Abstract
Gut-enriched Krüppel-like factor (GKLF) has been reported to partially inhibit α-smooth muscle actin (α-SMA) gene transcription by competing for binding to the TGF-β control element (TCE) with known activators such as Sp1 and other Krüppel-like factors. This incomplete inhibition via the TCE suggests an additional mechanism, which was evaluated in this study. The results showed that an α-SMA promoter mutated in the TCE remained susceptible to inhibition by GKLF in rat lung fibroblasts consistent with the existence of an additional TCE-independent mechanism. Since TGF-β– induced α-SMA expression is Smad3-dependent, potential interaction between GKLF and Smad3 was examined as a basis for this additional inhibitory mechanism. Co-immunoprecipitation and yeast two-hybrid assays revealed that GKLF could bind Smad3 through the Smad3 MH2 domain. Electrophoretic mobility shift assays and ChIP assay indicated that this GKLF–Smad3 interaction inhibited Smad3 binding to the Smad3-binding element (SBE) in the α-SMA promoter, and the activity of an SBE containing artificial promoter. Further analysis using smad3(−/−) fibroblasts confirmed that the TCE-independent inhibition by GKLF was dependent on Smad3. These data taken together suggest that in addition to inhibition via the TCE, GKLF represses α-SMA gene expression by interacting with Smad3 to prevent Smad3 binding to the SBE. It represents the first evidence to directly link GKLF with Smad3, a key intracellular mediator of TGF-β signaling, which should lead to a clearer understanding of the mechanism of how GKLF regulates TGF-β–induced gene expression.
Keywords: α-smooth muscle actin, GKLF, KLF4, Smad3, TGF-β
CLINICAL RELEVANCE
Our data taken together suggest that in addition to inhibition via the TCE, GKLF represses α-SMA gene expression by interacting with Smad3 to prevent Smad3 binding to the SBE. It represents the first evidence to directly link GKLF with Smad3.
De novo emergence of the myofibroblast is a characteristic of tissue repair, remodeling, and fibrosis. In pulmonary fibrosis the emergence and persistence of this cell is thought to be important for the pathogenesis of fibrosis and is a common feature of the fibroblastic foci encountered in idiopathic pulmonary fibrosis (1). This cell is commonly identified by expression of α-smooth muscle actin (α-SMA), which is expressed primarily in normal adult smooth muscle cells and transiently during development of cardiac and skeletal muscle (2, 3). Its expression, however, is not exclusive to smooth muscle cells and is a prominent feature of other cells such as myofibroblasts (4–7). TGF-β strongly activates its expression and induces the differentiation of fibroblast to myofibroblast (4, 8–10). In myofibroblast differentiation, Smad3 mediates the TGF-β induction of α-SMA gene expression by binding to a Smad-binding element (SBE) located at −552 to −513 from the transcriptional start site of the α-SMA gene promoter (4). In addition, another TGF-β control element (TCE) at −56 to −45 from the transcriptional start site of the α-SMA gene promoter is also important for both basal and TGF-β−induced α-SMA gene expression (4, 11–15).
Analysis of the TCE indicates it contains the minimal essential binding sequence of 5′-G/AG/AGGC/TGC/T-3′ for gut-enriched Krüppel-like factor (GKLF) and other Krüppel-like factors, such as Sp1 and BTEB2 (13–16). Transfection of a GKLF expression construct inhibits α-SMA gene expression, while transfection of Sp1 or BTEB2 activates its expression, thus indicating reciprocal regulation of the α-SMA promoter at the TCE by these inhibitory (e.g., GKLF) and stimulatory (e.g., BTEB2) Krüppel-like factors (13, 14). However, a significant portion of the inhibitory effect of GKLF on the α-SMA promoter is TCE-independent (15). This TCE-independent mechanism has not been identified. GKLF, also known as Krüppel-like factor 4 (KLF4), belongs to the family of Krüppel-like zinc finger transcription factors characterized by three CX2CX3FX5LX2HX3H zinc finger motifs separated by a highly conserved 7–amino acid spacer, TGEKP(Y/F)X (16). It plays important roles in regulation of the cell cycle, as well as cell growth and differentiation (17–20). Despite this mounting evidence of the importance of this transcription factor in regulating gene expression underlying many key cellular processes, the molecular mechanism responsible for this regulation remains incomplete.
Smad3 is a key molecule mediating TGF-β signaling in many cells (21, 22). It is composed of N-terminal Mad-homology domains 1 (MH1) and C-terminal Mad-homology domains 2 (MH2) connected by a proline-rich linker (23, 24). The MH1 domain of Smad3 mediates direct binding to DNA containing “CAGA” sequences, and in conjunction with other transcription factors, regulates promoter activity (24). The MH2 domain mediates interaction with other proteins to form homo- or heteromeric complexes, resulting in either activation or inhibition of target gene expression (25).
In this study, we hypothesized that the TCE-independent mechanism of GKLF inhibition of α-SMA transcription may be mediated by direct interaction between GKLF and Smad3, resulting in interference with Smad3 binding and activation of the α-SMA promoter. The results indicated that GKLF does bind Smad3 at its MH2 domain, which prevented Smad3 from binding to the SBE in the α-SMA promoter. This effectively abrogated the ability of Smad3 to activate α-SMA gene expression as well as an artificial SBE-containing promoter. Therefore, a novel mechanism for GKLF to regulate its target gene expression was suggested.
