Abstract
Candida albicans is an opportunistic fungal pathogen that is found in the normal gastrointestinal flora of most healthy humans. However, under certain environmental conditions, it can become a life-threatening pathogen. The shift from commensal organism to pathogen is often correlated with the capacity to undergo morphogenesis. Indeed, under certain conditions, including growth at ambient temperature, the presence of serum or N-acetylglucosamine, neutral pH, and nutrient starvation, C. albicans can undergo reversible transitions from the yeast form to the mycelial form. This morphological plasticity reflects the interplay of various signal transduction pathways, either stimulating or repressing hyphal formation. In this review, we provide an overview of the different sensing and signaling pathways involved in the morphogenesis and pathogenesis of C. albicans. Where appropriate, we compare the analogous pathways/genes in Saccharomyces cerevisiae in an attempt to highlight the evolution of the different components of the two organisms. The downstream components of these pathways, some of which may be interesting antifungal targets, are also discussed.
INTRODUCTION
Opportunistic fungal pathogens, such as Candida albicans, are found in the normal gastrointestinal flora and the oral mucosa of most healthy humans. However, in immunocompromised patients, bloodstream infections often cause death, despite the use of antifungal therapies (152). The underlying molecular mechanisms for survival inside the human body and adaptation to various environments are probably distinct but overlapping. Dietary factors, such as an excess of or deficiency in certain nutrients, may alter the endogenous microbial flora. Mechanical factors, such as trauma or occlusive injury, can also alter the microenvironment, deplete the system of “friendly bacteria,” and enable the pathogenic fungus to take over. Immunocompromised or immunosuppressed persons, including AIDS patients, neonates, and transplant recipients, are also particularly susceptible to fungal infections.
The most common systemic fungal infection is candidiasis, which accounts for well over half of these invasive mycoses. A single species, C. albicans, causes the majority of these infections. Its success stems in part from its capacity to live as a benign commensal in a variety of body locations, most notably the oral cavity, genitalia, and gastrointestinal tract (272). C. albicans expresses various traits critical for existence on mucosal surfaces, where a constant but dynamic interplay occurs between innate and acquired host defense mechanisms. The pathogenic Candida species also establish well-developed biofilms, which occur easily on various implants and are resistant to antifungal agents (76). The nature of disease resulting from tissue invasion by this organism is complex and depends on a variety of physical and physiological conditions in the host and on specific C. albicans traits. The capacity of C. albicans to rapidly acquire resistance to antifungal drugs, such as amphotericin B, flucytosine, and a series of azoles, means that continued development of new antifungals remains an important focus for clinicians and pharmaceutical companies.
An important feature of C. albicans, relevant to its pathogenesis, is its ability to switch between different morphological forms. C. albicans can grow in a single-celled, budding yeast form (blastospore) or in a filamentous form (including both pseudohyphae and true hyphae) (31). A crucial component of this versatility is the ability to survive as a commensal in several anatomically distinct sites, each with its own specific set of environmental pressures. Thus, C. albicans must be able to adapt its growth to a range of physiological extremes. To achieve adaptability, the fungus has evolved sophisticated mechanisms of sensing and responding to environmental cues by activating developmental switches that result in coordinated changes in cell physiology, morphology, and adherence. Progress in understanding many aspects of the biology of C. albicans has been hindered by the inability to carry out simple, large-scale genetic screens because of the diploid nature of this organism. A major breakthrough in assessing the contribution of specific genes to morphogenesis and virulence occurred with the development of transformation protocols and methods of deleting both alleles of a gene sequentially. Also, whole-genome microarray analysis has now become an important tool for probing signal transduction pathways during morphogenesis in C. albicans (102).
Additional interest in the molecular mechanisms of C. albicans morphopathogenic determinants originated from the necessity of identifying new drug targets due to increased drug resistance in clinical isolates. There is hope that recently developed techniques of manipulating C. albicans and the sequencing of its whole genome will lead to a thorough understanding of its virulence and biology, thus offering the possibility of a knowledge-based approach to the development of novel antifungal agents. A major strategy for determining virulence genes as molecular targets for antifungal drugs and vaccines is to identify a specific biochemical or structural target unique to C. albicans (or to fungi in general) in an attempt to specifically and selectively disrupt them and determine their effects on virulence.
In this review, we focus on recent advances in the environmental sensing and signal transduction pathways that mediate the morphogenesis and pathogenesis of C. albicans. Where possible, we compare the pathways of C. albicans with the analogous pathways/genes in Saccharomyces cerevisiae.
DIMORPHISM, AN IMPORTANT VIRULENCE FACTOR
The terms “dimorphism” and “dimorphic fungus” (i.e., existing in two morphological forms) are commonly accepted in reference to C. albicans. Strictly speaking, however, this fungus has the ability to adopt a spectrum of morphologies; thus, C. albicans can be considered a “polymorphic” or “pleomorphic” organism (71, 289). The production of germ tubes results in conversion to a filamentous growth phase or hypha, also called the mycelial form. The formation of pseudohyphae occurs by polarized cell division when yeast cells growing by budding have elongated without detaching from adjacent cells. Under certain nonoptimal growth conditions, C. albicans can undergo the formation of chlamydospores, which are round, retractile spores with a thick cell wall. These morphological transitions often represent a response of the fungus to changing environmental conditions and may permit adaptation to a different biological niche. The transition from a commensal to pathogenic lifestyle may also involve changes in environmental conditions and dispersion within the human host. Although progress has been achieved in recent years, the molecular mechanisms governing these morphogenetic conversions are still not fully understood, partly due to the difficulty of genetic manipulations with C. albicans (164), an issue we address briefly below.
GENETIC MANIPULATION WITH C. ALBICANS
Sequencing of the genome of C. albicans has recently been completed. C. albicans has a diploid genome consisting of eight pairs of chromosomes that can be separated by pulsed-field gel electrophoresis. With a size of ∼16 Mb, the haploid genome is slightly larger than that of the model yeast, S. cerevisiae. More information on the C. albicans genome can be found at www.candidagenome.org (68). Many genes are conserved between S. cerevisiae and C. albicans, and it is based on this similarity that the mechanisms of many biological processes in C. albicans have been discovered. As highlighted throughout this review, in many cases, although the specific components of relevant signaling pathways are conserved, molecular mechanisms and environmental signals have often diverged, most likely because of the coevolution of C. albicans and its human host.
C. albicans poses special problems for scientists interested in studying gene function because it is diploid and because CUG is translated into a serine instead of a leucine (164). To analyze the function of a gene, one must disrupt each of the two alleles by transformation. Although methods of transformation (spheroplast-polyethylene glycol, lithium acetate, and electroporation) are patterned after those used with S. cerevisiae, the transformation efficiency is poor. For the study of gene function in C. albicans, a number of disruption protocols and selectable markers are commonly utilized (300). The most widely used marker is the URA3 gene, which encodes orotidine-5′-phosphate decarboxylase and confers uracil prototrophy. However, this marker must be used with caution; URA3 expression levels can affect virulence and are susceptible to chromosome position effects, thereby complicating the analysis of strains constructed with URA3 as a selectable marker (62, 261, 281, 292). To avoid the use of the URA3 marker, various other auxotrophic markers, as well as dominant markers, have been developed and are currently used (206, 224, 241, 263). To study the functions of essential genes, the C. albicans MET3 promoter, which is regulated by the level of methionine and/or cysteine in the medium, or the tetracycline on/off system can be used. These systems allow conditional expression so that the consequences of depletion of a gene product may be investigated (57, 231, 254). In order to study the expression of certain genes at either the RNA level or the protein level, a number of C. albicans-optimized reporter constructs, or tags, have been generated (32). The development of these molecular tools has greatly accelerated the elucidation of morphogenesis and pathogenesis in C. albicans.
ENVIRONMENTAL SENSING PATHWAYS REQUIRED FOR MORPHOGENESIS AND PATHOGENESIS
All organisms, from bacteria and yeast to higher eukaryotes, respond to changes that occur in the environment. In C. albicans, the yeast-to-hypha transition is triggered by various environmental cues, such as serum, N-acetylglucosamine (GlcNAc), neutral pH, high temperature, starvation, CO2, and adherence. In recent years, receptors/sensors that may mediate environmental responses have been identified and partially characterized. Many in vivo and in vitro experiments point to a prominent role for amino acids as nitrogen sources and as ligands for membrane receptors involved in the regulation of cellular morphology and virulence (summarized in Fig. 1 and Table 1). In the following sections, we highlight work aimed at identifying receptors, ligands, and signaling pathways involved in the sensing of environmental signals in C. albicans.
FIG. 1.
Regulation of dimorphism in C. albicans by multiple signaling pathways. The Cph1-mediated MAPK pathway and the Efg1-mediated cAMP pathway are well-characterized signaling pathways in dimorphic regulation. In C. albicans, Ras1 is an important regulator of hyphal development and likely functions upstream of both pathways. In the cAMP-PKA pathway, two catalytic subunits or isoforms of PKA, Tpk1 and Tpk2, have differential effects on hyphal morphogenesis under different hypha-inducing conditions. The MAPK cascade includes Cst20 (PAK), Hst7 (MAPKK), Cek1 (MAPK), and the downstream transcription factor Cph1, which is a homolog of the S. cerevisiae transcription factor Ste12. Transcription of the hyphal regulator TEC1 is regulated by Efg1 and Cph2. Rim101 or Czf1 may function through Efg1 or act in parallel with Efg1. Tup1 is the negative regulator of the hyphal transition. Tup1, recruited by Rfg1, Nrg1, or Mig1, and Rbf1 are also implicated in dimorphic transitions. GlcNAc-inducible hexokinase, Hxk1, plays a negative role in hyphal development under certain conditions. Cell wall proteins (HWP1 and ECE1, etc., which are involved in adherence) are also regulated by Efg1. Transcription factors are shown in rectangular boxes.
TABLE 1.
Major morphopathogenic determinants in C. albicans
| C. albicans protein | S. cerevisiae homologa | Protein function | Reference(s) |
|---|---|---|---|
| Gpa2 | Gpa2 | α subunit of G protein | 193, 249 |
| Gpr1 | Gpr1 | G protein receptor | 193, 249 |
| Ras1 | Ras2 | GTPase | 92 |
| Cdc25 | Cdc25 | Ras-GEF | 112, 193 |
| Cdc35/Cyr1 | Cyr1 | Adenylate cyclase | 139, 243 |
| Srv2 | Cap1 | Adenylate cyclase-associated protein | 13 |
| Pde2 | Pde2 | High-affinity phosphodiesterase | 16, 143 |
| Flo8 | Flo8 | Transcription factor | 56 |
| Tpk1/Tpk2 | Tpk1/Tpk2 | Catalytic subunit of cAMP-dependent PKA | 37, 66, 276 |
| Bcy1 | Bcy1 | Regulatory subunit of cAMP-dependent PKA | 58 |
| Cst20 | Ste20 | PAK | 153, 171 |
| Hst7 | Ste7 | MAPKK | 171, 266 |
| Cek1 | Fus3/Kss1 | MAPK | 69 |
| Cpp1 | Cpp1 | MAPK phosphatase | 114, 256 |
| Tec1 | Tec1 | Transcription factor of TEA/ATTS family | 257 |
| Cph1 | Ste12 | Transcription factor | 181, 195 |
| Efg1 | Sok2/Phd1 | Transcription factor of HLH family | 36, 175, 285, 294 |
| Cph2 | Hms1 | Transcription factor of HLH family | 169, 170 |
| Fkh2 | Fkh2 | Forkhead transcription factor | 29 |
| Mcm1 | Mcm1 | Transcription factor of MADS box family | 245 |
| Cdc42 | Cdc42 | Rho G protein | 119, 305 |
| Cln2 | Cln2 | G1 cyclin | 185 |
| Tup1 | Tup1 | Transcriptional repressor | 41, 210 |
| Nrg1 | Nrg1 | Transcription factor with zinc finger domain | 43, 210, 211 |
| Mig1 | Mig1 | Transcription factor with zinc finger domain | 210 |
| Rfg1 | Rox1 | DNA-binding partner of Tup1 | 146, 148 |
| Rbf1 | Rbf1 | RPG box-binding factor | 137, 262 |
| Rap1 | Rap1 | Transcriptional silencer | 34 |
| Rad6 | Rad6 | Transcription-repressing factor | 176 |
| Hog1 | Hog1 | MAPK for oxidative stress | 2, 3, 227 |
| Ssn6 | Ssn6 | Forms complex with Tup1 | 136 |
| Sln1 | Sln1 | Histidine kinases | 212 |
| Hxk1 | No known homolog | GlcNAc kinase | 162, 267, 326 |
| Hwp1 | No known homolog | Hyphal wall protein | 262, 283, 303 |
| Int1 | Int1 | Surface glycoprotein | 7, 25, 98 |
| Als1 | Als1 | Adhesin factor | 95, 329 |
| Sap1 to Sap9 | No known homolog | Aspartyl proteinases | 134, 165, 255 |
| Plb1 | Plb1 | Phospholipase | 173, 208 |
| Gcn4 | Gcn4 | General amino acid control | 302 |
| Gcn2 | Gcn2 | General amino acid control | 301 |
| Gap1 | Gap1 | General amino acid permease | 35 |
| Csy1 | Csy1 | Amino acid sensor | 45 |
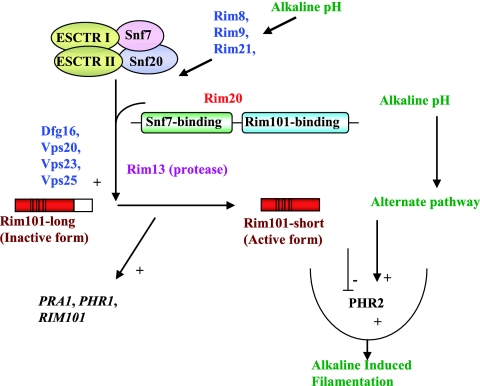
| Rim20 | PalA* | Proteolysis of Rim101 | 72, 73 |
| Rim8 | PalF* | Proteolysis of Rim101 | 72, 73 |
| Rim101 | PacC* | Zinc finger transcription factor | 72, 73, 87, 166 |
| Czf1 | No known homolog | Putative zinc finger transcription factor | 111 |
| Hgc1 | Cln21 | G1 cyclin-related protein | 332 |
| Sho1 | Sho1 | Adaptor protein | 244 |
| Crz1 | Crz1 | Calcineurin-regulated transcription factor | 250 |
| Mkc1 | Mkc1 | Mitogen-activated protein kinase | 80 |
| Rsr1 | Rsr1 | Ras-like GTPase | 118 |
| Bud2 | Bud2 | GTPase-activating protein | 118 |
| Cln3 | Cln3 | G1 cyclin | 59 |
| Clb2, Clb4 | Clb2, Clb4 | B-type cyclin | 28 |
| Erg1 | Erg1 | Epoxidase | 232 |
| Mep1, Mep2 | Mep1, Mep2 | Ammonium permease | 33 |
| Cdc4 | Cdc4 | Ubiquitin ligase | 8 |
| Pmt4 | Pmt4 | Mannosyltransferase | 236 |
| Hsl1 | Hsl1 | Ser/Thr protein kinase | 304 |
| Pld1 | Pld1 | Phospholipase D1 | 82 |
| Ecm33 | Ecm33 | GPI-anchored protein | 199 |
*, A. niger protein.
Amino Acid Availability Regulates Morphogenesis of C. albicans
Amino acid sensing by Csy1.
Serum- and amino acid-based media are known to induce filamentous growth in C. albicans. Although the mechanism by which amino acids induce filamentation is not well established, several genes have been implicated in the amino acid response. In S. cerevisiae, an amino acid-sensing complex (SPS sensor for Ssy1, Ptr3, and Ssy5) that is required for amino acid transporter expression through the induction of proteolytic cleavage of two latent transcription factors has been described (4, 94). Recently a similar sensing complex in C. albicans was described (45). Furthermore, it was shown that the amino acid sensor Csy1 (the homolog of the yeast protein Ssy1) plays an important role in filamentation. Loss of Csy1 results in a lack of amino acid-mediated activation of amino acid transport and a lack of induction of transcription of specific amino acid permease genes. Csy1 mutants also show altered colony morphology and hyphal formation in serum- and amino acid-based solid media but not in media that do not contain amino acids. As in S. cerevisiae, Csy1 appears to discharge its critical role in amino acid transport and filamentation by promoting proteolytic cleavage of two latent transcription factors, Stp1 and Stp2. Truncated Stp2 migrates to the nucleus and induces the expression of genes required for amino acid uptake, and truncated Stp1 induces genes required for the degradation of extracellular proteins (SAP2) and for uptake of peptides (OPT1) (198). The Ljungdahl group also discovered that Csh3, the functional homolog of S. cerevisiae Shr3, which is involved in endoplasmic reticulum exit after packaging, is essential for the proper uptake and sensing of extracellular amino acids and that a null mutant is unable to switch morphology in response to inducing amino acids (197).
Amino acid sensing (and transport) by the general amino acid permease Gap1.
In S. cerevisiae, the Gap1 protein functions not only as an amino acid transporter but also as an amino acid sensor for rapid activation of a fermentable growth medium-induced signaling pathway that controls protein kinase A (PKA) targets (83, 140). Receptors with dual roles as sensors have been dubbed “transceptors” (124). Our group has discovered that the C. albicans Gap1 transporter, which is a functional homolog of the budding yeast gene, may also be a transceptor important for initiating signal transduction pathways resulting in morphogenesis and virulence. GAP1 was isolated in a screen to identify and characterize genes that could be involved in the regulation of morphogenesis and virulence induced by GlcNAc, an important hyphal induction regulator of C. albicans (35) (see below). A null mutant of GAP1 has a defect in filamentation under nitrogen starvation conditions, as well as upon the addition of GlcNAc (35). Addition of serum, however, still resulted in the induction of filamentation in the gap1 mutant, suggesting that Gap1 is not required for serum-induced hyphal formation. As in S. cerevisiae, the expression of GAP1 is strongly repressed in medium containing ammonium as a good nitrogen source. How exactly amino acid transport (and/or sensing) results in hyphal formation remains to be determined. One interesting link is the fact that GlcNAc induces GAP1 expression at the yeast-to-germ-tube transition. This induction depends on Cph1, the transcription factor that is the downstream target of the mitogen-activated protein kinase (MAPK) pathway (see below). This may indicate that Gap1-mediated filamentation is regulated by this pathway.
Amino acid sensing by Gpr1.
In S. cerevisiae, ScGpr1 is a G protein-coupled receptor that functions upstream of the cyclic AMP (cAMP)-PKA pathway and promotes a rapid cAMP increase after the addition of glucose or sucrose (156). In contrast, C. albicans Gpr1 is not required for the glucose-induced increase in cAMP (193) but seems to directly or indirectly sense methionine (193). Addition of methionine to wild-type cells results in a rapid internalization of Gpr1, reminiscent of ligand-induced internalization, a typical feature of GPCR systems in higher eukaryotic cells. Gpr1 is also required for the methionine-induced yeast-to-hypha transition (193). Further research is required to determine the mechanism by which Gpr1 activates the PKA pathway and what the role of methionine is in this mechanism. Interestingly, a role for methionine in morphogenesis is not limited to C. albicans. Recently, Heitman and colleagues discovered that the addition of methionine results in the internalization of a G protein-coupled receptor (Gpr4) in Cryptococcus neoformans and that a gpr4 null mutant is still responsive to glucose (323). We discuss the Gpr1 receptor in more detail in “cAMP-PKA Pathway,” below.
Ammonium sensing by Mep2.
