Abstract
The tobacco N gene is a member of the Toll-interleukin-1 receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) class of plant resistance (R) genes and confers resistance to tobacco mosaic virus (TMV). We investigated the importance of specific domains of N in inducing TMV resistance, by examining various N deletion and point mutations that introduce single amino acid substitution mutants in vivo. Our deletion analysis suggests that the TIR, NBS, and LRR domains play an indispensable role in the induction of resistance responses against TMV. We show that amino acids conserved among the Toll/IL-1R/plant R gene TIR domain and NBS-containing proteins play a critical role in N-mediated TMV resistance. Some loss-of-function N alleles such as the TIR deletion and point mutations in the NBS (G216A/E/V/R, G218R, G219D, K222E/N, and T223A/N) interfere with the wild-type N function and behave like dominant negative mutations. These F1 plants mount a hypersensitive response (HR) that is indistinguishable from that of the wild-type N plants, yet TMV was able to move systemically, causing a systemic hypersensitive response (SHR). Many amino acid substitutions in the TIR, NBS, and LRR domains of N lead to a partial loss-of-function phenotype. These mutant plants mount delayed HR compared with the wild-type N plants and fail to contain the virus to the infection site. In addition, some partial loss-of-function alleles (W82S/A, W141S/A, G218V/S, and G219V) interfere with the wild-type N function, leading to SHR. The partial loss-of-function and dominant negative mutant alleles described in this report will be useful in furthering our understanding of the TIR-NBS-LRR class of R genes.
In plants, R genes are hypothesized to encode receptors that interact directly or indirectly with ligands produced by the corresponding invading pathogen avirulence (Avr) genes. This initial recognition event triggers defense responses that halt pathogen spread. Absence of an R gene in the plant or specific Avr gene in the invading pathogen results in successful colonization of the pathogen on the plant and further development of disease. This R-Avr-mediated defense response in plants is termed a “gene-for-gene” type of resistance (1).
The typical plant defense response includes the hypersensitive response (HR) or localized cell death at the site of pathogen ingress and containment of the pathogen to the infection site. The HR shares many characteristic features with nematode and animal-programmed cell apoptosis and may play a direct role in the restriction of a pathogen to the infection site (2, 3). A plethora of biochemical and physiological changes coincides with R gene-triggered resistance and host cell death (4). Some of the processes that are associated with gene-for-gene resistance are generation of reactive oxygen species and nitric oxide, production of antimicrobial compounds, lipid peroxidation, ion fluxes, cell wall strengthening, lignin deposition, and induction of defense genes. The local induced HR response often correlates with the induction of a nonspecific general defense response throughout the plant called systemic acquired resistance (SAR) (5). During SAR, salicylic acid (SA) levels increase throughout the plant, defense genes such as pathogenesis-related (PR) genes are expressed, and the plant becomes more resistant to further pathogen attack.
More than 20 R genes from diverse plant species conferring race-specific resistance to viral, bacterial, fungal, nematode, and insect pathogens have been cloned (6, 7). Most R genes contain an LRR domain (except Pto), and these domains are present in a wide variety of proteins and function largely in mediating protein–protein, protein–ligand, and protein–carbohydrate interactions (8). The LRR domain of R proteins has been implicated in playing a direct role in determining the specificity in gene-for-gene interactions (6, 9). In support of this view, LRRs have been shown to have considerable sequence variation among members within clustered R gene families (10–13). This variation in LRRs correlates directly with new specificities for pathogen recognition in the case of flax L alleles and tomato Cf genes (10, 13). However, no direct interaction between an LRR and pathogen ligand has been demonstrated to date for any LRR-containing R genes. Although the primary function of the LRR is assumed to be Avr protein recognition, indirect evidence suggests that it also plays a direct role in downstream signaling (14). A mutation in the C-terminal region of the RPS5 LRR specifically affects the recognition of Pseudomonas syringae AvrPphB. A mutation in the third LRR of the RPS5 suppresses resistance conferred by multiple R genes.
