Abstract
macroH2A histone variants have been implicated to function in gene silencing by several studies, including ones showing a preferential association of macroH2A on the inactive X chromosome. To examine macroH2A function in vivo, we knocked out macroH2A1. macroH2A1 knockout mice are viable and fertile. A broad screen of liver gene expression showed no evidence of defects in X inactivation but did identify genes that have increased expression levels in macroH2A1 knockouts. macroH2A1-containing nucleosomes are enriched on the coding and/or upstream regions of these genes, suggesting that their increased expression levels are a direct effect of the absence of macroH2A1. The concentrations of macroH2A1 nucleosomes on these genes are low in the livers of newborn mice, and the macroH2A1 knockout had little effect on the expression levels of these genes in newborn liver. Our results indicate that an increase in liver macroH2A1 during the transition from newborn to young-adult status contributes to a decrease in the expression levels of these genes. These genes cluster in the area of lipid metabolism, and we observed metabolic effects in macroH2A1 knockouts. Our results indicate that the function of macroH2A1 histones is not restricted to gene silencing but also involves fine tuning the expression of specific genes.
The nucleosome is an important target for modifying chromatin functions, including transcription. One mechanism for modifying nucleosome function is the substitution of variant histones for the major or canonical histones. Genetic studies showed that histone variants H2A.X, H2A.Z, CENPA, and H3.3 have important functional properties that cannot be provided by their conventional counterparts (reviewed in reference 24).
macroH2A histone variants have an unusual structure, consisting of a full-length H2A domain linked to a large nonhistone domain, producing proteins that are nearly three times the size of conventional core histones (Fig. 1A) (26). macroH2As are highly conserved among vertebrates (1, 27). They appear to be absent from most invertebrates but are present in the sea urchin. The H2A domain of macroH2As is ∼65% identical to conventional H2As. Most of the nonhistone region appears to be derived from a domain that is found in many contexts (3, 27). Recent studies showed that some “macrodomains,” including the one from macroH2A1.1, bind ADP ribose (17). The significance of this binding for macroH2A function is not known. There are three macroH2A variants. macroH2A1.1 and 1.2 are formed by alternate splicing of macroH2A1 (gene symbol H2afy), while macroH2A2 is encoded by a separate gene (4, 7, 26, 30). The distributions of macroH2A variants are different in different cell types and change during development (7, 25). The macroH2A1 (1.1 and 1.2) content of adult rat liver chromatin, a relatively rich source of macroH2A1, was estimated to be one for every 30 nucleosomes (26).
FIG. 1.
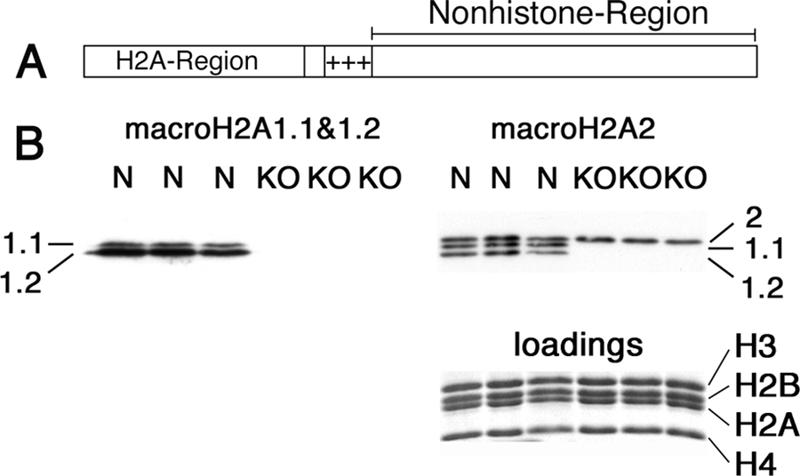
Absence of macroH2A1 proteins in the macroH2A1 knockout. (A) Diagram of macroH2A. The H2A region is ∼65% identical to conventional H2A. +++ indicates a highly basic region that may bind DNA. macroH2A1.1 and 1.2 differ in a single ∼30-amino-acid segment in the nonhistone region. (B) Western blots with antibodies against nonhistone regions of macroH2A1.1 and 1.2 (left) and macroH2A2 (right) are shown; note that this macroH2A2 serum has some cross-reaction with macroH2A1.1 and 1.2, so all three macroH2A variants show. macroH2A1 variants are much more abundant in mouse liver than macroH2A2 (7). Total nuclear extracts were prepared from normal mouse liver (lanes N) and macroH2A1 knockout mouse liver (lanes KO). Loadings were equalized using core histones (see stained gel below the macroH2A2 blot).