Ammonium transport in C. albicans is mediated by Mep1 and Mep2 (33). Recently it was shown that Mep2, but not Mep1, also functions as a receptor for the induction of filamentous growth under nitrogen starvation conditions, placing Mep2 in the transceptor family of proteins (33, 124). More detailed analysis has shown that, as with the budding yeast protein ScGap1, the C terminus of Mep2 is required for proper sensing but not for transport (33). Signaling by Mep2 functions through both the MAPK and the cAMP-PKA pathways and is Ras1 dependent (see below for more information on the Ras pathway). Under high ammonium conditions, when expression of Mep2 is repressed, filamentation is blocked. A role for Mep2 in polarized morphogenesis is supported by work with both budding yeast and Ustilago maydis. In S. cerevisiae, ScMep2 is required for pseudohyphal induction (186). The Mep2 homolog of U. maydis (Ump1) is able to complement an S. cerevisiae mep1 mep2 mep3 strain both for ammonium transport and for pseudohyphal induction (269). Interestingly, mutation of a putative PKA phosphorylation site in either Mep2 or Ump1 specifically blocked pseudohyphal induction, whereas transport capacity was not affected. This result suggests that phosphorylation by PKA is required for Mep2 to be able to exert its effect on pseudohyphal induction.
Role of Gcn4 in nitrogen-regulated morphogenesis in C. albicans.
Nitrogen availability has profound significance in fungal biology. In response to nitrogen limitation, fungi initiate morphological changes, sexual and asexual sporulation, and expression of virulence determinants (200). For instance, haploid MATα cells of C. neoformans, which typically grow as yeast, develop hyphae and fruiting bodies in nitrogen-deficient media; these processes are inhibited by ammonia (318). In S. cerevisiae, starvation for a single amino acid stimulates the expression of genes for all amino acid biosynthetic pathways in a phenomenon termed general amino acid control (GCN response) (122). Nitrogen catabolite repression, the regulation of gene expression in relation to nitrogen availability, has been demonstrated in numerous fungi (200). Though somewhat a misnomer (192), this global control mechanism prevents expression of the many genes required to utilize various secondary nitrogen sources when an adequate supply of preferred nitrogen sources is available, typically ammonium or glutamine (200). Nitrogen regulation is controlled by global positive (Gln3 and Gat1) and negative (Dal80 and Deh1) transcription factors as well as by pathway-specific transcription factors.
Tripathi et al. reported that C. albicans can respond to amino acid starvation in at least two ways: by stimulating cellular morphogenesis and by a GCN-like response (302). Both responses are dependent on Gcn4, a functional homolog of the S. cerevisiae transcription factor ScGcn4. Hence, Gcn4 plays a central role in coordinating morphogenetic and metabolic responses to amino acid starvation in C. albicans (302). In S. cerevisiae, ScGcn4 is mainly regulated at the translational level by the eIF-2α kinase ScGcn2 (122). In C. albicans, however, Gcn4 appears to be regulated mainly at the transcriptional level, with only a small contribution for Gcn2 (301). These differences between C. albicans and S. cerevisiae fit with the fact that ScGcn4 has not been implicated in pseudohyphal development (101), and transcript profiling of the GCN response has not revealed any obvious link with pseudohyphal development in S. cerevisiae (214). These results suggest a divergence in the cellular roles of Gcn4 between S. cerevisiae and C. albicans and underscore the importance of amino acids in the regulation of morphogenesis in C. albicans. In budding yeast, Gcn4 is targeted for degradation by the Cdc4 ubiquitin ligase complex, and deletion of CDC4 in C. albicans results in constitutive hyphal growth (8). This observation suggests that Cdc4-mediated protein degradation may be involved in the regulation of the dimorphic switch. Possible key targets of Cdc4 include Sol1, a homolog of ScSic1, a cyclin-dependent kinase inhibitor, and transcription factor Tec1 (63, 257), which are all targets of ScCdc4. So far, however, none of the candidates alone account for the hyperfilamentation phenotype of the cdc4 mutant.
Signal Transduction, Quorum Sensing, and the MAPK Cascade Module
In eukaryotic cells, MAPK cascades are key elements in mediating the transduction of many signals generated at the surface to the nucleus. As in S. cerevisiae, a MAPK pathway is involved in filamentation in C. albicans. The cascade consists of the kinases Cst20 (homologous to the p21-activated kinase [PAK] kinase Ste20), Hst7 (homologous to the MAPK kinase [MAPKK] Ste7), and Cek1 (homologous to the Fus3 and Kss1 MAPKs) (65, 69, 153, 171, 266, 317) (Fig. 1). The C. albicans MAPKK kinase (MAPKKK) Ste11 has not been characterized in detail, although its homolog in C. glabrata functionally complements an S. cerevisiae Ste11 mutant (51). The transcription factor Ste12 functions downstream of the pheromone-responsive MAPK cascade in budding yeast through a heptamer sequence, TGAAACA, called the pheromone response element (PRE). Genes involved in mating are regulated through the PRE by the Ste12 transcription factor and the regulatory proteins Dig1 and Dig2. By contrast, filamentation gene expression depends on Ste12 acting in concert with a filamentation-specific factor, Tec1, which binds cooperatively with Ste12 to the filamentation response element (64). The C. albicans homolog of Ste12 is called Cph1; expression of CPH1 in S. cerevisiae can complement both the mating defect in haploids and pseudohyphal formation in diploid cells (181, 195). Null mutations in any of the genes in the MAPK cascade (Cst20, Hst7, or Cek1) or the transcription factor Cph1 confer a hyphal defect on solid medium in response to many inducing conditions; however, all of these mutants filament normally in response to serum (69, 153, 171). Interestingly, although a cek1 MAPK mutant strain forms morphologically normal filaments in response to serum, it has a minor growth defect on serum-containing medium (69). The cek1 mutant strain also has a virulence defect that may be attributable to this growth defect (69). These results indicate that the Cek1 MAPK may function in more than one pathway or that deletion of the gene causes aberrant cross-talk between distinct MAPK cascades, similar to the altered signaling reported for MAPK mutants of S. cerevisiae. Recently, clear evidence of an interaction between a second MAPK module—the HOG1 pathway—and the CEK1 pathway has been obtained. Cek1 is activated in certain HOG pathway mutants, apparently to compensate for their defects in cell wall architecture (244). Under conditions that require active growth, Sho1, a sensor protein that links oxidative stress to morphogenesis, is essential for the activation of the Cek1 MAPK (244). Recently, a review of the interactions between the different MAPK pathways in C. albicans was published (205).
Other elements of the Cek1 pathway have small but varied effects on virulence. cst20 mutant strains have a modest virulence defect in a mouse model of systemic candidiasis (171). However, hst7 and cph1 mutant strains are able to cause lethal infection in mice at rates comparable to those of wild-type strains (171, 182). In addition to these components, a MAPK phosphatase, Cpp1, that regulates filamentous growth in C. albicans has been identified (69, 256). Disruption of both alleles of the CPP1 gene derepresses hyphal production and results in a hyperfilamentous phenotype. This hyperfilamentation is suppressed by deletion of the MAPK Cek1 (69), indicating that Cpp1 functions primarily in this MAPK pathway. cpp1 mutant strains are also reduced for virulence in both systemic and localized models of candidiasis (69, 114).
Activation of the MAPK pathway can occur through the Cdc42 small GTPase. Cdc42 binds with high affinity to the Cst20 kinase, as well as to a second PAK kinase, Cla4 (172, 287). Cdc42 and its exchange factor, Cdc24, are both required for hyphal growth (22, 305, 306). Certain point mutants of Cdc42 do not affect its mitotic functions but strongly affect morphogenesis upon serum stimulation. Consistent with this phenotype, the expression of hypha-specific genes is also reduced or transient in these mutants (23, 306). Addition of serum results in a transient increase in CDC24 expression, and this increased expression depends on Tec1, the second transcription factor (the homolog of ScTec1) downstream of the Cek1 MAPK module (23). Cdc24 is then recruited to the tip of the hypha. The regulatory relationship between Tec1 and Cdc24 suggests a positive feedback loop that contributes to the level of active Cdc42 at the tip of the germ tube.
Inputs into the Cek1 MAPK pathway may also occur by quorum sensing. Quorum-sensing molecules allow bacteria to monitor their growth and to control cell density-dependent phenomena. Recently, similar regulatory molecules, tyrosol and farnesol, were identified in C. albicans, and they are involved in morphogenesis. Studies of the morphological transition from a filamentous to a budding yeast form in C. albicans revealed excretion of an autoregulatory substance, 3,7,11-trimethyl-2,6,10-dodecatrienoate, or farnesol, into the medium. Farnesol inhibits filamentous growth and may be involved in developmental signaling (226). Whereas farnesol is a compound that prevents hyphal formation (it is produced during high-density growth), tyrosol stimulates the growth of Candida cells and, under the proper conditions, hyphal formation (61, 125). Recent work shows that farnesol may function through the Cek1 MAPK pathway to inhibit morphogenesis, as the addition of farnesol represses the expression of CPH1 and HST7 (253). How farnesol is sensed and how the signal is transmitted to this MAPK pathway remains to be determined. Interestingly, C. albicans mutants lacking the histidine kinase Chk1 (see below) are refractory to the inhibitory effect of farnesol both in cell suspension and during the formation of a biofilm (158). This indicates a role of two-component signal transduction proteins in quorum sensing and as upstream components of the Cek1 MAPK pathway.
cAMP-PKA Pathway
The cAMP-PKA pathway plays a very important role in filamentation in S. cerevisiae, C. albicans, and other fungi (177). Nitrogen starvation in S. cerevisiae results in the formation of elongated buds termed pseudohyphae, which is dependent on activation of the cAMP pathway (110, 157, 177). In C. albicans, an increase in cAMP levels accompanies the yeast-to-hypha transition, and inhibition of the cAMP phosphodiesterase induces this transition (247). Previous reports of cAMP levels during the yeast-to-hypha transition (13, 85, 193, 247) are difficult to compare because of differences in strains and experimental conditions. Nonetheless, it is clear is that the cAMP signal is less pronounced in C. albicans than in S. cerevisiae. Our understanding of the C. albicans cAMP-PKA pathway is based on considerable work with budding yeast, which we briefly review below. We then discuss the different components of the cAMP pathway in C. albicans and their roles in morphogenesis in more detail.
Upstream components of the cAMP-PKA pathway.
In S. cerevisiae, the cAMP-PKA pathway is activated by a G protein-coupled receptor system consisting of the G protein-coupled receptor ScGpr1 and the Gα protein ScGpa2. ScGpr1 was identified in two-hybrid screens with the Gα protein ScGpa2 as bait (156, 324) and in a screen for mutants deficient in glucose-induced loss of heat resistance, a property controlled by the cAMP-PKA pathway (156). Apart from the receptor, ScGpa2 also interacts with ScGbp1/ScKrh2 and ScGbp2/ScKrh1, two proteins that appear to act as Gβ-mimicking subunits, based on structural resemblance with classical Gβ proteins (24, 117). Recently, two different molecular mechanisms by which these kelch repeat proteins function were described (116, 233). It seems that activated ScGpa2 relieves the inhibition imposed by the kelch repeat proteins on PKA, thereby bypassing adenylate cyclase for direct regulation of PKA. It is also clear that both ScKrh1 and ScKrh2 may bind different components of the pathway, thereby functioning as scaffolding proteins. Another interaction partner of ScGpa2 is ScGpg1, which has predicted structural properties typical of a Gγ-like subunit, although ScGpg1 lacks apparent target sequences for posttranslational modifications, which are typical of Gγ subunits (117). The activity of ScGpa2 is also controlled by the RGS protein ScRgs2 (309). Hence, a G protein-coupled receptor (GPCR) system composed of ScGpr1, ScGpa2, and ScRgs2 has been proposed to act as a glucose-sensing system for control of the cAMP pathway (296, 310). Recently we obtained evidence that ScGpr1 is a sensor for sucrose and glucose and that mannose acts as an antagonist (174). This GPCR system is required for pseudohyphal and invasive growth induction, and ScGPR1 or ScGPA2 mutants, deficient in this morphogenesis, can be suppressed by the addition of cAMP (187, 188, 293).
Genes similar to ScGPA2 and ScGPR1 have been identified in C. albicans, but their precise functions remain unclear. Genetic evidence suggests that GPA2 may function upstream of both the Cek1 MAPK pathway (249) and the cAMP-PKA pathway (193, 204). Deletion of GPA2 or GPR1 produces defects in hyphal formation and morphogenesis in C. albicans, which are reversed by exogenous addition of (db)cAMP or by overexpression of downstream components in the pathway. As expected, epistasis analysis revealed that Gpa2 (the Gα protein) acts downstream of Gpr1 (the receptor) in the same signaling pathway, and a two-hybrid assay indicated that the carboxy terminus of Gpr1 interacts with Gpa2. Moreover, expression levels of HWP1 and ECE1, which are cAMP-dependent hypha-specific genes, are reduced in both mutants (204). Interestingly, the morphogenesis defect is present only when cells are cultured on solid media. A possible link between Gpa2 and the CEK1 MAPK pathway was recently solidified by the discovery that Gpa2 is involved in mating. Thus, Gpa2 seems to integrate the nutrient-sensing pathway with the pheromone response MAPK pathway, providing an explanation for why the function of the latter pathway strongly depends on nutritional conditions (27).
Conflicting data regarding the possible ligand for Gpr1 exist. Miwa et al. (204) showed that, as in S. cerevisiae, Gpr1 is required for the glucose-induced cAMP signal, but we found that the gpr1 strain showed the same glucose-induced increase in cAMP as did a wild-type strain (193). However, deletion of either CDC25 or RAS1 affected the glucose- and serum-induced cAMP signal, consistent with mediation of the glucose-induced cAMP increase in C. albicans by the Cdc25-Ras1 branch of the glucose response pathway and probably not via the Gpr1-Gpa2 branch (193). Addition of serum results in a rapid response, similar to that seen with glucose, supporting a recent finding that glucose is the main factor in serum affecting Candida morphogenesis (135). Two important gaps remain in our understanding of the upstream components of the cAMP-PKA pathway in C. albicans: (i) the identity of the receptor activating the Cdc25 guanine nucleotide exchange protein and (ii) the nature of the ligand for Gpr1. No data from budding yeast or C. albicans regarding the Cdc25 receptor are available, but, as mentioned above, preliminary investigations aimed at identifying the possible ligand for Gpr1 seem to point to amino acids, specifically methionine, although glucose or other sugars cannot be excluded (193, 194).
The concentration of glucose in the medium is a very important parameter for the hyphal response. Most research groups use normal yeast extract-peptone-dextrose or synthetic complete medium containing 2% glucose. Under these conditions, a wild-type strain displays smooth colonies. However, in the presence of more physiologically relevant concentrations (around 0.1% glucose in the blood), and when there is methionine present in the medium, a wild-type strain rapidly starts to form hyphae. This methionine-dependent, low glucose concentration-induced hyphal production is completely absent in the gpr1 mutant (194), as well as in an hgt12 mutant (189). Hgt12 is homologous to S. cerevisiae Snf3, which is the high-affinity glucose sensor required for the expression of glucose transporter genes. The pathway that is activated by Hgt12 in C. albicans remains to be investigated, but the Hgt12 receptor is required for hyphal development during macrophage infection. Hgt12 does not have the long C-terminal tail that is typical of the yeast Snf3 and Rgt2 glucose sensors. Recently, conflicting data regarding the role of Hgt12 as a sensor have been presented. Brown and colleagues constructed independent Hgt12 mutants and were unable to reproduce the growth or filamentation abnormalities. They showed that Hgt4, another homolog of the yeast Snf3 glucose sensor that does contain a long C-terminal tail, might be the sensor regulating the expression of Hgt12, which, according to these authors, is a normal glucose transporter (48).
Ras1, the master hyphal regulator.
Mutants of the single Ras homolog, Ras1, in C. albicans are viable but have a severe defect in hyphal growth in response to serum and other inducing conditions (92). In addition, while a dominant-negative Ras1 mutation [Ras1(A16)] causes a defect in filamentation, a dominant-active Ras1 mutation [Ras1(V13)] enhances the formation of hyphae (92). As mentioned above, the morphogenesis defect of a ras1 mutant can be suppressed by exogenous cAMP, and ras1 mutants are defective in cAMP induction in response to glucose and serum stimulation. Both phenotypes are shared with mutants lacking the guanine nucleotide exchange factor Cdc25 (193), suggesting that the Cdc25-Ras1 pathway functions upstream of the cAMP-PKA pathway. This is supported by a recent study in which a clear interaction between Ras1 and adenylate cyclase, Cyr1, is demonstrated (90). However, as with the situation in S. cerevisiae, Ras1 also appears to function upstream of the Cek1 MAPK pathway (Fig. 1), as the morphogenesis defect of a ras1 mutant can also be suppressed by overexpressing components of the filament-inducing MAPK cascade. Other evidence for a role in both pathways is that ras1 mutant strains exhibit a filamentation defect that is similar to that of the efg1 cph1 mutant strain (see below).
Cyr1, Srv2, and Pde2.
C. albicans has a single gene homologous to the S. cerevisiae adenylate cyclase gene (CYR1/CDC35). Like Ras1, the cyclase is not essential for growth in C. albicans but is required for hyphal development (243). Also as in budding yeast, genes encoding an adenylate cyclase-associated protein (SRV2) and two phosphodiesterases (PDE1 and PDE2) are present in the C. albicans genome. The Srv2 protein regulates adenylate cyclase activity and is required for wild-type germ tube formation and for virulence in a mouse model of systemic infection; it is the homolog of the S. cerevisiae Cap1 protein. Like other mutants in the pathway, srv2 mutants have defects in morphogenesis, but they can be rescued by the addition of cAMP (13). Interestingly, the basal level of cAMP is higher in the srv2 mutant than in the wild type (61.8 ± 6.5 versus 45.3 ± 4.6 pmol/mg protein), indicating a possible negative-feedback inhibition.
To characterize more fully the effects of hyperactivation of the cAMP pathway, two groups disrupted the PDE2 gene, which encodes the high-affinity phosphodiesterase (PDEase) (16, 143). An advantage of studying cAMP hyperactivation by disrupting PDE2 rather than overexpressing TPK1 and TPK2, which encode the catalytic subunits of PKA, is that the pde2 mutant displays phenotypes that result solely from an increase in the cAMP level, thereby preempting any unpredictable effects of inequitable overexpression of one cAMP signaling component relative to the others. ScPDE1 and ScPDE2 encode the low- and high-affinity cAMP PDEases, respectively, in S. cerevisiae (190, 221, 252). Budding yeast Scpde1 and Scpde2 mutants exhibit sensitivity to heat shock and nutrient starvation (190), and Scpde2 mutants also have cell wall-related phenotypes, such as lysis upon hypo-osmotic shock, suggesting a role for ScPde2 (and/or cAMP) in the maintenance of cell wall integrity in S. cerevisiae (299). Although C. albicans also has a low-affinity PDEase encoded by PDE1 (127), so far only PDE2 has been studied in detail. As with the situation in budding yeast, pde2 mutants show cell wall- and membrane-related phenotypes, such as increased sensitivity to membrane-perturbing agents such as sodium dodecyl sulfate or antifungals such as amphotericin B and a strong reduction in the thickness of the cell wall, caused mainly by alterations in the ergosterol and glucan composition (144). PDE2 mutants are also sensitive to nutrient deprivation and defective in entry into stationary phase and are avirulent in a mouse model of systemic candidiasis (16). Under conditions in which wild-type strains form smooth colonies composed of budding yeasts on agar media, the homozygous pde2 mutant displayed a wrinkled colony phenotype. The wrinkled colonies exhibited mixtures of elongated yeasts and filamentous forms (pseudohyphae and true hyphae). Germ tube formation by the homozygous pde2 mutant was accelerated in liquid media compared to wild-type strains. Thus, it is clear that Pde2 is required to regulate the level of cAMP in the cells and that disruption of this enzyme results in a constitutive activation of the PKA pathway.
