sole nitrogen source (see Materials and Methods). The GlnL (NtrB) protein is a histidine autokinase that donates phosphoryl groups to response regulator GlnG (NtrC) and thereby activates it (18). The autokinase is more active in glnL(Up) [NtrB(Con)] strains than in wild-type strains, and hence genes and operons under NtrC (GlnG) control are overexpressed even under conditions of nitrogen excess.
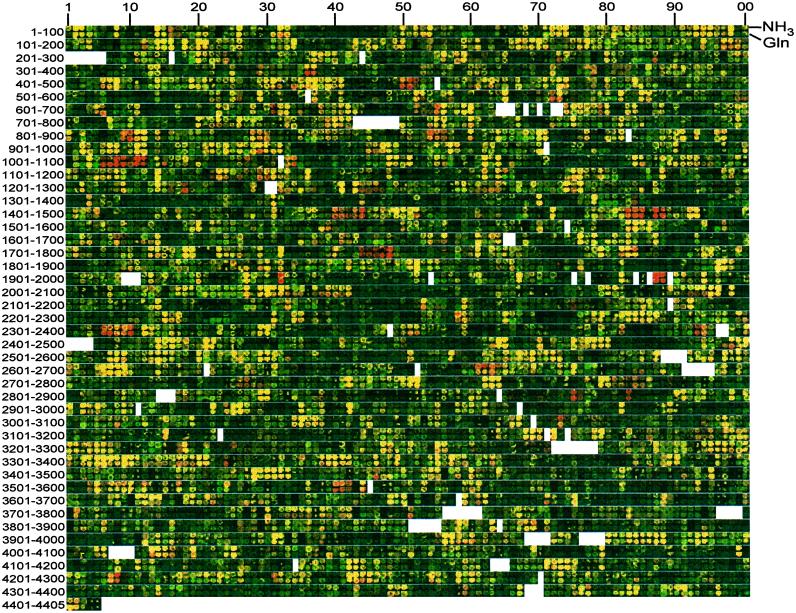
Data from DNA microarrays were displayed in genome order (Fig. 1), and genes for which mRNA levels in the glnL(Up) strain were at least 2.5-fold higher than those in the glnG strain were identified (Tables 1 and 2). The value of 2.5 was the minimum that yielded at least one gene in each operon known from previous biochemical and genetic studies to be under NtrC control in enteric bacteria [nine operons containing 27 genes (Table 1)]. The spots that met our first criterion often fell in groups of spots that appeared to be red by visual inspection (Fig. 1). With few exceptions (see Discussion), the boundaries of such groups corresponded to operon borders (6). Despite the relatively high ratio required in the first criterion, the induced operons included a number of new ones. Among those whose substrate specificities were easily rationalized were the following: three operons (b0854–57, b1243–47, b3540–44; 15 genes) specifying ATP-binding cassette transporters for putrescine, oligopeptides, and dipeptides, respectively, and two operons (b2393 and b4208) encoding secondary ion-coupled transporters for nucleosides, and d-alanine/d-serine/glycine, respectively. A mixed transport and catabolic operon for cytosine (codBA; b0336–37) and a gene encoding a probable ornithine aminotransferase (b3073) were also in this group.
Figure 1.
Two aligned genome images. Microarrays were probed with mixtures of cDNAs from the glnL(Up) and glnG strains grown on ammonium (upper row of each pair, experiment c) or glutamine (lower row, experiment e) as nitrogen source. Spots from fluorescence scanning images of microarrays were rearranged in genome order. The b number centuries (6) are indicated to the left. Blanks represent either b numbers that do not correspond to ORFs or that no longer exist. Red spots can be seen for most operons in Table 1. For some highly expressed genes—e.g., codBA (b0336–37) in the upper row and glnA (b3870)—spots appear intense yellow rather than red because of image saturation.
Putative NtrC-controlled operons included three specifying ATP-binding cassette transporters of unknown substrate specificity (b1440–44, b1483–88, and b3268–71) (6). Paulsen and colleagues (30) predicted that their substrates were spermidine, dipeptides, and an amino acid, respectively. Lessard and colleagues (10) implicated d-alanine-d-alanine as the dipeptide transported by the products of the b1483–88 operon, which also encodes a specific dipeptidase. Also putatively activated by NtrC were: cbl (b1987), which encodes a regulator for sulfur metabolism (see below); ompF (b0929), which encodes a large outer-membrane pore (see below); and three operons of unknown function (b1006–12, b1783–84, and b1932). Finally, NtrC appeared to activate a group of genes (b3504–17) around the hdeAB (H-NS-determined expression) operon (b3509–10). Additional experiments indicated that the “hdeAB group” was not under NtrC/Nac control (see below).