A large number of cloned R genes belong to the NBS-LRR class (15). These R genes contain a centrally located NBS and C-terminal LRR of various lengths. The NBS-LRR class of R genes can be further classified into TIR-NBS-LRR and LZ-NBS-LRR based on their N-terminal domain sequence. Members of the TIR-NBS-LRR class of R proteins contain an N-terminal domain that is similar to the cytoplasmic domains of the Toll, interleukin-1 receptor (IL-1R), and Myd88 (16,17). The TIR-NBS-LRR subclass includes the tobacco N (TMV resistance), flax L6 and M (rust resistance), Arabidopsis RPP5 and RPP1 (downy mildew resistance), and Arabidopsis RPS4 (bacterial resistance) (ref. 7 and references therein, and ref. 18). The LZ-NBS-LRR contains a leucine zipper sequence (LZ) at the N terminus. This class includes Arabidopsis RPM1, RPS2, RPS5, and tomato Prf (resistance to P. syringae pathovars), Arabidopsis RPP8 (downy mildew resistance), tomato Mi (root knot nematode resistance), and potato Rx1 (potato virus X resistance) (ref. 7 and references therein, and ref. 19). The tomato I2, maize Rp1-D, lettuce RGC2, and pepper Bs2 genes do not contain either TIR or LZ motifs (ref. 20 and references therein). In addition to the above-mentioned domains, these R genes contain a highly conserved domain of unknown function called GLPLAL between the NBS and LRR domains. The NBS region of the R genes discussed above shares sequence homology with the NBS region of cell death genes, CED4, from Caenorhabditis elegans and Apaf-1, FLASH, CARD4, and Nod1 from humans (21, 22).
NBS motifs are also found in many families of proteins, including the RAS group, ATPases, elongation factors, and G-proteins (23). These proteins serve as molecular switches and are critical for numerous fundamental eukaryotic cellular events such as cell growth, differentiation, cytoskeletal organization, vesicle transport, apoptosis, and defense (21, 22, 24). The P-loop, Kinase 2, and Kinase 3a are three structural motifs found in the NBS domain (25). The P-loop motif, [GXXXXGK(T/S)], is involved in interactions with phosphates and Mg2+ ions (23). The kinase 2 motif, which is believed to function in a phospho-transfer reaction, contains four consecutive hydrophobic amino acids followed by a conserved aspartate (D), which coordinates the divalent metal ion (Mg2+). The kinase 3a motif is involved in binding purine or ribose and contains a tyrosine (Y) or arginine (R). In addition to the above-discussed motifs, the NKXD, DXXG, and (C/S)AX motifs are highly conserved in the GTPase superfamily (24). The absence of these domains in the predicted plant R proteins and their homology with CED4/Apaf-1 suggests that R proteins may bind ATP and/or act as ATPases. However, to date there is no biochemical evidence on R protein NBS amino acids in the postulated nucleotide binding.
The presence of conserved TIR, NBS, and LRR structural motifs in different R proteins implies their involvement in protein complexes that recognize pathogen-derived ligands and trigger signal transduction leading to induction of defense responses. However, the precise function of the TIR-NBS-LRR domains in recognition and disease resistance signaling remains obscure. Some naturally occurring or induced alleles of R genes in the TIR, NBS, and LRR domains have been described (13, 14, 18, 26–31). In addition, site-directed mutagenesis work has been done in the GLPLAL region of the RPS2 (32). In this report we systematically investigated the role of the N-encoded TIR, NBS, and LRR domains in conferring TMV resistance by generation and analysis of a series of deletion and amino acid substitution mutant alleles of N.
Materials and Methods
Plant Materials and TMV Strains.
TMV-sensitive Nicotiana tabacum cv. petite Havana SR1 (SR1∷nn) and TMV-resistant Samsun NN were used in this study. All plants were grown in a virus-free greenhouse. W. O. Dawson (University of Florida, Citrus Research and Education Center, Lake Alfred, FL) kindly provided the TMV (U1 strain) infectious clone (pTMV004). Virus was propagated in TMV-sensitive tobacco, SR1∷nn.
Plasmid Constructions.
All DNA manipulations were performed essentially as described (33). Most constructs used in this study were derived from pGEM34 (34). Details of the constructions used in this report are available on request. TIR, NBS, GLPLAL, and LRR deletions were created by using flanking restriction enzyme sites. Reading frames were maintained in these deletions. Site-directed mutagenesis of TIR, NBS, and LRR regions was performed by two-step PCR as described (35). All constructions were confirmed by double-stranded DNA sequencing.
TMV Inoculation and Phenotypic Analysis.
Full-length infectious TMV RNA transcripts were generated, and inoculum was prepared by following the method as described (34). The leaf sap with virus and carborundum was rubbed onto 4- to 5-week-old T0, T1, and F1 plants with a sponge. Plants were scored for the development of a resistance or susceptible response to TMV from 3 to 20 days after infection.
Plant Transformation.
TMV-sensitive tobacco (SR1∷nn) plants were transformed with Agrobacterium tumefaciens AGL1 carrying various transgene constructions, using a leaf disk transformation procedure (36). Transformants were selected on 150 mg/liter kanamycin. At least 15 independent transformants were generated for each construction. Transformants were confirmed by PCR using N gene-specific primers. T0 transformants were transferred to soil and grown for 4 weeks before TMV inoculation. T0 transformants were selfed to generate T1 progeny plants. F1 progeny were generated by crossing individual T0 transformants, containing various mutations, to wild-type N-containing plants (Samsun NN).