MATERIALS AND METHODS
Cell Culture
Fisher 344 rats were purchased from Charles River Laboratories, Inc. (Wilmington, MA) while smad3 knockout mice (smad3(−/−)) (strain name: 129-smad3tm1Par/J) and corresponding control wild-type mice (smad3(+/+)) were from the Jackson Laboratory (Bar Harbor, ME). Fibroblasts were isolated from rat or mouse lungs and cultured as described previously (4, 12). For TGF-β1 or basic fibroblast growth factor (bFGF) treatment, the fibroblasts were cultured overnight in complete medium and then were deprived of serum by rinsing twice in PBS and incubating in Dulbecco's modified Eagle's medium (DMEM) containing 0.5% PDS for 8 h. This was followed by addition of 2 ng/ml TGF-β1 or 50 ng/ml bFGF (R&D Systems, Minneapolis, MN) for different times as indicated in the text before harvesting.
Promoter Constructs and cDNA Clones
The wild-type rat α-SMA promoter was cloned by PCR and inserted into promoterless vector pGL3-basic (Promega, Madison, WI) at the SmaI site to form the fusion plasmid pGL3-αSMAp (also named α-SMAp-luc) as previously described (4), resulting in a construct wherein the expression of luciferase was driven by the rat α-SMA promoter. To make TCE-mutated α-SMA promoter mutant–luciferase fusion plasmid, pGL3-α-SMAp-TCEm (also named α-SMAp-TCEm) primers A (5′-TGGGAAGCGAGCTGCAGGGGATCAGACCA-3′) and B (5′-TGGTCTGATCCCCTGCAGCTCGCTTCCCA-3′) corresponding to DNA sequence of α-SMA promoter at −64 to −36 with TCE binding site mutated were used, together with wild-type α-SMAp-luc fusion plasmid pGL3-αSMAp in a QuickChange mutagenesis following the protocol of Stratagene (La Jolla, CA) as before (4). The artificial pBV-luc promoter construct and the same vector containing four tandem SBEs, SBE4-luc, were generous gifts of Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD) (26).
The GKLF expression plasmid pMT3-GKLF was a generous gift from Dr. Vincent W. Yang (Emory University School of Medicine, Atlanta, GA) (16), and it was used in transient transfection studies. For production of recombinant Smad3 and GKLF, the coding regions of these respective molecules were amplified by PCR using plasmids pEGFPC2-RSmad3 (4) or pMT3-GKLF as templates. They are inserted into vector pGEX-4T-2 (Amersham Biosciences, Piscataway, NJ) in frame to form GST fusion protein expression plasmids pGEX-Smad3 and pGEX-GKLF, respectively. The respective GST-fusion proteins were produced in Escherichia coli and purified to homogeneity (a single band on gel electrophoresis) by chromatography on glutathione-conjugated agarose beads (Amersham Biosciences).
Western Blotting
Western blotting was done as before (4). Briefly, cells were treated as above with or without TGF-β for the indicated times and then harvested for SDS polyacrylamide gel electrophoresis. After transfer onto nitrocellulose membranes and blocking of nonspecific binding, α-SMA was detected using anti–α-SMA monoclonal antibody (Sigma, St Louis, MO) by chemiluminescence. Specificity of antibodies was confirmed using isoform-matched nonimmune immunoglobulins.
Transfection and Reporter Gene Assay
Transient transfections of cells were performed using either the FuGENE6 reagent (Roche Applied Science, Indianapolis, IN) or lipofectamine2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions as previously described (4). Supercoiled DNA was isolated with an endotoxin-free Qiagen column kit (Qiagen Inc, Valencia, CA). Unless otherwise indicated, 2 μg DNA of the construct of interest and 1 μg plasmid pRL-TK control vector (used for normalization) were co-transfected per culture into cells with or without 2 ng/ml TGF-β treatment. In experiments to examine the effects of Smad3 and/or GKLF on α-SMA promoter activity, 1 μg each of Smad3 expression plasmid and/or GKLF expression plasmid were co-transfected with the α-SMA promoter reporter gene construct. The cells were harvested 48 h after transfection, and the activity of firefly or Renilla luciferase was measured using the dual luciferase assay system from Promega. The relative luciferase activity was calculated by normalizing firefly luciferase activity to that of Renilla luciferase. Experiments with each construct were repeated two to four times, and relative activity (fold over promoterless control) was expressed as mean ± SE.
Yeast Two-Hybrid Assay
In the hybrid hunter two-hybrid system (Invitrogen), Smad3 and its truncated mutants, Smad3(MH1) with the MH2 domain deleted, Smad3(MH2) with the MH1 domain deleted, and full-length Smad3 with the phosphorylation site deleted were cloned into the “bait” vector pHybLex/Zeo to form pHyblex-Smad3, pHyblex-Smad3(MH1), pHyblex-Smad3(MH2), and pHyblex-Smad3-ΔP, respectively. They were expressed as fusions with the bacterial-derived LexA DBD. GKLF was cloned in frame with the B42 AD in the “prey” vector pYESTrp2 to form pYESTrp2-GKLF. In accordance with the manufacturer's protocol, the pHybLex/Zeo “bait” and pYESTrp2 “prey” fusion constructs were transformed sequentially by the lithium acetate method into yeast strain L40 (MATa his3 200 trp1–901 leu2–3112 ade2 LYS2∷(4lexAop-HIS3) URA3∷(8lexAop-lacZ) GAL4) hosting genomic LexAop-lacZ reporter. Co-transformants growing on both Trp− and Zeo+ selective media were assayed for β-galactosidase activity by liquid assay using ONPG (o-nitrophenol-β-D-galactoside) as substrate after lysis by freezing-thaw in liquid nitrogen in accordance with protocol of BD Biosciences Clontech (Palo Alto, CA).