The distribution of macroH2A in chromatin suggests a role in transcriptional repression. We recently mapped the distribution of macroH2A1, using DNA isolated from macroH2A1-containing nucleosomes (5). Active genes were depleted of macroH2A1, while the inactive X chromosome showed preferential enrichment. macroH2A1 was not enriched on four regions of the inactive X chromosome that escape inactivation (5). The enrichment of macroH2A on the inactive X chromosome was previously observed by immunofluorescence and by expression of macroH2A-green fluorescent protein fusion proteins (4, 6-8). A recent study showed that a macroH2A1 knockdown decreases the stability of X inactivation in cultured human cells, indicating that macroH2A1 has a role in maintenance of X inactivation (15). Immunofluorescence studies indicate that macroH2As are preferentially associated with other large domains of the transcriptional silent chromatin, including pericentromeric heterochromatin in some cell types (8, 11), the XY body of spermatocytes (16), and transcriptionally silent senescence-associated heterochromatic foci (36). A role for macroH2A1 in gene silencing is supported by a recent study that showed a dramatic derepression of interleukin-8 (IL-8) when macroH2A1 expression was knocked down in a cultured human B-cell line (2). In this paper, we investigate the in vivo functions of macroH2A by examining the effects of a macroH2A1 knockout mutation on gene expression in mouse liver.
MATERIALS AND METHODS
Mice.
All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Mice were maintained on a 12-h day/night cycle, with the lights coming on at 8 a.m. Mice used for organ harvest were killed between 8 and 11 a.m. For most studies, macroH2A1 knockout mice and their age- and sex-matched controls were raised in the same cage.
macroH2A1 knockout.
We based the targeting vector on an ∼10-kb ApaI fragment of mouse genomic DNA that contains exon 2 of macroH2A1 (H2afy) (see Fig. S1 in the supplemental material). We obtained this fragment from a mouse bacterial artificial chromosome clone (library CitbCJ7; Invitrogen) made from an embryonic stem (ES) cell line that has a 129Sv background. A cassette containing a promoterless internal ribosome entry site βgeo marker (22) was inserted into a PstI site in the first intron. This cassette was flanked by LoxP sites. A third LoxP site was inserted into a SalI site in the second intron (see Fig. S1 in the supplemental material).
Electroporation and drug selection were done by Incyte Genomics Inc., using 129SvJ ES cells. G418-resistant clones were screened by Southern blot analyses, with genomic fragments just 5′ or 3′ of the ApaI fragment used for the targeting vector (see Fig. S1 in the supplemental material). A clone that was positive for homologous recombination was transiently transfected with a plasmid that expressed the Cre recombinase (pMCCreN) (12), and G418-sensitive clones were screened by PCR for excision of the internal ribosome entry site βgeo cassette and exon 2.
ES cells were injected into C57BL/6 blastocysts, and the chimeric males were mated to obtain germ line transmission of the macroH2A1 knockout allele. The macroH2A1 knockout allele was backcrossed into the C57BL/6 background for 10 generations. macroH2A1 knockout and normal alleles were identified by PCR (see Fig. S1 in the supplemental material), using a small piece of the tail that was digested with proteinase K. The primers used were as follows: Pst forward, GTGAGACACTTGAGAAAAGTCATTGTCAGTATAAC; Pst reverse, AACAGCACAGCAGGCAGCTGCTGA; Sal reverse, CCTCCAGTCCTTGTTCAACATAACCACCAT. Reactions were run using Taq polymerase from New England Biolabs in its standard buffer with an annealing temperature of 55°C.
Microarrays.
The mice were raised in the same cage following weaning and were all killed over a period of approximately 1 h on the same morning. The large lobe of the liver was immediately dissected and frozen in liquid nitrogen. Total RNA was isolated using TRIzol (Invitrogen). RNA concentration was estimated by UV absorption and confirmed by gel electrophoresis.
All protocols were conducted as described in the Affymetrix GeneChip expression analysis technical manual. Briefly, total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 min, and hybridized for 16 h at 45°C to Affymetrix MOE430 v2.0 microarrays. The microarrays were then washed at low (6× SSPE) (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and high (100 mM MES [morpholineethanesulfonic acid], 0.1 M NaCl) stringencies and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect fluorescence signal after excitation at 570 nm.
Affymetrix Microarray Suite 5.0 was used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as the background and subtracted from all features in that sector. Probe intensities were then exported as .cel files and imported into GeneSpring (v7.2; Agilent Technologies), where probe set signals were calculated using Gene Chip robust multiarray. The expression values were log2 transformed in Excel and analyzed using statistical analysis of microarrays (SAM v2.1; Stanford University). This analysis gave us a list of the 111 most significant genes (see Table S1 in the supplemental material). This list had a maximum false-discovery rate of 47% based on 200 permutations.
Subtractive hybridization.