Results and Discussion
TIR, NBS, and LRR Domains Are Essential for N Function.
The presence of conserved TIR, NBS, GLPLAL, and LRR motifs among different R genes from distantly related plant species indicates that these motifs are structural and/or functional domains involved in determining resistance responses to diverse groups of plant pathogens (37, 38). To understand the importance of the TIR, NBS, GLPLAL, and LRR motifs in N-mediated signaling, we created an array of in-frame deletion mutations in the N genomic clone (Fig. 1). Deletions were created in the genomic clone, because alternative splicing is required for TMV resistance, and full-length N-cDNA alone is insufficient for TMV resistance (34). The N deletion mutants were cloned into a T-DNA vector and transformed into TMV-susceptible tobacco (SR1 nn) by Agrobacterium-mediated transformation. T0 transformants were inoculated with the U1 strain of TMV, and the development of resistance (localized HR and containment of virus to the inoculated leaf) or susceptible (mosaic and systemic spread of virus) response was observed. T1 progeny were analyzed to confirm the transmission of TMV-induced phenotypes. Dominance of the mutants was tested by analyzing F1 progeny derived from each mutant crossed with wild-type NN plants.
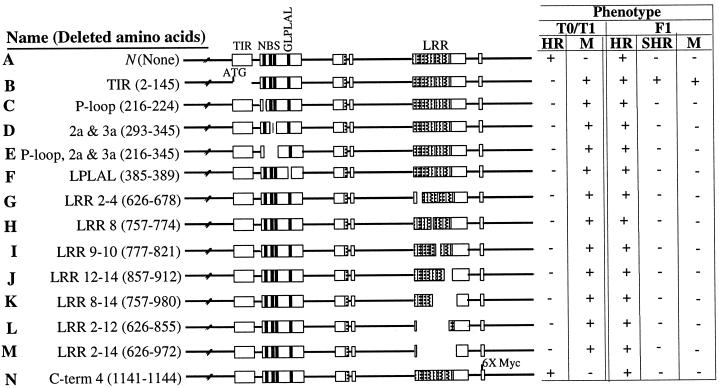
Figure 1.
Summary of the N gene deletion mutants and their phenotypes. The 13 deletion constructs (B to N) are depicted schematically. The phenotypes of primary transformants (T0), selfed T0 progeny (T1), and progeny derived from the cross between mutant T0 and wild-type NN plant (F1) are shown. TIR, NBS, and LRR are structural domains of the N gene. 6× Myc, six copies of Myc tag (EQKLSEEDLE) are inserted at the C terminus of the N gene. HR, hypersensitive response; SHR, systemic hypersensitive response; M, mosaic.
Plants bearing the N gene normally display localized HR at the site of virus infection within approximately 48 h, and lesions are discrete (<5 mm in diameter) (Fig. 1A). TMV becomes localized within the cells in and immediately surrounding the site of the HR lesion. Plants lacking the N gene allow TMV to replicate and spread systemically and develop a characteristic mosaic phenotype that is visible approximately 10 days after infection (Fig. 1B). Our analysis of T0 and T1 progeny bearing various deletion constructions of the N gene suggests that most of the deletions abolish the N-mediated resistance response to TMV (Fig. 1 B–M) compared with the N transgenic plants (Fig. 1A). It is interesting that even a single LRR repeat (amino acids 757–774) deletion fails to complement the resistant phenotype (Fig. 1H). The potential rigidity of the LRR domain, as shown by its inability to withstand minor amino acid alteration, implies that it may provide a site for interaction with the TMV ligand and/or other cellular proteins. In fact, in adenylate cyclase, a 600-amino acid LRR region is indispensable for its interaction with RAS protein (39). The N encoded protein tolerates modification at the C terminus (Fig. 1N). Insertion of six copies of a Myc tag (6 × EQKLSEEDLE) that resulted in the deletion of four C-terminus amino acids (amino acids 1141–1144) had no effect on the N function (Fig. 1N). Taken together, these results suggest that TIR, NBS, GLPLAL, and LRR domains are indispensable for the N-mediated TMV resistance response.