As is the case for operons known to be activated by NtrC, most differences in mRNA levels between the glnL(Up) strain and the glnG (ntrC) strain seen with ammonia as sole nitrogen source were also apparent with glutamine (Fig. 1; Tables 1 and 3). Note that the relative growth rates of the two strains were reversed on glutamine (Materials and Methods). Genes of the hdeAB group were an exception. Because these genes did not meet the criterion that mRNA levels in the glnL(Up) strain be ≥2.5 times higher than those in the glnG strain on glutamine as nitrogen source, they and others like them are not included in Table 1. Hierarchical clustering (17) also indicated that the hdeAB group was not under NtrC/Nac control (http://coli.berkeley.edu/∼zimmer/PNAS_NtrC_2000/). Additional differences between regulatory mutant strains seen only with glutamine as nitrogen source have not been analyzed in detail.
Most differences between mRNA levels in regulatory mutant strains also were seen in the congenic wild-type strain MG1655 when it was grown under two different conditions, namely with glutamine (Cy3) or ammonia (Cy5) as sole nitrogen source (Fig. 2; Tables 2 and 3). Although the reasons are not obvious, growth on glutamine is known to elevate expression of genes under NtrC control (50).
Assessment of Nac Involvement.
To determine which of the operons under NtrC control depended on the NtrC-Nac regulatory cascade, we compared mRNA levels in the glnL(Up) strain to those in a glnL(Up) nac strain on ammonia and on glutamine as nitrogen source (genes under Nac control detected). In addition, we compared mRNA levels in the glnL(Up) nac strain to those in the glnG (ntrC) strain under the same conditions (genes directly under NtrC control detected). These comparisons (Materials and Methods; Table 1) indicated that Nac activated transcription of the putative spermidine transport operon (b1440–44) (Fig. 2), the b1932 operon (ORF), the nucleoside transport operon nupC (b2393), the amino acid transport operon fklB-cycA (b4207–08), and the codBA operon (b0336–37). The latter was in accord with slow growth of a nac strain on cytosine as sole nitrogen source (2). Nac activation of other operons, e.g., ompF and opp, was less clear and may be caused by indirect effects. These operons are known to be subject to a plethora of other controls (Table 1). NtrC apparently activated transcription of the putrescine transport operon (b0854–57), the b1006–12 operon (ORFs), the transport and catabolic operon for d-alanine-d-alanine (b1483–88) (Fig. 2), and the probable ornithine aminotransferase operon (b3073), in addition to operons known or thought to be under its control (Table 1). Although we did not focus on repression, we noted that Nac apparently repressed transcription of the serA (b2913) and gltBDF (b3212–14) operons by ≥3-fold. Only repression of gltBDF was known (R.A.B., unpublished work).
NtrC Control of a ddp-lacZ Transcriptional Fusion.
Previous studies indicated that a ddp-lacZ transcriptional fusion placed at the λ attachment site of E. coli was transcribed in the late exponential phase of growth on enriched medium (to 150 Miller units) and that expression depended on σS (10). Having observed a large and reproducible difference between the mRNA levels for the ddp operon in a glnL(Up) and a glnG (ntrC) strain (up to 60-fold for ddpX; Figs. 1 and 2), we tested for NtrC control of the ddp-lacZ fusion. The differential rate of synthesis of β-galactosidase in a strain carrying the fusion and the glnL(Up) mutation was >230-fold higher than that in the congenic strain carrying the glnG (ntrC) mutation when they were grown on minimal medium with ammonium chloride as nitrogen source (2,300 versus <10 units). It was >390-fold higher with glutamine as nitrogen source (3,900 versus <10 units). In support of NtrC control, there is a putative σ54 promoter for the ddp operon located 53 bp upstream of the translational start for ddpX (6, 49) and a putative NtrC-binding site 140 bp upstream. Both are present in the fusion construct.
Analysis of Periplasmic Fractions.