We used PCR Select subtractive hybridization (Clontech Laboratories, Palo Alto, CA) to enrich for cDNAs with increased abundance in cDNA prepared from female macroH2A1 knockout liver in comparison to that for normal female mouse liver. Subtracted cDNA was cloned, and clones that were enriched in the subtracted cDNA were sequenced and tested by real-time PCR for enrichment in unsubtracted macroH2A1 knockout liver cDNA.
Real-time PCR.
Real-time PCR was performed using the LightCycler system (Roche Applied Science). We used Titanium Taq polymerase (BD Biosciences) with its standard buffer. Denaturation was for 1 s at 95°C, annealing for 3 s (variable temperatures) (see Table S3 in the supplemental material), and elongation for 5 s at 72°C for reactions with genomic DNA and 13 s at 72°C for reactions with cDNA. Detection was with SYBR green I, and quantification was done with standard LightCycler data analysis software, using the second derivative maximum to compare different samples. Using three standard primer pairs, we estimated a difference of approximately 1.9-fold per cycle and used this value for all primer pairs. Product purity was checked by melting curves and gel electrophoresis. We synthesized cDNA, using poly d(T)12-18 (Amersham Biosciences) as a primer and BD PowerScript reverse transcriptase (CloneTech) under standard conditions. All cDNAs were normalized using primers to glyceraldehyde phosphate dehydrogenase. Following normalization, normal and knockout cDNA samples were compared directly in the same run. Genomic DNAs were equalized by UV absorption and by ethidium bromide-stained agarose gels. Primer sequences are listed in Table S3 in the supplemental material.
Glucose tolerance test.
Mice were fasted overnight and the following morning were injected with 2 g glucose/kg of body weight intraperitoneally, using a solution of 20% glucose dissolved in phosphate-buffered saline. Blood glucose was measured at 0, 15, 30, 60, 90, and 120 min following the injection, using a One Touch Ultra glucometer. Blood was collected by cutting the tip of the tail. The mice were 2 to 3.5 months of age. Knockout and normal mice used in individual tests were closely age matched, usually within a week, and knockout and normal mice were caged together in most cases. We tested 63 different normal males, 49 knockout males, 44 normal females, and 47 knockout females. There was no difference in the average weights of knockout and normal males. The average weight of the knockout females was slightly higher than that for the normal females, 21.4 versus 20.7 g.
Purification of macroH2A1-containing nucleosomes.
macroH2A1-containing mono- and oligonucleosomes were purified from bulk chromatin by thiol affinity chromatography as described previously (5). Briefly, an H1-stripped S2 chromatin fraction was digested to mono- and oligonucleosomes and macroH2A1-containing nucleosomes were purified by binding them to thiopropyl Sepharose (Amersham Biosciences) and eluting them with mercaptoethanol. The chromatin was passed through a column of activated thiol Sepharose (Amersham Biosciences), which does not preferentially bind macroH2A1, prior to being passed through a thiopropyl Sepharose column. The thiopropyl column was washed with 0.5 M NaCl to remove nucleosomes bound by nonhistone proteins prior to elution of macroH2A1-containing nucleosomes with mercaptoethanol.
Northern blots.
RNA was run in formaldehyde-containing 1% agarose gels (NorthernMax; Ambion) under standard conditions. The transfers were done by downward elution with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)/10 mM NaOH (following the Ambion protocol) onto Zeta-Probe GT membranes (Bio-Rad). Membranes were washed in 2× SSC and UV cross-linked. Hybridization was done at 65°C, following the standard protocol for Zeta-Probe GT membranes. The Lpl probe was the insert from a mouse cDNA clone (gi 13097035). The Serpina7 probe was prepared by PCR amplification from mouse liver cDNA. The primers were as follows: forward, GTATCGGAGGCTCTCTGTGG; reverse, GGAGGTACTTCCTGCTGATCC. The product is 981 bp based on Serpina7 cDNA sequences. DNA probes were labeled by random priming.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate gel electrophoresis, transferred onto polyvinylidene difluoride membranes (7, 25), and probed with rabbit polyclonal antibodies raised against the nonhistone regions of rat macroH2A1.1, 1.2, and 2 (6). The blocked membranes were incubated overnight with the primary antibody, followed by a secondary peroxidase conjugated donkey anti-rabbit immunoglobulin G, and the signal was detected using chemiluminescence.
RESULTS
Knockout of macroH2A1.
