Abstract
Maize and a variety of other plant species release volatile compounds in response to herbivore attack that serve as chemical cues to signal natural enemies of the feeding herbivore. N-(17-hydroxylinolenoyl)-l-glutamine is an elicitor component that has been isolated and chemically characterized from the regurgitant of the herbivore-pest beet armyworm. This fatty acid derivative, referred to as volicitin, triggers the synthesis and release of volatile components, including terpenoids and indole in maize. Here we report on a previously unidentified enzyme, indole-3-glycerol phosphate lyase (IGL), that catalyzes the formation of free indole and is selectively activated by volicitin. IGL's enzymatic properties are similar to BX1, a maize enzyme that serves as the entry point to the secondary defense metabolites DIBOA and DIMBOA. Gene-sequence analysis indicates that Igl and Bx1 are evolutionarily related to the tryptophan synthase alpha subunit.
Many plant species elevate the release of volatile compounds in response to herbivore damage (1, 2). Herbivore predators and parasitic wasps exploit these chemical cues to locate their prey or hosts. Several such chemically mediated tritrophic interactions have been documented for agrarian systems including lima bean (1), cucumber (3), apple trees (4), cotton, and maize (5). Turlings et al. (2, 6, 7) have demonstrated that maize seedlings damaged by beet armyworm caterpillars release a specific blend of terpenoids and indole that is attractive to Cortesia marginiventris, a parasitic wasp that attacks larvae of several species of Lepidoptera.
Two elicitors have been identified in the oral secretions of insect herbivores that trigger the production of volatiles. Mattiacci et al. (8) found that a β-glucosidase in Pieris brassicae caterpillars elicits the release of volatiles from cabbage leaves. In maize, volatile production is activated by an elicitor present in the regurgitant of beet armyworm caterpillars (7). Volicitin [N-(17-hydroxylinolenoyl)-l-glutamine] was identified as the major active elicitor in the oral secretion of these larvae (9). Synthesized and natural volicitin induce maize seedlings to release the same blend of volatile terpenoids and indole as released when they are damaged by caterpillar feeding (9).
Maize seedlings contain indole as an intermediate in at least two biosynthetic pathways. The BX1 enzyme catalyzes the conversion of indole-3-glycerol phosphate (IGP) to indole, which is further converted to the defense-related secondary metabolite DIMBOA [2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one] (10, 11). Indole also serves as the penultimate intermediate in the formation of tryptophan by tryptophan synthase. The sequence similarity of BX1 and tryptophan synthase alpha (TSA), one subunit of the tryptophan synthase complex, suggests that BX1 has been evolutionarily modified to serve in secondary metabolism (11). Because the beet armyworm elicitor volicitin triggers increased release of indole in maize seedlings, the current study was undertaken to determine whether volicitin regulates expression of an enzyme catalyzing synthesis of free indole. Bx1 gene expression was not elicited by volicitin. However, a BX1 homologous enzyme, named here indole-3-glycerol phosphate lyase (IGL), catalyzes the same reaction. The corresponding gene Igl is shown to be regulated transcriptionally by volicitin. The induction pattern of Igl parallels the emission of free indole from the whole plant.
Materials and Methods
Plant Source.
Maize seeds, Zea mays, planted in potting soil were maintained in growth chambers illuminated with metal halide and high-pressure sodium lamps on a 14-h light/10-h dark photoperiod. Incubator temperature fluctuated between 20°C during the dark cycle and 30°C when the lights were on. The maize inbred-line CI31A was the source of genomic clones. Maize line B73 was the source for the Stratagene maize cDNA library (whole plant, 7 leaves stage, 1.5 million primary clones) used for the isolation of the cDNA clones. CI31A and bx1bx1 mutant lines (12) were used for the induction experiments. For all experiments, seedlings were harvested 11 days after planting.
Heterologous Expression and IGL Enzyme Assay.
The IGL protein was expressed in Escherichia coli by inserting the cDNA into a modified pET3a vector (13). The enzyme was purified by a six-nucleotide oligomeric COOH-terminal histidine tag (Qiagen Ni-NTA purification system) and was used for fluorometric enzyme assays (14) as described by Frey et al. (10).
Volicitin Incubations.
Maize seedlings were cut off above the root and placed in shell vials containing 500 μl of 50 mM sodium phosphate buffer adjusted to pH 8; seedlings were treated with buffer alone or with buffer containing 300 pmol synthetic volicitin. For RNA analysis, plantlets were incubated for 0, 1, 2, 4, and 8 h, and 3 plantlets were combined for each time point. When the seedlings were dissected, material from nine seedlings was combined. All tissue was frozen in liquid nitrogen and stored at −70°C. For volatile collections, seedlings were incubated first for 0, 1.5, 3, 4.5, 6, or 10 h.