Adenylate cyclase and CO2 sensing.
C. albicans is present in various parts of the body where the CO2 concentration is more than 150-fold higher (5%) than in atmospheric air (0.033%). In other words, cells growing on the skin face much lower CO2 concentrations than do cells present in the intestine or blood. The presence of 5% CO2 strongly induces pseudohyphal development and invasion of the underlying agar (151), a response that requires the catalytic domain of adenylate cyclase (Cyr1/Cdc35) but not Ras1. In many cases, cellular effects observed with CO2 can be mediated via its hydrated form, bicarbonate. C. albicans NCE103 encodes a carbonic anhydrase that greatly accelerates bicarbonate formation. Nce103 is required for C. albicans cells to grow in air but not under high CO2 concentrations (>0.5%). Nce103 is also required for tissue damage under atmospheric conditions but not under high CO2 conditions. Whether there is indeed a specific CO2 sensor protein on the cell membrane (e.g., through aquaporin water channels as in plant and mammalian cells) remains to be established. Klengel et al. have already excluded C. albicans Aqy1 as a possible candidate (151). A similar CO2-sensing pathway in Cryptococcus neoformans has also been described (14), and a review of this topic was recently published (15).
PKA.
Growth and cellular differentiation of eukaryotic cells depends to a large extent on the activity of cAMP-dependent protein kinases (PKA). PKAs are structurally conserved, consisting of two catalytic subunits that are inactivated by the binding of a homodimer of regulatory subunits. External cues elevate intracellular levels of cAMP, whose binding to the regulatory subunits liberates and thereby activates the catalytic subunits. A comparison of PKA subunit isoforms in C. albicans and S. cerevisiae reveals several significant differences. (i) In S. cerevisiae, PKA is encoded by three paralogues (ScTPK1 to ScTPK3) (298), whereas only two paralogues (TPK1 and TPK2) are present in the genome of C. albicans (66). (ii) In C. albicans, both isoforms have a positive, stimulatory function on the formation of true hyphae, whereas in S. cerevisiae, ScTpk2 has a positive function and ScTpk1 and ScTpk3 have negative functions in pseudohyphal development (229).
Although both C. albicans Tpk isoforms act positively to regulate hyphal formation, they have various phenotypic effects. For example, tpk1 mutants are defective in hyphal formation on solid media but are less affected in liquid media (276) (Fig. 1). In contrast, hyphal formation in tpk2 mutants is partially affected on solid media but is blocked in liquid medium. Because Tpk1 and Tpk2 mainly differ in their N-terminal domains (80 to 90 amino acids), hybrid Tpk proteins, with exchanged N-terminal domains, were tested by Bockmuhl et al. (37). Tests of hybrid proteins suggested that the catalytic domain mediates Tpk protein specificities in filamentation, whereas agar invasion is mediated by the N-terminal domain of Tpk2.
The regulatory subunits of PKA in S. cerevisiae and C. albicans are encoded by ScBCY1 and BCY1, respectively (58, 298). Cantore's group examined the morphogenetic behavior of C. albicans yeast cells lacking the PKA regulatory subunit and generated a bcy1 tpk2 double mutant strain, because a homozygous bcy1 mutant in a wild-type genetic background could not be obtained. In the bcy1 tpk2 mutant, PKA activity (due to the presence of the TPK1 gene) was cAMP independent, indicating that the cells harbored an unregulated phosphotransferase activity. This mutant has constitutive protein kinase activity and displays a defective germinative phenotype in GlcNAc and in serum-containing media. In addition, a Tpk1-green fluorescent protein (GFP) fusion is dispersed throughout the cell in the bcy1 tpk2 double mutant, while it is normally predominantly nuclear in wild-type cells. These genetic studies, together with biochemical evidence, suggest that C. albicans Bcy1 may tether the PKA catalytic subunit to the nucleus and thereby perform a pivotal role in regulating the enzymatic activity and availability of PKA in response to growth phase-related nutritional requirements (58).
Efg1 and Efh1, Major Transcriptional Regulators
The C. albicans EFG1 and EFH1 genes encode basic helix-loop-helix (bHLH) transcription factors that are members of the APSES family of fungus-specific regulators and involved in morphogenetic processes (81). This class includes StuAp of Aspergillus nidulans and Asm1 in Neurospora crassa, as well as the Phd1 and Sok2 proteins of S. cerevisiae. StuA is involved in the formation of conidiophores, which are important for asexual reproduction in A. nidulans (202, 264), while Asm1 of N. crassa has a role in ascospore maturation (6). Phd1 enhances and Sok2 suppresses pseudohyphal formation in S. cerevisiae (109, 314). In C. albicans, Efg1 plays a central role in morphogenesis. Below, we first discuss Efg1 as a downstream component of the PKA pathway. Next, we discuss target genes induced or repressed by Efg1, followed by a more general discussion of the genetic network regulating the expression of yeast- or hypha-specific genes. Finally, we discuss the role of Efg1 in morphogenesis under embedded conditions.
Efg1 is a downstream component in the cAMP-PKA pathway.
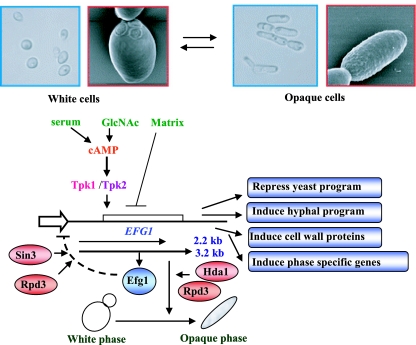
Several lines of evidence suggest that Efg1 functions downstream of PKA. Overexpression of TPK2 is unable to suppress the mutant phenotype of efg1, whereas overexpression of EFG1 can bypass the filamentation defect in a tpk2 mutant (276). The suppression activity of Efg1 depends on a threonine residue at position 206, a potential phosphorylation site for PKA (36). Induction of morphogenesis after the addition of serum, a process known to be regulated by the PKA pathway, requires Efg1. When EFG1 transcript levels were examined in transformants overexpressing TPK1 or TPK2, a reduction (about fourfold) of EFG1 mRNA was observed (294). This finding suggests that Efg1 is down-regulated by its immediate upstream regulator, PKA, but other observations point to autoregulation of EFG1 gene expression. Overexpression of EFG1 increases the levels of a smaller 2.2-kb transcript produced by the locus, while the levels of the endogenous 3.2-kb major EFG1 transcript decline dramatically (294) (Fig. 2). Serial deletion analysis of the promoter region revealed that the TATA box region is required for EFG1 autoregulation. Binding of Efg1 to the EFG1 transcriptional initiation region by chromatin immunoprecipitation was also shown. In summary, it appears that the levels and/or activities of Efg1, Tpk1, and Tpk2 are able to down-regulate the major EFG1 promoter. In addition, Efh1, the homolog of Efg1, also affects Efg1 down-regulation, indicating that at least four proteins can contribute to EFG1 expression. Negative autoregulation and PKA-mediated down-regulation are probably mediated though the Sin3-Rpd3 (279)-containing histone deacetylase complex (Fig. 2).
FIG. 2.
(Top) White-opaque switching in C. albicans. Shown are scanning electron micrographs at a magnification of ×1,000 and bright-field images of white and opaque cells. (Reprinted, with permission, from the Annual Review of Microbiology [26], volume 59, © 2005 by Annual Reviews.) (Bottom) Model of morphogenetic regulation by Efg1. Under hypha-inducing conditions (e.g., serum, GlcNAc), Efg1 is induced and activated; under microaerophilic conditions, however, it is repressed. The activated Efg1 (by PKA isoforms Tpk1 and Tpk2) initiates hyphal formation by inducing genes involved in hyphal formation and/or repressing genes directing the yeast form. Efg1 also induces the cell wall proteins (HWP1, HWP2, and RBE1) that are involved in adherence. The phase-specific genes (at the white-to-opaque-phase transition period) are also induced by Efg1. In parallel, Efg1, in conjunction with the Sin3-Rpd3 deacetylase complex, silences chromatin and thereby down-regulates EFG1 promoter activity (294). The 3.2-kb major transcript of EFG1 is expressed in the white phase, and the less-abundant 2.2-kb transcript is expressed in opaque cells. The Hda1-Rpd3 deacetylase complex regulates the white-to-opaque-phase transition as well as EFG1 down-regulation (279).
Roles of Efg1 and Efh1 in morphogenesis in C. albicans.
Overexpression of Efg1 or Efh1 induces pseudohyphae and opaque-white switching, a specific morphogenetic transition (81, 182, 285). An efg1 mutant strain has a moderate but not complete defect in hyphal growth in response to many environmental conditions (182). Addition of serum or GlcNAc as an inducer in liquid or on solid medium completely blocks hyphal formation in efg1 null mutants. In contrast, under microaerophilic or embedded conditions, hyphal formation is unaffected—and may be stimulated—in homozygous efg1 mutants (111). These results indicate that, depending on environmental cues (and depending on the genetic background), Efg1 may act as a transcriptional activator and repressor for different subsets of genes, whose balanced activities are essential for yeast, pseudohyphal, and hyphal morphogenesis of C. albicans. An efg1 cph1 double mutant strain has an extreme filamentous growth defect, with no detectable filamentation under almost any conditions tested, including the presence of serum (182). In addition, while the efg1 mutant strain has a minor reduction in virulence and the cph1 mutant has little or no defect, an efg1 cph1 double mutant strain is essentially avirulent in a mouse model of systemic infection (182). Recently, it was shown that this double mutant is still able to cause infections in the kidneys of infected mice. So, although this mutant is avirulent, local proliferation of C. albicans cells can occur in certain tissues (60). Thus, Cph1 and Efg1 define elements of two separate pathways that together are essential for both filamentation and virulence in C. albicans.
Efg1 and embedded growth.
As we have just described, efg1 mutants have a drastic block in true hyphal formation under most standard induction conditions, but considerable filamentation occurs in certain environments (46, 276). A limited supply of oxygen, as occurs under a coverslip during induction of chlamydospores, allows wild-type cells to form filaments, which is enhanced in efg1 mutants (275). Similarly, growth of wild-type colonies embedded in agar stimulates filamentation, which still occurs in homozygous efg1 cph1 mutants (111, 242). Thus, an Efg1-independent pathway of filamentation that is operative under microaerophilic/embedded conditions appears to exist in C. albicans. In wild-type strains, the filaments produced under microaerophilic conditions are mostly pseudohyphae; however, they are mostly true hyphae in efg1 mutants (276). Interestingly, the alternative filamentation pathway is not only independent of Efg1 but may be repressed by it to a certain degree. The enhanced filamentation in efg1 mutants does not depend on the Cph1 MAPK, because a homozygous efg1 cph1 strain is as hyperfilamentous as the homozygous efg1 mutant (276). It is possible that agar embedding generates microaerophilic conditions, which activate the same Efg1-independent pathway of morphogenesis under both conditions. The putative transcription factor Czf1 is probably an important element of the alternative pathway of filamentation in C. albicans (47). The central portion of Czf1 contains four clusters of glutamine residues, and the C terminus contains a cysteine-rich region similar to zinc finger elements. Overexpression of CZF1 stimulates filamentous growth but only under embedded conditions and in certain media lacking glucose. The expression of Czf1 is strongly up-regulated in a vps34 mutant; as a result, a vps34 mutant is derepressed in hyphal formation under microaerophilic conditions (150). Vps34 is phosphatidylinositol-3-kinase, which influences vesicular intracellular transport, filamentous growth, and virulence. The exact role of Vps34 in Czf1-mediated morphogenesis is not yet clear. Homozygous czf1 null mutants filament normally under standard induction conditions, but they are defective in hyphal development when embedded in agar. This defective phenotype occurs only during embedding in certain media, such as complex medium containing sucrose or galactose as carbon sources at 25°C, but not at 37°C, or in media containing strong inducers, including serum and GlcNAc. These characteristics suggest that factors other than Czf1 contribute to filamentation under embedded conditions. The defective phenotype of a czf1 mutant is exacerbated by the presence of a cph1 mutation, which by itself shows defects in the types of media used for monitoring the czf1 phenotype. Thus, although the cph1 mutant phenotype does not appear to be specific for embedded conditions, it worsens the filamentation defects caused by the czf1 mutation. Hyperfilamentation of efg1 single and efg1 cph double mutants suggests that Efg1 is a negative modulator of the Czf1 pathway under microaerophilic/embedded conditions. However, recent data show that this hyperfilamentation phenotype of efg1 mutants may be caused by a Czf1-independent pathway, as efg1 mutants do not express CZF1. Chromatin immunoprecipitation analysis has further indicated that Efg1 and Czf1 interact with the promoter of CZF1. This indicates that, like EFG1, CZF1 is autoregulated (312).
Other aspects of C. albicans morphogenesis are also dependent on Efg1. Chlamydospore formation requires Efg1 protein in PKA-dependent regulation, as T206 is required (275). Also, recent studies revealed the involvement of EFG1 in normal biofilm formation (237), as well as in normal filamentation under oxygen limitation conditions (260), which are probably similar to the microaerophilic conditions. Under these conditions, Efg1 seems to function as a repressor of filamentation. In contrast to efg1, an efh1 mutant does not give any clear phenotype except in an efg1 mutant background. In this case, an efh1 null mutant is hyperfilamentous under embedded or hypoxic conditions, indicating the cooperation of Efg1 and Efh1 in suppression of an alternative morphogenetic signaling pathway (81).
Efg1 and phenotype switching.
As noted briefly above, one form of phenotypic switching is the white-opaque transition, first described by Soll and colleagues in 1987 (268). C. albicans colonies of some strains (e.g., WO-1) can switch from the normal size, shape, and white color with high frequency (274) to larger, flatter, and gray colonies. White cells are similar in shape, size, and budding pattern to cells of common laboratory strains. Opaque cells, however, are bean shaped and exhibit three times the volume and twice the mass of white cells (Fig. 2) (268). Transcription of EFG1 is regulated in a unique fashion during the white-opaque transition (280); the more-abundant 3.2-kb transcription is expressed in the white phase, while the less-abundant 2.2-kb transcription is detected in the opaque phase (Fig. 2). Experiments using deletion or overexpression of EFG1 show that Efg1 functions downstream of the switch event in the regulation of a subset of white-phase-specific genes involved in the generation of the round white cell shape (278). Furthermore, a detailed promoter analysis suggests that the upstream region of EFG1 contains overlapping promoters for the expression of white-phase-specific and opaque-phase-specific transcripts (280). Overlapping promoters are also seen for the α and β mRNAs of the EFG1 homolog StuA in A. nidulans (320). It should be noted that EFG1 is not the only phase-regulated gene expressed as an alternative-molecular-weight transcript during the white-opaque transition in C. albicans. The deacetylase HOS3 is also transcribed as a 2.5-kb transcript in the white phase and as a less-abundant, lower-molecular-weight 2.3-kb transcript in the opaque phase (279). Recently, a key transcription factor required to establish and maintain the opaque growth phase was described by different groups. Like Efg1, this transcription factor, Wor1 (Tos9), binds to its own regulatory region, thereby activating its own expression. It is believed that high levels of Wor1 regulate the epigenetic inheritance of the opaque phase of growth (60, 131, 277).
Efg1 and cell wall dynamics.
Recently, several genes encoding cell wall components were shown to be regulated by Efg1, implying cell wall regulation in C. albicans morphogenesis. These genes include the cell wall mannoprotein Hwp1 (42, 262), the glycosylphosphatidylinositol (GPI)-anchored cell wall protein HYR1 (17), and ALS1, which encodes a cell surface glycoprotein (95). In order to analyze cell wall dynamics and the regulatory function of Cph1 and Efg1 in the transcriptional control of cell wall genes in a systematic manner, Sohn et al. used a DNA microarray to assay transcriptional profiles from wild-type cells and cph1, efg1, and cph1 efg1 double-mutant strains cultured under various yeast- or hypha-inducing conditions (271). Overall, their data demonstrate that Efg1 plays a major role in the regulation of cell wall genes analyzed under both yeast- and hypha-inducing conditions, while Cph1 plays a minor role. During induction of filamentation, many hypha-specific genes that are important for adhesion and virulence are expressed in wild-type cells (in an Efg1-dependent manner). Among those genes are the hyphal wall proteins HWP1 and HWP2. Concomitant with the up-regulation of hyphal wall proteins, the expression of yeast-specific genes such as YWP1 is down-regulated during the yeast-to-hypha transition. Thus, the reduced virulence of the efg1 mutant likely reflects both a lack of expression of hypha-specific genes and a change in the overall organization of the cell wall (155, 182). Microarray analysis has further demonstrated that apart from yeast- or hypha-specific genes, Efg1 is also important for the expression of metabolism genes, inducing glycolytic genes and repressing genes essential for oxidative metabolism (81). Finally, Erg3, which is involved in drug resistance, is also down-regulated by Efg1, and efg1 mutants are more resistant to antifungal agents (183).
Convergent Regulation of Cph1 and Efg1: Involvement of Tec1 and Cph2
Tec1, a member of the TEA/ATTS family of transcription factors, has been shown to regulate hyphal development and virulence in C. albicans (257). TEA/ATTS family members AbaA in A. nidulans and ScTec1 in S. cerevisiae are involved in the regulation of conidiophore formation and pseudohyphal growth, respectively (5, 106). As mentioned before, in S. cerevisiae ScTec1 and ScSte12 form a transcription factor complex to specifically activate genes involved in pseudohyphal growth (64, 191). Such complexes have not yet been identified in C. albicans. In C. albicans, TEC1 transcription is not regulated by the ScSte12 homolog Cph1 (171) but by Cph2 and Efg1. Cph2, an myc family bHLH protein, regulates hyphal development in C. albicans (169). cph2 mutant strains are impaired in hyphal development and in the induction of hypha-specific genes in liquid Lee's medium, and Cph2 is necessary for the transcriptional induction of TEC1 (Fig. 1). Cph2 binds directly to two sterol regulatory element 1-like elements upstream of TEC1. Furthermore, the ectopic expression of TEC1 suppresses the defect of cph2 in hyphal development. The function of Cph2 in hyphal transcription is therefore mediated, in part, through Tec1. tec1 mutants exhibit suppressed filamentation in liquid serum-containing media. EFG1 overexpression does not suppress the morphological defect of the tec1 mutant, whereas TEC1 overexpression has a partial phenotype in the efg1 mutant (257). These results, coupled with the fact that efg1 strains have a more severe defect in hyphal development than do tec1 strains, suggest that Tec1 is downstream of Efg1. In summary, then, it is clear that Tec1 is downstream of both Cph2 and Efg1.
The study of the C. albicans yeast-to-hypha transition has been informed by studies performed with budding yeast. Very recently, the group of Snyder performed a detailed analysis of the transcription factor network involved in the yeast-to-pseudohypha transition in S. cerevisiae. Predicted binding sites for two transcription factor-encoding genes, ScPHD1 and ScMGA, appear upstream of most genes involved in pseudohyphal growth, including Flo8 (38). Overexpression of either ScPHD1 or ScMGA1 induces pseudohyphal growth, even under noninducing conditions. This indicates that ScPhd1 and ScMga1 are the master regulators in this system. ScPhd1 is the homolog of C. albicans Efg1, and ScMga1 is homologous to orf19.3969, which has not yet been characterized in C. albicans (44). A common theme for regulation of gene expression during pseudohyphal development is the cooperative binding of transcription factors (probably in preformed complexes, as described above) to target promoters. Cooperative binding between ScTec1 and ScSte12 is well established (64, 191), and ScFlo8 and ScMss1 cooperate to activate ScFLO11 or ScSTA1 expression (149, 307). Other cooperating pairs include ScMga1 and ScFlo8 (38) and the C. albicans APSES proteins Efg1 and Efh1 (81). Recently the C. albicans FLO8 gene was characterized (56). As in S. cerevisiae, Flo8 is required for hyphal development and for hypha-specific gene expression. Cooperative binding of two different transcription factors may also occur in C. albicans, as Flo8 and Efg1 interact with each other and Flo8 controls a subset of the target genes of Efg1 (56).