We produced a knockout mutation of macroH2A1 by removing the second exon (see Fig. S1 in the supplemental material). This exon contains the initiation codon and encodes the first part of the H2A domain. The next methionine does not occur until the final 30 amino acids of the protein, making it very unlikely that any functional macroH2A-like protein could be encoded by the knockout allele. Western blot analysis confirmed that neither macroH2A1.1 nor macroH2A1.2 was present in nuclear extracts prepared from the livers of macroH2A1 knockout mice (Fig. 1B).
macroH2A1 knockout mice develop and grow with no obvious defects in their appearances, sizes, or fertility. The fertility of males suggests that macroH2A1 is not required for silencing of the XY body, although we have not directly addressed this question. We have not observed any obvious deficiency in the births of macroH2A1 knockout males or females. The viability of knockout females rules out any gross deficiency in X inactivation (21). We kept eight macroH2A1 knockout and eight control females for more than 1.5 years without the emergence of any consistent unusual pathology in the knockouts. Histological analysis of three knockout females showed no significant pathology at 8 weeks of age. In comparison to age-matched controls, the knockouts had mildly enlarged spleens, increased lymphocytic inflammation in a variety of tissues, and smaller Peyer's patches, but these differences were within normal limits.
We examined the possibility of up-regulation of macroH2A2 in macroH2A1 knockout mice by Western blot analyses (Fig. 1B). Scans of the Western blot did not detect a change in the overall level of macroH2A2 in the livers of macroH2A1 knockout mice. Immunofluorescence studies of macroH2A1 knockout liver did not reveal any obvious increase in the macroH2A2 content of hepatocytes (not shown). As expected, the immunofluorescence for macroH2A1, including staining of the inactive X chromosome, was lost in macroH2A1 knockout liver (not shown).
Gene expression in macroH2A1 knockout liver.
We used an Affymetrix MOE 430 v2.0 microarray to compare gene expression in macroH2A1 knockout liver to that in normal mouse liver. This array is based on the mouse UniGene database (build 107, June 2002) and was designed to probe the expression of more than 34,000 genes. The knockout mutation was inbred into the C57BL/6 background for 10 generations. Normal and knockout mice were caged together, and livers were collected at the same time of day; the mice were 2-month-old females. Livers from five normal mice were compared to livers from five macroH2A1 knockouts. We used significance analysis of microarrays (SAM v2.1; Stanford University) to analyze the results (32). With the maximum false-discovery rate set at 47%, significance analysis of microarrays identified 54 genes that had increased expression levels in macroH2A1 knockout liver and 50 genes that had decreased expression levels (see Table S1 in the supplemental material). Most of these genes showed <2-fold changes.
There is no evidence for reactivation of the inactive X chromosome in this microarray analysis. Only 3 of the 54 genes that showed increased expression levels are on the X chromosome, and 1 of these, Dlgh3, did not show increased expression levels when we analyzed it by real-time PCR in a larger group of females. The absence of any obvious effect on X inactivation is consistent with previous studies that showed that X inactivation is robustly maintained by multiple mechanisms (9, 15).
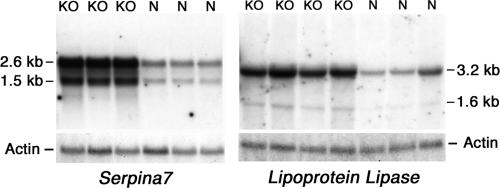
In order to identify genes that have consistently altered expression levels in macroH2A1 knockout mice, we used real-time PCR to examine the expression of selected genes in the livers of a large independent group of 2-month-old females. This analysis identified five genes that have significantly increased expression levels in the knockouts (Table 1). We used Northern blot analysis to examine the expression of two of these genes, the lipoprotein lipase (Lpl) and thyroxine binding globulin (Serpina7) genes. This analysis confirmed the PCR results and gave no evidence of unusual transcripts in the knockout mice (Fig. 2). Our PCR analysis identified three genes that showed small but significant decreases in expression in macroH2A1 knockout liver (Table 1).
TABLE 1.
Expression of selected genes in macroH2A1 knockout mice
| Gene | Value for indicated liver group
|
Function | Reference | ||
|---|---|---|---|---|---|
| Adulta
|
Newborn femaleb,c | ||||
| Femalec | Malec | ||||
| Serpina7 | 4.1f | 1.8f | 0.76g | Serum thyroxine binding protein | 31 |
| Lpl | 2.6f | 2.6f | 1.1d | Hydrolysis of circulating triacylglycerols | 29 |
| Krt1-23 | 2.4f | 5.6f | 1.1 | Type 1 keratin | 35 |
| ATP11a | 1.9f | 1.5d | 1.1 | Putative transbilayer amphipath transporter | 13 |
| Scd2 | 1.5d | 1.4d | 1.1 | Fatty acid desaturase | 23 |
| CD36 | 1.4e | 1.6f | 1.0 | Fatty acid translocase | 28 |
| Thrsp | 1 | 1.6e | ND | Regulates fatty acid synthesis | 37 |
| GTPbp4 | 0.78e | 0.9 | ND | Nuclear GTP binding protein | 20 |
| Sucnr1 | 0.77d | 1.1 | ND | G protein couple receptor for succinate | 14 |
| Ar | 0.65d | 1.2 | ND | Androgen-regulated transcription factor | 33 |
Numbers of adult livers: female, 13 normal and 12 knockout; male, 12 normal and 12 knockout.