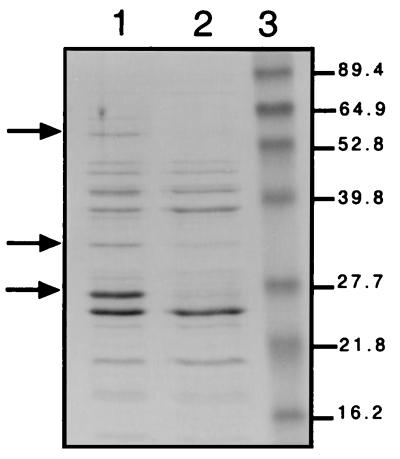
Analysis by SDS/PAGE indicated that the periplasmic fraction from the glnL(Up) strain grown on ammonia contained several proteins not present or present in much lower amounts in the same fraction from the glnG strain (Fig. 3). Analyses of periplasmic fractions by HPLC and MS indicated that binding protein components of the transport systems for d-alanine-d-alanine (ddpA) and arginine (argT) and of the putative transport system for polyamines (b1440) were among the proteins most highly elevated in the glnL(Up) strain over the glnG strain (R. W. Corbin and D. Hunt, personal communication).
Figure 3.
Periplasmic fractions of the glnL(Up) strain (lane 1) and the glnG strain (lane 2) on ammonia. After SDS polyacrylamide electrophoresis (2 μg protein; 12% gel), the gel was stained with Coommassie blue. Molecular weights of the standard proteins [GIBCO Benchmark Prestained Protein Ladder] in lane 3 are indicated to the right. Three bands of notably higher intensity in the glnL(Up) strain are indicated with arrows to the left.
Physiological Studies of Cbl.
Whereas many of the new operons/genes detected by probing for NtrC-activated genes on DNA microarrays were easily rationalized, the cbl (CysB-like) gene (b1987), which lies just downstream of nac (b1988), was not. The difference in mRNA levels—up to 30-fold higher in the glnL(Up) strain—was one of the largest we observed. Although we could not demonstrate a clear physiological role for Cbl in nitrogen metabolism, it activates transcription of operons whose products catabolize nitrogen- and sulfur-containing compounds (51). The cbl mRNA in glnL(Up) strains appeared to be full length (not shown), and we hope to determine whether it is translated to yield protein.
Discussion
The Extended NtrC/Nac Regulon.
NtrC appears to control almost 2% of E. coli genes (∼75) (Table 1). Transcription of most is apparently activated by NtrC in conjunction with σ54-holoenzyme, but transcription of some (≈9 operons; 25 genes) appears to depend on the adapter Nac in conjunction with σ70-holoenzyme.
The most striking observation about the extended NtrC/Nac regulon is the number of transport operons it contains. Almost 2/3 (45 genes) of the genes activated by NtrC/Nac are members of transport operons, which were expressed even in the absence of specific inducing compounds in the medium. Clearly, E. coli uses its capacity to scavenge for nitrogen-containing compounds as a first line of defense against nitrogen starvation. It uses both multicomponent ATP-binding cassette transporters, which often have high affinity for their substrates, and lower affinity ion-coupled transporters, usually encoded by a single gene. Not only does E. coli scavenge for compounds that may be found in the human intestine or other environments, but also for compounds it releases into the periplasmic space during cell growth and division. The latter include d-alanine (d-ala) and the d-alanine-d-alanine dipeptide (d-ala-d-ala) released during murein synthesis. The cycA gene (b4208), which encodes a transporter for d-ala, is under Nac control and the ddp operon (b1483–88), which encodes a transporter and a specific dipeptidase for d-ala-d-ala (10), is under NtrC control.
Although there has been no investigation under nitrogen-limiting conditions, under other conditions of limitation/starvation, E. coli increases cross-links between diaminopimelate (DAP) moieties in the murein, otherwise a minor proportion of the total (10, 52). This increases release of the d-ala-d-ala dipeptide (as opposed to d-ala, which is released when the more common d-ala-DAP cross-links are formed) and could enhance recovery of nitrogen. To the degree that preexisting murein can be remodeled to convert the common d-ala-DAP cross-links to DAP–DAP cross-links, additional d-ala would be released. Interestingly, increased DAP–DAP cross-bridges and other changes in murein composition that occur upon nutrient limitation are correlated with greater resistance of this layer to destruction (10, 52). Presumably, both scavenging and toughening of the murein would enhance survival under nitrogen-limiting conditions.
Data Analysis.