Other Positive Regulators: Cdc5, G1 Cyclins, Int1, Mcm1, and Fkh2
Several other genes in C. albicans may contribute to morphogenesis, but information is limited or not well focused on pathogenicity relationships. These include genes encoding the polo-like kinase Cdc5, three major G1 cyclins, and MADS box and forkhead transcription factor families, among others (Table 1). C. albicans cells lacking the polo-like kinase CDC5 are blocked early in nuclear division. Cell cycle arrest is characterized by the formation of hypha-like filaments under yeast growth conditions. The filaments resemble serum-induced hyphae, and filament formation is independent of Cph1 or Efg1 but requires Cyr1/Cdc35 (11). Microarray experiments confirmed the resemblance of cell cycle-arrested filaments to serum-induced hyphae, as in both cases several targets of hypha-signaling pathways were expressed (10).
Cyclin-dependent protein kinases (Cdks) regulate major cell cycle transitions in eukaryotic cells. In S. cerevisiae, three G1 cyclins, ScCln1, ScCln2, and ScCln3, activate the Cdc28 Cdk to promote G1-phase progression (30). C. albicans contains homologs of the S. cerevisiae G1 cyclins, including Ccn1 (= ScCln1), Hgc1, and Cln3 (www.candidagenome.org). Deletion of CCN1 results in the inability to maintain hyphal growth under certain conditions (185), while deletion of HGC1 prevents hyphal growth under all hypha-inducing conditions (332). Ccn1 and Hgc1 are not essential for progression through the cell cycle (they have a slow-growth phenotype), suggesting that these G1 cyclin homologs in C. albicans have evolved important roles in hyphal morphogenesis as opposed to cell cycle progression. The induction of three known hypha-specific genes, HYR1, ECE1, and HWP1, in ccn1 mutant cells in response to serum is slightly reduced (about 50%) in comparison to that in wild-type cells. On the other hand, in liquid amino acid-based Lee's medium, the ccn1 strain shows a profound defect in the transcriptional activation of all three hypha-specific genes. Thus, Ccn1 may coordinately regulate hyphal development with signal transduction pathways in response to various environmental cues (185). Cln3, however, is essential for the C. albicans yeast cell cycle. When unbudded cells are depleted for Cln3, they increase in size but do not bud (12, 59). Eventually, however, these enlarged cells spontaneously form true hyphae. Cln3 seems to be less important for the hyphal cell cycle, although morphological abnormalities are observed in Cln3-depleted cells when they are treated with hypha-inducing signals such as serum (59). Apart from the G1 cyclins, C. albicans also contains two B-type cyclins, Clb2 and Clb4, that are important for morphogenesis but affect polarized growth negatively (28).
Other kinases have also been investigated for their role in pseudohyphal development. The Nim1 kinases, Gin4 and Hsl1, are important for the formation of septin structures, and both gin4 and hsl1 mutants form pseudohyphae constitutively (319). In S. cerevisiae, ScHsl1 regulates the tyrosine phosphorylation of the cyclin-dependent protein kinase Cdc28 by deactivating the protein kinase ScSwe1, which regulates the G2/M transition (295). Interestingly, in C. albicans, the Hsl1-Swe1-Cdc28 pathway appears to be important for cell elongation of both the yeast and hyphal forms and for virulence (304). Gin4-depleted pseudohyphae are unable to form hyphae when challenged with serum, but this can be overcome by ectopic expression of Gin4 from the MET3 promoter. Thus, Gin4 plays an important role in regulating the developmental switch from pseudohyphae to hyphae (Fig. 3) (319).
FIG. 3.
Multiple environmental conditions activate the kinase cascade, resulting in coordination of the stress response with morphogenesis. Arrows indicate activation; lines with bars indicate inhibition. See the text for details.
Another morphogenesis factor, Int1, was originally cloned by virtue of its limited homology to vertebrate leukocyte integrins (98). Like cph1 mutants, C. albicans int1 strains have a reduced ability to form hyphae in response to most hypha-inducing conditions but form apparently normal hyphae in the presence of serum (100). The C terminus of Int1 has homology to S. cerevisiae Bud4, which is localized to the septin rings at the mother-bud neck (7). C. albicans INT1 induces filamentous growth in S. cerevisiae, and this property has been used to explore the cytoskeleton components required for INT1-induced filamentous growth. Sla2, a cytoskeleton protein, may interact with Int1 to mediate morphogenesis by modulating the actin skeleton (7). In C. albicans, Int1 is important in axial budding pattern and colocalizes with the Cdc3 septin in a ring at the mother-bud neck of yeast and pseudohyphal cells. Under conditions that induce hyphae, both Cdc3 and Int1 localize to a ring distal to the junction of the mother cell and germ tube. Thus, placement of the Int1/septin ring with respect to the mother-daughter cell junction distinguishes yeast/pseudohyphal growth from hyphal growth (99).
Finally, several cell cycle-regulated transcription factors, such as the MADS box and forkhead transcription factors, are also required for morphogenesis. The C. albicans homolog of the MADS box transcription factor MCM1 was identified in a screen for genes that could activate pFLO11::lacZ expression in S. cerevisiae (245). Either overexpression or repression of MCM1 induces hyphae in C. albicans, indicating that correct timing of expression is important for normal morphogenesis.
In S. cerevisiae, two forkhead transcription factors, ScFKH1 and ScFKH2, regulate the expression of genes whose transcription peaks in early M phase (163, 333), including mitotic cyclins. In addition, Scfkh1 Scfkh2 mutants display constitutive pseudohyphal growth. C. albicans has only one forkhead homolog, FKH2. Cells lacking this gene also form constitutive pseudohyphae under all yeast and hyphal growth conditions tested. fkh2 mutants exhibit reduced expression of hypha-specific mRNAs in the presence of serum, while under yeast growth conditions expression of several genes encoding proteins likely to be important for cell wall separation is reduced. Together, these results imply that Fkh2 is required for morphogenesis of true hyphae as well as of yeast cells. Cph1 and Efg1 are not required for pseudohyphal morphology of fkh2 mutants, implying that Fkh2 acts in a parallel or downstream pathway. Cells lacking Fkh2 do not damage human epithelial and endothelial cells in vitro, suggesting that Fkh2 contributes to C. albicans virulence (29).
Transcriptional Repressor Tup1
As highlighted throughout this review, activation of signal transduction pathways causes transcription factors to induce global expression programs that direct cells among distinct developmental pathways. The Tup1 transcription factor is required to keep cells in the yeast form in the presence of glucose and other noninducing conditions. In S. cerevisiae, the ScTup1 protein regulates about 60 genes involved in glucose regulation, oxygen stress response, and DNA damage (79). The C. albicans Tup1 homolog (41) contains seven conserved WD40 repeats at the C terminus, which likely promote interaction with DNA-binding proteins (DBPs), and an N-terminal domain that promotes interaction with the Ssn6 corepressor, as in budding yeast (147, 154). A homozygous C. albicans tup1 mutant grows in filamentous form in all media tested; filaments on most media have characteristics of pseudohyphae, but in some media true hyphae appear. Pseudohyphae of a tup1 mutant, unlike pseudohyphae produced by EFG1 overexpression (285), cannot be induced to form germ tubes or true hyphae by the addition of serum (41). Tup1 has activities besides repression of filamentation, because tup1 mutants exhibit pleiotropic phenotypes, including a failure to grow at 42°C and misshapen cell walls. In epistasis experiments, most of the filamentation phenotype induced by the tup1 mutation is abolished by the presence of an efg1 mutation, while a cph1 mutation has very little effect. These and other genetic results indicate that Efg1 is the main contributor to the tup1 hyphal phenotype. Genes repressed by Tup1 have been identified recently, of which some are expressed in a filament-specific manner (42). Braun et al. used subtractive hybridization to identify six genes termed “repressed by TUP1” (RBT), whose expression is regulated by Tup1 (40). One of the genes (HWP1) has previously been characterized, and a seventh TUP1-repressed gene (WAP1) was recovered due to its high similarity to RBT5 (Fig. 1). These genes all encode secreted or cell surface proteins, and four out of the seven (HWP1, RBT1, RBT5, and WAP1) encode putatively GPI-modified cell wall proteins. The remaining three, RBT2, RBT4, and RBT7, encode, respectively, an apparent ferric reductase, a plant pathogen-related protein (PR-1), and a putative secreted RNase T2.
In S. cerevisiae, ScTup1 forms a transcriptional corepressor complex in concert with ScSsn6 and regulates a diverse set of genes controlled by mating type, glucose, oxygen, and DNA damage (270). Although neither ScTup1 nor ScSsn6 itself has any DNA-binding activity, each is recruited to a specific promoter through interaction with distinct upstream DBPs, such as ScMig1, ScRox1, ScCtr1, and ScNrg1 (270). C. albicans Ssn6 encodes a putative global transcriptional corepressor (136). Its expression level declines significantly in response to strong hyphal inducers, such as serum. Overexpression of Ssn6 causes increased filamentation and decreased virulence (136). Mutants lacking Ssn6 display a stubby pseudohyphal growth pattern, derepressed expression of filament-specific genes in response to increased temperature, and failure to develop true hyphae. Such morphological defects are not rescued by overexpression of TUP1, CPH1, or EFG1. Recent data indicate that Ssn6 is not essential for repression of hypha-specific genes, at least under a few conditions (Fig. 4), and ssn6 mutants form filaments in response to serum in the absence of Cph1 and Efg1. These results suggest that filamentation of ssn6 mutants is not dependent on the MAPK or cAMP protein kinase pathways (104).
FIG. 4.
(Top) Ssn6 is not directly required for Tup1-mediated repression of hypha-specific genes (HSG's), and Tup1 may interact directly with the DBP. The DBP-responsive element (RE) may affect Ssn6 dependency. (Bottom) Model summarizing the transcriptional repression mediated by Tup1, Nrg1, Mig1, and Rfg1 in C. albicans.
Tup1 repression with Nrg1, Mig1, and Rfg1.
The DBP Nrg1 contains a zinc finger domain that is conserved in transcriptional regulators from fungi to humans. It is most closely related to ScNrg1, which represses transcription in a Tup1-dependent fashion. nrg1 mutant cells are predominantly filamentous under non-filament-inducing conditions and show attenuated virulence. Nrg1 represses several filament-specific genes, such as ECE1 and HWP1. Most of these genes contain an Nrg1 response element in their promoters. These genes constitute a subset of those under Tup1 control, providing further evidence that Nrg1 acts by recruiting Tup1 to target genes (43, 104, 210, 211).
In S. cerevisiae, another transcriptional repressor, ScMig1, targets the ScTup1-ScSsn6 complex to the promoters of glucose-repressed genes to repress their transcription. Murad et al. (210) provided new insights into the regulatory functions of Tup1, Nrg1, and Mig1 in C. albicans. Nrg1 and Tup1 regulate a set of genes different from those regulated by Mig1 and Tup1. This is consistent with the idea that Mig1 and Nrg1 target the Tup1 repressor to a specific subset of C. albicans genes (210) (Fig. 4). However, Mig1 and Nrg1 repress other C. albicans genes in a Tup1-independent fashion (210, 211). An HMG protein, Rfg1, homolog of S. cerevisiae ScRox1, is also an important DBP that recruits Tup1 to the promoters of hyphal growth genes. Recently, Kadosh and Johnson identified 61 hypha-specific genes that are induced in response to growth in serum at 37°C by using whole-genome microarray analysis, and they showed that approximately half of the genes are under the negative control of the Rfg1, Nrg1, and/or Tup1 transcriptional repressors (145).
Other Negative Regulators: Rap1, Rbf1, and Rad6
In S. cerevisiae, the ScRap1 protein acts as both a transcriptional silencer and a structural protein at telomeres by binding to a sequence designated the RPG box (84). The C. albicans Rap1 homolog is not essential for survival but is required to repress pseudohypha formation under conditions favoring yeast growth (34). A second C. albicans protein, Rbf1, is not homologous to Rap1 but binds to the RPG box of S. cerevisiae (137). Rbf1 contains two glutamine-rich regions embedded within a region with weak similarity to bHLH domains, which binds to RPG sequences. Homozygous rbf1 null mutants are constitutively filamentous; the filaments formed are characteristic of pseudohyphae rather than true hyphae (137). Virulence of the rbf1 mutant in the mouse model of systemic infection was significantly attenuated (137). Recently, by screening for sequences that mediate Rbf1-dependent transcriptional regulation, target genes were identified in the heterologous host S. cerevisiae. Among the genes identified as Rbf1 targets was the white-phase-specific gene WH11 (273); the level of WH11 transcripts is reduced in homozygous rbf1 mutants compared to wild-type cells (137). Another repressing factor is the Rad6 protein, which, besides contributing to UV protection, represses hyphal growth under inducing conditions by an unknown pathway; its deficiency under noninducing conditions generates a pseudohyphal phenotype (176). All of these repressors have clear homologs in S. cerevisiae, and this may be the reason that they seem to be more involved in the repression of pseudohyphae than of true hyphae. In general, it also becomes clear that the regulation of morphogenesis is tightly controlled by a whole group of activators and repressors.
Other MAPK Pathways Involved in Morphogenesis
The HOG MAPK pathway: response to oxidative stress.
The recent annotation of the C. albicans genome sequence revealed four putative osmosensor proteins: Chk1, Nik1 (Cos1), Sln1, and Ssu81 (Sho1). As for other signaling pathways, the organization of the osmosensing pathways in C. albicans has been informed by studies with budding yeast. In S. cerevisiae, the ScHog1 MAPK pathway is involved in osmotic stress signaling. The pathway starts with the activation of cell membrane-bound receptor proteins (ScSho1, ScMsb2, and ScSln1) that act to sense external osmolarity, feeds through the HOG MAPK pathway, and ends with the synthesis of osmoprotectants such as glycerol (70, 123). The ScSln1 branch is required to induce expression of several genes in response to very high solute levels and operates over a broader range of osmolarities than does the ScSho1/ScMsb2 branch. Under osmotic stress, ScSln1 promotes phosphorylation of a downstream target protein, ScYpd1, which continuously transfers a phosphate group to the response regulator protein ScSsk1. This pathway activates two partially redundant MAPKKKs, ScSsk2 and ScSsk22. The ScSho1/ScMsb2 branch requires ScCdc42, ScSte20, and ScSte50 to activate the MAPKKK ScSte11. Any of the three MAPKKKs is able to activate ScPbs2, which then phosphorylates ScHog1. Phosphorylation of ScHog1 by the MAPKK ScPbs2 results in its nuclear localization and causes regulation of gene expression through several transcription factors—ScHot1, ScSko1, and ScSmp1—and probably also through ScMsn1, ScMns2, and ScMsn4 (105, 228, 240, 248, 315).
Although most components of these pathways are conserved in C. albicans, it seems that, instead of osmolarity, the sensors are activated by oxidative stress. Mechanisms of defense against oxidative stress are especially relevant for many pathogenic fungi, such as C. albicans, since the neutrophil-macrophage system controls infection (308) through an oxidative killing mechanism. The Hog1 MAPK mediates an adaptive response to both osmotic (as in yeast) and oxidative stress in C. albicans. Hog1 also participates in two distinct morphogenetic processes, either as a repressor (yeast-to-hypha transition) or as an inducer (chlamydospore formation) (3). The HOG pathway appears to be a major virulence determinant: the pathway represses the serum-induced yeast-to-hypha transition in C. albicans and also represses filamentous growth under other partially inducing conditions, such as low temperature, low pH, or nitrogen starvation (86) (Fig. 3). Hog-mediated repression of the yeast-to-hypha switch is independent of the Efg1 and Cph1 transcription factors. A triple deletion mutant, efg1 cph1 hog1, is able to form filaments under subinducing conditions, similar to a hog1 mutant. In contrast, the function of Hog1 in chlamydospore formation is dependent on the CEK1 MAPK pathway (86). The exact mechanism by which Hog1 represses hypha formation is still unclear but may involve activation of the transcriptional repressor Rbf1.
The upstream sensors of the Hog1 MAPK pathway, Ssu81 (Sho1) and Sln1, have clear roles in oxidative stress tolerance and morphogenesis. Although sho1 mutants are sensitive to oxidative stress, the signal appears to travel mainly through the other branch of the pathway via the Sln1-Ssk1 components (244). Sho1 is essential for the activation of the Cek1 MAPK under conditions that require active cell growth and/or cell wall remodeling, and cek1 mutants are defective in morphogenesis on different hypha-inducing media. So it seems that in C. albicans, Sho1 links oxidative stress, cell wall biogenesis, and morphogenesis. The SLN1 gene encodes a two-component histidine kinase containing a sensor and regulator domain. In vitro autophosphorylation activity has been shown for the Sln1 protein (325). In S. cerevisiae, activation of Sln1 occurs at normal osmolarity and leads to phosphorylation (and thereby inactivation) of the Ssk1 regulator. Although the C. albicans Sln1 and Ssk1 proteins are the direct homologs of the S. cerevisiae Sln1 and Ssk1 proteins, they are not essential in sensing hyperosmolarity. However, hyphal development of sln1 and ssk1 null mutants is blocked on starvation-type media and is severely impaired on serum agar, while filamentation is normal in all liquid media (54, 212, 325). Interestingly, while growth of the ssk1 mutant under nitrogen starvation conditions does not allow formation of hyphae, invasive growth is stimulated significantly compared to the wild-type strains (54). Apart from Sln1 and Sho1, two other two-component histidine kinases have been described, Nik1 (Cos1) and Chk1 (54, 325). The Cos1 protein is a homolog of the Neurospora crassa Nik1 histidine kinase, which regulates hyphal growth and protection against osmotic stress. In C. albicans, null mutants lacking NIK1 or CHK1 have no defect in osmoprotection but are significantly defective in hyphal formation on solid media (starvation-type or medium containing serum) but not in liquid media (1, 325). In addition, the Chk1 histidine kinase is needed to prevent flocculation of hyphae (53). Interestingly, deletions of SLN1 or NIK1 alleles in a chk1 mutant restored filamentation and virulence, suggesting that Sln1 and Nik1 act upstream of Chk1 via a negative regulator (Fig. 3) (325). Expression of CHK1 is down-regulated in an sln1 mutant but is not changed in a nik1 mutant, under most conditions tested (179). Thus, histidine kinase pathways, including Sln1 and Nik1, are possibly down-regulated during hyphal development on agar surfaces and within agar.
Role of the PKC pathway in C. albicans morphology.
In S. cerevisiae, the protein kinase C (PKC)-MAPK “cell integrity pathway” is involved in responses to a wide variety of stresses, including nutritional (311) and osmotic (103) stresses. These environmental signals are sensed by a family of sensor proteins: ScMtl1, ScWsc1, ScWsc2, ScWsc3, and ScMid2. The cell surface sensors transmit the signal to ScRom2, a guanine nucleotide exchange factor protein of the GTP-binding protein ScRho1. ScRho1 then activates the ScPkc1 protein kinase, which in turn activates a MAPK module consisting of ScBck1; two MAPKKs, ScMkk1 and ScMkk2; and the MAPK ScSlt2 (Mpk1). ScSlt2 then activates downstream transcription factors such as ScRlm1, involved in the activation of cell wall genes, and SBF, a heterodimeric transcription factor required for G1-specific gene expression (120, 123, 178). Based on these data with budding yeast, mutants in the C. albicans PKC pathway homologs have been tested for defects in morphogenesis (Fig. 5). A pkc1 null strain has a cell lysis defect, which is osmotically remediable; however, normal hyphal morphogenesis occurs in stabilized liquid serum media (230). The downstream target of Pkc1, the MAPK Mkc1 (homolog of yeast Slt2), is also affected in morphogenesis under certain conditions (215, 217). Homozygous mkc1 mutant cells have cell wall defects and are recovered in lower counts with shorter hyphae in the target tissues of infected mice. These data show that Mkc1 is clearly necessary for virulence in a mouse model of systemic infection (80).
