Abstract
Many organisms use circadian clocks to keep temporal order and anticipate daily environmental changes. In Drosophila, the master clock gene Clock promotes the transcription of several key target genes. Two of these gene products, PER and TIM, repress CLK–CYC-mediated transcription. To recognize additional direct CLK target genes, we designed a genome-wide approach and identified clockwork orange (cwo) as a new core clock component. cwo encodes a transcriptional repressor that synergizes with PER and inhibits CLK-mediated activation. Consistent with this function, the mRNA profiles of CLK direct target genes in cwo mutant flies manifest high trough values and low amplitude oscillations. Because behavioral rhythmicity fails to persist in constant darkness (DD) with little or no effect on average mRNA levels in flies lacking cwo, transcriptional oscillation amplitude appears to be linked to rhythmicity. Moreover, the mutant flies are long period, consistent with the late repression indicated by the RNA profiles. These findings suggest that CWO acts preferentially in the late night to help terminate CLK–CYC-mediated transcription of direct target genes including cwo itself. The presence of mammalian homologs with circadian expression features (Dec1 and Dec2) suggests that a similar feedback mechanism exists in mammalian clocks.
Keywords: Circadian, clk, Drosophila, transcriptional oscillations, chromatin
Most living organisms use circadian rhythms for maintaining internal temporal order and anticipating daily environmental changes (Panda et al. 2002). Clocks use self-sustained biochemical oscillators and are manifest at molecular, physiological, and behavioral levels (Dunlap 1999; Roenneberg and Merrow 2005). In higher eukaryotes, these rhythms in physiology and behavior rely on intracellular oscillations in abundance, post-translational modifications, and nuclear localization of several clock proteins, proposed to be generated by interconnected transcription and post-transcriptional feedback loops (Hall 2003).
In Drosophila, the genes Clock (Clk) and cycle (cyc) may sit at the top of a genetic hierarchy governing circadian rhythms (Zhao et al. 2003) and promote the rhythmic transcription of several key clock genes (Allada et al. 1998; Rutila et al. 1998b). The protein products of two of these target genes, PER (period) and TIM (timeless), repress CLK–CYC-mediated transcription toward the end of every cycle and thereby repress their own synthesis (Hardin et al. 1990; Sehgal et al. 1994, 1995). The comparable event in mammals involves cryptochromes (CRYs) as well as PERs as the major transcriptional repressors (Panda et al. 2002). A second transcriptional feedback loop in flies affects Clk mRNA cycling and involves vri (vrille) and Pdp1 (Par domain protein 1), two other direct targets of the CLK–CYC heterodimer (Cyran et al. 2003; Glossop et al. 2003).
The mechanism by which PER represses CLK-driven transcription is still uncertain. Recent reports indicate that cyclical CLK target gene expression may be the result of cyclical DNA binding, both in Drosophila and mammals (Ripperger and Schibler 2006; Yu et al. 2006). Moreover, recent evidence suggests that Drosophila PER may deliver the kinase DBT (doubletime) to the CLK–CYC dimer. CLK phosphorylation likely ensues, with the subsequent disassociation of CLK–CYC from target E-boxes (Lee et al. 1999; Yu et al. 2006). Although comparable biochemical detail is lacking for mammalian clocks, activation–repression cycles generate high-amplitude mRNA oscillations in both systems and are proposed to be important for behavioral oscillations. At least in the fly system, there is good genetic evidence that this is the case (Allada et al. 2003).
A recent report suggests an additional “active” repression mechanism, as important changes in chromatin structure escort circadian transcriptional oscillations in mammals and Neurospora (Ripperger and Schibler 2006; Belden et al. 2007). In mammals, these modifications appear to follow the CRY-PER repression events and may enhance the oscillation amplitude of various cycling mRNAs. It is likely that similar phenomena take place in the Drosophila system.
Although this view advances transcriptional oscillations as an important feature of circadian timekeeping, recent evidence strongly indicates that they might be dispensable in the cyanobacterial system (Tomita et al. 2005). Moreover, there are transgenic fly strains in which behavioral and molecular (protein) rhythmicity can persist despite the absence of per and tim transcriptional oscillations (Yang and Sehgal 2001). Consequently, the role of cyclic transcription in generating the molecular and behavioral rhythms characteristic of wild-type flies is still uncertain.
Nonetheless, it is curious that all known bona fide direct targets of the master gene Clk are involved in transcriptional regulation (per, vri, tim, Pdp1). We therefore reasoned that finding additional CLK direct targets might identify new biochemical pathways relevant to central clock function or perhaps reinforce the centrality of transcriptional regulation. We report here the isolation and characterization of a new core clock component: clockwork orange (cwo). cwo transcription is activated by CLK–CYC and repressed by PER–TIM. As a consequence, cwo mRNA oscillates with an amplitude and phase comparable with other CLK direct targets; for example, vri, Pdp1, per, and tim. cwo is prominently expressed in circadian brain neurons and cooperates with PER to repress CLK–CYC-mediated transcription. Characterization of flies deficient in cwo activity demonstrates that the protein is essential for robust oscillations of core clock mRNAs as well as persistent behavioral rhythms with wild-type periods. Because the oscillation amplitude of direct clock transcripts is specifically affected in cwo-deficient strains, the core transcriptional feedback loop appears essential for circadian rhythms in Drosophila. As cwo orthologs (Dec1 and Dec2) are possible pacemaker components in mammals (Honma et al. 2002), this view may also extend to other animal systems.
Results
Identification of direct CLK targets in flies
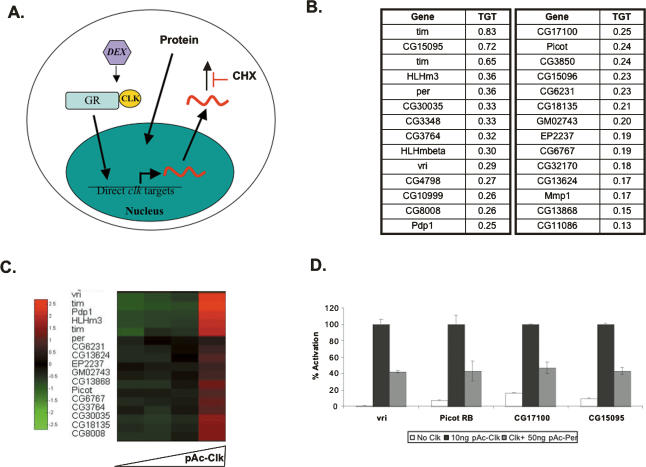
To find additional CLK direct targets, we extended to flies an approach previously used in Drosophila S2 cells (Fig. 1A; McDonald et al. 2001). Briefly, we generated a transgenic fly line expressing a CLK–glucocorticoid receptor fusion protein (CLKGR) and expressed it in clock neurons (tim-gal4; UAS-CLKGR fly strain). To identify direct CLK targets, fly heads were cultured and stimulated with dexamethasone in the presence of the protein synthesis inhibitor cycloheximide. A parallel experiment was carried out in S2 cells.