Numbers of newborn female (3-day) livers: seven normal and eight knockout. Similar results were obtained with a smaller group of newborn males.
Values were determined in duplicate by real-time PCR and are given as ratios of expression levels for macroH2A1 knockout mice to those for normal controls. ND, not determined.
P < 0.05 (one-tailed t test).
P < 0.01 (one-tailed t test).
P < 10−4 (one-tailed t test).
P < 0.05 (two-tailed t test).
FIG. 2.
Northern blot analysis of Serpina7 and Lpl expression in macroH2A1 knockout mice. Total mouse liver RNA was analyzed on Northern blots for Serpina7 and Lpl expression. The blots were rehybridized with β actin to show equal loadings. KO, RNA from female macroH2A1 knockout mouse liver; N, RNA from normal female mouse liver.
The five genes that were significantly increased in female knockouts also had increased expression levels in male knockout liver (Table 1). There were two notable differences. One was Serpina7, which was 1.8-fold higher in knockout males compared to more than 4-fold higher in knockout females. The other was Krt1-23, which was 5.6-fold higher in knockout males compared to 2.4-fold higher in knockout females. In contrast to those in female liver, expression levels of Ar, Sucnr1, and GTPbp4 were not significantly decreased in male knockout liver. The sexual dimorphic effect of the knockout on Serpina7 expression may be related to the sexual dimorphic expression of this gene in normal mice, which we found to be nearly four times higher in normal adult males than in normal adult females. Krt1-23 expression levels were about 30% higher in normal adult females than in normal adult males.
We identified two additional genes that have increased expression levels in macroH2A1 knockout mice. The thyroid hormone responsive SPOT14 homolog gene (Thrsp) was identified in a microarray analysis of gene expression in male macroH2A1 knockout liver, and CD36 was identified by subtractive hybridization. Both of these genes were confirmed by real-time PCR, but Thrsp showed increased expression levels only in males (Table 1). Interestingly, four of the seven genes that have increased expression levels in knockouts encode proteins directly involved in fatty acid metabolism (Table 1).
Distribution of macroH2A1 on genes with altered expression levels.
We examined the distribution of macroH2A1-containing nucleosomes on the genes that had altered expression levels in order to assess whether the effect of the knockout on these genes was likely to be direct. We purified macroH2A1-containing mono- and oligonucleosomes with a high degree of specificity, using our previously described thiol affinity procedure (Fig. 3) (5). We used real-time PCR to compare the distribution of specific sequences in DNA isolated from macroH2A1-containing nucleosomes to that in DNA from the nucleosomes applied to the columns. The results presented in Fig. 4 are the averages for four preparations, two from male liver and two from female liver. The male and female results are presented separately in Table S2 in the supplemental material and in most cases were very similar to each other.
FIG. 3.
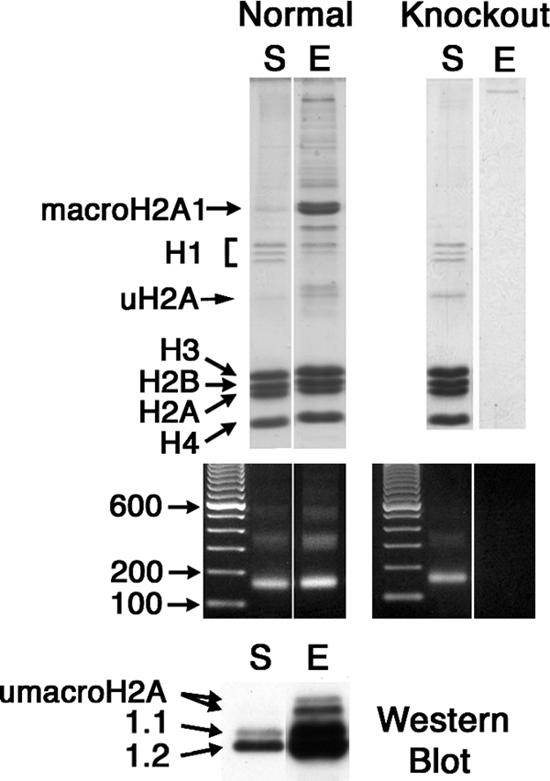
Purification of macroH2A1-containing nucleosomes by thiol-affinity chromatography. Mouse liver mono- and oligonucleosomes were prepared, and macroH2A1-containing chromatin fragments were purified by selective thiol affinity chromatography (5). S, starting material; E, material eluted from thiopropyl Sepharose with β-mercaptoethanol. The mercaptoethanol-eluted chromatin is highly enriched for macroH2A1-containing nucleosomes. Results for normal mouse liver are shown on the left, and results for macroH2A1 knockout liver are on the right. Protein compositions were analyzed by sodium dodecyl sulfate gel electrophoresis (top panels). DNA was analyzed in agarose gels (middle panels). The bottom panel is a Western blot of the starting material and a pool of the eluted fractions stained with antibodies against macroH2A1.1 and 1.2; these lanes were loaded for equal amounts of H3 and H4.