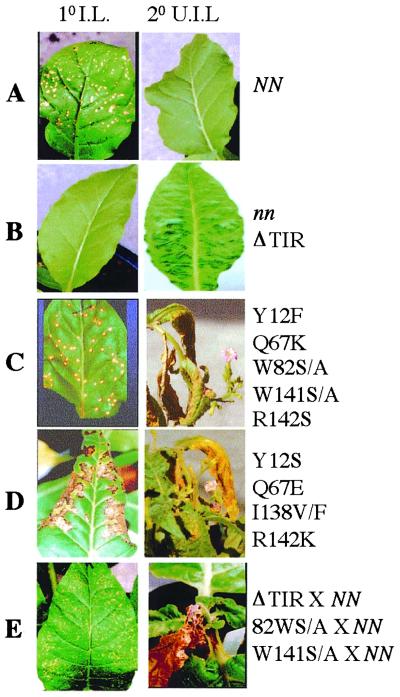
Analysis of F1 progeny derived from crosses between deletion mutant plants and N tobacco plants suggests that most of the deletion mutations are recessive to the wild-type N function (Fig. 1 C–N). F1 progeny plants exhibit HR and resistance responses similar to those observed for wild-type N-containing plants with respect to timing and containment of TMV to the inoculation site. F1 progeny derived from TIR deletion mutant (amino acids 2–145) and N tobacco crosses, however, show a dominant change-of-function phenotype (Figs. 1B and 2E). These plants mount localized HR on the inoculated leaves (Fig. 1E) similar to that of wild-type N-containing plants (Fig. 1A). However, at 10 days after infection, upper uninoculated leaves displayed SHR (Fig. 1E). This SHR phenotype is virus dependent. We believe that the observed SHR phenotype in the upper uninoculated leaves is due to virus escape from the infection site, because plant sap from upper uninoculated leaves was able to induce HR on TMV-resistant Samsun NN plants. Taken together, these results indicate that the TIR deletion allele may interfere with the action of the wild-type N by affecting dimerization or multimerization or interaction with other cellular proteins. These results are interesting because in the case of IL-1R and Myd88, deletion of the TIR domain leads to loss of IL-1R-mediated cellular response to its ligand, IL-1 (40, 41). Furthermore, IL-1R and Myd88 TIR deletion mutants act as dominant negative mutations and interfere with the wild-type function (41, 42).
Figure 2.
Comparison of TIR domains of Toll, human IL-1R, and plant resistance proteins. Alignment was performed with the clustal w program (60). Amino acids conserved in all of these proteins are represented by an asterisk (*). Two dots represent conservative changes, and one dot represents semiconservative changes. Amino acids that are subjected to mutagenesis in this study are highlighted.
TIR Amino Acids Conserved Among Plant R Genes, Toll, and IL-1R Are Required for N-Mediated Resistance to TMV.
To further characterize amino acids of the TIR domain that are crucial for N function, we introduced 17 substitutions in nine TIR amino acids by site-directed mutagenesis (Table 1 and Fig. 2). We targeted these nine amino acids because they are essential for the activation of intracellular signaling events in Toll and IL-1R pathways (17, 43, 44) and are conserved among Toll, IL-1R, and TIR-containing plant R genes (Fig. 2). Some single amino acid substitutions lead to a wild-type or complete loss-of-resistance response to TMV (Table 1). Substitution I63V in N-TIR, a conserved amino acid present in both dToll and hIL-1R, gives rise to TMV-resistant plants. In contrast, transgenic plants bearing amino acid substitution I63 M are TMV susceptible. Interestingly, in dToll, V911 M (I63 is the corresponding position in the N gene) mutation leads to a dorsalizing phenotype (43). Another dorsalizing recessive allele in dToll is H893Y (D46 is the corresponding position in the N gene). In fact, Y is present at this position in hIL-1R (Fig. 2). Interestingly, substitution mutation D46Y has no effect on the N-mediated resistance to TMV; however, substitution to H results in nonfunctional N (Table 1).
Table 1.
Site-directed mutagenesis analysis of the N gene TIR, NBS, and LRR domains
| Domain | Name of mutation | Phenotype
|
|||||
|---|---|---|---|---|---|---|---|
| T0/T1
|
F1
|
||||||
| HR | SHR | M | HR | SHR | M | ||
| NN | + | − | − | + | − | − | |
| nn | − | − | + | + | − | − | |
| TIR | D46Y | + | − | − | + | − | − |
| I63V | + | − | − | + | − | − | |
| S66A | + | − | − | + | − | − | |
| D46H | − | − | + | + | − | − | |
| I63M | − | − | + | + | − | − | |
| Y12S*/F | + | + | + | + | − | − | |
| Q67E*/K | + | + | + | + | − | − | |
| I138V*/F* | + | + | + | + | − | − | |
| R142K*/S | + | + | + | + | − | − | |
| W82S/A | + | + | + | + | + | + | |
| W141S/A | + | + | + | + | + | + | |
| NBS | G218P | + | − | − | + | − | − |
| G216A/E/V/R | − | − | + | + | + | + | |
| G218R | − | − | + | + | + | + | |
| G219D | − | − | + | + | + | + | |
| K222E/N | − | − | + | + | + | + | |
| T223A/N | − | − | + | + | + | + | |
| G218V*/S | + | + | + | + | + | + | |
| G219V* | + | + | + | + | + | + | |
| T223S | + | + | + | + | − | − | |
| D301H/N/Y | − | − | + | + | − | − | |
| R325Y/G | − | − | + | + | − | − | |
| LRR | P619S | − | − | + | + | − | − |
| P619A/T | + | + | + | + | − | − | |
T0: 15 independent transformants were tested for TMV response. T1: Analysis of selfed progenies from T0 plant. F1: Analysis of progenies from T0 mutant crossed to Samsun NN. M, mosaic.