Figure 1.
Identification of direct CLK targets in Drosophila S2 cells and fly heads. (A) Diagram illustrating the approach for the identification of direct CLK targets from Drosophila S2 cells and fly heads. (Dex) Dexamethasone; (GR) ligand-binding domain of the glucocorticoid receptor; (CHX) cycloheximide. Head or cells were treated for 6 h with CHX + vehicle or CHX + Dex. Heads or cells were harvested, and RNA was extracted and hybridized to oligonucleotide microarrays. Both the new Affymetrix Drosophila 2.0 Chips and the first generation of chips for Drosophila (Affymetrix Drosophila Genome) were used. (B) Top 28 direct CLK targets identified by the approach described in A. (TGT) Targetness. This index was obtained by averaging the relative stimulation by dexamethasone from S2 cells and fly heads. For example, for tim, TGT = Average{(36.75/38.3), (5.47/7.65)} = 0.83, where 36.75 is the stimulation ratio obtained for the tim probe, 38.3 is the ratio for the most stimulated probe in fly heads, and 5.47 and 7.65 are the equivalent values from S2 cells. (C) CLK protein expression activates most of the direct CLK-GR targets. Transient transfections were performed with varying amounts of pAc-Clk plasmid in S2 cells (0, 10, 30, and 100 ng). After 48 h, cells were harvested and total RNA was isolated. Microarray analysis was performed as described in Materials and Methods using the Drosophila 2.0 genomic Affymetrix chips. A heat map was generated using a normalized value as indicated in Materials and Methods. (D) Effect of CLK and PER expression on vri-Luc (Blau and Young 1999), picot-Luc, CG15095-Luc, and CG17100-Luc reporters on S2 cells. pAc-Clk and pAc-per refers to CLK- and PER-expressing plasmids, respectively (Nawathean and Rosbash 2004). In all cases, cotransfection with pCopia-Renilla Luciferase was performed to normalize for cell number, transfection efficiency, and general effects on transcription. For each condition, a normalized firefly/Renilla Luciferase value was obtained by setting the ratio obtained with the addition of pActin-Clk to 100%. The graph shows the results of a representative experiment (in which duplicates for each condition were performed). Error bars indicate the standard error of the mean (SEM). The experiment was performed three times with similar results.
This approach identified candidate direct CLK targets, which we ranked according to targetness (TGT) as described in Materials and Methods. Among the 28 genes that passed a stringent cutoff criterion (Fig. 1B) were the four known direct targets as well as other genes described as cyclically expressed or affected in the Clk mutant Jrk (Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Ceriani et al. 2002; Lin et al. 2002; Ueda et al. 2002). However, 60% were not previously connected to Drosophila rhythms, and many of these were also activated by CLK in S2 cells (Fig. 1C). Since overexpression studies can reveal nonphysiological targets, we tested some of these candidates in an independent assay. To this end, we constructed luciferase reporter constructs from the promoters of three new targets and tested them in S2 cells with vri-luciferase as a positive control (Blau and Young 1999). All were strongly activated when cotransfected with a CLK-expressing plasmid (pAc-Clk) and repressed by PER cotransfection (Fig. 1D).
cwo is expressed cyclically and in pacemaker neurons
Among putative direct CLK targets was a gene encoding a transcription factor with an ORANGE domain, CG17100, which we call clockwork orange (cwo). (This gene was previously misannotated as stich1 [Salzberg et al. 1994; Dubruille et al. 2002].) cwo belongs to a family of transcriptional repressors (basic helix–loop–helix-O [bHLH-O]) involved in various aspects of cell physiology and metabolism (Davis and Turner 2001). Importantly, two close mouse relatives, dec1 and dec2, are circadianly expressed in the suprachiasmatic nucleus (SCN). However, there is no evidence that they contribute to the generation or maintenance of circadian rhythms (Honma et al. 2002). We previously reported that cwo mRNA oscillates in a circadian manner by microarray and RNase protection assays (McDonald and Rosbash 2001). We verified this observation with RT–PCR as well as with new 2.0 Drosophila Affymetrix microarrays. By both criteria, cwo mRNA cycles with a phase (peak around 14 h Zeitgeber time [ZT14]) (Fig. 2A; data not shown) that resembles those of per, tim, Pdp1, and vri mRNAs (Hardin et al. 1990; Sehgal et al. 1995; Blau and Young 1999; Cyran et al. 2003). These CLK direct targets contain numerous CLK–CYC-binding elements (E-boxes) in their promoters. E-boxes are necessary for transcriptional oscillations and have been shown in some cases to mediate CLK activation followed by PER repression (McDonald et al. 2001; Wang et al. 2001). There are six E-boxes within the promoter of cwo, 2 kb upstream of the transcriptional start site, and 15 E-boxes within the first intron (data not shown). Moreover, cwo mRNA was regulated in clock mutant strains like characterized direct target genes (Fig. 2B), namely, low and high mRNA levels in the Clk mutant Jrk and the per01 mutant, respectively (Allada et al. 1998; Rutila et al. 1998b; Blau and Young 1999).
Figure 2.
cwo gene expression is regulated in a circadian manner. (A) Expression profile for cwo and tim across six time points in wild-type (yw) flies. Data were obtained by Q-PCR. Expression values for each transcript and time point were generated by dividing the cwo or tim mRNA signal by the expression value for a control noncircadian mRNA (ribosomal protein 49, RP49). A value of 1 was assigned to the maximum expression across the six time points; the values in the other time points were calculated as a fraction of this maximum value. The data are the average of the normalized cwo or tim expression values for three independent RNA samples. The error bars indicate the SEM. (B) cwo mRNA levels in control (yw), ClkJrk, and per01 fly strains. Flies were entrained for 4 d in LD conditions and then transferred to DD. After 2 d in DD conditions, flies were harvested at CT3 and CT15. RNA from both time points for each strain was combined, and Q-PCR was performed using primers for cwo and RP49. Expression values are reported as a ratio of cwo/RP49 expression. We assigned a value of 1 to the ratio obtained for control flies and proceed as in A. The experiment was performed twice and the results were averaged. The error bars indicate the SEM. (C) cwo localization in the fly brain examined with GFP. p{cwo-gal4 (NP7442-gal4)}; UAS-GFP flies were stained with PDF antiserum. Strong overlap between the PDF staining and GFP expression can be observed in the region corresponding to the position of the pacemaker neurons (sLNv). An amplified picture of the overlap in the sLNv region is shown in the bottom right panels.