Collection and Analysis of Volatiles.
Treated seedlings or freshly cut plantlets were placed into individual volatile-collection chambers, and volatiles were collected for 1.5 h as described (9). All treatments were run in triplicate. Volatiles were analyzed by capillary GC (Hewlett–Packard 6890) on a 15-m × 0.25-mm (i.d.) fused silica column with a 0.1-mm-thick bonded methyl-silicone mode for 0.3 min with an injector temperature of 230°C and detector temperature of 250°C. The column was held at 40°C for 0.5 min, increased 12°C⋅min−1 to 180°C, and then increased 40°C⋅min−1 to 220°C and held for 2 min. Helium was used as a carrier gas at a linear flow velocity of 35 cm⋅sec−1. Plant volatile components were identified by comparison of GC retention times with those of authentic standards. Compounds were quantified via flame-ionization detection by using the peak area of nonyl acetate added as an internal standard. Select samples were also analyzed by mass spectroscopy (HP 5973) interfaced to the GC and operated in the electron impact mode. Oven conditions were the same as listed above except that a DB-5MS column was used.
Transcript Analysis.
RNA was isolated as depicted by Logemann et al. (15), and poly(A)+ RNA was purified with OligoTex beads (Qiagen, Germany) as directed by the supplier. For Northern blot analysis, the poly(A)+ RNA was separated, transferred to Hybond N membrane (Amersham Pharmacia), and hybridized with 32P-labeled DNA probes as described by Frey et al. (16). All membranes were hybridized by using the Igl and glycerol-phosphate dehydrogenase, cytosolic form (GAP C) cDNAs (17). Poly(A)+ RNA equivalent to 1.5 plantlets was applied per lane. To normalize the differences in loading, all values of the GAP C signals were divided by the value of the highest GAP C hybridization signal. Every Igl signal value was then divided by the so-calculated fraction of the respective GAP C signal. The Storm 860 PhosphorImager (Amersham Pharmacia) and the program IMAGE QUANT were used for quantification. One hybridization unit is defined as 103 IMAGE QUANT volumes. For the determination of transcript quantities by the real-time PCR experiment shown in Fig. 2, gene-specific primers were designed: Igl, 5′-ATGGCCTCCGCGATCAAGGCTGCATC-3′ and 5′-CTGAGAGTGAGAGCACACGAGTTCC-3′; Bx1, 5′-ATGGCTTTCGCGCCCAAAACGTCCTC-3′ and 5′-CGTGGACCCCCGCCTCTTTCATCTCG-3′; TSA, 5′-CCACAAAGGCAGCGCTCGGAGGTG-3′ and 5′-GCCTCGCTCCTCAGCAACGTCGTCT-3′; GAP C, 5′-GCTAGCTGCACCACAAACTGCCT-3′ and 5′-TAGCCCCACTCGTTGTCGTACCA-3′. The experiment was performed on the LightCycler instrument with the FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals). Two independent incubation experiments were analyzed in triplicate. Linearized plasmids containing the different cDNAs were quantified by agarose gel electrophoresis and ethidium-bromide staining, and used as quantification standards. Single-stranded template cDNA was synthesized by using 0.5 μg of poly(A)+ RNA and the TaqMan Reverse Transcription Reagents kit (Perkin–Elmer) by priming with random hexamers as directed by the supplier. After quantification, the final PCR products were checked for homogeneity and absence of intron sequences by agarose gel electrophoresis.
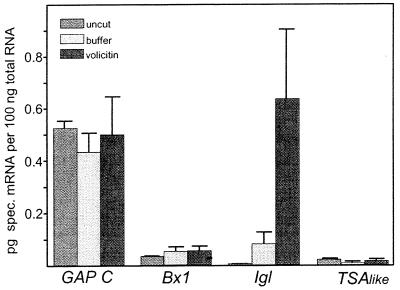
Figure 2.
Quantification of mRNA levels in the maize inbred-line CI31A. Incubation in buffer or volicitin solution was for 2 h. The standard deviation is indicated by the bar.
Molecular Biology Methods.