Co-Immunoprecipitation Assays
Cell lysates and immunoprecipitations were done using the Co-IP kit (Roche Applied Science, Indianapolis, IN) in accordance with the manufacturer's protocol. Anti-GKLF antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and anti-Smad3 antibodies were from Upstate USA, Inc. (Charlottesville, VA). Immunoblots were performed as described above in the section on Western blotting.
Electrophoretic Mobility Shift Assay
GST-Smad3 fusion proteins were tested for its ability to bind SBE by electrophoretic mobility shift assay (EMSA). A double-stranded SBE oligonucleotide probe bearing nucleotide sequences corresponding to the region between −552 and −513 of the α-SMA promoter with the sense strand sequence 5′-TACAGACTTCATTGATACTACACA CAGACTCCAGACTAC-3′ was synthesized and used for the EMSA. It was labeled with γ-32P by T4 polynucleotide kinase and incubated with 1 μg of protein(s) of interest, 1.0 μg of Poly dI-dC, 0.1 μg poly-L-lysine in a final volume of 25 μl in Dignam's Buffer C. Where indicated, the EMSA reaction mixtures were pre-incubated with GKLF or GST only on ice for 30 min before probe addition and incubation for another 20 min at room temperature. Samples were then analyzed by electrophoresis on 4% nondenaturing polyacrylamide gels at 150 V in 1× Tris-borate/EDTA electrophoresis buffer (TBE) for ∼ 2 h. After electrophoresis the gels were dried and exposed to X-ray film for 24–48 h.
Chromatin Immunoprecipitation Assay
This was performed using a kit from Upstate USA Inc. (Charlottesville, VA), using the manufacturer's protocol as previously described (27). Lung fibroblasts were transfected with either a GKLF expression plasmid (pMT3-GKLF) or the expression vector only (pMT3). Four hours after transfection, the cells were treated with TGF-β for 24 h. They were then fixed with 1% formaldehyde in DMEM to crosslink bound transcription factors to DNA, and the samples were then washed twice with PBS and then suspended in SDS lysis buffer. After sonication to shear the DNA to an average of 1,000 base pairs, the lysates were centrifuged and the supernatants collected. After preclearing with protein A agarose, each sample was aliquoted (20 μl) separately for use as “input DNA” in PCR analysis. The remainder of each sample was then divided equally into three aliquots for incubation with (1) anti-Smad3 antibody (anti-Smad3 bound DNA fraction), (2) nonimmune rabbit IgG (nonspecific antibody background DNA fraction), or (3) PBS-buffer (no antibody background fraction), respectively. Any immune complexes formed were affinity-adsorbed with protein A agarose and collected by centrifugation. The bound DNA was washed extensively and eluted from the protein A agarose with freshly made elution buffer (1% SDS, 0.1 M NaHCO3). The eluates and the original “input DNA” sample were incubated at 65°C for 4 h to reverse the crosslink and then used as templates for PCR analysis along with the oligonucleotide primers A (5′-ACGGTCCTTAAGCATGATAT-3′) and B (5′-CTTACCCTGATGGCGACTGGCTGG-3′). These primers were designed to amplify a 764-bp α-SMA promoter region containing the SBE. The PCR products were then analyzed by gel electrophoresis in 1.3% agarose, along with 100 bp DNA ladder from New England Biolabs, Inc. (Ipswich, MA).
RESULTS
Effects of GKLF on Wild-Type and TCE-Mutated α-SMA Promoters
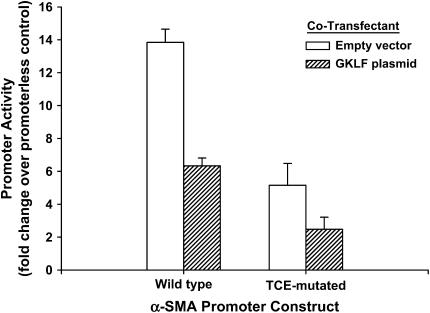
De novo induction of myofibroblast differentiation is a key characteristic of wound healing, tissue repair, and fibrosis/remodeling, and which is subject to regulation by TGF-β (4, 8–12). α-SMA is the most commonly used marker gene for myofibroblast differentiation (4–7), whose expression in TGF-β–stimulated rat lung fibroblasts is dependent on Smad3 binding to the SBE in the α-SMA promoter (4). Its expression is also suppressed by GKLF in part by binding to a TCE in the α-SMA promoter (13–15). However, GKLF inhibition of α-SMA gene expression is not entirely dependent on the TCE, and specifically its interaction with Smad3-mediated activation of the promoter has not been explored. To examine this possibility, the effects of a GKLF expression plasmid on wild-type or a TCE-mutated α-SMA gene promoter activity were analyzed in lung fibroblasts. As shown in Figure 1, co-transfection with the GKLF plasmid caused marked inhibition (> 50%) of α-SMA promoter activity in TGF-β–treated lung fibroblasts. The TCE-mutated promoter activity was lower than the activity of the wild-type promoter, but showed comparable inhibition (> 50%) by GKLF. These results, in addition to confirming the importance of the TCE for α-SMA gene expression in response to TGF-β, clearly showed that the TCE-mutated α-SMA promoter activity could still be inhibited by GKLF in lung fibroblast. This finding is consistent with the existence of a TCE-independent mechanism of GKLF suppression of the α-SMA promoter.