To make it more likely that the effects we observed were due directly to NtrC or Nac, we analyzed experiments done with pairs of regulatory mutant strains grown under the same conditions rather than the wild-type strain grown under two different conditions. The latter would be complicated by less direct responses. In fact, NtrC/Nac-controlled operons (Table 1) constitute a subset of those whose expression changed when the wild-type strain was grown on glutamine rather than ammonium. Obtaining stronger evidence that effects of NtrC and Nac on the newly identified operons in Table 1 are direct—or demonstrating that they are not—will require in vitro analysis and/or genetic dissection of promoter-regulatory regions. Although we have detected some putative NtrC binding sites by sequence analysis, we have not yet identified the pairs of sites that usually constitute functional enhancers (53).
Analysis of data from E. coli microarrays was facilitated by the use of genome images, i.e., the display of spot images in genome order (Fig. 1). This allowed easy recognition of differences in expression of multigenic operons, groups of contiguous genes that are transcribed together. We note, however, that differences in mRNA levels between our pairs of regulatory mutant strains were not uniform across operons (Fig. 2, Tables 2 and 3, and http://coli.berkeley.edu/∼zimmer/PNAS_NtrC_2000/). In some instances lack of coordinacy may have a biological explanation—e.g., there are large differences in stability for portions of mRNA encoding periplasmic and membrane components of ATP-binding cassette transporters (54, 55). In other cases, however, we know or suspect that apparent lack of coordinacy was an artifact of differences in the quantity or quality of PCR products (e.g., see b1486 in Figs. 1 and 2). Use of genome images also assisted in eliminating false signals caused by antisense RNA [e.g., see tesB (b0452) in Fig. 1] and in recognizing supraoperonic organization [e.g., see nac-cbl (b1987-88) and the hdeAB group (b3504-17)]. For all of the reasons discussed above, visualization of microarray data in genome order should be generally useful for bacteria, archaea, and their viruses. In addition, it should facilitate detecting differences in expression of large regions of eukaryotic chromosomes (56).
Supplementary Material
Acknowledgments
We are very grateful to D. Botstein and P. Brown for their help in initiating experiments with DNA microarrays and N. Cozzarelli and C. Gross for sharing essential materials. We thank M. Hryniewicz, I. Lessard, and C. Walsh for advice and generous gifts of materials. We thank our colleagues L. Csonka, J. Ingraham, R. LaRossa, B. Magasanik, W. van Heeswijk, N. Wingreen, and C. Yanofsky for thoughtful criticisms of the manuscript and O. Carmi for help in its preparation. This work was supported by National Institutes of Health Fellowship GM19862 to D.P.Z., a fellowship from the Deutsche Forschungsgemeinschaft to V.F.W., a Program in Mathematics and Molecular Biology/National Science Foundation postdoctoral fellowship to A.B.K., National Institutes of Health Grant GM47156 to R.A.B., National Institutes of Health Grant GM31657 to N. Cozzarelli, National Institutes of Health Grant HG00983 and support from the Howard Hughes Medical Institute to P. Brown, and National Institutes of Health Grant GM38361 and a grant from the Novartis Agricultural Discovery Institute, San Diego (to S.K.).
Abbreviations
- NtrC
nitrogen regulatory protein C
- Nac
nitrogen assimilation control
- DAP
diaminopimelate
Footnotes
See commentary on page 14044.