To examine cwo spatial expression, we crossed a UAS-GFP line with an enhancer trap fly line (NP7492; NP Consortium, Kyoto, Japan) containing a GAL4-coding sequence in the promoter region of cwo. Costaining with anti-PDF antisera (PDF is a neuropeptide specific for pacemaker cells) showed strong GFP expression in brain pacemaker neurons (Fig. 2C). Recent independent reports confirm this observation: A LacZ enhancer trap in this same gene is prominently expressed in clock cells—that is, in all PER-expressing cells (Shafer et al. 2006)—and comparable data with an anti-CWO antibody are presented in the accompanying paper by Matsumoto et al. (2007).
cwo is a new core clock component
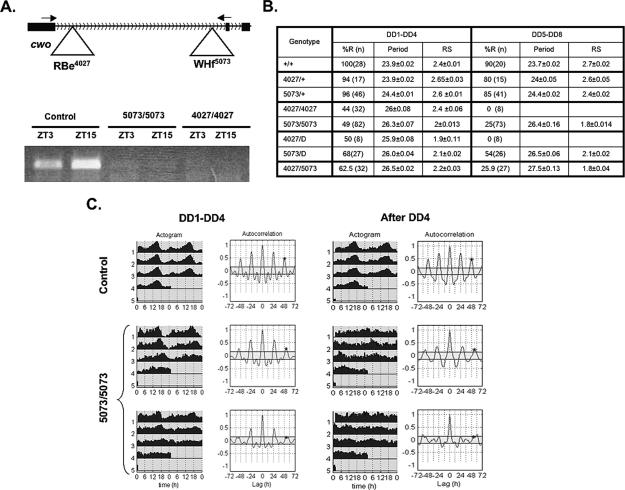
We next characterized two cwo-deficient fly strains (e4027 and f5073; Exelixis collection, Harvard University), which contain unique transposon insertions at the beginning and at the end of the first intron (Fig. 3A, top). RT–PCR shows that both insertions reduce cwo mature mRNA levels below the level of detection, probably as a consequence of deficient splicing (Fig. 3A, bottom). Moreover, no cwo protein is detectable in the 5073 strain as shown in Matsumoto et al. (2007).
Figure 3.
cwo is a new clock component. (A, top) Schematic of e4027 and f5073 insertions in the first intron of cwo. Arrows indicate the position of primer pairs for Q-PCR. (Bottom) Q-PCR from control (5073/+), 5073/5073, and 4027/4027 fly heads at ZT3 and ZT15 using the primer pair shown in the top part. (B) Behavioral analysis of fly strains deficient for cwo and control flies. “D” refers to a deletion that includes the cwo locus (see text). (C) Comparison of circadian locomotor behavior of control (5073/+; top panels) and cwo-deficient flies (5073/5073; bottom panels). In each case, the behavior is shown in average actograms (left) and autocorrelation analysis (right) (Levine et al. 2002). (DD1–DD4) The first 4 d in DD; (after DD4) the locomotor activity after 4 d in DD. For each interval of time (DD1–DD4 and DD5–DD8), 5073/5073 flies can be divided in two groups according to their behavioral rhythms (top for rhythmic flies and bottom for arrhythmic flies). The results illustrate a model experiment (n = 28 for control flies and n = 24 for 5073 flies).
These strains have strong circadian locomotor activity phenotypes (Fig. 3B,C; Supplementary Fig. 1). During the first 4 d in free running conditions (constant darkness, DD), more than half of these flies were arrhythmic (56% and 51% for strains 4027 and 5073, respectively); the remaining flies had weaker and longer rhythms than control strains, which have nearly 0 arrhythmic flies. After 4 d in DD, most mutant flies were arrhythmic (75% or 100% for 5073 and 4027, respectively) (Fig. 3B,C; Supplementary Fig. 1). An indistinguishable phenotype, long and weak rhythms, was obtained when either of the two chromosomes was heterozygous over a deficiency that includes the cwo gene, and an identical phenotype was observed with the trans-heterozygous strain; that is, 5073/4027 (Fig. 3B; Supplementary Fig. 1). Moreover, expression of a cwo RNA interference (RNAi) construct in circadian neurons produces a similar phenotype (Matsumoto et al. 2007).
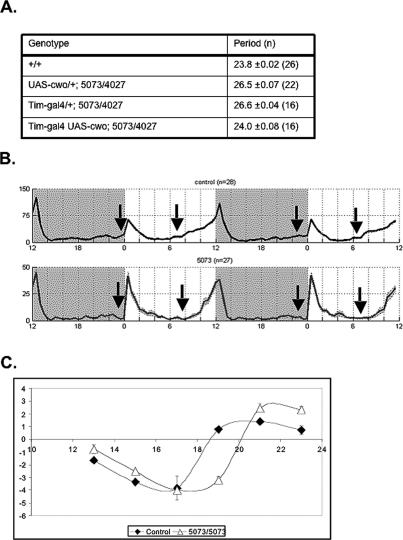
The long period phenotype of the cwo-deficient genotypes is a consequence of lack of cwo expression in circadian cells, since expression of a UAS-cwo transgene with a timeless driver restores the circadian period to 24 h in the trans-heterozygous mutant background (Fig. 4A). A slow clock is also apparent under light–dark (LD) conditions in 5073 flies, as there is a clear delay in morning anticipation (Fig. 4B). Moreover, 5073 flies also have an aberrant phase-response curve (PRC) (Fig. 4C), when subjected to a light pulse during the last night of LD entrainment. The mutant PRC is similar in the early night but then lags behind the wild-type PRC, which we interpret to indicate a slow clock even in these LD conditions (Fig. 4B,C).
Figure 4.
cwo is responsible for the slow clock in LD and DD conditions. (A) Expression of cwo in the circadian neurons can rescue the long rhythms present in cwo-deficient flies. (B) Comparison of the circadian locomotor behavior of control (5073/+; top panels) and cwo-deficient flies (5073/5073; bottom panels) in LD conditions. Two standard days are shown, with timing indicated by alternating white and gray background areas, with white representing the illuminated interval of LD (ZT0–ZT12), and gray representing the dark period (ZT12–ZT24). The arrows indicate the phase of morning or evening anticipation for each fly strain. (Control) 5073/+ flies. (C) PRC for control and 5073 flies. The time onset of the photic stimuli was plotted on the X-axis (ZT, in hours). The phase response was plotted on the Y-axis as the difference (in hours) from the phase of untreated flies. The experiment was repeated twice, and the error bars represent the SEM.