Figure 1.
Inhibition property of α-SMA promoter activity by GKLF. Wild-type and TCE-mutated α-SMA promoters mutant constructs were co-transfected with GKLF expression plasmid or empty expression vectors into rat lung fibroblasts as indicated. Cells were then treated with TGF-β and analyzed for firefly and Renilla luciferase activities, respectively. After normalization to the Renilla luciferase activity, the data were expressed as fold over promoterless control and shown as means ± SE. Experiments with each combination were repeated two to four times and yielded similar results.
Smad3-GKLF Binding Interactions
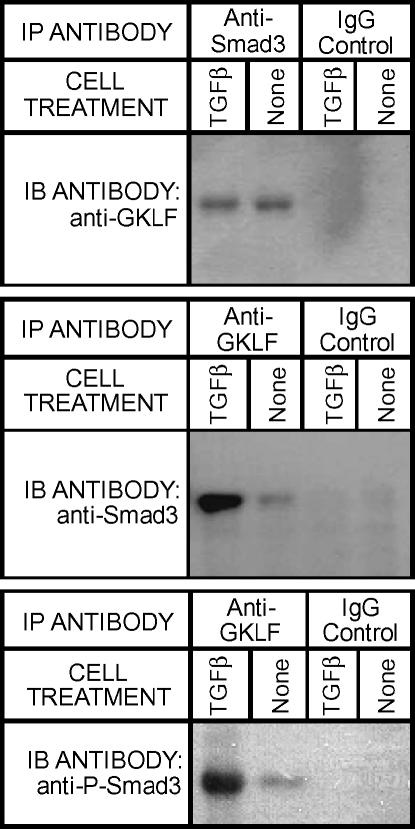
Smad3 is a key intracellular mediator of TGF-β signaling and found to be essential for TGF-β induction of α-SMA expression by binding to an SBE identified in the α-SMA promoter (4). In view of this essential role of Smad3, the possibility that GKLF could interact directly with Smad3 to interfere with its activating function was evaluated using co-immunoprecipitation (Co-IP) analysis. Immunoprecipitation of fibroblast lysates with anti-Smad3 antibodies followed by immunoblotting with anti-GKLF antibodies yielded a positive band that migrated with the expected molecular weight for GKLF (Figure 2, upper panel). A positive signal migrating with the expected molecular weight for Smad3 was also generated when the immunoprecipitation was done with anti-GKLF antibodies followed by immunoblotting with anti-Smad3 antibodies (Figure 2, middle panel). In both instances, use of control IgG instead of the specific immunoprecipitating antibodies failed to generate a signal upon immunoblotting. These results indicated the ability of GKLF to bind Smad3 in fibroblast extracts. Since Smad3 is phosphorylated and translocates to the nucleus in response to TGF-β (24), the effect of phosporylation on Smad3 interaction with GKLF was also examined by Co-IP analysis. Upon immunoprecipitation of the lysates with anti-GKLF antibody and immunobloting with anti-phosphorylated-Smad3 antibody, a strong signal was observed, indicating that phosporylation of Smad3 does not affect its interaction with GKLF (Figure 2, bottom panel).
Figure 2.
Co-immunoprecipitition analysis of GKLF–Smad3 interaction. Fibroblasts were treated with either TGF-β or buffer only (None) for 48 h. The total proteins were then extracted and, after preclearing with protein A beads, were incubated with anti-Smad3 (upper panel) or anti-GKLF antibodies (middle and lower panels) (IP ANTIBODY) or their corresponding IgG control. After stringent washing, the proteins bound on the respective beads were eluted and equal volume of the elution was subjected to Western blot analysis using the indicated IB ANTIBODY.
To confirm and extend this finding, a deletion analysis of Smad3 in a yeast two-hybrid system was performed. Full-length Smad3, Smad3(MH1) with the MH2 domain deleted, Smad3(MH2) with the MH1 domain deleted, or full-length Smad3 with the phosphorylation site deleted were fused with LexA DBD as “baits,” respectively. Each were co-transfected with B42 AD fused GKLF “preys” into yeast strain L40 containing a genome-integrated LexAop-lacZ reporter. After confirmation of the co-transfection in Trp− and Zeo+ selective media, the β-galactosidase activity was tested to show the interaction between the “baits” and “preys.” As shown in Table 1, significant interaction was observed between the full-length Smad3 and GKLF when compared with the corresponding vector only control. Deletion of the MH1 domain of Smad3 resulted in a > 8-fold enhancement in GKLF-Smad3 interaction. In contrast, deletion of the MH2 domain caused a > 75% reduction in the interaction between GKLF and Smad3. Consistent with the Co-IP assay, deletion of the Smad3 phosphorylation site does not affect the GKLF-Smad3 interaction. These findings confirmed direct binding interaction between GKLF and Smad3, which appeared to be mainly via the MH2 domain of Smad3.
TABLE 1.
YEAST TWO-HYBRID ANALYSIS OF GUT-ENRICHED KRÜPPEL-LIKE FACTOR –Smad3 INTERACTION
| “Prey”
|
||
|---|---|---|
| “Bait” | pYEStrp2 | pYEStrp2-GKLF |
| pHyblex-Zeo | 0.45 ± 0.03 | 0.41 ± 0.04 |
| pHyblex-Smad3 | 0.43 ± 0.17 | 5.43 ± 0.16 |
| pHyblex-Smad3(MH1) | 0.42 ± 0.12 | 1.32 ± 0.35 |
| pHyblex-Smad3(MH2) | 0.97 ± 0.06 | 43.89 ± 0.11 |
| pHyblex-Smad3–ΔP | 0.047 ± 0.05 | 4.78 ± 0.13 |
Definition of abbreviation: GKLF, gut-enriched Krüppel-like factor.