Cloning was performed as described by Frey et al. (16). The Stratagen maize cDNA library (106 plaque forming units) was screened, and four cDNA clones were identified for Igl and five were identified for TSAlike. cDNA and genomic sequences were cloned into the pBluescript KS(+) vector (Stratagene). Sequencing was done by primer walking on an Applied Biosystems PRISM 377 DNA sequencer with oligomeric primers purchased from Eurogentec (Brussels) and GIBCO/BRL. The 5′ end of the transcripts was isolated with the 5′-3′ rapid amplification of cDNA ends kit (Roche Molecular Biochemicals). DMSO at a final concentration of 5% (vol/vol) was included in the first strand cDNA transcription mix.
Mapping.
Data were generated by Southern blot analysis with the internal genomic 1.2-kb PstI fragment of Igl as a probe. A unique restriction fragment length polymorphism was revealed for the CM37xT232 recombinant inbred-mapping population (18).
Results
Isolation of the Igl Gene and Enzymatic Properties.
To probe for genes that are involved in the synthesis of indole, a pivotal intermediate for both primary and secondary metabolism, the BX1-cDNA probe, was used for the screening of a genomic library (16). A genomic clone that encoded a protein with homology to TSAs was isolated (Fig. 1B). Because of a close homology to Bx1, the IGL of the DIMBOA biosynthetic pathway, the gene was termed Igl.
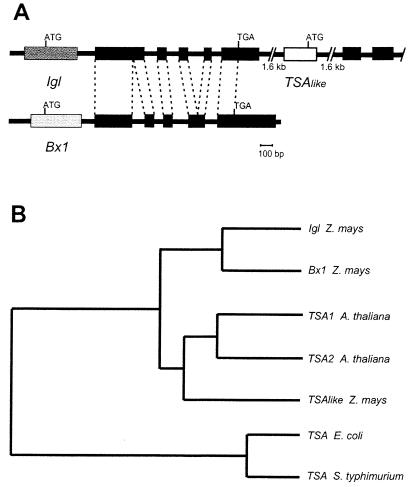
Figure 1.
Genes related to TSA in maize. (A) Gene structure of Igl, Bx1, and TSAlike. Exons are represented by filled boxes, and corresponding exons are indicated by dashed lines. A putative TSA gene (TSAlike) is located downstream of Igl (1,615 bp separate the first polyadenylation site of Igl from the transcription start site of TSAlike). The first exons of IGL, BX1, and TSAlike are only 45% identical with respect to each other. Homology with bacterial TSA enzymes starts with the amino acids encoded by exon 2 of the three genes. The exon-intron structure of BX1 and IGL is not absolutely conserved. IGL and BX1 have 82% identity in the amino acids encoded by exons 2–6. Comparison to TSAlike reveals a 66% identity for IGL and 67% identical amino acids for BX1 in this fraction of the protein. Exons 2 and 3 of TSAlike comprise the second exon of Bx1. The genomic sequence of TSAlike was not fully determined. (B) Phylogenetic tree. The relationship is calculated for the conserved region of the proteins that begins with exon 2 for the plant enzymes. The program PILEUP of the GCG package, version 8.1 for UNIX, was used for the calculation.
Several cDNA clones of Igl were identified by screening a whole plant cDNA library with the 1.2-kb internal PstI fragment of the genomic clone. Because the cDNA clones proved to be incomplete at the 5′ end, a 5′-rapid amplification of cDNA ends experiment was performed to get the full-size sequence of the gene. The reading frame of Igl encodes a 339-aa peptide and is interrupted by five intervening sequences as revealed by comparison with the cDNA clones (Fig. 1A). The exon-intron structure of Igl and Bx1 is not fully conserved. Whereas the amino acids encoded by exon 2 to the C terminus display 82% identity to BX1 and 63% to TSA of Arabidopsis thaliana, the amino acids encoded by the first exon are <45% identical between these protein species. In the case of Bx1 (11) and TSA of A. thaliana (19), exon 1 encodes a transit peptide.
A third gene with homology to TSA was isolated from maize as a full-size cDNA clone and as a partial genomic clone (Fig. 1). A 5′ rapid amplification of cDNA ends experiment was performed to verify the 5′-end. The product of this gene is more related to TSA from A. thaliana (70% identity) than to Igl and Bx1 (65% and 64% identity, respectively; Fig. 1B). This product represents a candidate for the maize TSA and was termed provisionally TSAlike. Sequence analyses of the genomic Igl clone revealed that Igl and TSAlike are separated by 1.6 kb (Fig. 1). The genes map to the long arm of chromosome one (1L260, id. tum11).