The timing and appearance of HR in these mutants was different compared with the wild-type N-containing plants.
Conservative or nonconservative substitution at positions 12, 67, 82, 138, 141, and 142 in the N-TIR domain leads to partial loss-of-function phenotypes (Table 1 and Fig. 3). The timing and appearance of HR in the plants bearing Y12F, Q67K, W82S/A, W141S/A, and R142S alleles is similar to that of the wild-type N-containing plants (compare Figs. 3C and 3A). However, in plants bearing Y12S, Q67E, I138F/V, and R142K alleles, the occurrence of HR was delayed (5–7 days vs. 2 days), and the appearance (nondiscrete 20 mm vs. discrete 5 mm) of HR is different compared with that of the wild-type N plants (compare Figs. 3D and 3A). Irrespective of the timing of the HR occurrence and the size of the HR lesions, all plants bearing the above-discussed mutant alleles developed the SHR phenotype at approximately 10 days after infection. It is possible that the HR (wild-type appearing or delayed) leading to discrete cell death lesions in these mutants may not be sufficient to contain virus to the infection site. Among these mutant N alleles, W82S/A and W141S/A alleles are dominant and interfere with the wild-type N function (Fig. 3E). Taken together, these results suggest that some amino acid substitutions in the N-TIR domain lead to partial loss-of-function phenotype. In addition, some of the loss-of-function and partial loss-of-function alleles act as dominant negative and interfere with the wild-type N function.
Figure 3.
Response to TMV of plants expressing the N gene TIR domain deletion and various amino acid substitution mutants. Primary TMV-U1-inoculated (1o I.L.) and secondary uninoculated upper leaves (2o U.I.L.) of mutant plants and progeny derived from mutants crossed to wild-type NN plants are shown.
Conserved amino acids in the TIR family of proteins among the distantly related species of Drosophila, humans, and plants underscore the possible importance of the TIR domain in innate immune responses. In agreement with this notion, our results indicate that mutations that affect dToll and hIL-1R signaling also affect N-mediated signaling events leading to TMV resistance. The plant R-TIR domain may also function in pathogen recognition (13). In flax, the L6 and L7 alleles differ only in the TIR region but show different specificities against fungal pathogens. In any case, the results described here provide no direct insight into how the TIR domain of the N and other TIR-containing R products participate in pathogen recognition and the resistance signaling cascade compared with Toll and IL-1R pathways. Further biochemical and molecular studies of the N-TIR domain will elucidate the precise role of this domain in the N signal transduction pathway.
Conserved Amino Acids in the NBS Domain Play an Important Role in the N Function.
Our analysis of the N-NBS domain deletion mutations confirms their importance in N-mediated signal transduction leading to TMV resistance. To further determine whether conserved amino acids critical for NBS function in other proteins are required for N-mediated TMV resistance, we introduced 20 point mutations in seven conserved amino acids in the three subdomains of the NBS region of the N (Table 1). In the N gene, the P-loop sequence includes 216GMGGVGKT223. Any substitution in the invariant G216 and K222 in the P-loop of the N leads to loss of resistance to TMV (Table 1). However, these alleles interfere with the wild-type N function (Table 1). Interestingly, mutations in the conserved lysine residue in the P-loop of the CED4 and Apaf1 abolish binding of ATP and activation of CED3/procaspase-9 and cell death (45–47). The presence of conserved domains between CED4 and Apaf1 and the N protein and the disruption of the function of the three corresponding genes by mutations in the conserved ATPase domain suggest that these proteins may activate similar cell death machinery and mechanisms.