FIG. 5.
Downstream targets of the PKC pathway. Under environmental stress conditions, the MAPK Mkc1 is activated in a Pck1-dependent manner. Then, it activates transcription factors Efg1, Czf1, and Bcr1 under different conditions for morphogenesis in C. albicans. CZF1 expression requires Efg1 and is negatively regulated by Czf1. Tec1 regulates the expression of BCR1.
Mkc1 is phosphorylated in the presence of oxidative stress, changes in osmotic pressure, cell wall damage, and a decrease in the growth temperature. The phosphorylation of Mkc1 largely depends on Pkc1, but under certain conditions Hog1 is required for Mkc1 phosphorylation, illustrating the interconnection between different MAPK pathways in C. albicans. One possibility is that Hog1 may be required for the expression of genes that are important for the glycosylation of the receptors (Wsc or Mid2 homologs) that activate the Mkc1 pathway (216). Kumamoto has further investigated the role of Mkc1 as a kinase that is specifically activated upon contact with a solid support (160, 161). This is important from a clinical point of view, as biofilm formation on various implants is currently the biggest problem in hospitals. Mkc1 is required for invasive hyphal growth and normal biofilm development (Fig. 5). The transcription factors that are activated (or indirectly induced) by Mkc1 remain to be identified, but possible candidates are Efg1 and Czf1. As described previously, Efg1 is required for biofilm development (237). Czf1 is required for filamentous growth under embedded and semisolid conditions (47), and ectopic expression of the Czf1 gene restores the filamentation defect of the mkc1 mutant. Another potential downstream target of Mkc1 is the zinc finger transcription factor Bcr1. Bcr1 is required for biofilm formation but not for hyphal growth. The expression of BCR1 depends on the hyphal regulator Tec1, and the downstream effectors of Bcr1 are genes encoding cell surface proteins and adhesins (Fig. 5) (223).
GlcNAc Catabolic Pathway of C. albicans
GlcNAc is one of the known in vitro inducers of the yeast-to-hypha transition in C. albicans and promotes germ tube formation in C. albicans within 3 h. The response of C. albicans to GlcNAc involves dramatic changes in the enzyme levels of the aminosugar catabolic pathway. The most interesting feature of this pathway is that all of the genes involved in the catabolism of GlcNAc exist in a cluster (162) housing six genes (Fig. 6). Three of the cluster genes encode known enzymes—glucosamine-6-phosphate deaminase (Nag1), GlcNAc-6-phosphate deacetylase (Dac1/Nag2), and GlcNAc kinase (Hxk1/Nag5)—that act sequentially on GlcNAc to generate fructose-6-phosphate, which is fed into the glycolytic pathway (Fig. 6). The notable traits of this metabolic pathway are that it is inducible by GlcNAc and that GlcNAc can be utilized as a sole carbon source. The transcripts for these genes are induced within minutes of each other (162), indicating the activation of a trans-acting factor(s) that enables the coordinated expression of the catabolic pathway genes. GlcNAc was unable to induce the other three genes in the cluster, which encode predicted transmembrane proteins (Nag3/Tmp1 and Nag4/Tmp2) and a cytosolic protein with a possible GTP-binding site (Nag6) (258, 326, 327). Disruption of the Nag regulon cluster, including deletion of DAC1, NAG1, and HXK1, results in a mutant that is unable to grow on aminosugars and which is avirulent in a murine model of systemic candidiasis (267). Hyphal formation under GlcNAc-inducible conditions is attenuated in the mutant, but it shows hyperfilamentation and a change in colony morphology under other filament-inducing conditions, such as Spider (where mannitol is used as the carbon source) or SLAD (nitrogen starvation) medium. When the genes (NAG1, DAC1, and HXK1) are disrupted individually, the growth pattern remains the same, i.e., all three mutants show impaired growth in GlcNAc but normal growth in glucose. The hyperfilamentation phenotype on Spider and SLAD media appears attributable to disruption of the NAG1 gene. Virulence in a murine model of systemic candidiasis is variably affected in the three individual mutants, with the strongest attenuation for the hxk1 mutant, followed by the dac1 and nag1 mutants (267, 326). In summary, these three genes are involved not only in metabolism of GlcNAc but also in morphogenesis and virulence. These processes do not seem to be related to one another. Experiments with immobilized GlcNAc, unavailable to the cells, show that GlcNAc utilization by the cells is not required for germ tube induction (290). This clearly points toward sensing of GlcNAc at the cell surface, but which receptor is involved is not yet understood.
FIG. 6.
(Top) Chromosomal organization of the NAG gene cluster in C. albicans. The organization of the cluster of genes on chromosome 6 and their direction of transcription are illustrated. NAG1 and DAC1 are transcribed in opposite directions from a bidirectional promoter. (Bottom) Metabolism of GlcNAc in C. albicans. GlcNAc is synthesized by C. albicans from fructose-6-phosphate by sequential action of glutamine: Frc 6-P amidotransferase (Gfp1), GlcN6-P acetyltransferase (Gna1), GlcNAc6P mutase (Agm1), UDP-GlcNAc pyrophosphorylase (Uap1). On the other hand, GlcNAc permease, GlcNAc kinase, GlcNAc-6-phosphate deacetylase, and GlcN-6-phosphate deaminase act sequentially on GlcNAc to generate the fructose-6-phosphate that is fed into the glycolytic pathway. N-Acetylmannosamine epimerase converts N-acetylmannosamine, which is imported in the cell by an unknown permease (?), into GlcNAc. Glucosamine can directly enter the cell via a general sugar permease and is converted to glucosamine-6-phosphate by the action of a GlcN-kinase.
The capacity to metabolize GlcNAc is present in various bacteria, such as Escherichia coli (313), Klebsiella pneumoniae, and Vibrio species (328), but, surprisingly, is absent in S. cerevisiae. The main reason for this seems to be the absence of genes homologous to NAG1, DAC1, and HXK1. A notable exception is S. cerevisiae strain 3059, a clinical isolate, which grew well on both glucose- and GlcNAc-containing media (265). How this GlcNAc is metabolized in this particular strain is not yet known. The functions of the other genes in the Nag regulon are not yet clear. NAG3 and NAG4 have more than 80% homology to each other and have significant sequence homology to S. cerevisiae Ypr156C and Ygr138C (both transporters of spermine) (162, 327). NAG3 and NAG4 are not required for GlcNAc uptake, as there was no difference between the growth of a nag3 nag4 mutant in GlcNAc-containing medium and the growth of the wild-type strain (258). The nag3 nag4 mutant became sensitive to cycloheximide, 4-nitroquinoline 1-oxide, and 1,10-phenanthroline. The above observations indicate that though NAG3 and NAG4 are present in the same gene cluster as the catabolic pathway genes, they are not indispensable for GlcNAc metabolism or GlcNAc-mediated signaling (258). NAG6 encodes a protein involved in drug sensitivity that has no human or murine homolog (327). This, together with the fact that Nag6 is required for virulence, makes it an interesting target for antifungals.
β-N-Acetylglucosaminidase and virulence.
β-N-Acetylglucosaminidase is a chitobiase that likely acts together with chitinase to release GlcNAc from chitin by hydrolysis (20). This process could have two biological effects: first, it may provide an appropriate carbon source to the fungus; second, it may facilitate adhesion of Candida cells to host tissues by bringing about conformational changes in the cell wall (55). The enzyme is reported to be induced by GlcNAc and repressed by other sugars, such as glucose (94-fold lower), fructose, galactose, and mannose (220). Thus, GlcNAc not only regulates its own catabolic pathway but also regulates the availability of GlcNAc to the cell. β-N-Acetylglucosaminidase is secreted and deposited into various parts of the cell wall of both the yeast and hyphal forms. A mutant deficient in β-N-acetylglucosaminidase exhibits attenuated virulence in a mouse model of candidiasis (141), although it shows normal growth. The HEX1 gene encodes β-N-acetylglucosaminidase and produces a transcript that is 200 nucleotides longer when isolated from GlcNAc-grown cells than from glucose-grown cells, implying that alternate transcription termination sites are used depending on the growth conditions (55). Temperature and pH changes also affect the level of HEX1 transcripts and β-N-acetylglucosaminidase activity.
pH Regulation in C. albicans
C. albicans causes infections in a broad range of host niches, which show significant differences in ambient pH. For example, the mouse systemic pH is 7.3, whereas the pH in the rat vagina is 4.5. The ability of Candida to react appropriately to (among other environmental variables) rather different pH environments is crucial for its pathogenicity. The environmental pH acts as a manipulator for many physiological functions, including morphogenesis (93). Under optimal (37°C) temperature conditions, filamentation is favored by ambient pH values close to neutral and is considerably reduced at pH values lower than 6. In contrast, the yeast form predominates almost exclusively at pH 4 (50).
Genes involved in pH regulation.
The pathway controlling pH-responsive gene expression has been most extensively dissected for the ascomycete Aspergillus nidulans (234). Central to the pH response is the pH-dependent activation of the zinc finger transcription factor encoded by pacC (297). PacC is synthesized in an inactive form, which is activated at alkaline pH by proteolytic removal of the carboxy terminus (203). Proteolysis is dependent on six genes: palA, palB, palC, palF, palH, and palI (297). The activated form of PacC induces the expression of alkaline pH-expressed genes and represses acidic pH-expressed genes (297). This regulatory pathway is apparently conserved, as various elements have been identified in Yarrowia lipolytica and S. cerevisiae. The Y. lipolytica RIM101 gene product is activated by carboxy-terminal truncation and is required for alkaline pH-dependent expression of XPR2-encoded protease (168). In S. cerevisiae, the PacC homolog, ScRIM101, was initially identified as controlling meiosis and haploid invasiveness (180, 286). ScRIM101 and the yeast homolog of PalB, ScCpl1, have been implicated in a pH-dependent growth response of yeast (97). Genes homologous to palA and palI are also present in S. cerevisiae (77, 218). Saporito-Irwin et al. used differential screening techniques to isolate genes with a pH-dependent expression profile and identified PHR1, a prototypical alkaline pH-expressed gene encoding a protein of 548 residues, carrying a glycosylphosphatidylinositol lipid anchor, which is required for apical cell growth and morphogenesis (251). PHR1 is homologous to ScGAS1, which encodes a GPI-anchored protein of the S. cerevisiae cell surface (225). PHR1 is strongly expressed under conditions of alkaline pH but not at any pH below 5.5 (251). A second pH-regulated gene, designated PHR2 (207), is expressed at an ambient pH below 5.5 and also plays a role in morphogenesis (251). Deletion of either one or both genes has a strong effect on virulence. Deletion of PHR1 results in pH-conditional defects in growth, morphogenesis, and virulence, evident at neutral to alkaline pHs. Conversely, a phr2 null mutant exhibits similar pH-conditional defects in growth and morphogenesis that manifest at acidic rather than alkaline pH values. Engineered expression of PHR1 at acidic pH in a phr2 mutant strain and PHR2 at alkaline pH in a phr1 mutant strain complements the defects in the opposing mutant. A strain lacking both PHR1 and PHR2 shows pH-independent constitutive growth and morphological defects (108, 207). When such strains were tested for pathogenicity in various niches of the host, the virulence phenotype paralleled the pH dependence of the in vitro phenotypes. The phr1 mutant was avirulent in a mouse model of systemic infection but uncompromised in its ability to cause vaginal infection in rats. The virulence phenotype of a phr2 mutant was the inverse. Heterozygous mutants exhibited partial reductions in their pathogenic potential, suggesting a gene dosage effect (75). Another pH-regulated gene, PRA1, a homolog of surface antigens of Aspergillus species, with maximal expression at neutral pH, has an important role in host-pathogen interactions during candidal infection (259).
To investigate the pH-dependent regulation of PHR1 and PHR2 in C. albicans, Fonzi et al. isolated and characterized PRR1 (pH response regulator), which is the C. albicans homolog of palF (now renamed RIM8). Expression of RIM8 is pH dependent, and a mutant lacking RIM8 is defective in pH-dependent regulation of gene expression. PHR1 is no longer induced at alkaline pH, and PHR2 is no longer repressed in both alkaline and acidic pH. Thus, Rim8 is a component of the pH response pathway in C. albicans. A rim8 mutant exhibited no morphological abnormalities at either pH; however, it cannot form hyphae on serum-containing medium (235). The ability to form hyphae on serum is not restored by forced expression of PHR1, indicating that additional RIM8-dependent genes are required for hyphal development. As discussed above, HWP1 is an Efg1 target gene required for normal filamentation, but it also appears to be downstream of Rim8. Expression of HWP1 is induced in the absence of RIM8, indicating that the pH response pathway is not required for the expression of EFG1-dependent genes. This suggests that the pH response pathway is distinct from the EFG1-dependent signaling pathway (235). In this regard it might be noted that EFG1 lies downstream of TPK2, which encodes a cAMP-dependent protein kinase (276). Rim101 of S. cerevisiae contains a functionally significant recognition site for phosphorylation by cAMP-dependent protein kinases (286). Although this site is not conserved in the C. albicans homolog, two potential phosphorylation sites are present, and this could provide a regulatory connection between Tpk2, Rim101, and Efg1.
Rim101-dependent and -independent pathways.
Fonzi's group characterized the full-length homolog of pacC, which they designated PRR2 (for “pH response regulator,” now renamed RIM101). C. albicans RIM101 encodes a protein of 604 amino acids that shares two structural features with other members of the Rim101 family. First, the zinc finger domain of the Rim101 family is highly conserved; C. albicans Rim101 has 57% identity and 87% similarity to the S. cerevisiae homolog in this region. Second, C. albicans Rim101 has an 84-amino-acid C-terminal region with 32% D/E residues, which is conserved in other members of this family (in S. cerevisiae, Y. lipolytica, and A. nidulans).
As with its homologs in other species (see above), C. albicans Rim101 activity is controlled by proteolytic processing. Under acidic conditions, Rim101 exists primarily in a full-length “long” form (180). Under alkaline conditions, a carboxyl-terminal portion is cleaved to yield the active “short” form. Proteolysis is controlled by pH through the action of a number of gene products, including Rim20, Rim8, Rim13, and Rim9 (78, 180) (Fig. 7). Together with two other pH-regulatory genes, RIM8 and RIM20, Rim101 acts to induce expression of PRA1, PHR1, and RIM101 itself under alkaline conditions (238). In the absence of RIM101, alkaline and acidic pH-expressed genes, such as PHR2, are no longer repressed (239). The same phenotype is observed in the rim8 mutant (235). PacC homologues from three filamentous fungi bind the consensus sequence 5′-GCCARG-3′ in vitro (251, 288, 297), and a similar DNA sequence (5′-NCCAAG-3′) is recognized by Rim101 of C. albicans.
FIG. 7.
Alkaline responses in Rim101 regulation. Alkaline pH stimulates Rim101 activity through increased expression and proteolytic activation. The ESCRT-I, ESCRT-II, and Snf7-Vps20 complexes are required for Rim101 activation. The Rim13p-dependent C-terminal proteolytic processing event also depends on Rim20, Rim8, and other transmembrane proteins. Full-length Rim101-long does not have a known activity. Processed Rim101-short is required for the alkaline response, which includes activation of alkaline pH-induced genes, repression of alkaline pH-repressed genes, and filamentation. Since Rim101 is an alkaline pH-induced gene, its expression may depend on autoregulation by Rim101-short. Alkaline pH also stimulates a RIM101-independent pathway. This pathway activates PHR2 expression and stimulates filamentation in conjugation with Rim101.
Unlike Aspergillus and Yarrowia spp., C. albicans remains pH responsive in the absence of the RIM101 pathway. For example, a dominant RIM101-405 allele, which complements the filamentation defect of the rim101 null mutant, promotes filamentation very weakly at acidic pH. Thus, the uncoupling of Rim101 processing from the upstream regulators does not completely bypass the control of filamentation by external pH. In addition, PHR2 becomes an alkaline pH-induced gene in cells that lack RIM101 pathway functions. Thus, both morphological and gene expression responses suggest the existence of a RIM101-independent pH response pathway (Fig. 7). This pathway has two roles: to stimulate PHR2 expression at alkaline pH and to act in conjunction with Rim101 to activate filamentation (72). Davis et al. also found that the RIM101 pathway is necessary in vivo for pathogenesis. Using the mouse model of hematogenously disseminated systemic candidiasis, they showed that both rim101 and rim8 mutants have significant reductions in virulence. The virulence defect of the rim8 mutant is not due solely to a failure to process Rim101, since the rim8 mutant has a more pronounced virulence defect than does a rim101 mutant. These observations suggest that both processed and unprocessed Rim101 may have a function during infection. Thus, it seems also that unprocessed Rim101 has a functional role in target gene regulation (73).
Participation of endocytic machinery in pH signaling.
Rim20 is a Bro1-domain protein; these are proteins that normally participate in endosome metabolism, but in C. albicans Rim20 is required for alkaline pH-stimulated cleavage of Rim101 (321). Rim101 cleavage removes a C-terminal PEST-like sequence allowing the N-terminal zinc finger domain to regulate transcription (167, 180). Rim20 is thought to function as an adaptor (similar to the yeast Bro1 gene), binding Snf7, to bring the protease Rim13 in close proximity with Rim101, thus promoting Rim101 cleavage and activation (Fig. 7) (322). Snf7 is a component of machinery designated ESCRT (for “endosomal sorting complexes required for transport”), a group of three protein complexes that sort endocytic cargo into multivesicular bodies (9, 159). A connection between ESCRT machinery and Rim101 signaling was predicted based on two-hybrid assay-based interactions between Rim20 and Snf7 (138). Also, mutants with defects in the ESCRT subunits fail to produce cleaved Rim101 (67, 322). A GFP-Rim20 fusion is located in the cytoplasm under acidic conditions; under alkaline conditions, it is located to punctate foci (39). All known proteins required for Rim101 cleavage, i.e., the arrestin homolog Rim8; the transmembrane proteins Rim9 and Rim21; and Dfg16, Snf7, Vps20, Vps23, and Vps25, are required for the appearance of Rim20-GFP to punctuate foci. Dfg16 and Rim21 are seven-transmembrane domain-containing proteins and therefore may constitute the sensors for external pH (21, 234). Sensing of external pH may actually be triggered by the Rim8-directed endocytosis of these two receptor proteins (21). Evidence for this hypothesis comes again from work performed with Aspergillus. The Rim8 homolog PalF behaves as a pH-responsive arrestin: alkaline growth conditions induce its phosphorylation and ubiquitination. This is the first example of an upstream component of the Rim101 pathway that is directly controlled by pH (121).
DOWNSTREAM TARGETS OF THE DIFFERENT ENVIRONMENTAL SENSING PATHWAYS INVOLVED IN MORPHOGENESIS
Adhesins
The virulence factors expressed or required by Candida species, and in particular by C. albicans, vary depending on the type of the infection (i.e., mucosal or systemic), the site and stage of infection, and the nature of the host response (89). One of the important virulence attributes is adhesion. Most studies focus on the role of two well-characterized adhesins (Hwp1 and the ALS family) in the morphogenesis, pathogenicity, and phenotype switching of C. albicans (291).
Role of Hwp1 in morphogenesis and pathogenicity.