Phylogenetic analysis of the maize and A. thaliana TSA homologous proteins defines two branches: one is comprised of the A. thaliana TSAs and the TSAlike sequence from maize, and the second consists of BX1 and IGL (Fig. 1B). Association with the tryptophan synthase beta subunit (TSB) has been shown to be basic for enzymatic activity of TSA in bacteria and in plants (19–21). In contrast, BX1 activity is independent of TSB (10). To address the question of whether the intimate structural similarity of IGL and BX1 reflects the same enzymatic mechanism, i.e., the conversion of IGP to indole, Igl was expressed in E. coli, purified, and used for the determination of the steady-state kinetic constants as described for BX1 (10). The Michaelis constant KmIGP of 0.1 mM is ≈8-fold higher compared with BX1, but the catalytic rate constant kcat of 2.3 s−1 is similar for both enzymes. The kcat/KmIGP of 23 mM−1⋅s−1 proves that IGL is catalyzing the formation of indole, and is ≈3 times more efficient in this reaction as bacterial TSA in its active form in the α2β2 complex (Table 1; ref. 22). Hence, two maize enzymes, BX1 and IGL, are capable of producing indole efficiently.
Table 1.
Comparison of catalytic properties
|
E. coli
|
Zea
mays
|
|||
|---|---|---|---|---|
| α | α2β2 | BX1 | IGL | |
| KmIGP | 0.5 mM | 0.03 mM | 0.013 mM | 0.1 mM |
| Kcat | 0.002 s−1 | 0.2 s−1 | 2.8 s−1 | 2.3 s−1 |
| Kcat/KmIGP | 0.004 mM−1⋅s−1 | 7.4 mM−1⋅s−1 | 215 mM−1⋅s−1 | 23 mM−1⋅s−1 |
Igl Transcript Level Is Elevated by Volicitin.
Real-time PCR with gene-specific primer pairs was used to reveal a possible causal relationship between the biosynthesis of volatile indole induced by volicitin and expression of the homologous maize genes (Fig. 2). Whereas the steady-state transcript level of Bx1 is not influenced by volicitin treatment of the seedlings, for Igl an ≈8-fold increase compared with the buffer control was observed. This finding indicates that Igl is a putative target gene in the signaling cascade that starts with the perception of volicitin and leads to the synthesis and emission of a defined blend of volatiles. The amount of Igl transcript at 2 h after volicitin treatment exceeds the level of GAP C and represents a major transcript under this condition. TSAlike is not influenced by volicitin treatment and is expressed at a lower level compared with Bx1 and Igl (Fig. 2).
Emission of Volatile Indole Is Independent of a Functional Bx1 Gene.
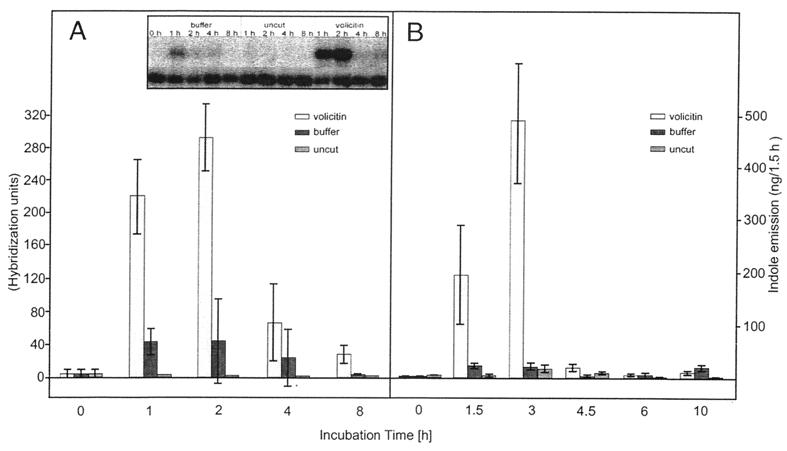
To examine how the enzymes BX1 and IGL catalyze free indole emission in maize, a mutant line defective in the BX1 gene product was examined. The bx1bx1 line carries a deletion of the Bx1 gene that removes ≈0.5 kb of the 5′ part of the mRNA (10), allowing the hybridization signals derived from Igl and bx1 to be distinguished unambiguously by the size difference. Elevated levels of indole were detected for volicitin treatment in the mutant line within 1.5 h, and the maximum response was observed at the 3-h collection, with indole emissions of 326 ± 77 ng⋅h−1 compared with 13 ± 4 and 6 ± 7 for buffer and untreated controls, respectively (Fig. 3B).
Figure 3.