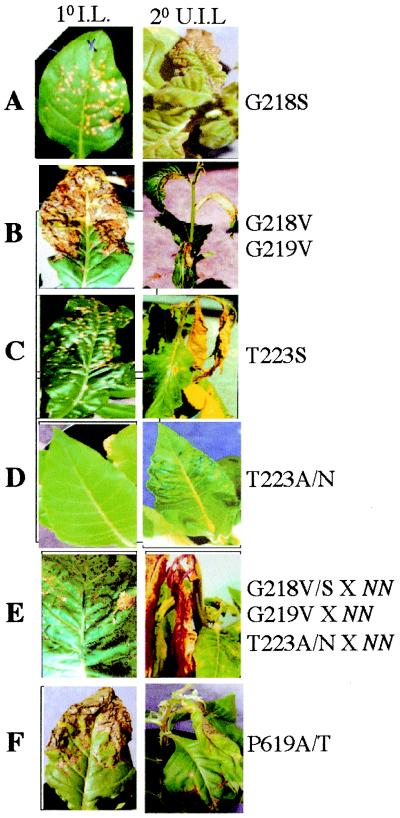
The G12 and G13 amino acids in RAS play an important role in oncogenic transformation, and these mutants have reduced GTPase activity (48). Therefore, we made several substitutions at residue G218 and 219 of N (Table 1). All tested substitutions in N except proline at G218 lead to a loss-of-function or a partial loss-of-function phenotype. Plants bearing the G218S allele display a normal HR phenotype. Plants bearing the G218V or G219V alleles display a massive HR phenotype compared with the wild-type N-containing plants (Fig. 4B vs. Fig. 3A). The initiation of HR is delayed (5 days vs. 2 days), and the lesion size is greater (25 mm vs. 5 mm in the wild type). The G218V/S and G219V alleles are dominant over the wild-type N allele (Fig. 4E). These phenotypic results are in agreement with the RAS studies (48); however, precise biochemical analysis of N and other NBS-containing R proteins is necessary to draw such an analogy.
Figure 4.
Response of plants expressing various amino acid substitution mutations in the N gene NBS and LRR domains to TMV. Primary TMV-U1-inoculated (1o I.L.) and secondary uninoculated upper leaves (2o U.I.L.) from mutant plants and progeny derived from mutants crossed to wild-type NN plants are shown.
Structural studies of NBS proteins indicate that a hydroxyl group of Ser or Thr in the P-loop is involved in binding of Mg2+ associated with bound nucleotides (49). Mutations in this amino acid position result in reduced NTPase activity (49). Substitution of T223S in the N leads to partial loss-of-N-function phenotype (Table 1 and Fig. 4C). The appearance of HR lesion size and timing is similar to that of the wild-type N-containing plants. However, HR continues to spread throughout the plant and results in death within 5–7 days. The death of the plant because of virus-induced SHR in plants bearing this allele is the fastest compared with any other partial loss-of-function alleles described in this report. These results are surprising, given the fact that the T223S substitution is a conservative substitution.
Substitution of serine in the P-loop with alanine or asparagine in NBS-containing proteins leads to dominant inhibitory biological function (50–52). Therefore, we replaced T223 of the N gene with asparagine and alanine. These N alleles fail to initiate a defense response against TMV infection and exhibit mosaic symptoms similar to those of plants with no N gene (Table 1 and Fig. 4D). However, these alleles interfere with wild-type N function (Fig. 4E). Therefore these alleles act as dominant interfering mutants similar to those that have been described in the other NBS-containing proteins (51–53).
The aspartate at position 301 in the kinase 2 (297LIVLD301) and arginine at position 325 in the kinase 3a (320FGNGSR325) domains are highly conserved among NBS proteins (25). In agreement with this notion, any mutations in these two residues of the N gene lead to loss of N function (Table 1). It is interesting that in CED4, mutation in the highly conserved aspartate residue of the kinase 2 domain disrupts the oligomerization of CED4 and inhibits activation of CED3 protease and cell death (54).
Taken together, our analysis of the N-NBS domain suggests that binding of nucleotide may be required for N protein function. It will be interesting to determine whether the mutant N proteins described in this report display altered properties at the level of nucleotide binding and hydrolysis processes. Biochemical analysis using purified wild-type N and mutant proteins may shed light on the role of the NBS domain in NBS-LRR R protein function.
Proline-619 in the Second LRR of N Protein Plays an Important Role in TMV Resistance.