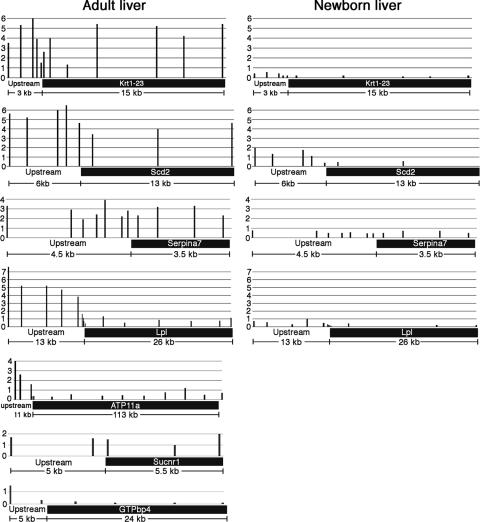
FIG. 4.
Distribution of macroH2A1 on genes that have altered expression levels in macroH2A1 knockout mouse liver. Results for adult liver (2-month-old mice) are on the left, and those for newborn liver (3-day-old mice) are on the right. macroH2A1-containing chromatin fragments were purified from normal mouse liver (Fig. 3). The relative macroH2A1 concentration on a sequence was estimated by using real-time PCR to calculate the relative concentration of the sequence in the macroH2A1-enriched, thiopropyl-eluted fraction in comparison to that of the starting material. A value of 1 on the scales on the left of the diagrams indicates a macroH2A1 concentration equal to that of the starting material prepared from adult liver. A few sites tested with the genes of the adult samples were not tested with the newborn samples. The horizontal bars on the bottom of the charts indicate the transcribed regions of the genes.
macroH2A1 was enriched on the transcribed and upstream regions of Scd2, Krt1-23, and Serpina7 (Fig. 4, adult liver), consistent with the possibility that the increased expression levels of these genes in knockouts are a direct effect of the absence of macroH2A1. Serpina7 is located on the X chromosome. In contrast to what we have seen with other active X-linked genes (5), Serpina7 showed nearly equal enrichment levels for macroH2A1 in male samples and female samples at most sites (see Table S2 in the supplemental material). macroH2A1 was enriched on the upstream regions of Lpl and ATP11a (Fig. 4, adult liver). This suggests that the increased expression levels of these genes in macroH2A1 knockout liver may be a direct effect of the knockout on the regulatory regions of these genes. This enrichment was especially striking for Lpl, where it started within 450 bp of the start of the gene and extended to more than 10 kb upstream. macroH2A1 was depleted on most of the transcribed regions that we probed for both of these genes, consistent with what we have observed for other active genes (5).
We examined the distribution of macroH2A1 on two of the genes that had decreased expression levels in female macroH2A1 knockout liver (Fig. 4). As with most active genes that we have examined, macroH2A1 was depleted on the transcribed and nearby upstream regions of GTPbp4. There was a small enrichment for macroH2A1 on both the transcribed and the upstream regions of Sucnr1. The low concentrations of macroH2A1 on these genes indicate that the effect of a macroH2A1 knockout on these genes is indirect.
Gene expression in newborn liver.
The macroH2A1 content of liver nuclei from 3-day-old mice is much lower than that of liver nuclei from 2-month-old mice (Fig. 5) (25). Based on scans of Western blots, the macroH2A1.1 content of liver nuclei isolated from 3-day-old-mice was 16-fold lower and the macroH2A1.2 content was 2.7-fold lower. This suggested that the effect of a macroH2A1 knockout on gene expression might be different in the livers of newborn mice. Indeed, none of the genes that showed increased expression levels in adult knockout liver showed increased expression levels in newborn knockout liver (Table 1). The expression levels of Serpina7, Lpl, Scd2, Krt1-23, and CD36 are higher in newborn liver than in adult liver, but ATP11a expression levels were similar in newborn and adult livers (Table 2). These findings suggest that an increase in liver macroH2A1 during this period contributes in part to a decrease in the expression levels of Serpina7, Lpl, Scd2, Krt1-23, and CD36. Consistent with this idea, we found that the concentrations of macroH2A1 on Serpina7, Lpl, Scd2, and Krt1-23 were much lower in the livers of newborn mice than in adult livers (Fig. 4).