“Prey” and “Bait” vectors were as indicated, and resulting co-transformants were assayed for β-galactosidase activity. The numbers showed the β-galactosidase activities (in Miller units) of each combination of constructs (between respective columns and rows) in yeast and were expressed as means ± SE of three individual experiments.
Effect of GKLF on Smad3 Binding to SBE
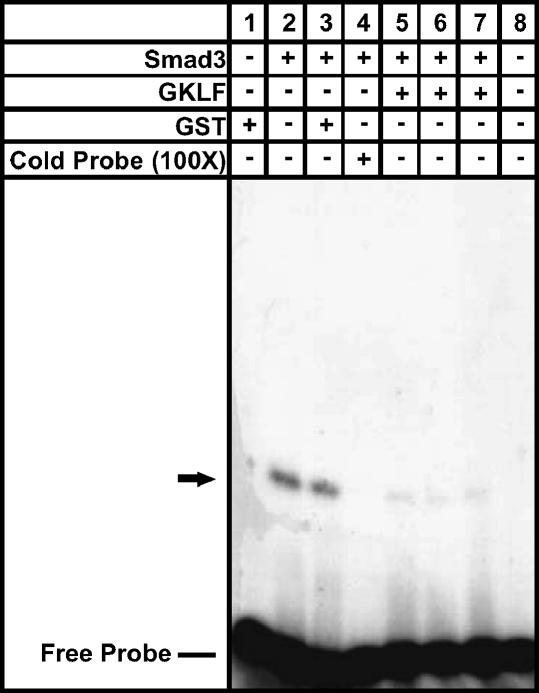
Since Smad3 binds to the SBE of the α-SMA gene promoter to regulate its expression (4), GKLF may inhibit α-SMA expression by affecting Smad3 binding to the SBE on this promoter. This possibility was examined by EMSA using recombinant GKLF and Smad3 in conjunction with a 32P-labeled synthetic oligonucleotide probe containing the SBE binding site of the α-SMA promoter. Where indicated, Smad3 was pre-incubated with GKLF or GST only before probe addition. As shown in Figure 3, incubation of GST with the SBE probe failed to generate a shifted or retarded band (lane 1), but incubation of Smad3 with the SBE probe caused the appearance of a major shifted (retarded) band indicating binding of Smad3 (but not GST) to the hot probe (lane 2). Binding of Smad3 to the SBE probe was not significantly affected by preincubation with GST, but was abolished by addition of 100-fold excess of cold probe (lane 4), thus confirming specificity of binding to the probe. Interestingly, preincubation of Smad3 with GKLF before addition to the hot SBE probe significantly reduced Smad3 binding to this probe (lanes 5–7). The specificity of this GKLF inhibitory effect was supported by the lack of effect of preincubation with GST (lane 3). These results suggested that the GKLF–Smad3 interaction inhibited the binding of Smad3 to SBE in the α-SMA promoter.
Figure 3.
Effect of GKLF on Smad3 binding to SBE. 32P-labeled double-stranded oligonuleotide probe corresponding to the sequence of the SBE in the rat α-SMA promoter was incubated with either recombinant Smad3 protein or GST protein only, and then analyzed by gel electrophoresis. Selected samples were pretreated with the indicated substances before addition of radiolabeled probe. A shifted band indicative of DNA–protein complex formation was indicated with an arrow. Addition of a 100-fold excess of unlabeled probe, as indicated, was used to document the specificity of the binding. A film was shown from a representative experiment, which was repeated twice, yielding similar results.
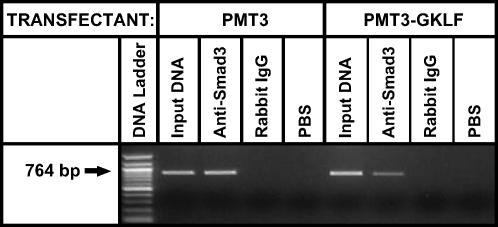
To further confirm that GKLF inhibits Smad3 binding to SBE in the α-SMA promoter, a GKLF expression plasmid or the expression vector only was transfected into rat lung fibroblasts and then the cells were treated with TGF-β for 24 h. The cells were then fixed and lysed for ChIP assay to determine the effect of GKLF on Smad3 binding to DNA. As shown in Figure 4, Smad3 binding to α-SMA promoter DNA was reduced in cells transfected with GKLF expression plasmid, relative to that seen in cells transfected with the empty vector only. This result further confirmed that GKLF inhibited Smad3 binding to the SBE in the α-SMA promoter.
Figure 4.
ChIP analysis of Smad3 binding to the α-SMA promoter. Rat lung fibroblasts were transfected with GKLF expression plasmid or the empty vector only and then treated with TGF-β. After fixation with formaldehyde and sonication, the lysates were incubated with anti-Smad3 antibody (Anti-Smad3), nonimmune rabbit IgG (Rabbit IgG), or PBS as indicated. These precipitated DNA samples, along with untreated whole samples (Input DNA) were analyzed by PCR using primers spanning the SBE region of the α-SMA promoter. The PCR products and a 100-bp DNA ladder were analyzed by agarose gel electrophoresis. A representative gel is shown.