Analysis of Igl transcript levels and indole emission in the bx1bx1 mutant line. The seedlings were harvested at the indicated time points after incubation in buffer or buffer with volicitin. Uncut seedlings were taken as a control. The standard deviation is given by the vertical bars. (A) RNA quantification. The hybridization signals of the Igl and GAP C probe were quantified with the PhosphorImager. GAP C values were taken to normalize the loading of the lanes. Five individual experiments were made for each time point except for 8 h, at which time only two repetitions were made. An example of a Northern blot is given (Inset). The Igl signal is shown in the upper part, and hybridization of GAP C is shown in the lower part. (B) Determination of indole emission. Indole produced within 1.5 h was collected at the indicated time points and quantified as described in Materials and Methods.
Volicitin Induction of Igl Expression Is Fast and Transient.
The kinetics of Igl induction were investigated by Northern blot analysis with the bx1bx1 mutant maize line. GAP C, a gene not influenced by volicitin treatment (Fig. 2), was used to normalize for loading differences on the gels. The low level of Igl transcript in untreated plants was increased by wounding (buffer control), and more dramatically by wounding in combination with volicitin treatment (Fig. 3A). The increase induced by cutting and incubation in buffer was 10-fold, and the maximum value was reached 1 h after treatment and dropped slowly to resume the noninduced level after 8 h. In contrast, a 50- to 60-fold increase was achieved with volicitin incubation. The principal augmentation of Igl transcript was found within the first hour of treatment, but there was still an increase during the next hour. The decrease within the next 6 h was more pronounced as compared with that of the buffer treatment. Thus, the rise of Igl transcript levels induced by volicitin is clearly transient. The emission of indole showed a similar transient release that peaked about 2 h after the maximum level of Igl transcript was reached (Fig. 3). The effect of wounding on indole emission is relatively small. This response could indicate that a certain threshold of Igl induction has to be exceeded for notable indole production to occur.
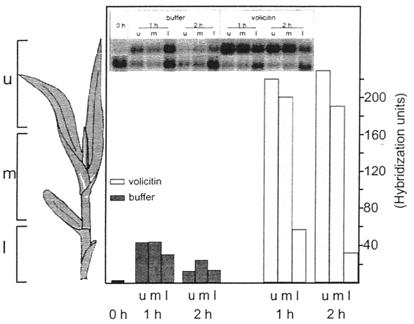
Igl Is Expressed Highest in Upper Parts of the Plantlet.
To determine whether Igl transcript responds differently in distinct plant parts to volicitin, plants were dissected into three fractions after incubation (Fig. 4). For both incubation times (1 and 2 h), the highest level of Igl transcript was detected in the upper part of the seedling. The buffer-control seedlings displayed a lower level of Igl transcript for all parts, and there was no pronounced variation between the different fractions.
Figure 4.
Analysis of Igl mRNA levels in different parts of the seedling. Seedlings of the bx1bx1 mutant line were cut and incubated for 1 and 2 h in buffer or buffer containing volicitin. The lower (l), middle (m), and upper (u) parts of the seedling were assayed individually. The Inset displays the Northern blot analysis with the Igl probe (u) and GAP C probe (l). Analysis was performed as described in Fig. 3.
Discussion
An Enzymatic Mechanism to Produce Indole.
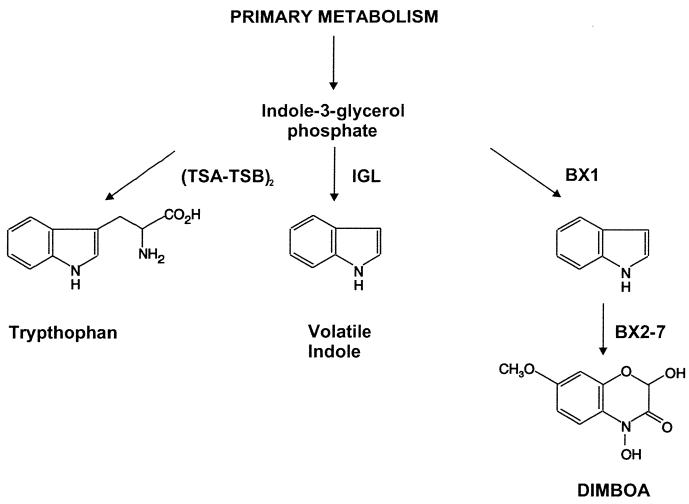
A crucial step in the evolution of secondary metabolic pathways is the establishment of the branch reactions that channel metabolites out of primary metabolism. In maize, the first step in the synthesis of the benzoxazinoid DIMBOA (Fig. 5) is the conversion of IGP to indole. This reaction is catalyzed by the BX1 enzyme (10) and defines the branch point from tryptophan biosynthesis. We have characterized here a second maize enzyme, IGL, with similar properties. Both enzymes act efficiently in vitro as IGP lyases (Table 1). The sequence similarity suggests that IGL and BX1 originated from TSA, the enzyme responsible for the penultimate reaction in tryptophan biosynthesis (Fig. 5). Tryptophan synthase is typically a tetrameric heterosubunit complex that is formed by two TSA–TSB complexes that are linked via TSB (20). These complexes were originally described in bacteria; however, an analogous heterosubunit tryptophan synthase complex exists in plants (19, 23). Indole is usually not released from the tryptophan synthase complex but is directly converted to tryptophan by TSB. TSA activity depends almost completely on complex formation.