Proadhesive and proinvasive factors of C. albicans contribute to disease by mediating the penetration of host tissues. Filamentous forms, particularly true hyphae, embed themselves within the superficial, keratinized layer of stratified squamous epithelium and grow by apical extension (283). The true hyphae are extensions of germ tubes that emerge from yeasts. To explore the role of surface proteins in tissue invasion, the function of HWP1, a developmentally regulated gene in germ tubes and true hyphae, was studied (283). HWP1 encodes an outer mannoprotein, with a cell surface-exposed NH2-terminal domain and COOH-terminal features conferring covalent integration into cell wall β-glucan. It may belong to a unique subset of GPI-anchored proteins characterized by the presence of a conserved structural motif, suggesting that it imparts a general property, e.g., interaction with specific surface proteins or wall polysaccharides. The surface localization of Hwp1 is compatible with diverse functions, from cell wall assembly to cell signaling. The evidence for the common occurrence of both membrane- and cell wall-anchored forms of cell wall-localized proteins comes from studies using fusion proteins consisting of α-galactosidase fused to C-terminal sequences from GPI-anchored cell wall proteins (196). Staab et al. showed that a 325-kDa GPI-anchored membrane species of Hwp1 is the precursor of a 301-kDa intermediate, which becomes covalently attached to β-glucan in the cell wall. This intermediate form may be released from membranes by phosphatidylinositol-specific phospholipase C, and it is found in the soluble fraction in cytoplasmic extracts and in small amounts in culture supernatants (282). HWP1 has no known S. cerevisiae homolog.
HWP1 was originally isolated as a hypha- and germ tube-specific gene from a differential screen (283, 291). HWP1 is conditionally required for hyphal formation: the ability to form hyphae on solid media is severely reduced in an HWP1 heterozygous mutant and eliminated in the null mutant. In the presence of serum, colonies of the null mutant produce peripheral hyphae but at reduced levels compared to the wild type. All mutants of HWP1 maintain the ability to invade agar directly beneath the colony and to form germ tubes in liquid serum-containing cultures. An hwp1 null mutant has reduced virulence in a hematogenously disseminated murine model (262, 283), germinates less readily in the kidneys of infected mice, and causes less endothelial cell damage (303). When tested in vitro, the hwp1 mutants germinate normally in liquid media containing 10% serum but exhibit reduced hyphal development when grown on serum-containing agar (262). In the murine kidney, the organisms are almost certainly exposed to serum constituents. The finding that the null mutants germinated very poorly in vivo suggests that the signal transduction pathway(s) that induces germination on solid media may also regulate germination in the murine kidney. The transcriptional and functional regulations of Hwp1 are summarized in Fig. 8. Staab et al. reported that the Hwp1 protein might act as an adhesin by serving as a substrate for host cell transglutaminases (283). More recently they have shown that Hwp1 participates in cross-links with proteins on the mammalian mucosa (292). In a model system of oroesophageal candidiasis, which is caused by the combined action of fungal virulence factors and host inflammatory responses when protective immunity is absent (18), the wild-type strain resulted in extensive alterations of the lingual and esophageal mucosa, whereas mice treated with the hwp1 mutant were not affected. Thus, HWP1 is a promising target for the development of antifungal drugs for treatment of oroesophageal candidiasis.
FIG. 8.
Transcriptional and functional regulation of HWP1. Transcription of HWP1 is regulated by Efg1. Interestingly, the hyphal gene repressors Tup1 and Rbf1 are positive regulators of Hwp1, whereas the MADS box transcription factor Mcm1 (245) negatively regulates HWP1 transcription. The pH regulators Mds3 and Dfg5 are required for expression of HWP1 in alkaline medium (74). HWP1 is expressed in conjugation tubes of a/a cells by α pheromone of opposite-mating-type cells (184). The cell surface protein Hwp1 is involved in adherence and plays a role in mating.
ALS family and adhesion.
The ALS gene family of C. albicans encodes large cell surface glycoproteins with a three-domain structure that are implicated in the process of adhesion to host surfaces (126, 128). The ALS (for “agglutinin-like sequence”) family includes eight genes, ALS1 to ALS7 and ALS9. Recent work showed that ALS3 and ALS8, which were presumed to represent separate loci, are the same gene, eliminating ALS8 from the family nomenclature (331). In C. albicans, ALS genes are found on three different chromosomes: chromosomes 3 (ALS6 and ALS7), R (ALS3), and 6 (ALS1, ALS2, ALS4, ALS5) (96, 128). Each Als protein has a relatively conserved N-terminal domain, a central domain consisting of tandem copies of a highly conserved 108-bp unit of a repeated motif, and a serine-threonine-rich C-terminal domain that is relatively variable across the family. Within a given ALS gene, the size of tandemly repeated regions varies considerably between alleles due to differences in the numbers of copies of the 108-bp sequence. The different ALS family members are differentially regulated and exhibit variability in size and expression. Additionally, examination of various C. albicans isolates showed that certain strains lack ALS genes that are present in others (128, 129). Finally, allelic sequence polymorphisms can be found outside of repeated regions. These sequence differences are more pronounced within the 3′ end of the gene, which encodes the heavily glycosylated portion of the mature protein (128). Als1 was the first cell surface protein identified that functions as a downstream effector of filamentation in C. albicans. Overexpression of ALS1 causes extensive flocculation and the formation of large aggregates of cells, a phenotype similar to that seen for S. cerevisiae Flo11. Both proteins function as effectors of filamentation and mediate adherence and flocculation (96, 115). Als1 functions as an effector of the cAMP-PKA pathway via Efg1 but does not appear to be an effector of the MAPK pathway (95). Als1 appears to be essential for virulence in hematogenously disseminated infection in a murine model (52). Recently, a function for Als3 in biofilm formation was found by performing genome-wide expression analysis of the bcr1 mutant, which is affected in biofilm formation. Als3 expression is strongly down-regulated in a bcr1 mutant (223). ALS3 overexpression rescues the bcr1 mutant biofilm defect in vivo, arguing for an important role for Als3 under these in vivo conditions (222, 330).
Extracellular Hydrolytic Enzymes
In susceptible hosts, C. albicans enters the bloodstream and causes deep-seated infection in target organs. Because the organisms must cross the endothelial cells of the blood vessels to enter these organs, the interaction between C. albicans and vascular endothelial cells is likely to be a critical step in the initiation of a disseminated infection. One factor that contributes to this process of virulence is hydrolytic enzyme production. The three most significant extracellular hydrolytic enzymes produced by C. albicans are the secreted aspartyl proteinases (Sap), phospholipase B enzymes, and lipases. All 10 SAP genes of C. albicans encode preproenzymes approximately 60 amino acids longer than the mature enzymes, which are processed when transported via the secretory pathway (213).
SAP gene expression in the yeast-to-hypha transition.
The hypha-deficient efg1 mutant is, in contrast to the cph1 mutant, not able to invade or damage tissue (see above). However, whether this could be explained solely by a block in dimorphism was not clear. Hube's group investigated the expression of the 10 SAP genes under hypha-inducing conditions (5% serum, different pH values, and different temperatures) in different genetic backgrounds (wild type, efg1 mutant, and cph1 mutant) (133). Expression of SAP4 to SAP6 was strongly reduced in efg1 mutants in serum-containing medium, which is a strong environmental signal for hyphal growth of wild-type cells (316). However, expression of all three genes was enhanced in both efg1 and cph1 mutants when hyphal formation was induced via a pH and temperature shift protocol, suggesting that the transcription factors Efg1 and Cph1 can be either positive or negative regulators of SAP4 to SAP6 expression. Since efg1 mutants do not produce hyphal cells under these conditions, the expression of SAP4 to SAP6 is not strictly linked to hyphal morphology but is regulated by another factor that regulates hyphal formation. Expression of Sap antigen during systemic infections has long been recognized in antigen-antibody studies (246). Staib et al. demonstrated high-level expression of SAP5 in all phases of infection with wild-type cells and a late induction of SAP2 and SAP6 using in vivo expression technology. With Sap-specific antibodies, Sap1 to Sap3 proteinases were detected on both yeast and hyphal wild-type cells, while the Sap4- to Sap6-specific antigen was identified mostly on penetrating hyphal cells, conferring hypha-specific expression of Sap4 and Sap6. The transcription factors Cph1 and Efg1 are involved in the expression of SAP5, as the avirulent cph1 efg1 mutant does not express this gene (284). Although expression of SAP2 was high in all in vivo samples investigated and large amounts of Sap1 to Sap3 antigen were found on all types of wild-type Candida cells, the level of expression did not correlate with the importance of the corresponding gene for invasion. Only mutants lacking SAP6 had strongly reduced abilities to invade and damage parenchymal organs, despite the fact that hyphal production was normal and all other proteinase genes were still expressed (91). It can be concluded that the reduced virulence of hypha-deficient mutants is not only due to the inability to form hyphae but also due to modified expression of SAP genes normally associated with hyphal morphology.
Phospholipase B.
Phospholipases are important pathogenicity determinants in C. albicans. They play a significant role in damaging cell membranes and invading host cells. High phospholipase production is correlated with increased adherence and a higher mortality rate in animal models (201). Various phospholipases reported from C. albicans include phospholipases A, B, C, and D (219). Phospholipase A and lysophospholipase activities are found in the cell wall of yeast cells and hyphae. Enzyme activity is higher in the walls of older yeast cells than of younger cells and is more prominent at the tip of growing hyphae (113). A C. albicans strain deleted for the phospholipase B gene PLB1 has significantly attenuated virulence (107, 173). Although deletion of PLB1 does not produce any detectable effects on Candida adherence to human endothelial or epithelial cells, the ability of the plb1 null mutants to penetrate host cells is dramatically reduced (209). Thus, phospholipase B may well contribute to the pathogenicity of C. albicans by abetting the fungus in damaging and traversing host cell membranes, processes which likely increase the rapidity of disseminated infection (173). Expression of PLB1 is regulated by nutritional supplementation, environmental factors, and the growth phase of the C. albicans cells, as well as by physiological conditions. The differential expression of PLB1 in response to environmental factors may be correlated to host-specific components available to C. albicans (208). These data prove that phospholipase B is essential for Candida virulence and pave the way for studies directed at determining the mechanism(s) through which phospholipases modulate virulence in this organism. The phospholipase D1 has been reported to be important for morphological transition under certain conditions (132).
FUTURE CHALLENGES
The mechanism of the yeast-to-hypha transition and identification of potential virulence attributes of C. albicans is a critical problem that requires further investigation for the development and improvement of targeted antifungal therapy. In the past, progress in C. albicans research was slow due to the lack of effective molecular tools, such as stable plasmid expression systems, limited mating ability, and the absence of known sporulation. The recent annotation of the C. albicans genome (68), together with the development of a whole range of molecular tools (e.g., codon-optimized reporter constructs), has greatly accelerated the pace of genetic research with this organism. In this review, we have highlighted studies that reveal the dynamics of cellular signaling during the yeast-to-hypha transition in C. albicans. Morphogenesis in C. albicans is regulated by a limited number of transcription factors, such as Efg1, Cph1, Rim101, Tup1, Bcr1, Czf1, Tec1, Nrg1, and Rbf1. These transcription factors are induced and/or activated by a whole array of environmental signals through a number of signaling pathways. Many of these transcription factors have common downstream targets, which are the real triggers of morphogenesis. In the classical view, we see environmental sensing as a system in which a membrane protein is the upstream component of a signal transduction pathway. Recent studies have shown that this is not always the case. The sensing of environmental CO2 concentrations seems to occur directly at the level of carbonic anhydrase and adenylate cyclase and does not seem to involve a membrane receptor. Similar systems may exist for the sensing of ammonia or other gases or some of the small quorum-sensing molecules.
Several important questions remain unanswered. (i) How do the different signals leading to morphogenesis and virulence affect the transcription of specific subsets of genes? What are the transcription factors that mediate the responses? The major transcription factor Efg1 is a key regulator of the yeast and hyphal program, but it is not required, or may be even inhibitory, for the later phase of filament formation. These observations suggest that additional regulators are involved in the control of morphogenesis and that there is a strong interplay between different signal transduction pathways. Alternatively, more feedback loops of gene regulation, such as for EFG1 and WOR1, will probably be discovered. A future challenge will be to identify all transcription factors involved in morphogenesis and how they interact with each other or how they are interdependent. (ii) How are morphogenesis and virulence regulated in vivo? Mutants that are either nonfilamentous (e.g., cph1 efg1 mutants) or hyperfilamentous (e.g., pde2 or hxk1 mutants) in vitro have strongly reduced virulence. Thus, it seems that a balance in filamentation is critical at the time of invasion and establishment of virulence. (iii) A third important issue that needs to be clarified is how the aminosugar GlcNAc induces cellular morphogenesis. Aminosugar utilization is an attribute of pathogenic yeast, and knockout of this catabolic pathway abolishes virulence. (iv) Finally, although several membrane receptors have been described, our knowledge of ligands remains limited. In this area of research, comparison with S. cerevisiae may be of limited use, as C. albicans has coevolved with its host. For instance, whereas in S. cerevisiae we have clearly shown that glucose and sucrose are the ligands of the G protein-coupled receptor Gpr1, the ligand of the homologous receptor in C. albicans remains to be identified. In addition, we will have to look into specific host components as potential ligands for certain receptors.
FINAL OUTLOOK
C. albicans is a suitable organism for studying dimorphism and pathogenicity. Major challenges still remain in determining how cells sense different sets of specific environmental signals and how the various signaling cascades process and integrate these signals. The phrase “many components but few connections” can characterize the current knowledge of the mechanisms of yeast-to-hypha transitions in C. albicans. Therefore, efforts should be directed toward analyses of functional linkages in addition to an expansion of the list of components affecting morphogenesis and virulence. A human-curated assembly of the C. albicans genome sequence has been released (www.candidagenome.org/) (44, 142). Bruno and Mitchell describe different strategies for overcoming the limitation of C. albicans gene functional analysis (49), and Noble and Johnson describe the use of new “lab” strains (avoiding the use of the URA3 marker) and “high-throughput” gene deletion strategies (224). Comparative genome analysis provides important clues about the evolution of the species and its mechanisms of pathogenesis (142). Comparative analysis of genetic and biochemical pathways in S. cerevisiae and C. albicans has also been extremely fruitful in revealing pathways regulating virulence in C. albicans. With the current speed of identification of genomes of other pathogens (e.g., C. glabrata and C. dubliniensis), comparative genomics will be an important tool in the future to identify broad-spectrum antifungal targets and to understand fungal pathogenesis. The ability of C. albicans to pass through the diverse body tract requires niche-specific gene expression (130). Recently, tools for monitoring the expression of specific genes in different locations of the host have become available (19). The availability of new functional genomic technologies, including DNA microarrays for high-throughput gene expression profiling and knock-out mutant collections, should enable functional characterization of novel genes. As has been achieved for S. cerevisiae, the C. albicans community should put together a consortium to generate a deletion strain collection, as well as conditional mutant collections for essential genes. While technologies for rapid gene deletion construction exist (88, 224), such collections should be constructed with the use of dominant and reusable markers in order to avoid marker problems and for the generation of multiple deletions. At the signaling level, new strategies for identifying nonlinear complexities in signal transduction, such as cross-talks and feedback loops, must be developed. As a result, our view and appreciation of Candida biology and pathogenesis should undergo something just short of a revolution in the very near future.
Acknowledgments
We are grateful to Brenda Andrews, K. Natarajan, and C. W. Schweinfest for valuable suggestions during preparation of the manuscript. S. Ghosh and K. H. Rao are acknowledged for critical reading of the manuscript.
Recent studies of the GlcNAc signaling pathway in the laboratory of A.D. have been supported by the Department of Biotechnology, India. Original work performed in the laboratory of P.V.D. was supported by grants from the Research Fund of Flanders (G.0242.04) and from the European Commission, Marie-Curie RTN Project CanTrain (MRTN-CT-2004-512841).