References
- 1.Ikeda T P, Shauger A E, Kustu S. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 2.Muse W B, Bender R A. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomposiello P J, Janes B K, Bender R A. J Bacteriol. 1998;180:578–585. doi: 10.1128/jb.180.3.578-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. . (First Published October 10, 2000; 10.1073/pnas.220414297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.He L, Soupene E, Kustu S. J Bacteriol. 1997;179:7446–7455. doi: 10.1128/jb.179.23.7446-7455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y M, Backman K, Magasanik B. J Bacteriol. 1982;150:214–220. doi: 10.1128/jb.150.1.214-220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backman K, Chen Y M, Magasanik B. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lessard I A, Pratt S D, McCafferty D G, Bussiere D E, Hutchins C, Wanner B L, Katz L, Walsh C T. Chem Biol. 1998;5:489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- 11.Kustu S G, McFarland N C, Hui S P, Esmon B, Ames G F. J Bacteriol. 1979;138:218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob F, Monod J. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 13.Soupene E, He L, Yan D, Kustu S. Proc Natl Acad Sci USA. 1998;95:7030–7034. doi: 10.1073/pnas.95.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ames G F, Prody C, Kustu S. J Bacteriol. 1984;160:1181–1183. doi: 10.1128/jb.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang S E, Daniels D L, Blattner F R. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin-Chao S, Cohen S N. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 17.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi F, Turnbough C L., Jr J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- 20.Urbanowski M L, Stauffer L T, Stauffer G V. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 21.Claverie-Martin F, Magasanik B. Proc Natl Acad Sci USA. 1991;88:1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nohno T, Saito T. Nucleic Acids Res. 1987;15:2777. doi: 10.1093/nar/15.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schellhorn H E, Audia J P, Wei L I, Chang L. J Bacteriol. 1998;180:6283–6291. doi: 10.1128/jb.180.23.6283-6291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 25.Pratt L A, Hsing W, Gibson K E, Silhavy T J. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 26.Andrews J C, Blevins T C, Short S A. J Bacteriol. 1986;165:428–433. doi: 10.1128/jb.165.2.428-433.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews J C, Short S A. J Bacteriol. 1986;165:434–442. doi: 10.1128/jb.165.2.434-442.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvo J M, Matthews R G. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith M W, Payne J W. FEMS Microbiol Lett. 1992;79:183–190. doi: 10.1111/j.1574-6968.1992.tb14038.x. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen I T, Sliwinski M K, Saier M H., Jr J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 31.Lessard I A, Walsh C T. Proc Natl Acad Sci USA. 1999;96:11028–11032. doi: 10.1073/pnas.96.20.11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraley C D, Kim J H, McCann M P, Matin A. J Bacteriol. 1998;180:4287–4290. doi: 10.1128/jb.180.16.4287-4290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C D, Abdelal A T. J Bacteriol. 1999;181:1934–1938. doi: 10.1128/jb.181.6.1934-1938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider B L, Kiupakis A K, Reitzer L J. J Bacteriol. 1998;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwanicka-Nowicka R, Hryniewicz M M. Gene. 1995;166:11–17. doi: 10.1016/0378-1119(95)00606-8. [DOI] [PubMed] [Google Scholar]
- 36.Feng J, Goss T J, Bender R A, Ninfa A J. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferro-Luzzi Ames G, Nikaido K. EMBO J. 1985;4:539–547. doi: 10.1002/j.1460-2075.1985.tb03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz G, Dürre P, Mullenbach G, Ames G F. Mol Gen Genet. 1987;209:403–407. doi: 10.1007/BF00329673. [DOI] [PubMed] [Google Scholar]
- 39.Schmitz G, Nikaido K, Ames G F. Mol Gen Genet. 1988;215:107–117. doi: 10.1007/BF00331311. [DOI] [PubMed] [Google Scholar]
- 40.Craig J E, Zhang Y, Gallagher M P. Mol Microbiol. 1994;11:1159–1168. doi: 10.1111/j.1365-2958.1994.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 41.Kahane S, Levitz R, Halpern Y S. J Bacteriol. 1978;135:295–299. doi: 10.1128/jb.135.2.295-299.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzer E, Halpern Y S. J Bacteriol. 1990;172:3250–3256. doi: 10.1128/jb.172.6.3250-3256.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niegemann E, Schulz A, Bartsch K. Arch Microbiol. 1993;160:454–460. doi: 10.1007/BF00245306. [DOI] [PubMed] [Google Scholar]
- 44.Abouhamad W N, Manson M D. Mol Microbiol. 1994;14:1077–1092. doi: 10.1111/j.1365-2958.1994.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 45.Hirschman J, Wong P K, Sei K, Keener J, Kustu S. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt T P, Magasanik B. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reitzer L J, Magasanik B. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi H, Schneider B L, Reitzer L. J Bacteriol. 2000;182:5332–5341. doi: 10.1128/jb.182.19.5332-5341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thieffry D, Salgado H, Huerta A M, Collado-Vides J. Bioinformatics. 1998;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- 50.Reitzer L J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. I. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 380–390. [Google Scholar]
- 51.van der Ploeg J R, Iwanicka-Nowicka R, Kertesz M A, Leisinger T, Hryniewicz M M. J Bacteriol. 1997;179:7671–7678. doi: 10.1128/jb.179.24.7671-7678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuomanen E, Markiewicz Z, Tomasz A. J Bacteriol. 1988;170:1373–1376. doi: 10.1128/jb.170.3.1373-1376.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 54.Newbury S F, Smith N H, Higgins C F. Cell. 1987;51:1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- 55.Higgins C F, McLaren R S, Newbury S F. Gene. 1988;72:3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- 56.Blumenthal T. BioEssays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.