Figure 5.
Branching of primary and secondary metabolic pathways in maize. (TSA-TSB)2: tryptophan synthase complex; BX2–7: enzymes necessary for the conversion of indole into DIMBOA. IGL and BX1 are TSA-homologous proteins that catalyze indole formation. TSA is active only in a complex with TSB. Indole formed transiently in this complex is immediately converted to tryptophan.
For both IGL and BX1, changes relative to the TSA-consensus sequence are observed and located in two domains. In IGL and BX1, an unusual tyrosine substitutes a conserved aliphatic residue (position 58 of Salmonella typhimurium TSA). Two conserved glutamine residues (position 134, 135 of S. typhimurium TSA) are substituted by valine and alanine in the case of BX1, and by glycine and asparagine in the case of IGL. A 4-aa deletion (BX1) and an addition of 2 aa (IGL) are found in close proximity to these substitutions; hence the architecture of this part of the protein might be altered. Both modified regions are part of the interaction domains of TSA and TSB based on the crystal structure of the S. typhimurium tryptophan synthase complex (24, 25). Whether these alterations are significant for the establishment of the different enzymatic properties of IGL and BX1 versus TSA remains to be shown. No deviation from the TSA consensus is found for TSAlike.
The synthesis of several other secondary metabolites in plants, such as the indole glucosinolates, anthranilate-derived alkaloids, and tryptamine derivatives (19, 26, 27), could depend on indole as an intermediate. IGP was proposed as a branch point from the tryptophan pathway for the synthesis of the indolic phytoalexin camalexin (3-thiazol-2′-yl-indole) in A. thaliana (28, 29). It would not be surprising if BX1 and IGL would represent the preferred route for indole production.
Function of Igl in Maize.
Indole has been identified as one of the blend of volatile chemicals emitted from maize in response to herbivore damage (2). The production and release of indole in plants has been shown to be an active process in which de novo synthesis is triggered in response to insect feeding (30). Because volicitin (9) has been identified as an elicitor molecule that can mimic larvae feeding in that it triggers indole emissions, induction of an indole-biosynthetic pathway that responds directly or indirectly to volicitin application is predicted. The data presented here suggest that the Igl gene product is a key player in this process: IGL is an efficient indole-3-glycerol phosphate lyase in vitro (see above). Igl transcript accumulation (Fig. 3) closely resembles the temporary fashion of indole emission induced by volicitin application. Igl transcript abundance is ahead of the rise of indole production by about 2 h; this delay could result from translation of Igl mRNA into a functional enzyme. A similar time interval was recognized for the sesquiterpene cyclase stc1 transcript and emission of a naphthalene-based sesquiterpene [see accompanying paper (ref. 31)]. Neither Bx1 nor TSAlike transcript levels are influenced by volicitin, and, hence, neither are expected to contribute directly to volicitin-induced volatile indole production.
A further correlation of Igl induction and volatile emission is found in the requirement of a certain developmental stage (two to three expanded leaves) for volatile emission and Igl induction as response to herbivory and volicitin treatment. Application in earlier stages of plant development (4 days) results in stimulation of neither Igl transcription nor indole formation (data not shown). The same developmental stage is necessary for the induction of stc1 [see accompanying paper (ref. 31)]. The results implicate that Igl is a target gene in the volicitin signaling cascade; however, ultimate prooof will require an Igl mutant.
The cell types that are responsible for volatile production remain to be determined. Whether there is substrate channeling and indole produced by IGL is used exclusively as a volatile, or whether it is available for other biosynthetic pathways, for example DIMBOA formation, can be addressed in future experiments.
Elevated Igl Transcript Level with Elicitor Application.