TCE-Independent Inhibition by GKLF and its Dependence on Smad3
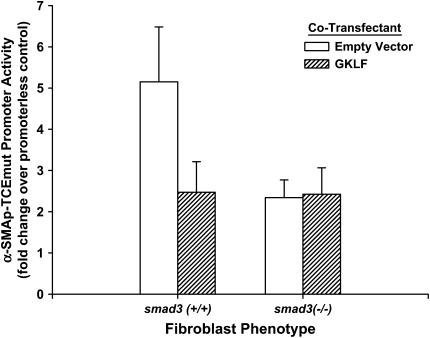
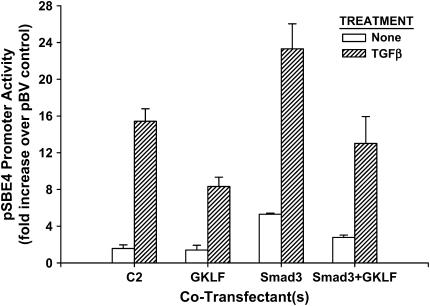
To further test if the TCE-independent inhibition by GKLF was dependent on Smad3, the effects of GKLF on the TCE-mutated α-SMA gene promoter activity were analyzed in smad3(+/+) or smad3(−/−) lung fibroblasts. The results showed that co-transfection with the GKLF expression plasmid caused marked inhibition (> 50%) of TCE-mutated promoter activity in TGF-β–treated smad3(+/+) cells (Figure 5). However, GKLF failed to inhibit the same mutant promoter construct activity in smad3(−/−) cells. This result suggested that the TCE-independent inhibition by GKLF was dependent on Smad3. However, this lack of detectable inhibition by GKLF may be due to the already low level of mutant promoter activity in smad3(−/−) cells, although it remained ∼ 2-fold above the promoterless control. Hence, to confirm the ability of GKLF to inhibit SBE-dependent promoter activity independent of any effect on the TCE, a Smad3-regulated artificial promoter (pSBE4-luc) was used in co-transfection studies with the Smad3 and GKLF expression plasmids. No TCE site was identified in this artificial promoter or its control (lacking SBE) construct. As expected, this artificial promoter was markedly induced by treating the cells with TGF-β, and/or by co-transfection with the Smad3 plasmid (Figure 6). When the GKLF expression plasmid only was co-transfected with the pSBE4-luc artificial promoter construct, the promoter activity was significantly inhibited in TGF-β–treated cells, but not in cells without TGF-β treatment. The increased activity due to Smad3 overexpression was significantly inhibited by co-transfection with the GKLF plasmid in the absence or presence of TGF-β treatment. These results confirmed the ability of GKLF to inhibit Smad3-dependent (and thus SBE-dependent) promoter activity independent of the TCE.
Figure 5.
TCE-independent inhibition by GKLF depended on Smad3. TCE-mutated α-SMA promoters were co-transfected with GKLF expression plasmid or empty expression vectors into either smad3(+/+) or smad3(−/−) fibroblasts as indicated. Cells were then treated with TGF-β, and then analyzed for firefly and Renilla luciferase activities. After normalization to the Renilla luciferase activity, the data were expressed as fold over promoterless control and shown as means ± SE. Experiments with each combination were repeated two to four times and yielded similar results.
Figure 6.
GKLF inhibited Smad3-regulated artificial promoter. Artificial pBV-luc luciferase fusion construct (pBV) and the same vector with four tandem SBE repeats (pSBE4) were transfected into NIH3T3 cells, and co-transfected with the indicated expression plasmid(s) or empty vector control (C2). After treatment with TGF-β or buffer only (None), the cells were analyzed for firefly and Renilla luciferase activities. After normalization to the Renilla luciferase activity, the results were expressed as fold increase over the pBV control promoter activity and shown as means ± SE. Experiments with each combination were repeated two to four times, with similar results.
DISCUSSION
α-SMA is one of the most abundant proteins in the cell and is transiently expressed during development of cardiac and skeletal muscle (2, 3). It is also an important marker for myofibroblasts, which appear de novo at the early fibroproliferative stages of wound healing, desmoplastic responses in cancers, tissue repair/remodeling, and fibrosis in multiple organs (4–8). Myofibroblasts are key sources of extracellular matrix and cytokines important for the development and progression of fibrosis (28). Hence, understanding the regulatory mechanisms of α-SMA gene expression as they relate to emergence and persistence of the myofibroblast could provide critical insight into the pathogenesis of fibrotic diseases.
GKLF is a Krüppel-like transcription factor that is found to be involved in regulation of a wide variety of biological processes, including inhibition of cell proliferation by blocking G1/S progression of the cell cycle (18), acceleration of the formation of the epidermal permeability barrier (29), inhibition of the proliferation–differentiation switch, and initiation of squamous epithelial dysplasia (30). Therefore, investigation of the GKLF regulatory mechanism on myofibroblast differentiation also helps in understanding its role in these processes as well.
Previous studies have amply documented the inhibitory effect of GKLF on gene expression as mediated by interaction with the TCE present in the promoter of certain genes in smooth muscle cells. Several trans-acting factors, either activators such as Sp1 and BTEB2 or repressors such as GKLF, have been shown to compete for binding to the TCE and to reciprocally regulate gene expression (13, 14). However, a significant proportion of the inhibition by GKLF cannot be accounted for by this interaction with the TCE, since a TCE-mutated construct remains susceptible to inhibition by GKLF (15). Based on this finding, the authors of that study suggest the existence of additional TCE-independent GKLF inhibitory mechanisms (15). In this study, we investigated such a potential mechanism and focus it on the possible interaction of GKLF with the previously established Smad3-dependent activation of the α-SMA promoter (4). The results of studies using a TCE-mutated α-SMA promoter construct confirmed the existence of such a TCE-independent mechanism of GKLF inhibition of TGF-β–induced α-SMA expression in rat lung fibroblasts.