REFERENCES
- 1.Alex, L. A., C. Korch, C. P. Selitrennikoff, and M. I. Simon. 1998. COS1, a two-component histidine kinase that is involved in hyphal development in the opportunistic pathogen Candida albicans. Proc. Natl. Acad. Sci. USA 95:7069-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., F. Navarro-Garcia, G. Molero, R. Diez-Orejas, M. Gustin, J. Pla, M. Sanches, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso-Monge, R., F. Navarro-Garcia, E. Roman, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreasson, C., S. Heessen, and P. O. Ljungdahl. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aramayo, R., Y. Peleg, R. Addison, and R. Metzenberg. 1996. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144:991-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asleson, C. M., E. S. Bensen, C. A. Gale, A. S. Melms, C. Kurischko, and J. Berman. 2001. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol. Cell. Biol. 21:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atir-Lande, A., T. Gildor, and D. Kornitzer. 2005. Role of the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16:2772-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 10.Bachewich, C., A. Nantel, and M. Whiteway. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57:942-959. [DOI] [PubMed] [Google Scholar]
- 11.Bachewich, C., D. Y. Thomas, and M. Whiteway. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14:2163-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachewich, C., and M. Whiteway. 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell. 4:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahn, Y.-S., and P. Sundstrom. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahn, Y. S., G. M. Cox, J. R. Perfect, and J. Heitman. 2005. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 15:2013-2020. [DOI] [PubMed] [Google Scholar]
- 15.Bahn, Y. S., and F. A. Muhlschlegel. 2006. CO2 sensing in fungi and beyond. Curr. Opin. Microbiol. 9:572-578. [DOI] [PubMed] [Google Scholar]
- 16.Bahn, Y. S., J. Staab, and P. Sundstrom. 2003. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50:391-409. [DOI] [PubMed] [Google Scholar]
- 17.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balish, E., T. F. Warner, C. J. Pierson, D. M. Bock, and R. D. Wagner. 2001. Oroesophageal candidiasis is lethal for transgenic mice with combined natural killer and T-cell defects. Med. Mycol. 39:261-268. [DOI] [PubMed] [Google Scholar]
- 19.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett-Bee, K., and M. Hamilton. 1984. The detection and analysis of chitinase activity from the yeast form of Candida albicans. J. Gen. Microbiol. 130:1857-1861. [DOI] [PubMed] [Google Scholar]
- 21.Barwell, K. J., J. H. Boysen, W. Xu, and A. P. Mitchell. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4:890-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassilana, M., J. Hopkins, and R. A. Arkowitz. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4:588-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battle, M., A. Lu, D. Green, Y. Xue, and J. P. Hirsch. 2003. Krh1p and Krh2p act downstream of the Gpa2p G alpha subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116:701-711. [DOI] [PubMed] [Google Scholar]
- 25.Bendel, C. M., K. M. Kinneberg, R. P. Jechorek, C. A. Gale, S. L. Erlandsen, M. K. Hostetter, and C. L. Wells. 1999. Systemic infection following intravenous inoculation of mice with Candida albicans int1 mutant strains. Mol. Genet. Metab. 67:343-351. [DOI] [PubMed] [Google Scholar]
- 26.Bennett, R. J., and A. D. Johnson. 2005. Mating in Candida albicans and the search. Annu. Rev. Microbiol. 59:233-255. [DOI] [PubMed] [Google Scholar]
- 27.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62:100-119. [DOI] [PubMed] [Google Scholar]
- 28.Bensen, E. S., A. Clemente-Blanco, K. R. Finley, J. Correa-Bordes, and J. Berman. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benton, B. K., A. H. Tinkelenberg, D. Jean, S. D. Plump, and F. R. Cross. 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12:5267-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman, J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman, J., and P. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 33.Biswas, K., and J. Morchhauser. 2005. The Mep2 ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649-669. [DOI] [PubMed] [Google Scholar]
- 34.Biswas, K., K. J. Rieger, and J. Morschhauser. 2003. Functional analysis of CaRAP1, encoding the repressor/activator protein 1 of Candida albicans. Gene 307:151-158. [DOI] [PubMed] [Google Scholar]
- 35.Biswas, S., M. Roy, and A. Datta. 2003. N-Acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 36.Bockmühl, D. P., and J. F. Ernst. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bockmühl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1 and Tpk2 in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 38.Borneman, A. R., J. A. Leigh-Bell, H. Yu, P. Bertone, M. Gerstein, and M. Snyder. 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20:435-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boysen, J. H., and A. P. Mitchell. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the S. cerevisiae RIM101 pathway. Mol. Biol. Cell 17:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor Tup1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 42.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun, B. R., M. Van het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morchhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brega, E., R. Zufferey, and B. C. Mamoun. 2004. Candida albicans Csy1 is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown, A. J. P., and N. A. R. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 47.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues, and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 48.Brown, V., J. A. Sexton, and M. Johnston. 2006. A glucose sensor in Candida albicans. Eukaryot. Cell 5:1726-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruno, V. M., and A. P. Mitchell. 2004. Large-scale gene function analysis in Candida albicans. Trends Microbiol. 12:157-161. [DOI] [PubMed] [Google Scholar]
- 50.Buffo, J., M. A. Herman, and D. R. Soll. 1984. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 85:21-30. [DOI] [PubMed] [Google Scholar]
- 51.Calcagno, A. M., E. Bignell, T. R. Rogers, M. Canedo, F. A. Muhlschlegel, and K. Haynes. 2004. Candida glabrata Ste20 is involved in maintaining cell wall integrity and adaptation to hypertonic stress, and is required for wild-type levels of virulence. Yeast 21:557-568. [DOI] [PubMed] [Google Scholar]
- 52.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 53.Calera, J. A., and R. Calderone. 1999. Flocculation of hyphae is associated with a deletion in the putative CaHK1 two-component histidine kinase gene from Candida albicans. Microbiology 145:1431-1442. [DOI] [PubMed] [Google Scholar]
- 54.Calera, J. A., X. J. Zhao, F. De Bernardis, M. Sheridan, and R. Calderone. 1999. Avirulence of Candida albicans CaHK1 mutants in a murine model of hematogenously dissemeinated candidiasis. Infect. Immun. 67:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cannon, R. D., K. Niimi, H. F. Jenkinson, and M. G. Shepherd. 1994. Molecular cloning and expression of the Candida albicans β-N-acetylglucosaminidase (HEX1) gene. J. Bacteriol. 176:2640-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Liu. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 58.Cassola, A., M. Parrot, S. Silberstein, B. B. Magee, S. Passeron, L. Giasson, and M. L. Cantore. 2004. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chapa y Lazo, B., S. Bates, and P. Sudbery. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, C. G., Y. L. Yang, H. H. Chen, C. L. Su, S. F. Huang, C. T. Chen, Y. T. Liu, I. J. Su, and H. J. Lo. 2006. Non-lethal Candida albicans cph1/cph1 efg1/efg1 transcription factor mutant establishing restricted zone of infection in a mouse model of systemic infection. Int. J. Immunopathol. Pharmacol. 19:561-565. [DOI] [PubMed] [Google Scholar]
- 61.Chen, H., M. Fujita, Q. Feng, J. Clardy, and G. R. Fink. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 101:5048-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng, S., M. H. Nguyen, Z. Zhang, H. Jia, M. Handfield, and C. J. Clancy. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71:6101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chou, S., L. Huang, and H. Liu. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981-990. [DOI] [PubMed] [Google Scholar]
- 64.Chou, S., S. Lane, and H. Liu. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clark, K. L., P. J. Feldmann, D. Dignard, R. Larocque, A. J. Brown, M. G. Lee, D. Y. Thomas, and M. Whiteway. 1995. Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol. Gen. Genet. 249:609-621. [DOI] [PubMed] [Google Scholar]
- 66.Cloutier, M., R. Castilla, N. Bolduc, A. Zelada, P. Martineau, M. Bouillon, B. B. Magee, S. Passeron, L. Giasson, and M. L. Cantore. 2003. The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Gen. Biol. 38:133-141. [DOI] [PubMed] [Google Scholar]
- 67.Cornet, M., F. Bidard, P. Schwarz, G. Da Costa, S. Blanchin-Roland, F. Dromer, and C. Gaillardin. 2005. Deletions of edocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73:7977-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costanzo, M. C., M. B. Arnaud, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2006. The Candida Genome Database: faciliating research on Candida albicans molecular biology. FEMS Yeast Res. 6:671-684. [DOI] [PubMed] [Google Scholar]
- 69.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cullen, P. J., W. J. Sabbagh, E. Graham, M. M. Irick, E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. J. Sprague. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 72.Davis, D., J. E. Edwards, Jr., A. P. Mitchell, and A. S. Ibrahim. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Bernardis, F., F. A. Mühlchlegel, A. Cassone, and W. A. Fonzi. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.d'Enfert, C. 2006. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Curr. Drug Targets 7:465-470. [DOI] [PubMed] [Google Scholar]
- 77.Denison, S. H., S. Negrete-Urtasun, J. M. Mingot, J. Tilburn, W. A. Mayer, A. Goel, E. A. Espeso, M. A. Penalva, and H. N. Arst, Jr. 1998. Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol. Microbiol. 30:259-264. [DOI] [PubMed] [Google Scholar]
- 78.Denison, S. H., M. Orejas, and H. N. Arst, Jr. 1995. Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 270:28519-28522. [DOI] [PubMed] [Google Scholar]
- 79.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 80.Diez-Orejas, R., G. Molero, F. Navarro-Garcia, J. Pla, C. Nombela, and M. Sanchez-Perez. 1997. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect. Immun. 65:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doedt, T., S. Krishnamurthy, D. P. Bockmühl, B. Tebarth, C. Stempel, C. L. Russell, A. J. P. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dolan, J. W., A. C. Bell, B. Hube, M. Schaller, T. F. Warner, and E. Balish. 2004. Candida albicans PLD1 activity is required for full virulence. Med. Mycol. 42:439-447. [DOI] [PubMed] [Google Scholar]
- 83.Donaton, M. C., I. Holsbeeks, O. Lagatie, G. Van Zeebroeck, M. Crauwels, J. Winderickx, and J. M. Thevelein. 2003. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50:911-929. [DOI] [PubMed] [Google Scholar]
- 84.Drazinic, C. M., J. B. Smerage, M. C. Lopez, and H. V. Baker. 1996. Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p). Mol. Cell. Biol. 16:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Egidy, G., C. Paveto, S. Passeron, and M. A. Galvagno. 1990. cAMP levels and in situ measurement of cAMP related enzymes during yeast-to-hyphae transition in Candida albicans. Cell. Biol. Int. Rep. 14:59-68. [DOI] [PubMed] [Google Scholar]
- 86.Eisman, B., R. Alonso-Monge, E. Roman, D. Arana, C. Nombela, and J. Pla. 2006. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 5:347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.El Barkani, A., O. Kurzai, W. A. Fonzi, A. Ramon, A. Porta, M. Frosch, and F. A. Muhlschlegel. 2000. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol. Cell. Biol. 20:4635-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Enloe, B., A. Diamond, and A. P. Mitchell. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fallon, K., K. Bausch, J. Noonan, E. Huguenel, and P. Tamburini. 1997. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect. Immun. 65:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fang, H. M., and Y. Wang. 2006. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 61:484-496. [DOI] [PubMed] [Google Scholar]
- 91.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schafer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng, Q., E. Summers, B. Guo, and G. R. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fonzi, W. A. 2002. Role of pH response in Candida albicans virulence. Mycoses 45(Suppl. 1):16-21. [DOI] [PubMed] [Google Scholar]
- 94.Forsberg, H., F. Gilstring, A. Zargari, P. Martinez, and P. O. Ljungdahl. 2001. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42:215-228. [DOI] [PubMed] [Google Scholar]
- 95.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y. C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61-72. [DOI] [PubMed] [Google Scholar]
- 96.Fu, Y., G. Rieg, W. A. Fonzi, P. H. Belanger, J. E. Edwards, Jr., and S. G. Filler. 1998. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 66:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Futai, E., T. Maeda, H. Sorimachi, K. Kitamoto, S. Ishiura, and K. Suzuki. 1999. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260:559-568. [DOI] [PubMed] [Google Scholar]
- 98.Gale, C., D. Finkel, N. Tao, M. Meinke, M. McClellan, J. Olson, K. Kendrick, and M. Hostetter. 1996. Cloning and expression of a gene encoding an integrin-like protein in Candida albicans. Proc. Natl. Acad. Sci. USA 93:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Becker, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 101.Gancedo, J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107-123. [DOI] [PubMed] [Google Scholar]
- 102.Garaizar, J., S. Brena, J. Bikandi, A. Rementeria, and J. Ponton. 2006. Use of DNA microarray technology and gene expression profiles to investigate the pathogenesis, cell biology, antifungal susceptibility and diagnosis of Candida albicans. FEMS Yeast Res. 6:987-998. [DOI] [PubMed] [Google Scholar]
- 103.Garcia-Rodriguez, L. J., R. Valle, A. Duran, and C. Roncero. 2005. Cell integrity signaling activation in response to hyperosmotic shock in yeast. FEBS Lett. 579:6186-6190. [DOI] [PubMed] [Google Scholar]
- 104.Garcia-Sanchez, S., A. L. Mavor, C. L. Russell, S. Argimon, P. Dennison, B. Enjalbert, and A. J. Brown. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 16:2913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gavrias, V., A. Andrianopoulos, C. J. Gimeno, and W. E. Timberlake. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255-1263. [DOI] [PubMed] [Google Scholar]
- 107.Ghannoum, M. A. 1998. Extracellular phospholipases as universal virulence factor in pathogenic fungi. Nippon Ishinkin Gakkai Zasshi 39:55-59. [DOI] [PubMed] [Google Scholar]
- 108.Ghannoum, M. A., B. Spellberg, S. M. Saporito-Irwin, and W. A. Fonzi. 1995. Reduced virulence of Candida albicans PHR1 mutants. Infect. Immun. 63:4528-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 111.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentatous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goldberg, D., M. Segal, and A. Levitzki. 1994. Cdc25 is not the signal receiver for glucose induced cAMP response in S. cerevisiae. FEBS Lett. 356:249-254. [DOI] [PubMed] [Google Scholar]
- 113.Goyal, S., and G. K. Khuller. 1992. Phospholipid composition and subcellular distribution in yeast and mycelial forms of Candida albicans. J. Med. Vet. Mycol. 30:355-362. [DOI] [PubMed] [Google Scholar]
- 114.Guhad, F. A., C. Csank, H. E. Jensen, D. Y. Thomas, M. Whiteway, and J. Hau. 1998. Reduced pathogenicity of a Candida albicans MAP kinase phosphatase (CPP1) mutant in the murine mastitis model. APMIS 106:1049-1055. [DOI] [PubMed] [Google Scholar]
- 115.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harashima, T., S. Anderson, J. R. R. Yates, and J. Heitman. 2006. The Kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 22:819-830. [DOI] [PubMed] [Google Scholar]
- 117.Harashima, T., and J. Heitman. 2002. The G alpha protein Gpa2 controls yeast differentiation by interacting with Kelch repeat proteins that mimic G beta subunits. Mol. Cell 10:163-173. [DOI] [PubMed] [Google Scholar]
- 118.Hausauer, D., M. Gerami-Nejad, C. Kistler-Anderson, and C. Gale. 2005. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot. Cell 4:1273-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hazan, I., and H. Liu. 2002. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot. Cell 1:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heinisch, J. J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32:671-680. [DOI] [PubMed] [Google Scholar]
- 121.Herranz, S., J. M. Rodriguez, H. J. Bussink, J. C. Sanchez-Ferrero, H. N. Arst, Jr., M. A. Penalva, and O. Vincent. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 102:12141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van De Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29:556-564. [DOI] [PubMed] [Google Scholar]
- 125.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 127.Hoyer, L. L., L. B. Cieslinski, M. M. McLaughlin, T. J. Torphy, A. R. Shatzman, and G. P. Livi. 1994. A Candida albicans cyclic nucleotide phosphodiesterase: cloning and expression in Saccharomyces cerevisiae and biochemical characterization of the recombinant enzyme. Microbiology 140:1533-1542. [DOI] [PubMed] [Google Scholar]
- 128.Hoyer, L. L., and J. E. Hecht. 2001. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast 18:49-60. [DOI] [PubMed] [Google Scholar]
- 129.Hoyer, L. L., and J. E. Hecht. 2000. The ALS6 and ALS7 genes of Candida albicans. Yeast 16:847-855. [DOI] [PubMed] [Google Scholar]
- 130.Hromatka, B. S., S. M. Noble, and A. D. Johnson. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress. Mol. Biol. Cell 16:4814-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang, G., H. Wang, S. Chou, X. Nie, J. Chen, and H. Liu. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. USA 103:12813-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hube, B., D. Hess, C. A. Baker, M. Schaller, W. Schafer, and J. W. Dolan. 2001. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147:879-889. [DOI] [PubMed] [Google Scholar]
- 133.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 134.Hube, B., D. Sanglard, F. C. Odds, D. Hess, M. Monod, W. Schafer, A. J. Brown, and N. A. R. Gow. 1997. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hudson, D. A., Q. L. Sciascia, R. J. Sanders, G. E. Norris, P. J. Edwards, P. A. Sullivan, and P. C. Farley. 2004. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150:3041-3049. [DOI] [PubMed] [Google Scholar]
- 136.Hwang, C. S., J. H. Oh, W. K. Huh, H. S. Yim, and S. O. Kang. 2003. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 47:1029-1043. [DOI] [PubMed] [Google Scholar]
- 137.Ishii, N., M. Yamamoto, F. Yoshihara, M. Arisawa, and Y. Aoki. 1997. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology 143:429-435. [DOI] [PubMed] [Google Scholar]
- 138.Ito, T., T. Chiba, and M. Yoshida. 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. 19:S23-S27. [DOI] [PubMed] [Google Scholar]
- 139.Jain, P., I. Akula, and T. Edlind. 2003. Cyclic AMP signaling pathway modulates susceptibility of Candida species and Saccharomyces cerevisiae to antifungal azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 47:3195-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jauniaux, J. C., and M. Grenson. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39-44. [DOI] [PubMed] [Google Scholar]
- 141.Jenkinson, H. F., and M. G. Shepherd. 1987. A mutant of Candida albicans deficient in beta-N-acetylglucosaminidase (chitobiase). J. Gen. Microbiol. 133:2097-2106. [DOI] [PubMed] [Google Scholar]
- 142.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jung, W. H., and L. I. Stateva. 2003. The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 149:2961-2976. [DOI] [PubMed] [Google Scholar]
- 144.Jung, W. H., P. Warn, E. Ragni, L. Popolo, C. D. Nunn, M. P. Turner, and L. I. Stateva. 2005. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in C. albicans. Yeast 22:285-294. [DOI] [PubMed] [Google Scholar]
- 145.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709-719. [DOI] [PubMed] [Google Scholar]
- 148.Khalaf, R. A., and R. S. Zitomer. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim, T. S., H. Y. Kim, J. H. Yoon, and H. S. Kang. 2004. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol. Cell. Biol. 24:9542-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kitanovic, A., M. Nguyen, G. Vogl, A. Hartmann, J. Gunther, R. Wurzner, W. Kunkel, S. Wolfl, and R. Eck. 2005. Phospatidylinositol 3 kinase VPS34 of Candida albicans is involved in filamentous growth, secretion of aspartic proteases, and intracellular detoxification. FEMS Yeast Res. 5:431-439. [DOI] [PubMed] [Google Scholar]
- 151.Klengel, T., W. J. Liang, J. Chaloupka, C. Ruoff, K. Schroppel, J. R. Naglik, S. E. Eckert, E. G. Mogensen, K. Haynes, M. F. Tuite, L. R. Levin, J. Buck, and F. A. Muhlschlegel. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15:2021-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klepser, M. 2006. Candida resistance and its clinical relevance. Pharmacotherapy 26:68S-75S. [DOI] [PubMed] [Google Scholar]
- 153.Köhler, J. R., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Komachi, K., and A. D. Johnson. 1997. Residues in the WD repeats of Tup1 required for interaction with α2. Mol. Cell. Biol. 17:6023-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Korting, H. C., B. Hube, S. Oberhauser, E. Januschke, G. Hamm, A. Albrecht, C. Borelli, and M. Schaller. 2003. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 52:623-632. [DOI] [PubMed] [Google Scholar]
- 156.Kraakman, L., K. Lemaire, P. Ma, A. W. R. H. Teunissen, M. C. V. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 157.Kronstad, J., A. D. De Maria, D. Funnell, R. D. Laidlaw, N. Lee, M. M. de Sa, and M. Ramesh. 1998. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch. Microbiol. 170:395-404. [DOI] [PubMed] [Google Scholar]
- 158.Kruppa, M., B. P. Krom, N. Chauhan, A. V. Bambach, R. L. Cihlar, and R. A. Calderone. 2004. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3:1062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kullas, A. L., M. Li, and D. A. Davis. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 162.Kumar, M. J., M. S. Jamaluddin, K. Natarajan, D. Kaur, and A. Datta. 2000. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. USA 97:14218-14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kumar, R., D. M. Reynolds, A. Shevchenko, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 164.Kurtz, M. B., D. R. Kirsch, and R. Kelly. 1988. The molecular genetics of Candida albicans. Microbiol. Sci. 5:58-63. [PubMed] [Google Scholar]
- 165.Kvaal, C., S. A. Lachke, T. Srikantha, K. Daniels, J. McCoy, and D. R. Soll. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 168.Lambert, M., S. Blanchin-Roland, F. Le Louedec, A. Lepingle, and C. Gaillardin. 1997. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17:3966-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 170.Lane, S., S. Zhou, T. Pan, Q. Dai, and H. Liu. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21:6418-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. Gow, A. J. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Leberer, E., K. Ziegelbauer, A. Schmidt, D. Harcus, D. Dignard, J. Ash, L. Johnson, and D. Y. Thomas. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7:539-546. [DOI] [PubMed] [Google Scholar]