An altered clock is also apparent from the 5073 six-time-point microarray data, which show a striking decrease in mRNA oscillation amplitude for some clock components (per, tim, and vri) (Fig. 5A,B). Normalization of the data facilitates visualizing the delayed repression of these mRNAs in the 5073 strain (Fig. 5C). The mRNA trough is two- to threefold elevated for direct CLK target genes in the cwo mutant strain (Fig. 5D), which fits with “incomplete” repression, even in LD conditions when behavioral rhythms are robust. However, lower peak levels also contribute to the low amplitude of the cwo-deficient strain. This effect on oscillation amplitude is accompanied by little or no effect on average mRNA levels in the 5073 strain (Fig. 5E). Similar mRNA profiles for tim, vri, and per mRNAs were observed using RT–PCR (Supplementary Fig. 2; data not shown). It is intriguing that an oscillation amplitude effect was less apparent on other cycling genes, including core oscillator genes that are not direct CLK targets; for example, Clk and cry (Fig. 5A; data not shown). All of these data indicate that cwo has central pacemaker component characteristics. Similar data and a parallel conclusion are presented in Matsumoto et al. (2007).
Figure 5.
mRNA oscillation of the direct CLK targets is selectively affected in cwo-deficient flies. (A) Amplitude of oscillation for the core clock genes in 5073 flies. The data were obtained by Affymetrix oligonucleotide microarray. The maximum and minimum values were selected, and the amplitude of oscillation was defined as the ratio of these two values (N = 3 for ZT3 and N = 2 for ZT15). The amplitude is expressed as the percentage of control (yw). Error bars indicate the SEM. (B) Profile for tim, Pdp1, vri, and per mRNAs. Results were obtained by microarrays. (C) Normalized profile for vri and tim mRNA. Results were obtained by microarray, and the value for each time point was normalized to the maximum value across the six time points. (D) Comparison of trough values for tim, vri, and per for control and 5073 flies as measured by oligonucleotide microarray (N = 3). Error bars indicate the SEM. (E) Average mRNA levels for clock-relevant mRNAs. Data were obtained by oligonucleotide microarrays. The six time-point values were averaged for control and cwo-deficient flies and expressed as a percentage of control expression.
cwo cooperates with PER in repressing CLK-mediated transcription
Is the function of CWO to repress CLK direct target gene transcription, as its bHLH-O motif and the mutant data suggest? To this end, we used direct target promoter-luciferase reporters and transfected them into Drosophila S2 cells with a cwo expression vector (pAc-cwo). Even without CLK coexpression, cwo expression reduced basal transcription from the vri, tim, cwo, and CG15095 promoters but had no effect on the actin promoter (Fig. 6A). This is consistent with the notion that cwo encodes a transcriptional repressor.
Figure 6.
CWO binds to E-boxes and functionally cooperates with PER in inhibiting CLK–CYC-mediated transcription. (A) Effect of cwo expression (150 ng of cwo-expressing plasmid, pAc-cwo) on the transcription of pActin-Luc, vri-Luc, tim-Luc, CG15095-Luc, and cwo-Luc. Expression was measured and normalized as indicated for Figure 1D. A representative experiment is shown. Two experiments with duplicates for each condition were performed with similar results. Error bars represent the SEM. (B) Comparison of CYC and CWO amino acid composition in the bHLH domain. (Red) Zones with identical amino acid composition; (green) areas with similar charge amino acids. (C) Effect of cwoVP16 overexpression (0, 25, or 100 ng of pAc-cwoVP16) on the transcription driven by tim-Luc and timΔTer1ΔTer2m-Luc (McDonald et al. 2001). A representative experiment is shown. Two experiments with duplicates for each condition were performed with similar results. Error bars represent the SEM. (D) Effects of PER and CWO expression on CLK-mediated expression. “Clk, No Per” indicates that 10 ng of pAc-Clk was cotransfected with 0, 100, or 200 ng of pAc-cwo and the correspondent reporters. “Clk + Per” indicates that the transfection was performed as before, except 50 ng of pAc-per were also cotransfected. In both cases, the expression was obtained after normalization to the control Copia-Renilla Luciferase value. A value of 1 was assigned to the maximum value in this condition. A representative experiment is shown. Three experiments with duplicates for each condition were performed with similar results. Error bars represent the SEM. (E) PER repression of CLK-mediated transcription in the presence (100 ng of pAc-cwo) or absence of CWO. Expression was calculated and expressed as in Figure 1D. A representative experiment is shown. Two experiments with duplicates for each condition were performed with similar results.
Does cwo interact directly with CLK target genes? Close inspection of the cwo bHLH region reveals striking similarity with the CYC bHLH domain, suggesting that cwo could recognize clock gene E-boxes like the CLK partner CYC (Fig. 6B). To assess this possibility, we first added the VP16 domain to the C terminus of cwo (Wysocka and Herr 2003). cwo-VP16 is a potent transcriptional activator of CLK direct target genes, as it increased transcription of the vri-Luc and tim-luc reporters by 60- and 150-fold, respectively (Fig. 6C, left panel; Supplementary Fig. 3B). Deletion of the three E-boxes in the tim promoter (McDonald et al. 2001) prevented activation (Fig. 6C, right panels), and similar data were obtained with vri-Ebox and vri-Ebox mutant promoters (Supplementary Fig. 3C; Blau and Young 1999). Addition of cyc double-stranded RNA (dsRNA) had no effect (Supplementary Fig. 3D), indicating that cwo-VP16 activity does not require CYC, and ChIP data from S2 cells indicate direct binding of cwo to target gene E-boxes (Matsumoto et al. 2007; data not shown). All of these considerations indicate that cwo functions by interacting directly with promoter E-boxes of CLK direct target genes.
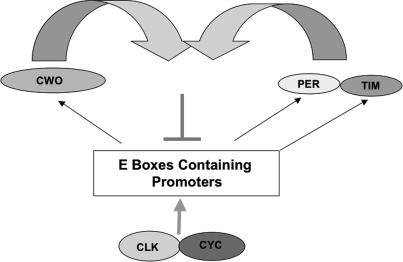
However, cwo overexpression only mildly repressed CLK-mediated transcription in S2 cells (Fig. 6D, left). This is probably not a consequence of CLK levels, since CWO expression has the same effect with lower amounts of pAc-Clk (Supplementary Fig. 3A). Based in part on the similar mRNA profiles of cwo (Fig. 2A) and per, we assayed CWO-mediated repression in the presence of PER. Indeed, CWO now repressed CLK-mediated transcription fivefold to 10-fold (Fig. 6D, right; data not shown). In a reciprocal manner, expression of a fixed amount of cwo plasmid strongly enhanced PER-mediated repression (Fig. 6E). For example, low doses of pAc-per (e.g., 25 ng) are ineffective in reducing CLK-mediated expression without CWO coexpression. The two complementary experiments demonstrate that CWO and PER can cooperate to repress CLK-mediated transcription, and we suggest that they work through different mechanisms (Fig. 7).