FIG. 5.
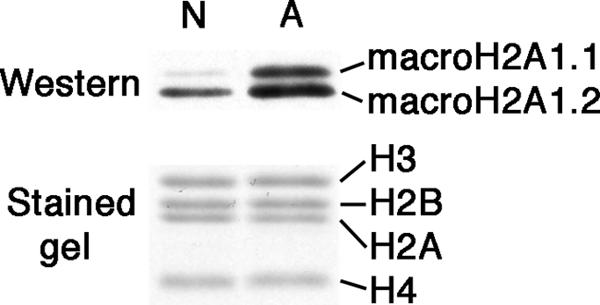
Developmental changes in the macroH2A1 content of mouse liver nuclei. Nuclei were isolated from the livers of newborn (3-day-old) and adult (2-month-old) mice. Total nuclear extracts were analyzed on Western blots using antibodies against macroH2A1.1 and 1.2. Gels were loaded for equal content of core histones; see Coomassie blue-stained lanes below the blot. Lane N, extract from newborn livers; lane A, extract from adult livers.
TABLE 2.
Expression in newborn and adult livers
| Gene | Expression levela |
|---|---|
| Serpina7 | 20 |
| Lpl | 12.5 |
| Krt1-23 | 17.9 |
| Scd2 | 5.9 |
| ATP11a | 1 |
| CD36 | 2.2 |
Expression levels in six newborn and six adult females were determined by real-time PCR and were normalized to glyceraldehyde phosphate dehydrogenase expression levels.
Glucose metabolism.
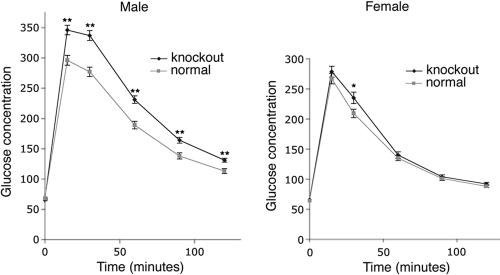
Lipoprotein lipase hydrolyzes triacylglycerols circulating in lipoprotein complexes in the blood and has a crucial role in the delivery of fatty acids to the tissues (29). It is expressed in neonatal mouse liver, but its expression levels in adult mouse liver are low (29). Mice that carry a liver-specific Lpl transgene that increases liver lipoprotein lipase expression levels fourfold develop insulin resistance in the liver and reduced glucose tolerance secondary to the increased delivery of fatty acids to the liver (18). The livers of adult macroH2A1 knockouts have increased expression levels of Lpl and the fatty acid transporter CD36, which should lead to increased fatty acid delivery and reduced glucose tolerance. As expected, macroH2A1 knockout males had significantly higher concentrations of blood glucose in glucose tolerance tests at all times except zero time (Fig. 6). Female knockouts showed only a small increase in blood glucose at the 30-min time point. We believe that this sexual dimorphism may reflect a difference in the responses of females and males to increased delivery of fatty acids to the liver. Previous studies of the effects of increased fatty acid delivery on liver insulin resistance have been done on male mice and rats (18, 19, 34), suggesting that the responses of females may be different.
FIG. 6.
Glucose tolerance is altered in macroH2A1 knockout mice. Mice were fasted overnight and injected with 2 g glucose/kg of body weight intraperitoneally, and blood glucose concentrations were measured at 0, 15, 30, 60, 90, and 120 min. The error bars indicate the standard errors of the means. *, P < 0.05 (two-tailed t test); **, P < 0.001 (two-tailed t test). Numbers of mice: 63 normal males, 49 knockout males, 44 normal females, and 47 knockout females.
DISCUSSION
macroH2A histone variants have been implicated to function in gene silencing. Interestingly, our broad-based examination of gene expression in macroH2A1 knockout mouse liver did not discover any genes showing dramatic derepression. Instead, we discovered several genes that showed moderate but consistent increases in expression. macroH2A1-containing nucleosomes were preferentially present in the upstream and/or transcribed regions of these genes, indicating that macroH2A1 in the regulatory and/or transcribed regions of these genes has a direct repressive effect on their expression levels. The distribution of macroH2A1 that we observed did not provide evidence for the localization of macroH2A1 to a particular site or small region of these genes. Instead, we found relatively large domains of similar macroH2A1 enrichment in the upstream and/or transcribed regions. We believe that these domains exert a repressive effect on these genes. The case for a direct effect on ATP11a is weaker, because the domain of macroH2A1 enrichment begins rather far upstream. Recent studies using in vitro transcription indicate that macroH2A1.2 nucleosomes can directly inhibit initiation of transcription (10). It is not known how close they need to be to the promoter region to exert a repressive effect or whether their presence on transcribed regions inhibits elongation. We discovered a few genes that have reduced expression levels in macroH2A1 knockout mice. macroH2A1 nucleosomes showed little or no enrichment on these genes, suggesting that their reduced expression levels are an indirect effect of the knockout.