Our data indicated that GKLF could bind Smad3 via its MH2 domain. Removal of the Smad3 MH1 domain enhances the GKLF-Smad3 interaction. The Smad3 MH1 domain was known to be responsible for the binding of Smad3 to SBE in the promoter. The GKLF–Smad3 MH2 interaction may induce conformational (secondary/tertiary structure) changes in Smad3 resulting in reduction of the ability of the Smad3 MH1 domain to bind to the SBE. This is the case for other Smad3 interacting proteins, such as the zinc finger transcription factor Evi-1 (31). In our study, a similar effect was observed with GKLF as the interacting protein. Binding of GKLF to Smad3 significantly reduced binding of Smad3 to the SBE in the α-SMA promoter, which was associated with reduced promoter activity. Since the interaction between full-length Smad3 and GKLF was weaker than the interaction between Smad3 MH2 domain only and GKLF, the conformation of the MH2 domain may be less favorable for binding to GKLF when the MH1 domain is intact. Conversely, it may be that GKLF binding to the MH2 domain constrains the MH1 domain from binding to the SBE.
Smad2 is another potential transacting factor that mediates TGF-β signaling in many cells (23–25). It is quite similar in structure and function to Smad3 but does not bind to DNA (or the SBE) directly (32). In contrast to more than 3-fold increase in Smad3 expression after TGF-β treatment, Smad2 is constitutively expressed in fibroblasts (4). Therefore, the induction of α-SMA by TGF-β was thought to occur mainly through Smad3 (4). Nevertheless, Smad2 is also found to activate α-SMA gene expression in rat hepatic stellate cells (33) and is possibly also responsible for the basal expression of α-SMA in rat lung fibroblasts. Interestingly, this basal level of expression mediated by Smad2 was not inhibited by GKLF, since no significant repression was observed for the promoter activity of TCE-mutated α-SMA promoter mutant in smad3(−/−) cells after transfection with GKLF expression plasmid.
Although the data presented in our study have identified a TCE-independent mechanism for GKLF inhibition of α-SMA expression by inhibiting Smad3 binding to the SBE, additional mechanisms have not been ruled out. Thus potential effects of GKLF on alternative pathways of TGF-β signaling, and on other as yet undiscovered pathways unrelated to the TCE, remain to be explored. For example, MAP kinase phosporylation of Smad3 (34) may represent an additional potential target for GKLF regulation. However, our data indicated that the Smad3–GKLF interaction was not affected by phosphorylation status of Smad3. However, members of the MAP kinase family, such as ERK1/2, are known to be required for the expression of GKLF (35). Since FGF-2 induces the phosphorylation of ERK1/2 (36) and antagonizes the stimulatory effect of TGF-β on α-SMA gene expression (37), an additional mechanism may be mediated by the induction of this growth factor. It may be that FGF-2 could inhibit α-SMA expression independent of the TCE as well. Further studies are necessary to evaluate these additional potential mechanisms.
Acknowledgments
The authors are grateful for the gifts of mouse GKLF eukaryotic expression plasmid from Dr. Vincent W. Yang, Emory University School of Medicine, and the artificial SBE containing promoter and control constructs from Dr. Bert Vogelstein, Johns Hopkins University.
This work was supported by grants HL28737, HL31963, HL52285, and HL77297 from the National Institutes of Health, and by an Award to S.H.P. from the Sandler Program in Asthma Research.
Originally Published in Press as DOI: 10.1165/rcmb.2006-0043OC on July 20, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Pardo A, Selman M. Idiopathic pulmonary fibrosis: new insights in its pathogenesis. Int J Biochem Cell Biol 2002;34:1534–1538. [DOI] [PubMed] [Google Scholar]
- 2.Campbell JH, Kocher O, Skalli O, Gabbiani G, Campbell GR. Cytodifferentiation and expression of alpha-smooth muscle actin mRNA and protein during primary culture of aortic smooth muscle cells: correlation with cell density and proliferative state. Arteriosclerosis 1989;9: 633–643. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock-Mitchell J, Mitchell JJ, Low RB, Kieny M, Sengel P, Rubbia L, Skalli O, Jackson B, Gabbiani G. Alpha-smooth muscle actin is transiently expressed in embryonic rat cardiac and skeletal muscles. Differentiation 1988;39:161–166. [DOI] [PubMed] [Google Scholar]
- 4.Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-β–induced alpha-smooth muscle actin expression. Am J Respir Cell Mol Biol 2003;29:397–404. [DOI] [PubMed] [Google Scholar]
- 5.Darby I, Skalli O, Gabbiani G. Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Lab Invest 1990;63:21–29. [PubMed] [Google Scholar]
- 6.Hu B, Wu Z, Jin H, Hashimoto N, Liu T, Phan SH. CCAAT/enhancer-binding protein beta isoforms and the regulation of alpha-smooth muscle actin gene expression by IL-1 beta. J Immunol 2004;173:4661–4668. [DOI] [PubMed] [Google Scholar]
- 7.Skalli O, Schurch W, Seemayer T, Lagace R, Montandon D, Pittet B, Gabbiani G. Myofibroblasts from diverse pathologic settings are heterogeneous in their content of actin isoforms and intermediate filament proteins. Lab Invest 1989;60:275–285. [PubMed] [Google Scholar]