The N was isolated by using Maize activator (Ac) transposon (55). Sequence analysis of excision sites using genomic DNA derived from TMV-susceptible plants lacking the Ac transposon suggested that these plants contained frameshift mutations that resulted in truncation of the N protein (56). Analysis of the sequence from the seven independent germinal revertants showed no changes in sequence compared with the wild-type N (56). These results suggested that reversion to the wild-type resistance phenotype is possible only if there is a precise excision event. Interestingly, the Ac was inserted adjacent to the proline residue at the beginning of the second LRR of N at position 619. Prolines in LRR proteins that cause kinks in the peptide backbone have been speculated to function in positioning the conserved core motifs [LXXLXLXX(N/C/T)XL] of LRR (9). Based on this observation, we hypothesized that mutation of the proline residue may disrupt the structure of ligand binding and therefore lead to a change in N function. To test this possibility, we introduced several substitutions at position 619 in the N protein. Substitution of serine for proline at position 619 leads to loss of N function (Table 1). Plants bearing this mutant allele exhibit mosaic symptoms after infection with TMV. Substitution of alanine or threonine for proline at position 619 leads to a partial loss-of-function phenotype (Table 1 and Fig. 4F). These plants mount delayed HR compared with the wild-type N-containing plants, and they develop SHR because virus escapes from the infection site. These results suggest that any change at position 619 of the N gene is not tolerable. It is possible that the P619S change may lead to failure of the N protein interaction with TMV ligand, thereby resulting in a complete loss-of-resistance response to TMV. However, P619A/T may recognize the TMV ligand and initiate HR but fail to activate signaling events that are required to restrict TMV spread. Similarly, in the case of the Arabidopsis RPS5, a proline-to-serine substitution at position 799 leads to loss of resistance to P. syringae carrying avrPphB (14).
Conclusions
In summary, our analysis suggests that the TIR, NBS, GLPLAL, and LRR domains of N play an important role in N-mediated recognition and signal transduction leading to TMV resistance. We cannot be certain that some of the observed phenotypes (especially loss of function) are due to lack of production of N mutant proteins. However, about 56% (10/18) of our loss-of-function alleles interfere with wild-type N function, suggesting that at least plants bearing these alleles make N mutant protein in vivo. Furthermore, our RT-PCR analysis of T1 and F1 plants bearing the TIR deletion, as well as the W82S/A, T223S/A/N, and G218V/S mutant alleles, suggests that the N gene is transcribed in these plants (data not shown). However, it is possible that mutant N proteins may be unstable or mislocalized or both. Some of these questions have yet to be addressed. The partial loss-of-function and dominant interfering alleles of N will be useful in determining the domain structure of the N protein and components of N-mediated signal transduction. Furthermore, these mutant alleles will be helpful in the next step of N analysis, including dimerization or multimerization, binding and hydrolysis of nucleotides, and interaction with the TMV ligand and other N signaling proteins.
The induction of HR in a race-specific resistance response has been implicated in halting the pathogen (2, 3). However, several R-Avr-mediated resistance responses do not involve HR. The potato Rx1 gene confers resistance to potato virus X (PVX) without the induction of visible HR lesions at the site of infection (19). In addition, the dnd1 mutation in Arabidopsis is able to elicit race-specific resistance responses in the absence of cell death (57). The HR induced in plants bearing some of the partial loss-of-function N alleles (Y12F, Q67K, W82S/A, W141S/A, R142S, G218S, and T223S) described in this report is indistinguishable with respect to size and timing of occurrence of HR compared with the wild-type N-containing plants. At least phenotypically the induction of normal HR in these mutant plants suggests that the N protein may be able to recognize the TMV ligand directly or indirectly and initiates HR. However, this cell death alone is not sufficient to inhibit TMV replication and systemic movement. One possible explanation for why these mutant N plants show normal HR but fail to inhibit virus movement may be the delayed occurrence of biochemical and physiological events that are associated with HR, like generation of reactive oxygen species, induction of salicylic acid, defense genes, etc. (58, 59). In any case, the N mutant alleles described in this report will provide us with important tools for understanding the molecular and biochemical basis of the role of cell death in TMV restriction. Interestingly, the T223S allele of the tobacco N gene induces a similar phenotype in tomato (Hwi-Hwa Cheng and B.J.B., unpublished observations) and Nicotiana benthamiana (S.P.D.-K., unpublished observations). These plants are used to isolate T223S suppressors.
Acknowledgments
We thank Shirleko Dai and Marianne Dutton for critical reading of the manuscript. We thank the Plant Gene Expression Center greenhouse manager David Hantz for his excellent help and advice. We thank Jack Lio, Michael Schiff, Sarah Hantz, and Amy Vittor for excellent technical help. S.P.D.-K. was supported by a Life Science Research Foundation Fellowship sponsored by the Department of Energy during most of this study.