Igl mRNA is hardly detectable in maize plantlets (Figs. 3 and 4). However, cutting and incubation in volicitin solution leads to a drastic increase of the Igl transcript steady-state level (Figs. 3 and 4), with peak amounts even exceeding the transcript levels of the housekeeping gene GAP C. Accumulation is achieved within 1 to 2 h after treatment with volicitin, and, although application is at the base of the plantlet, the top part displays the highest Igl transcript level by volicitin treatment (Fig. 4). This response demonstrates a relatively rapid movement of the signal. The induction kinetics are comparable to the examples of induced transcription of certain mitogen-activated protein kinases by the fungal elicitor Avr9 and wounding; here, maximal transcript accumulation is reached 1 to 2 h after elicitation of cell cultures or plants (32). For stc1 in maize [see accompanying paper (ref. 31)], induction by insect regurgitant or volicitin reaches maximum transcript levels at 4 h. Peak levels at 8 h or 12 h after caterpillar chewing are reported for the protease inhibitor II and 3-hydroxy-3-methylglutaryl-CoA reductase genes of potato (33). Interestingly, in these cases, induction is delayed for wounding compared with wounding, in conjunction with inductor application. In contrast, for Igl, both kinetics are comparable, but the maximum transcript levels are different (Fig. 3).
Evolution of Secondary Metabolic Pathways.
Duplication of genes of the primary metabolism and subsequent modification of gene function may be a driving force for the evolution of secondary metabolic pathways. Bx1 and Igl seem to be derived from TSA by gene-duplication events and were modified during evolution to obtain their distinct function in secondary metabolism. Interestingly, both enzymes efficiently catalyze indole formation independently of a TSB-like subunit. Gene duplication is indicated by the fact that Igl and TSAlike are separated by only 1.6 kb in the maize genome. Another example for the recruitment of a gene of the primary metabolism is represented by homospermidine synthase from Senecio vernalis. This enzyme is derived from deoxyhypusine synthase, an enzyme required for activation of translation initiation factor 5A (34).
The coexistence of IGL and BX1 demonstrates that modified genes can catalyze the same reaction directed toward different biochemical routes. Such parallel secondary metabolic pathways, at least in maize, seem to augment the chemical-defense capacity of the plant. Bx1 is expressed constitutively in the young seedlings and catalyzes the accumulation of the indole-derived antifeedant components DIBOA and DIMBOA. In contrast, Igl is induced later in plant development in response to herbivore damage, is able to catalyze the synthesis of free indole that is released from the leaves, and is part of the blend of odors that attracts beneficial insects. An inducible indole-producing function is also required for cotton (35) and several other plants (36) that respond to different chemical signals with emission of volatile indole. Because the efficiency of indole formation depends on the amount of available substrate (IGP) delivered by the primary metabolism, regulative interactions with the primary pathway as demonstrated for camalexin biosynthesis (37) are also anticipated for volatile indole production.
Acknowledgments
We thank B. Burr for processing the mapping data, H. Alborn for providing volicitin, and Verena Goretzki for sequence analysis of TSAlike. The excellent technical assistance of Regina Hüttl is acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 369) and by Fonds der Chemischen Industrie.
Abbreviations
- GAP C
glycerol-phosphate dehydrogenase, cytosolic form
- IGP
indole-3-glycerol phosphate
- TSA
tryptophan synthase alpha subunit
- TSB
tryptophan synthase beta subunit
- IGL
IGP lyase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF271383 and AF271384).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260499897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260499897
References
- 1.Dicke M, Sabelis M W, Takabayashi J, Bruin J, Posthumus M A. J Chem Ecol. 1990;16:3091–3118. doi: 10.1007/BF00979614. [DOI] [PubMed] [Google Scholar]