- 8.Desmouliere A. Factors influencing myofibroblast differentiation during wound healing and fibrosis. Cell Biol Int 1995;19:471–476. [DOI] [PubMed] [Google Scholar]
- 9.Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol 1993;122:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronnov-Jessen L, Petersen OW. Induction of alpha-smooth muscle actin by transforming growth factor-beta 1 in quiescent human breast gland fibroblasts: implications for myofibroblast generation in breast neoplasia. Lab Invest 1993;68:696–707. [PubMed] [Google Scholar]
- 11.Hautmann MB, Madsen CS, Owens GK. A transforming growth factor beta (TGFbeta) control element drives TGFbeta-induced stimulation of smooth muscle alpha-actin gene expression in concert with two CArG elements. J Biol Chem 1997;272:10948–10956. [DOI] [PubMed] [Google Scholar]
- 12.Roy SG, Nozaki Y, Phan SH. Regulation of alpha-smooth muscle actin gene expression in myofibroblast differentiation from rat lung fibroblasts. Int J Biochem Cell Biol 2001;33:723–734. [DOI] [PubMed] [Google Scholar]
- 13.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem 2000;275:37798–37806. [DOI] [PubMed] [Google Scholar]
- 14.Cogan JG, Subramanian SV, Polikandriotis JA, Kelm RJ Jr, Strauch AR. Vascular smooth muscle alpha-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer. J Biol Chem 2002;277:36433–36442. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Sinha S, Owens G. A transforming growth factor-β control element required for SM α-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem 2003;278: 48004–48011. [DOI] [PubMed] [Google Scholar]
- 16.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res 1998;26: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem 1996;271:20009–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem 2001;276:30423–30428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickenig G, Baudler S, Muller C, Werner C, Werner N, Welzel H, Strehlow K, Bohm M. Redox-sensitive vascular smooth muscle cell proliferation is mediated by GKLF and Id3 in vitro and in vivo. FASEB J 2002;16:1077–1086. [DOI] [PubMed] [Google Scholar]
- 20.Siddique A, Malo MS, Ocuin LM, Hinnebusch BF, Abedrapo MA, Henderson JW, Zhang W, Mozumder M, Yang VW, Hodin RA. Convergence of the thyroid hormone and gut-enriched Kruppel-like factor pathways in the context of enterocyte differentiation. J Gastrointest Surg 2003;7:1053–1061. [DOI] [PubMed] [Google Scholar]
- 21.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 1997;390:465–471. [DOI] [PubMed] [Google Scholar]
- 22.ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci 2004;29:265–273. [DOI] [PubMed] [Google Scholar]
- 23.Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell 1998;95:737–740. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature 1996; 383:168–172. [DOI] [PubMed] [Google Scholar]
- 25.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massague J, Shi Y. Crystal structure of a phosphorylated Smad2: recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell 2001;8:1277–1289. [DOI] [PubMed] [Google Scholar]
- 26.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1998;1:611–617. [DOI] [PubMed] [Google Scholar]
- 27.Hu B, Tack DC, Liu T, Wu Z, Ullenbruch MR, Phan SH. Role of Smad3 in the regulation of rat telomerase reverse transcriptase by TGFbeta. Oncogene 2006;25:1030–1041. [DOI] [PubMed] [Google Scholar]
- 28.Rogler G, Gelbmann CM, Vogl D, Brunner M, Scholmerich J, Falk W, Andus T, Brand K. Differential activation of cytokine secretion in primary human colonic fibroblast/myofibroblast cultures. Scand J Gastroenterol 2001;36:389–398. [DOI] [PubMed] [Google Scholar]
- 29.Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development 2003;130:2767–2777. [DOI] [PubMed] [Google Scholar]
- 30.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson AJ, Kudlow JE, Lobo-Ruppert SM, Ruppert JM. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 2005;24:1491–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature 1998;394:92–96. [DOI] [PubMed] [Google Scholar]
- 32.Derynck, R., Y. Zhang and X. H. Feng. Smads: transcriptional activators of TGF-beta responses. Cell 1998;95:737–740. [DOI] [PubMed] [Google Scholar]
- 33.Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell 2005; 16:4214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furukawa F, Matsuzaki K, Mori S, Tahashi Y, Yoshida K, Sugano Y, Yamagata H, Matsushita M, Seki T, Inagaki Y, et al. p38 MAPK mediates fibrogenic signal through Smad3 phosphorylation in rat myofibroblasts. Hepatology 2003;38:879–889. [DOI] [PubMed] [Google Scholar]
- 35.Fujita Y, Maruyama S, Kogo H, Matsuo S, Fujimoto T. Caveolin-1 in mesangial cells suppresses MAP kinase activation and cell proliferation induced by bFGF and PDGF. Kidney Int 2004;66:1794–1804. [DOI] [PubMed] [Google Scholar]
- 36.Shi YH, Wang YX, You JF, Heng WJ, Zhong HH, Fang WG. Activation of HIF-1 by bFGF in breast cancer: role of PI-3K and MEK1/ERK pathways. Zhonghua Yi Xue Za Zhi 2004;84:1899–1903. [PubMed] [Google Scholar]
- 37.Liu T, Hu B, Chung MJ, Ullenbruch M, Jin H, Phan SH. Telomerase regulation of myofibroblast differentiation. Am J Respir Cell Mol Biol 2006;34:625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]