Figure 7.
Proposed role of CWO in the circadian molecular circuit.
Discussion
We have identified cwo as a new core clock component in Drosophila. It encodes a transcriptional repressor, which synergizes with PER and inhibits CLK-mediated activation. Consistent with this function, the mRNA profiles of CLK direct target genes manifest high trough values and low amplitude oscillations in mutant flies. Because rhythmicity fails to persist in DD and there is little or no effect on average mRNA levels in the 5073 strain, transcriptional oscillation amplitude appears linked to rhythmicity. Moreover, the mutant flies are long period, consistent with the late repression indicated by the RNA profiles. These findings suggest that CWO acts preferentially in the late night to help terminate CLK–CYC-mediated transcription of direct target genes including cwo itself. The presence of cwo homologs (Dec1 and Dec2) in mammals suggests that a similar feedback mechanism exists in mammals.
cwo is a new core clock component
We used a genome-wide approach to identify candidate CLK targets from fly heads (Fig. 1A). Intriguingly, a significant fraction of these genes are nonoscillating. Because the S2 cell assays predict that most of these genes are probably bona fide CLK targets, they may reflect a noncircadian role of CLK (Fig. 1C,D). Accordingly, a recent study reported that CLK expression is not restricted to circadian neurons in the fly brain (Houl et al. 2006). In contrast, cwo mRNA cycles and is expressed in circadian neurons (Fig. 2A,C; Matsumoto et al. 2007). Moreover, the cwo mRNA profile is similar to that of the other core clock components, as the gene is activated by CLK–CYC and repressed by PER (Figs. 1D, 2A,B). We suggest that cyclical transcription of cwo probably contributes to circadian changes in the level of CWO similar to other direct CLK–CYC targets.
The two cwo insertion strains have no detectable cwo mRNA. Both also have long periods, which fail to persist after 4–5 d in DD. The penetrance of these two phenotypes, however, is not identical: 100% of flies have long periods, whereas 50%–75% are arrhythmic after 4 d in DD (Fig. 3B). This suggests that these two phenomena are separable and that the long periods are not due to the weak rhythms. The circadian phenotype is slightly more severe in the 4027 strain but is accompanied by a high mortality of flies (data not shown). In contrast, 5073 homozygous flies and 5073/4027 trans-heterozygous flies show no life-span effect (data not shown) and have comparable phenotypes; that is, long rhythms and 75% arrhythmic flies after 4 d in DD (Fig. 3B,C; Supplementary Fig. 1). This indicates that both rhythm features are determined by the absence of cwo expression.
The slow clock is not only manifest by a period phenotype in DD but also by a late activity phase in LD. More specifically, 5073/5073 flies have delayed anticipation of the lights-on transition (Fig. 4B). This is consistent with cwo acting in the pdf-expressing neurons, since these cells are both responsible for the morning anticipation in LD and period determination in DD (Grima et al. 2004; Stoleru et al. 2004, 2005). We therefore prefer the parsimonious interpretation that the delayed phase is caused by a slow central molecular oscillator rather than an output defect. More support for this hypothesis comes from the delayed mRNA profiles as well as the delayed PRC. Although aberrant PRCs often reflect defects in light perception, we have previously suggested that they can also reflect a fast or slow central oscillator (Rutila et al. 1998a). In this view, the wider PRC delay zone reflects a slower clock and in particular the broader transcriptional peak (Fig. 4C). Taken together with the 5073 mRNA curves, CWO may preferentially function to repress transcription at the end of each cycle (see below). In contrast, the more potent advance zone of the 5073 PRC may reflect an underlying weaker circadian oscillator (Vitaterna et al. 2006).
Expression of a cwo transgene in tim-expressing cells restored a 24-h period to the mutant genotype (Fig. 4A). In contrast, cwo overexpression using the pdf-gal4 as well as the tim-gal4 driver had no effect on the period of an otherwise wild-type strain (data not shown). This adds to the evidence that the long period phenotype is due to the absence of functional cwo. Although the rescue also improves the rhythm strength of the cwo-deficient host strain, it is not as strong as that of wild-type flies (data not shown). Moreover, cwo overexpression combined with heterozygosity for the 5073 or 4027 chromosomes also gives rise to weak rhythms (data not shown). We suspect that rhythm strength is sensitive to the levels and timing of cwo expression. We note that the UAS transgene lacks the cwo 5′ and 3′ untranslated (UTR) regions, which are unusually long (2.6 and 1.5 kb, respectively) and probably contribute to post-transcriptional regulation of CWO expression.
Model for cwo function
In the current model, high-amplitude oscillations of tim, per, vri, and Pdp1 mRNAs levels are due to cyclical activation and repression of the CLK–CYC heterodimer. Recent reports from mammals suggest that there is a daily change in chromatin structure, which parallels the CLK–BMAL (CLK–CYC equivalent) activation cycle. Moreover, circadian chromatin remodeling has recently been reported in the Neurospora system (Belden et al. 2007). How these changes are generated and/or linked to the activation–repression cycle is not known.
However, based on the link between bHLH-O proteins and histone deacetylase recruitment (Davis and Turner 2001), we suggest that CWO helps build a repressive chromatin structure during the end of a cycle not unlike the one observed at mammalian circadian promoters (Ripperger and Schibler 2006). This explains the higher trough values as well as the long period and delayed mRNA decline in the 5073 strain (Fig. 5C,D). PER probably recruits the kinase doubletime (DBT) to the CLK–CYC dimer, resulting in diminished CLK–CYC affinity for DNA (Lee et al. 1999; Yu et al. 2006); this should favor CWO binding to E-boxes and corepressor recruitment. Similarly, CWO activity may aid CLK–CYC inactivation by PER–DBT, as observed in the S2 cells experiments (Fig. 6F). As closed chromatin structures are necessary for full activation by several transcription factors (Beato and Eisfeld 1997), this may also help explain the lower mRNA peak of most CLK direct targets in the 5073 strain (Fig. 5B). This lower mRNA peak could also be an indirect or “system” effect, as peak direct CLK-target mRNA levels in this strain are comparable (∼60%) with the levels observed in other repressor mutant strains, namely, per01 and tim01.