Abbreviations
- TIR
Toll/interleukin-1 receptor homology region
- NBS
nucleotide-binding site
- LRR
leucine-rich repeat
- HR
hypersensitive response
- SHR
systemic hypersensitive response
- TMV
tobacco mosaic virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Flor H. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 2.Dangl J L, Dietrich R A, Richberg M H. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg J T. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:525–545. doi: 10.1146/annurev.arplant.48.1.525. [DOI] [PubMed] [Google Scholar]
- 4.Lamb C J. Cell. 1994;76:419–422. doi: 10.1016/0092-8674(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 5.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bent A. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis J, Jones D. Curr Opin Plant Biol. 1998;1:288–293. doi: 10.1016/1369-5266(88)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Kobe B, Deisenhofer J. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 9.Jones D A, Jones J D G. Adv Bot Res Incorp Adv Plant Pathol. 1997;24:90–167. [Google Scholar]
- 10.Parniske M, Hammond-Kosack K E, Golstein C, Thomas C M, Jones D A, Harrison K, Wulff B B, Jones J D. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- 11.McDowell J M, Dhandaydham M, Long T A, Aarts M G, Goff S, Holub E B, Dangl J L. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers B C, Shen K A, Rohani P, Gaut B S, Michelmore R W. Plant Cell. 1998;11:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis J G, Lawrence G J, Luck J E, Dodds P N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren R F, Henk A, Mowery P, Holub E, Innes R W. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyers B C, Dickerman A W, Michelmore R W, Sivaramakrishnan S, Sobral B W, Young N D. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 16.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slack J L, Schooley K, Bonnert T P, Mitcham J L, Qwarnstrom E E, Sims J E, Dower S K. J Biol Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- 18.Gassman W, Hinsch M E, Staskawicz B J. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 19.Bendahmane A, Kanyuka K, Baulcombe D C. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tai T H, Dahlbeck D, Clark E T, Gajiwala P, Pasion R, Whalen M C, Stall R E, Staskawicz B J. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Biezen E A, Jones J D. Curr Biol. 1998;8:R226–R227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 22.Arvind L, Dixit V M, Koonin E V. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 23.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 24.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 25.Traut T W. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 26.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 27.Mindrinos M, Katagiri F, Yu G-L, Ausubel F M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 28.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 29.Salmeron J M, Oldroyd G E, Rommens C M, Scofield S R, Kim H S, Lavelle D T, Dahlbeck D, Staskawicz B J. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 30.Parker J E, Coleman M J, Szabo V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botella M A, Parker J E, Frost L N, Bittner-Eddy P D, Beynon J L, Daniels M J, Holub E B, Jones J D. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley Interscience; 1998. [Google Scholar]
- 34.Dinesh-Kumar S P, Baker B. Proc Natl Acad Sci USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. . (First Published January 31, 2000; 10.1073/pnas.020367497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinesh-Kumar S P, Miller W A. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horsch R B, Fry J E, Hoffmann N L, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 37.Dangl J L. Cell. 1995;80:363–366. doi: 10.1016/0092-8674(95)90485-9. [DOI] [PubMed] [Google Scholar]
- 38.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki N, Choe H R, Nishida Y, Yamawaki-Kataoka Y, Ohnishi S, Tamaoki T, Kataoka T. Proc Natl Acad Sci USA. 1990;87:8711–8715. doi: 10.1073/pnas.87.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heguy A, Baldari C, Bush K, Nagele R, Newton R C, Robb R J, Horuk R, Telford J L, Melli M. Cell Growth Differ. 1991;2:311–315. [PubMed] [Google Scholar]
- 41.Leung K, Betts J C, Xu L, Nabel G J. J Biol Chem. 1994;269:1579–1582. [PubMed] [Google Scholar]
- 42.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 43.Schneider D S, Hudson K L, Lin T Y, Anderson K V. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 44.Heguy A, Baldari C T, Macchia G, Telford J L, Melli M. J Biol Chem. 1992;267:2605–2609. [PubMed] [Google Scholar]
- 45.Seshagiri S, Miller L. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 46.Chinnaiyan A M, Chaudhary D, O'Rourke K, Koonin E V, Dixit V M. Nature (London) 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 47.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 48.Seeburg P H, Colby W W, Capon D J, Goeddel D V, Levinson A D. Nature (London) 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 49.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilderbrandt J D, Day R, Farnsworth C L, Feig L A. Mol Cell Biol. 1991;11:4830–4838. doi: 10.1128/mcb.11.10.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen S-Y, Huff S Y, Lai C-C, Der C J, Powers S. Oncogene. 1994;9:2691–2698. [PubMed] [Google Scholar]
- 52.Wu T H, Marinus M G. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feig L A, Cooper G M. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Chang H Y, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 55.Whitham S, Dinesh-Kumar S P, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 56.Dinesh-Kumar S P, Whitham S, Choi D, Hehl R, Corr C, Baker B. Proc Natl Acad Sci USA. 1995;92:4175–4180. doi: 10.1073/pnas.92.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu I C, Parker J, Bent A F. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doke N, Ohashi Y. Physiol Mol Plant Biol. 1988;32:163–175. [Google Scholar]
- 59.Delaney T P, Ukness S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 60.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]