- 2.Turlings T C J, Tumlinson J H, Lewis W J. Science. 1990;250:1251–1253. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- 3.Takabayashi J, Dicke M, Posthumus M A. J Chem Ecol. 1994;20:1329–1354. doi: 10.1007/BF02059811. [DOI] [PubMed] [Google Scholar]
- 4.Takabayashi J, Dicke M, Posthumus M A. Phytochemistry. 1991;30:1459–1462. [Google Scholar]
- 5.De Moraes C M, Lewis W J, Paré P W, Alborn H T, Tumlinson J H. Nature (London) 1998;393:570–573. [Google Scholar]
- 6.Turlings T C, Tumlinson J H, Heath J H, Proveau A T, Doolittle R E. J Chem Ecol. 1991;17:2235–2251. doi: 10.1007/BF00988004. [DOI] [PubMed] [Google Scholar]
- 7.Turlings T C, McCall P J, Alborn H T, Tumlinson J H. J Chem Ecol. 1993;19:411–425. doi: 10.1007/BF00994314. [DOI] [PubMed] [Google Scholar]
- 8.Mattiacci L, Dicke M, Posthumus M A. Proc Natl Acad Sci USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alborn H T, Turlings T C, Jones T H, Stenhagen G, Loughrin J H, Tumlinson J H. Science. 1997;276:945–949. [Google Scholar]
- 10.Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley R B, Briggs S P, et al. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 11.Sicker D, Frey M, Schulz M, Gierl A. Int Rev Cytol. 2000;198:319–346. doi: 10.1016/s0074-7696(00)98008-2. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton R H. Weeds. 1964;12:27–30. [Google Scholar]
- 13.Rosenberg A H, Lade B N, Chui D S, Lin S W, Dunn J J, Studier F W. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 14.Creighton T E. Eur J Biochem. 1970;13:1–10. doi: 10.1111/j.1432-1033.1970.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 15.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 16.Frey M, Kliem R, Saedler H, Gierl A. Mol Gen Genet. 1995;246:100–109. doi: 10.1007/BF00290138. [DOI] [PubMed] [Google Scholar]
- 17.Brinkmann H, Martinez P, Quigley F, Martin W, Cerff R. J Mol Evol. 1987;26:320–328. doi: 10.1007/BF02101150. [DOI] [PubMed] [Google Scholar]
- 18.Burr B, Burr F. Trends Genet. 1991;7:55–60. doi: 10.1016/0168-9525(91)90232-F. [DOI] [PubMed] [Google Scholar]
- 19.Radwanski E R, Zhao J, Last R L. Mol Gen Genet. 1995;248:657–667. doi: 10.1007/BF02191705. [DOI] [PubMed] [Google Scholar]
- 20.Creighton T E, Yanofsky C. J Biol Chem. 1966;241:980–990. [PubMed] [Google Scholar]
- 21.Radwanski E R, Barczak A J, Last R L. Mol Gen Genet. 1996;253:353–361. doi: 10.1007/pl00008602. [DOI] [PubMed] [Google Scholar]
- 22.Weischet W O, Kirschner K. Eur J Biochem. 1976;65:365–373. doi: 10.1111/j.1432-1033.1976.tb10350.x. [DOI] [PubMed] [Google Scholar]
- 23.Nagao R T, Moore T C. Arch Biochem Biophys. 1972;149:402–413. doi: 10.1016/0003-9861(72)90338-4. [DOI] [PubMed] [Google Scholar]
- 24.Hyde C C, Ahmed S A, Padlan E A, Miles E W, Davies D R. J Biol Chem. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 25.Weyand M, Schlichting I. Biochemistry. 1999;38:16469–16480. doi: 10.1021/bi9920533. [DOI] [PubMed] [Google Scholar]
- 26.Kutchan T M. Plant Cell. 1995;7:1059–1070. doi: 10.1105/tpc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bak S, Nielsen H L, Halkier B A. Plant Mol Biol. 1998;38:725–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- 28.Tsuji J, Zook M, Somerville S C, Last R L, Hammerschmidt R. Physiol Mol Plant Pathol. 1993;43:221–229. [Google Scholar]
- 29.Zook M. Plant Physiol. 1998;118:1389–1393. doi: 10.1104/pp.118.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paré P W, Tumlinson J H. Nature (London) 1997;385:30–31. [Google Scholar]
- 31.Shen B, Zheng Z, Dooner H K. Proc. Natl. Acad. Sci. USA. 2000. http://www.pnas.org/cgi/doi/10.1073/pnas.240284097 10.1073/pnas.240284097. http://www.pnas.org/cgi/doi/10.1073/pnas.240284097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeis T, Piedras P, Zhang S, Klessig D F, Hirt H, Jones J D. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korth K L, Dixon R A. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ober D, Hartmann T. Proc Natl Acad Sci USA. 1999;96:14777–14782. doi: 10.1073/pnas.96.26.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughrin J H, Manukian A, Heath J H, Turlings T C, Tumlinson J H. Proc Natl Acad Sci USA. 1994;91:11836–11840. doi: 10.1073/pnas.91.25.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boland W, Hopke J, Donath J, Nüske J, Bublitz F. Angew Chem Int Ed Engl. 1995;34:1600–1602. [Google Scholar]
- 37.Zhao J, Williams C C, Last R L. Plant Cell. 1998;10:359–370. doi: 10.1105/tpc.10.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]