Role of transcriptional oscillations in generating circadian rhythms
Although mRNA and transcriptional oscillations were proposed long ago to be essential for circadian clock function, recent evidence strongly indicates that they are dispensable in cyanobacteria (Tomita et al. 2005). Consistent with this notion, there is evidence in the fly system that some rhythmicity persists without per and tim transcriptional cycling (Yang and Sehgal 2001). Importantly, behavioral rhythms and probably other CLK direct target genes still undergo oscillations in these strains. We suggest that the CWO feedback system contributes to this residual rhythmicity.
As shown above, the absence of CWO has two effects on the mRNA profiles of tim, per, and vri: late repression and low-amplitude oscillations. We propose that the former is responsible for the phase change in LD and period change in DD, whereas the latter causes the weak rhythmicity phenotype. Because mRNA oscillation amplitude is affected with little or no effect on average mRNA levels in the 5073 strain (Fig. 5E), transcriptional regulation appears essential for persistent DD rhythms, which fail after several days in the cwo mutant genotypes. In this view, we interpret the weak mRNA amplitude to be the cause of the weak rhythms. This adds to the evidence supporting the direct involvement of transcriptional oscillations in the timekeeping process (Allada et al. 2003). We note that we cannot rule out the possibility that the weak rhythmicity is a consequence of an additional role of cwo in the output pathway. Although there are no comparable genetic results in mammalian systems, the similar expression profiles as well as the conservation of Dec1 and Dec2 with cwo suggest that a comparable feedback mechanism with behavioral effects exists in mammals.
Materials and methods
Plasmids
pAc-clk, pAc-per, Copia-Renilla luciferase, tim-Luciferase, timΔ1Δ2m-Luciferase, vri-Luciferase, vriEbox-Luciferase, and vrimutEbox-Luciferase have been previously described (Blau and Young 1999; McDonald et al. 2001). pAc-cwo was constructed by amplifying the coding region of cwo by PCR and ligating in-frame into pAcB V5/His6 (Invitrogen). The construction of reporters CG15095-Luciferase, picot-Luciferase, and CG15095-Luciferase was done in two steps. First, a PCR fragment containing the Luciferase gene was amplified from the tim-Luciferase construct (McDonald et al. 2001) and ligated into pBS-KS (Stratagene). The corresponding promoters were added upstream of the Luciferase gene by ligation of the appropriate PCR fragments. pAc-Luciferase was constructed by ligation of the PCR product containing the Luciferase gene and an artificial poly-linker sequence into pAcB V5/His6 (Invitrogen). pAc-cwoVP16 was constructed by ligation of the PCR product containing cwo in-frame into pAc-VP16 (a kind gift from Rebecca Schoer, Columbia University, New York).
S2 cell transfection
S2 cells were maintained in 10% fetal bovine serum (FBS) (Invitrogen) insect tissue culture medium (HyClone). Cells were seeded in a six-well plate. Transfection was performed at 70%–90% confluence according to the manufacturer’s recommendations (12 μL of cellfectin [Invitrogen] and 2 μg of total DNA). In all experiments, 50 ng of pCopia-Renilla Luciferase plus 50 ng of the Luciferase firefly reporter were used. pBS-KS+ (Stratagene) was used to bring the total amount of DNA to 2 μg.
Luciferase activity assay
Forty-eight hours after transfection, cells were assayed using the Dual Luciferase Assay Kit (Promega) following the manufacturer’s instructions.
dsRNA synthesis and RNAi treatment
For both procedures, we followed the RNAi protocol in S2 cells previously described (Nawathean et al. 2005). Two different dsRNAs were synthesized for each gene.
Analysis of gene expression by RT–PCR
Total RNA was prepared from adults from the indicated strains using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA derived from this RNA (using Invitrogen SuperScript II) was used as template for quantitative real-time PCR performed with the Corbett Research Rotor-Gene 3000 real-time cycler. The PCR mixture contained Platinum Taq polymerase (Invitrogen), optimized concentrations of SYBR-green, and the corresponding primers: tim, 5′-CCTTTTCGTA CACAGATGCC-3′ and 5′-GGTCCGTCTGGTGATCCCAG 3′; vri, 5′-GCGCTCGCGATAAGTCTCTA-3′ and 5′-CTTT GTTGTGGCTGTTGGTG-3′; RP49, 5′-ATCCGCCCAGCA TACAG-3′ and 5′-TCCGACCAGGTTACAAGAA-3′; cwo, 5′-GTCTGTGGATCGAGGAGCAG-3′ and 5′-GGCATSTTCAG CACTGTCCT-3′.
Cycling parameters were 3 min at 95°C, followed by 40 cycles of 30 sec at 95°C, 45 sec at 55°C, and 45 sec at 72°C. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer. mRNA levels were quantified using a calibration curve based on dilution of concentrated cDNA. mRNA values from heads were normalized to that from ribosomal protein 49 (RP49).
Microarrays
Probe preparation
Total RNA was extracted from S2 cells or fly heads, using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. For heads, cDNA synthesis was carried out as described in the Expression Analysis Technical Manual (Affymetrix). The cRNA reactions were carried out using the BioArray High-Yield Transcript Labeling Kit (Enzo) or IVT (Affymetrix) depending on whether the oligonucleotide microarrays to be used were first or second generation, respectively. Affymetrix high-density arrays for Drosophila melanogaster were probed, hybridized, stained, and washed according to the manufacturer’s protocol.
Data analysis
GeneChip.CEL files were analyzed using R (http://www.r-project.org) and the bioconductor package (gcrma algorithm; http://www.bioconductor.org). Anti-logarithm (base 2) was applied to the data to obtain the expression values. These data have been deposited at NCBI GEO as series GSE7644, GSE7646, GSE7651, GSE7652, and GSE7653. Heat maps were done normalizing the gene expression value to the maximum for each gene across the different conditions in the following way: Normalized value = (Value − Maximum)/Maximum.
Determination of direct CLK targets
Experiment layout
For the experiments in S2 cells, duplicate experiments using Affymetrix Drosophila Genome oligonucleotide microarrays and Drosophila Genome 2.0 oligonucleotide microarrays were performed. In all cases, we performed two conditions: cycloheximide only (CHX) or CHX plus dexamethasone (Dex). In the case of microarrays from cultured fly heads, we performed two sets of Drosophila Genome microarrays (stimulation with Dex or vehicle in presence of CHX at ZT16) and four sets of Drosophila Genome 2.0 microarrays (stimulation with Dex or vehicle in presence of CHX at ZT4 and ZT16).