- 173.Leidich, S. D., A. S. Ibrahim, Y. Fu, A. Koul, C. Jessup, J. Vitullo, W. Fonzi, F. Mirbod, S. Nakashima, Y. Nozawa, and M. A. Ghannoum. 1998. Cloning and disruption of CaPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273:26078-26086. [DOI] [PubMed] [Google Scholar]
- 174.Lemaire, K., S. Van De Velde, P. Van Dijck, and J. M. Thevelein. 2004. Nutrients as ligand for the G protein coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293-299. [DOI] [PubMed] [Google Scholar]
- 175.Leng, P., P. R. Lee, H. Wu, and A. J. Brown. 2001. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 183:4090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Leng, P., P. E. Sudbery, and A. J. Brown. 2000. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol. Microbiol. 35:1264-1275. [DOI] [PubMed] [Google Scholar]
- 177.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li, D., V. Gurkovska, M. Sheridan, R. Calderone, and N. Chauhan. 2004. Studies on the regulation of the two-component histidine kinase gene CHK1 in Candida albicans using the heterologous LacZ reporter gene. Microbiology 150:3305-3313. [DOI] [PubMed] [Google Scholar]
- 180.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1736. [DOI] [PubMed] [Google Scholar]
- 182.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 183.Lo, H. J., J. S. Wang, C. Y. Lin, C. G. Chen, T. Y. Hsiao, C. T. Hsu, C. L. Su, M. J. Fann, Y. T. Ching, and Y. L. Yang. 2005. Efg1 involved in drug resistance by regulating the expression of ERG3 in Candida albicans. Antimicrob. Agents Chemother. 49:1213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. α-Pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Loeb, J. D., M. Sepulveda-Becerra, I. Hazan, and H. Liu. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19:4019-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luongo, M., A. Porta, and B. Maresca. 2005. Homology, disruption and phenotypic analysis of CaGS Candida albicans gene induced during macrophage infection. FEMS Immunol. Med. Micriobiol. 45:471-478. [DOI] [PubMed] [Google Scholar]
- 190.Ma, P., S. Wera, P. Van Dijck, and J. M. Thevelein. 1999. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol. Biol. Cell 10:91-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 192.Magasanik, B. 2003. Ammonia assimilation by Saccharomyces cerevisiae. Eukaryot. Cell 2:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Maidan, M. M., L. De Rop, J. Serneels, S. Exler, S. Rupp, H. Tournu, J. M. Thevelein, and P. Van Dijck. 2005. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-PKA pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maidan, M. M., J. M. Thevelein, and P. Van Dijck. 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G protein coupled receptor Gpr1. Biochem. Soc. Trans. 33:291-293. [DOI] [PubMed] [Google Scholar]
- 195.Malathi, K., K. Ganesan, and A. Datta. 1994. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J. Biol. Chem. 269:22945-22951. [PubMed] [Google Scholar]
- 196.Mao, Y., Z. Zhang, and B. Wong. 2003. Use of green fluorescent protein fusions to analyse the N- and C-terminal signal peptides of GPI-anchored cell wall proteins in Candida albicans. Mol. Microbiol. 50:1617-1628. [DOI] [PubMed] [Google Scholar]
- 197.Martinez, P., and P. O. Ljungdahl. 2004. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol. Microbiol. 51:371-384. [DOI] [PubMed] [Google Scholar]
- 198.Martinez, P., and P. O. Ljungdahl. 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25:9435-9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Martinez-Lopez, R., L. Monteoliva, R. Diez-Orejas, C. Nombela, and C. Gil. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence. Microbiology 150:3341-3354. [DOI] [PubMed] [Google Scholar]
- 200.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mayser, P., S. Laabs, K. U. Heuer, and K. Grunder. 1996. Detection of extracellular phospholipase activity in Candida albicans and Rhodotorula rubra. Mycopathologia 135:149-155. [DOI] [PubMed] [Google Scholar]
- 202.Miller, K. Y., J. Wu, and B. L. Miller. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6:1770-1782. [DOI] [PubMed] [Google Scholar]
- 203.Mingot, J. M., J. Tilburn, E. Diez, E. Bignell, M. Orejas, D. A. Widdick, S. Sarkar, C. V. Brown, M. X. Caddick, E. A. Espeso, H. N. Arst, Jr., and M. A. Penalva. 1999. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol. Cell. Biol. 19:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Miwa, T., Y. Takagi, M. Shinozaki, C.-W. Yun, W. A. Schell, J. R. Perfect, H. Kumagai, and H. Tamaki. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Monge, R. A., E. Roman, C. Nombela, and J. Pla. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905-912. [DOI] [PubMed] [Google Scholar]
- 206.Morschhauser, J., P. Staib, and G. Kohler. 2005. Targeted gene deletion in Candida albicans wild-type strains by MPAR flipping. Methods Mol. Med. 118:35-44. [DOI] [PubMed] [Google Scholar]
- 207.Muhlschlegel, F. A., and W. A. Fonzi. 1997. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17:5960-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mukherjee, P. K., J. Chandra, D. M. Kuhn, and M. A. Ghannoum. 2003. Differential expression of Candida albicans phospholipase B (PLB1) under various environmental and physiological conditions. Microbiology 149:261-267. [DOI] [PubMed] [Google Scholar]
- 209.Mukherjee, P. K., K. R. Seshan, S. D. Leidich, J. Chandra, G. T. Cole, and M. A. Ghannoum. 2001. Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology 147:2585-2597. [DOI] [PubMed] [Google Scholar]
- 210.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 211.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. P. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nagahashi, S., T. Mio, N. Ono, T. Yamada-Okabe, M. Arisawa, H. Bussey, and H. Yamada-Okabe. 1998. Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology 144:425-432. [DOI] [PubMed] [Google Scholar]
- 213.Naglik, J. R., S. J. Challacombe, and B. Hube. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Navarro-Garcia, F., R. Alonso-Monge, H. Rico, J. Pla, R. Sentandreu, and C. Nombela. 1998. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 144:411-424. [DOI] [PubMed] [Google Scholar]
- 216.Navarro-Garcia, F., B. Eisman, S. M. Fiuza, C. Nombela, and J. Pla. 2005. The MAP kinase Mkc1 is activated under different stress conditions in Candida albicans. Microbiology 151:2737-2749. [DOI] [PubMed] [Google Scholar]
- 217.Navarro-Garcia, F., M. Sanchez, J. Pla, and C. Nombela. 1995. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 15:2197-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Negrete-Urtasun, S., S. H. Denison, and H. N. Arst, Jr. 1997. Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J. Bacteriol. 179:1832-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Niewerth, M., and H. C. Korting. 2001. Phospholipases of Candida albicans. Mycoses 44:361-367. [DOI] [PubMed] [Google Scholar]
- 220.Niimi, K., M. Niimi, M. G. Shepherd, and R. D. Cannon. 1997. Regulation of N-acetylglucosaminidase production in Candida albicans. Arch. Microbiol. 168:464-472. [DOI] [PubMed] [Google Scholar]
- 221.Nikawa, J., P. Sass, and M. Wigler. 1987. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nobile, C. J., D. R. Andes, J. E. Nett, F. J. Smith, F. Yue, Q. T. Phan, J. E. Edwards, Jr., S. G. Filler, and A. P. Mitchell. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150-1155. [DOI] [PubMed] [Google Scholar]
- 224.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nuoffer, C., P. Jeno, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Oh, K. B., H. Miyazawa, T. Naito, and H. Matsuoka. 2001. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc. Natl. Acad. Sci. USA 98:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.O'Rourke, S. M., and I. Herskowitz. 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Paravicini, G., A. Mendoza, B. Antonsson, M. Cooper, C. Losberger, and M. A. Payton. 1996. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity but not dimorphism. Yeast 12:741-756. [DOI] [PubMed] [Google Scholar]
- 231.Park, Y. N., and J. Morschhauser. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4:1328-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pasrija, R., S. Krishnamurthy, T. Prasad, J. F. Ernst, and R. Prasad. 2005. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J. Antimicrob. Chemother. 55:905-913. [DOI] [PubMed] [Google Scholar]
- 233.Peeters, T., W. Louwet, R. Gelade, D. Nauwelaers, J. M. Thevelein, and M. Versele. 2006. Kelch-repeat proteins interacting with the G alpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 103:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Penalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Porta, A., A. M. Ramon, and W. A. Fonzi. 1999. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181:7516-7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Prill, S. K., B. Klinkert, C. Timpel, C. A. Gale, K. Schroppel, and J. F. Ernst. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546-560. [DOI] [PubMed] [Google Scholar]
- 237.Ramage, G., K. VandeWalle, J. L. Lopez-Ribot, and B. L. Wickes. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214:95-100. [DOI] [PubMed] [Google Scholar]
- 238.Ramon, A. M., and W. A. Fonzi. 2003. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell 2:718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290-8300. [DOI] [PubMed] [Google Scholar]
- 241.Reuß, O., A. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 242.Riggle, P. J., K. A. Andrutis, X. Chen, S. R. Tzipori, and C. A. Kumamoto. 1999. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect. Immun. 67:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rocha, C. R. C., K. Schröppel, D. Harcus, A. Marcil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signalling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Roman, E., C. Nombela, and J. Pla. 2005. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25:10611-10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rottmann, M., S. Dieter, H. Brunner, and S. Rupp. 2003. A screen in Saccharomyces cerevisiae identified CaMCM1, an essential gene in Candida albicans crucial for morphogenesis. Mol. Microbiol. 47:943-959. [DOI] [PubMed] [Google Scholar]
- 246.Ruchel, R. 1986. Cleavage of immunoglobulins by pathogenic yeasts of the genus Candida. Microbiol. Sci. 3:316-319. [PubMed] [Google Scholar]
- 247.Sabie, F. T., and G. M. Gadd. 1992. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia 119:147-156. [DOI] [PubMed] [Google Scholar]
- 248.Saito, H., and K. Tatebayashi. 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. (Tokyo) 136:267-272. [DOI] [PubMed] [Google Scholar]
- 249.Sanchez-Martinez, C., and J. Pérez-Martin. 2002. Gpa2, a G-protein α subunit required for hyphal development in Candida albicans. Eukaryot. Cell 1:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Santos, M., and I. F. de Larrinoa. 2005. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr. Genet. 48:88-100. [DOI] [PubMed] [Google Scholar]
- 251.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sass, P., J. Field, J. Nikawa, T. Toda, and M. Wigler. 1986. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83:9303-9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sato, T., T. Watanabe, T. Mikami, and T. Matsumoto. 2004. Farnesol, a morphogenetic autoregularoty substance in the dimorphic fungus Candida albicans, inhibits growth through suppression of a mitogen-activated protein kinase cascade. Biol. Pharm. Bull. 27:751-752. [DOI] [PubMed] [Google Scholar]
- 254.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentatous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Schaller, M., M. Bein, H. C. Korting, S. Baur, G. Hamm, M. Monod, S. Beinhauer, and B. Hube. 2003. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect. Immun. 71:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Schroppel, K., K. Sprosser, M. Whiteway, D. Y. Thomas, M. Rollinghoff, and C. Csank. 2000. Repression of hyphal proteinase expression by the mitogen-activated protein (MAP) kinase phosphatase Cpp1p of Candida albicans is independent of the MAP kinase Cek1p. Infect. Immun. 68:7159-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Schweizer, A., S. Rupp, B. N. Taylor, M. Röllinghoff, and K. Schröppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 258.Sengupta, M., and A. Datta. 2003. Two membrane proteins located in the Nag regulon of Candida albicans confer multidrug resistance. Biochem. Biophys. Res. Commun. 301:1099-1108. [DOI] [PubMed] [Google Scholar]
- 259.Sentandreu, M., M. V. Elorza, R. Sentandreu, and W. A. Fonzi. 1998. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J. Bacteriol. 180:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 361:399-411. [DOI] [PubMed] [Google Scholar]
- 261.Sharkey, L. L., W. L. Liao, A. K. Ghosh, and W. A. Fonzi. 2005. Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology 151:1061-1071. [DOI] [PubMed] [Google Scholar]
- 262.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Shen, J., W. Guo, and J. R. Köhler. 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 73:1239-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sheppard, D. C., T. Doedt, L. Y. Chiang, H. S. Kim, D. Chen, W. C. Nierman, and S. G. Filler. 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16:5866-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Singh, B., and A. Datta. 1979. Regulation of N-acetylglucosamine uptake in yeast. Biochim. Biophys. Acta 557:248-258. [DOI] [PubMed] [Google Scholar]
- 266.Singh, P., S. Ghosh, and A. Datta. 1997. A novel MAP-kinase kinase from Candida albicans. Gene 190:99-104. [DOI] [PubMed] [Google Scholar]
- 267.Singh, P., S. Ghosh, and A. Datta. 2001. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 69:7898-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Smith, D. G., M. D. Garcia-Pedrajas, S. E. Gold, and M. H. Perlin. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50:259-275. [DOI] [PubMed] [Google Scholar]
- 270.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 271.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 272.Soll, D. R. 2002. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 81:101-110. [DOI] [PubMed] [Google Scholar]
- 273.Soll, D. R. 1997. Gene regulation during high-frequency switching in Candida albicans. Microbiology 143:279-288. [DOI] [PubMed] [Google Scholar]
- 274.Soll, D. R. 1992. High-frequency switching in Candida albicans. Clin. Microbiol. Rev. 5:183-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Sonneborn, A., D. P. Bockmühl, M. Gerads, K. Kurpanek, D. Sanglard, and J. F. Ernst. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386-396. [DOI] [PubMed] [Google Scholar]
- 277.Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu, M. R. Seringhaus, M. Gerstein, S. Yi, M. Snyder, and D. R. Soll. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Srikantha, T., L. Tsai, K. Daniels, A. J. Klar, and D. R. Soll. 2001. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Srikantha, T., L. K. Tsai, K. Daniels, and D. R. Soll. 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 182:1580-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Staab, J., and P. Sundstrom. 2003. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 11:69-73. [DOI] [PubMed] [Google Scholar]
- 282.Staab, J. F., Y. S. Bahn, C. H. Tai, P. F. Cook, and P. Sundstrom. 2004. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 279:40737-40747. [DOI] [PubMed] [Google Scholar]
- 283.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 284.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 285.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Su, S. S., and A. P. Mitchell. 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21:3789-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Su, Z., M. J. Osborne, P. Xu, X. Xu, Y. Li, and F. Ni. 2005. A bivalent dissectional analysis of the high-affinity interactions between Cdc42 and the Cdc42/Rac interactive binding domains of signaling kinases in Candida albicans. Biochemistry 44:16461-16474. [DOI] [PubMed] [Google Scholar]
- 288.Suarez, T., and M. A. Penalva. 1996. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol. Microbiol. 20:529-540. [DOI] [PubMed] [Google Scholar]
- 289.Sudbery, P. E., N. A. Gow, and J. Berman. 2004. The distinct morphogenetic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 290.Sullivan, P. A., and M. G. Shepherd. 1982. Gratuitous induction by N-acetylmannosamine of germ tube formation and enzymes for N-acetylglucosamine utilization in Candida albicans. J. Bacteriol. 151:1118-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sundstrom, P. 1999. Adhesins in Candida albicans. Curr. Opin. Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 292.Sundstrom, P., E. Balish, and C. M. Allen. 2002. Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J. Infect. Dis. 185:521-530. [DOI] [PubMed] [Google Scholar]
- 293.Tamaki, H., T. Miwa, M. Shinozaki, M. Saito, C.-W. Yun, K. Yamamoto, and H. Kumagai. 2000. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267:164-168. [DOI] [PubMed] [Google Scholar]
- 294.Tebarth, B., T. Doedt, S. Krishnamurthy, M. Weide, F. Monterola, A. Dominguez, and J. F. Ernst. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329:949-962. [DOI] [PubMed] [Google Scholar]
- 295.Theesfeld, C. L., T. R. Zyla, E. G. Bardes, and D. J. Lew. 2003. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell 14:3280-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 297.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Tomlin, G. C., G. E. Hamilton, D. C. Gardner, R. M. Walmsley, L. I. Stateva, and S. G. Oliver. 2000. Suppression of sorbitol dependence in a strain bearing a mutation in the SRB1/PSA1/VIG9 gene encoding GDP-mannose pyrophosphorylase by PDE2 overexpression suggests a role for the Ras/cAMP signal-transduction pathway in the control of yeast cell-wall biogenesis. Microbiology 146:2133-2146. [DOI] [PubMed] [Google Scholar]
- 300.Tournu, H., J. Serneels, and P. Van Dijck. 2005. Fungal pathogens research: novel and improved molecular approaches for the discovery of antifungal drug targets. Curr. Drug Targets 6:909-922. [DOI] [PubMed] [Google Scholar]
- 301.Tournu, H., G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F. C. Odds, N. A. Gow, and A. J. Brown. 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 4:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. P. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tsuchimori, N., L. L. Sharkey, W. A. Fonzi, S. W. French, J. E. Edwards, Jr., and S. G. Filler. 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Umeyama, T., A. Kaneko, Y. Nagai, N. Hanaoka, K. Tanabe, Y. Takano, M. Niimi, and Y. Uehara. 2005. Candida albicans protein kinase CaHsl1p regulates cell elongation and virulence. Mol. Microbiol. 55:381-395. [DOI] [PubMed] [Google Scholar]
- 305.Ushinsky, S. C., D. Harcus, J. Ash, D. Dignard, A. Marcil, J. Morchhauser, D. Y. Thomas, M. Whiteway, and E. Leberer. 2002. CDC42 is required for polarized growth in the human pathogen Candida albicans. Eukaryot. Cell 1:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Vandenberg, A. L., A. S. Ibrahim, J. E. Edwards, Jr., K. A. Toenjes, and D. L. Johnson. 2004. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell 3:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Van Dyk, D., I. S. Pretorius, and F. F. Bauer. 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169:91-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Vazquez-Torres, A., and E. Balish. 1997. Macrophages in resistance to candidiasis. Microbiol. Mol. Biol. Rev. 61:170-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Versele, M., J. H. de Winde, and J. M. Thevelein. 1999. A novel regulator of G-protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 18:5577-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Versele, M., K. Lemaire, and J. M. Thevelein. 2001. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2:574-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Villela, F., E. Herrero, J. Torres, and M. A. de la Torre-Ruiz. 2005. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 280:9149-9159. [DOI] [PubMed] [Google Scholar]
- 312.Vinces, M. D., C. Haas, and C. A. Kumamoto. 2006. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot. Cell 5:825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Vogler, A. P., and J. W. Lengeler. 1989. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol. Gen. Genet. 219:97-105. [DOI] [PubMed] [Google Scholar]
- 314.Ward, M. P., C. J. Gimeno, G. R. Fink, and S. Garrett. 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15:6854-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Westfall, P. J., D. R. Ballon, and J. Thorner. 2004. When the stress of your environment makes you go HOG wild. Science 306:1511-1512. [DOI] [PubMed] [Google Scholar]
- 316.White, T. C., and N. Agabian. 1995. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J. Bacteriol. 177:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Whiteway, M., D. Dignard, and D. Y. Thomas. 1992. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc. Natl. Acad. Sci. USA 89:9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wightman, R., S. Bates, P. Amornrrattanapan, and P. Sudbery. 2004. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 164:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Wu, J., and B. L. Miller. 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17:6191-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Xu, W., F. J. J. Smith, R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Xue, C., Y. S. Bahn, G. M. Cox, and J. Heitman. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p G subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yamada-Okabe, T., T. Mio, N. Ono, Y. Kashima, M. Matsui, M. Arisawa, and H. Yamada-Okabe. 1999. Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J. Bacteriol. 181:7243-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Yamada-Okabe, T., Y. Sakamori, T. Mio, and H. Yamada-Okabe. 2001. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 268:2498-2505. [DOI] [PubMed] [Google Scholar]
- 327.Yamada-Okabe, T., and H. Yamada-Okabe. 2002. Characterization of the CaNAG3, CaNAG4, and CaNAG6 genes of the pathogenic fungus Candida albicans: possible involvement of these genes in the susceptibilities of cytotoxic agents. FEMS Microbiol. Lett. 212:15-21. [DOI] [PubMed] [Google Scholar]
- 328.Yamano, N., N. Oura, J. Wang, and S. Fujishima. 1997. Cloning and sequencing of the genes for N-acetylglucosamine use that construct divergent operons (nagE-nagAC) from Vibrio cholerae non-O1. Biosci. Biotechnol. Biochem. 61:1349-1353. [DOI] [PubMed] [Google Scholar]
- 329.Zhao, G. Q., Y. Zhang, M. A. Hoon, J. Chandrashekar, I. Erlenbach, N. J. P. Ryba, and C. S. Zuker. 2003. The receptors for mammalian sweet and umami taste. Cell 115:255-266. [DOI] [PubMed] [Google Scholar]
- 330.Zhao, X., K. J. Daniels, S. H. Oh, C. B. Green, K. M. Yeater, D. R. Soll, and L. L. Hoyer. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Zhao, X., C. Pujol, D. R. Soll, and L. L. Hoyer. 2003. Allelic variation in the contiguous loci encoding Candida albicans ALS5, ALS1 and ALS9. Microbiology 149:2947-2960. [DOI] [PubMed] [Google Scholar]
- 332.Zheng, X., Y. Wang, and Y. Wang. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]