Head culture
Heads were dissected and cultured in the 12:12 LD condition as described before (Levine et al. 2002). After 3 d of LD entrainment, CHX (0.1 mg/mL) and either dexamethasone (15 μM) or vehicle was added for 6 h.
Data analysis
The samples were obtained and processed as described above. Expression values were obtained as described above. For each probe set, fold changes between the control and Dex plus samples were calculated as follows: ratio = Min(Dex)/Max(No Dex), where Min(Dex) is the minimum value for each probe across the samples with Dex, and Max(No Dex) is the maximum value for each probe set across the control samples. This calculation was performed on data obtained from Affymetrix first- and second-generation microarrays from S2 cells and fly heads. The direct CLK targets from S2 cells and heads were determined separately. Those probes that pass one or more of the following criteria were selected as direct-CLK targets in S2 cells: (1) The ratio on both sets of chips (first and second generation) is >1.5. (2) The ratio on one of the sets is >2.
With these criteria, we found 46 probes (corresponding to 43 genes). To obtain the CLK targets from fly heads, we chose the probe sets that passed one or more of the following criteria: (1) The ratio is >1.5 for first-generation (one time point) and second-generation (both time points) oligonucleotide microarrays. (2) The ratio is >3 for one of the three sets of chips (one first generation or one of the time points from the second generation). Using these criteria, we found 73 probes sets (corresponding to 72 genes). To obtain the final targets, we selected probe sets that passed at least one of the following criteria: (1) They are on both the S2 and fly head lists. (2) They are in the S2 cell list and the ratio from fly head microarrays is >1.2, or they are in the fly head list and the ratio from the S2 cell microarray is >1.2. The Per gene is not expressed in S2 cells upon CLK addition. However, we decided to included it in the final list, since it is a known CLK direct target and the second most-induced gene upon addition of dexamethasone from fly heads (22-fold). For ranking the obtained targets, we built an index: Targetness (TGT) = Average{[(ratioHeads)/Max ratio Heads], [(ratioCell)/Max ratio Cell]}.
Fly strains
The insertion lines f5073 and e4027 were obtained from the Exelixis Consortium, Harvard University. The insertion line NP7492 was obtained from the NP Consortium, Kyoto, Japan.
Construction of UAS-cwo transgenic lines
UAS-cwo plasmids were generated by cloning a PCR fragment from pAc-cwo into pUAST (Brand and Perrimon 1993). This construct was used to generate germline transformants by injecting yw; Ki pp P[ry+Δ2–3]/+.
PRC
Flies were entrained to a 12:12 LD cycle for 4 d. During the fifth dark phase of the cycle, flies were given a 10-min saturating white light pulse (2000 lux) at 13, 15, 17, 19, 21, and 23 h after the last light-on event. A separate control group of flies was not given a light pulse. Flies were then put into DD. The phase of the locomotor activity peaks after the light pulse was determined and compared with the no-light-pulse control.
Locomotor behavior
Male flies were monitored for 4 d in (LD) conditions, followed by 4–5 d in DD using Trikinetics Drosophila Activity Monitors. Analyses were performed with a signal-processing toolbox (Levine et al. 2002). We used autocorrelation and spectral analysis to estimate behavioral cycle durations (periods) and the Rhythm Index to assess rhythm strength (Levine et al. 2002).
In situ hybridization and GFP detection
In situ hybridization and GFP detection were performed as described previously (Zhao et al. 2003).
Acknowledgments
We thank R. Allada for sharing unpublished results, critically reading the manuscript, suggesting the name (cwo) for CG17100, and exchanging a manuscript with similar conclusions. The accompanying manuscript from Ueda and colleagues (Matsumoto et al. 2007) also identifies cwo as a clock component with an essentially identical conclusion. In addition, we thank P. Hardin, C. Green, D.Tardiff, and E. Nagoshi for comments on the manuscript; A. Villella for assistance with confocal microscopy; and K. Palm and C. Tang for help with the PRC experiments. We also thank Heather Felton for administrative assistance. S.K. is a recipient of a HFSP post-doctoral fellowship. This work was supported in part by NIH grants NS44232, NS45713, and GM66778 to M.R.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1552607
References
- Allada R., White N.E., So W.V., Hall J.C., Rosbash M., White N.E., So W.V., Hall J.C., Rosbash M., So W.V., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Allada R., Kadener S., Nandakumar N., Rosbash M., Kadener S., Nandakumar N., Rosbash M., Nandakumar N., Rosbash M., Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–3375. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M., Eisfeld K., Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W.J., Loros J.J., Dunlap J.C., Loros J.J., Dunlap J.C., Dunlap J.C. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Blau J., Young M.W., Young M.W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Ceriani M.F., Hogenesch J.B., Yanovsky M., Panda S., Straume M., Kay S.A., Hogenesch J.B., Yanovsky M., Panda S., Straume M., Kay S.A., Yanovsky M., Panda S., Straume M., Kay S.A., Panda S., Straume M., Kay S.A., Straume M., Kay S.A., Kay S.A. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M.W., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M.W., Naef F., Boothroyd C., Rajewsky N., Young M.W., Boothroyd C., Rajewsky N., Young M.W., Rajewsky N., Young M.W., Young M.W. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Cyran S., Buchsbaum A., Reddy K., Lin M., Glossop N., Hardin P., Young M., Storti R., Blau J., Buchsbaum A., Reddy K., Lin M., Glossop N., Hardin P., Young M., Storti R., Blau J., Reddy K., Lin M., Glossop N., Hardin P., Young M., Storti R., Blau J., Lin M., Glossop N., Hardin P., Young M., Storti R., Blau J., Glossop N., Hardin P., Young M., Storti R., Blau J., Hardin P., Young M., Storti R., Blau J., Young M., Storti R., Blau J., Storti R., Blau J., Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Davis R.L., Turner D.L., Turner D.L. Vertebrate hairy and Enhancer of split related proteins: Transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Dubruille R., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B., Swoboda P., Couble P., Kernan M., Durand B., Couble P., Kernan M., Durand B., Kernan M., Durand B., Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Houl J.H., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Houl J.H., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Ng F.S., Dudek S.M., Hardin P.E., Dudek S.M., Hardin P.E., Hardin P.E. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Grima B., Chelot E., Xia R., Rouyer F., Chelot E., Xia R., Rouyer F., Xia R., Rouyer F., Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hall J. Adv. Genet. Vol. 48. 2003. Genetics and molecular biology of rhythms in Drosophila and other insects; pp. 1–280. [DOI] [PubMed] [Google Scholar]
- Hardin P.E., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K., Sato F., Noshiro M., Kato Y., Honma K., Noshiro M., Kato Y., Honma K., Kato Y., Honma K., Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Houl J.H., Yu W., Dudek S.M., Hardin P.E., Yu W., Dudek S.M., Hardin P.E., Dudek S.M., Hardin P.E., Hardin P.E. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J. Biol. Rhythms. 2006;21:93–103. doi: 10.1177/0748730405283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I., Bae K., Edery I., Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK–CYC/ dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol. Cell. Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J., Funes P., Dowse H., Hall J., Funes P., Dowse H., Hall J., Dowse H., Hall J., Hall J. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002 doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Han M., Shimada B., Wang L., Gibler T.M., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Han M., Shimada B., Wang L., Gibler T.M., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Shimada B., Wang L., Gibler T.M., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Wang L., Gibler T.M., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Gibler T.M., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Amarakone A., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Awad T.A., Stormo G.D., Van Gelder R.N., Taghert P.H., Stormo G.D., Van Gelder R.N., Taghert P.H., Van Gelder R.N., Taghert P.H., Taghert P.H. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Ukai-Tadenuma M., Yamada R.G., Houl J., Uno K.D., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Ukai-Tadenuma M., Yamada R.G., Houl J., Uno K.D., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Yamada R.G., Houl J., Uno K.D., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Houl J., Uno K.D., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Uno K.D., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Kasukawa T., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Dauwalder B., Ito T.Q., Takahashi K., Ueda R., Ito T.Q., Takahashi K., Ueda R., Takahashi K., Ueda R., Ueda R., et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator Drosophila circadian clock. Genes & Dev. 2007 doi: 10.1101/gad.1552207. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash M., Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash M., Emery P., Rosbash M., Emery P., Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol. Cell. Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P., Rosbash M., Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nawathean P., Menet J.S., Rosbash M., Menet J.S., Rosbash M., Rosbash M. Assaying the Drosophila negative feedback loop with RNA interference in S2 cells. Methods Enzymol. 2005;393:610–622. doi: 10.1016/S0076-6879(05)93032-2. [DOI] [PubMed] [Google Scholar]
- Panda S., Hogenesch J., Kay S., Hogenesch J., Kay S., Kay S. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A., Schibler U., Schibler U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Roenneberg T., Merrow M., Merrow M. Timeline: Circadian clocks—The fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 2005;6:965–971. doi: 10.1038/nrm1766. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Maltseva O., Rosbash M., Maltseva O., Rosbash M., Rosbash M. The timSL mutant affects a restricted portion of the Drosophila melanogaster circadian cycle. J. Biol. Rhythms. 1998a;13:380–392. doi: 10.1177/074873098129000200. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri V., Le M., So W.V., Rosbash M., Hall J.C., Suri V., Le M., So W.V., Rosbash M., Hall J.C., Le M., So W.V., Rosbash M., Hall J.C., So W.V., Rosbash M., Hall J.C., Rosbash M., Hall J.C., Hall J.C. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998b;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Salzberg A., D’Evelyn D., Schulze K.L., Lee J.K., Strumpf D., Tsai L., Bellen H.J., D’Evelyn D., Schulze K.L., Lee J.K., Strumpf D., Tsai L., Bellen H.J., Schulze K.L., Lee J.K., Strumpf D., Tsai L., Bellen H.J., Lee J.K., Strumpf D., Tsai L., Bellen H.J., Strumpf D., Tsai L., Bellen H.J., Tsai L., Bellen H.J., Bellen H.J. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Price J., Man B., Young M., Price J., Man B., Young M., Man B., Young M., Young M. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M., Young M.W., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M., Young M.W., Hunter-Ensor M., Chen Y., Myers M., Young M.W., Chen Y., Myers M., Young M.W., Myers M., Young M.W., Young M.W. Circadian oscillations and autoregulation of timeless RNA. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- Shafer O.T., Helfrich-Forster C., Renn S.C., Taghert P.H., Helfrich-Forster C., Renn S.C., Taghert P.H., Renn S.C., Taghert P.H., Taghert P.H. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J. Comp. Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M., Peng Y., Agosto J., Rosbash M., Agosto J., Rosbash M., Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Nawathean P., Rosbash M., Peng Y., Nawathean P., Rosbash M., Nawathean P., Rosbash M., Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Tomita J., Nakajima M., Kondo T., Iwasaki H., Nakajima M., Kondo T., Iwasaki H., Kondo T., Iwasaki H., Iwasaki H. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Matsumoto A., Kawamura M., Iino M., Tanimura T., Hashimoto S., Matsumoto A., Kawamura M., Iino M., Tanimura T., Hashimoto S., Kawamura M., Iino M., Tanimura T., Hashimoto S., Iino M., Tanimura T., Hashimoto S., Tanimura T., Hashimoto S., Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H., Ko C.H., Chang A.M., Buhr E.D., Fruechte E.M., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Ko C.H., Chang A.M., Buhr E.D., Fruechte E.M., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Chang A.M., Buhr E.D., Fruechte E.M., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Buhr E.D., Fruechte E.M., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Fruechte E.M., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Schook A., Antoch M.P., Turek F.W., Takahashi J.S., Antoch M.P., Turek F.W., Takahashi J.S., Turek F.W., Takahashi J.S., Takahashi J.S. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.K., Ousley A., Darlington T.K., Chen D., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A., Ousley A., Darlington T.K., Chen D., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A., Darlington T.K., Chen D., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A., Chen D., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A., Fu W., Hickman L.J., Kay S.A., Sehgal A., Hickman L.J., Kay S.A., Sehgal A., Kay S.A., Sehgal A., Sehgal A. Regulation of the cycling of timeless (tim) RNA. J. Neurobiol. 2001;47:161–175. doi: 10.1002/neu.1024. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Herr W., Herr W. The herpes simplex virus VP16-induced complex: The makings of a regulatory switch. Trends Biochem. Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Yang Z., Sehgal A., Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yu W., Zheng H., Houl J.H., Dauwalder B., Hardin P.E., Zheng H., Houl J.H., Dauwalder B., Hardin P.E., Houl J.H., Dauwalder B., Hardin P.E., Dauwalder B., Hardin P.E., Hardin P.E. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes & Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kilman V.L., Keegan K.P., Peng Y., Emery P., Rosbash M., Allada R., Kilman V.L., Keegan K.P., Peng Y., Emery P., Rosbash M., Allada R., Keegan K.P., Peng Y., Emery P., Rosbash M., Allada R., Peng Y., Emery P., Rosbash M., Allada R., Emery P., Rosbash M., Allada R., Rosbash M., Allada R., Allada R. Drosophilaclock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]