Four of the seven genes that we identified as having increased expression levels in macroH2A1 knockouts are directly involved in fatty acid metabolism: Lpl, Scd2, Thrsp, and CD36 (Table 1). This suggests that macroH2A1-mediated regulation of gene expression may be a mechanism for bringing about coordinated adaptive changes to the metabolic state of liver cells. Our studies with newborn and young-adult mouse liver support this idea. There is a substantial increase in the macroH2A1 content of liver as mice mature from newborns to young adults (Fig. 5). Our results indicate that this increase contributes to the down-regulation of specific genes that influence the metabolic state of the liver. The increase of macroH2A1.1 during this period is especially marked, suggesting that it may be important for these effects. We have observed a similar developmental change in macroH2A1 composition in rat liver and kidney (25), suggesting that this may be an evolutionarily conserved mechanism used in a variety of tissues to adapt gene expression to different functional states.
Why is our list of genes that have altered expression levels in a macroH2A1 knockout short, when macroH2A1 histones are widely distributed in the chromatin? The microarray that we used was designed to examine the expression of all well-described genes as of 2002. Some genes were likely missed due to technical limitations of the arrays or technical problems with our analyses. We believe that genes were missed due to the relatively small effects of the knockout on the level of expression. Normal variation in gene expression or technical variations in our experiments will confound the detection of genes with small changes.
Most of the macroH2A may be involved in silencing regions of the genome, such as the inactive X chromosome, that are controlled by multiple repressive mechanisms. Detecting the effects of a macroH2A knockout on many of these regions may require special treatments and single-cell assays, such as those used for detecting X reactivation in macroH2A1 knockdown cells (15). A recent study showed that a knockdown of macroH2A1 produced a several-hundredfold increase in IL-8 expression in a cultured human lymphoid cell line (2). We were not able to examine whether macroH2A1 is essential for silencing IL-8 in mouse B cells, because mice do not have an IL-8 gene or any obvious homologue. Our gene expression analyses and the mild phenotype of macroH2A1 knockout mice suggest that dramatic derepression of genes is rare in macroH2A1 knockout mice. While our microarray analysis did not identify any genes showing dramatic derepression, we have detected significant derepression of specific endogenous retroviruses in macroH2A1 knockout mice (unpublished data). Overall, we believe that there is good evidence that macroH2A1 nucleosomes contribute to maintaining gene silencing in many regions of the genome. Our current study indicates that macroH2A function is not restricted to suppressing inappropriate or unwanted gene expression but also includes fine tuning the expression of specific genes.
Supplementary Material
Acknowledgments
We thank Austin Smith for providing the plasmid pGTiresbgeo, Don Baldwin and John Tobias for assistance with the microarrays, Katherine McKeown for work on the cDNA subtraction experiment, Linden Craig for assistance with histopathology, Mitchell Lazar and Rexford Ahima for suggestions related to metabolic tests, Lionel Larue for assistance with the knockout, Rachel Weinstein for assistance with statistical analyses, and Mike Atchison and Narayan Avadhani for comments on the manuscript.
This work was supported by Public Health Service grant GM49351 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 22 January 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbott, D. W., M. Laszczak, J. D. Lewis, H. Su, S. C. Moore, M. Hills, S. Dimitrov, and J. Ausio. 2004. Structural characterization of macroH2A containing chromatin. Biochemistry 43:1352-1359. [DOI] [PubMed] [Google Scholar]
- 2.Agelopoulos, M., and D. Thanos. 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 25:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. [DOI] [PubMed] [Google Scholar]
- 4.Chadwick, B. P., and H. F. Willard. 2001. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 10:1101-1113. [DOI] [PubMed] [Google Scholar]
- 5.Changolkar, L. N., and J. R. Pehrson. 2006. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol. Cell. Biol. 26:4410-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 7.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 276:21776-21784. [DOI] [PubMed] [Google Scholar]
- 8.Costanzi, C., P. Stein, D. M. Worrad, R. M. Schultz, and J. R. Pehrson. 2000. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation embryos. Development 127:2283-2289. [DOI] [PubMed] [Google Scholar]
- 9.Csankovszki, G., A. Nagy, and R. Jaenisch. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153:773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyen, C. M., W. An, D. Angelov, V. Bondarenko, F. Mietton, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2006. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 26:1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoryev, S. A., T. Nikitina, J. R. Pehrson, P. B. Singh, and C. L. Woodcock. 2004. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 117:6153-6162. [DOI] [PubMed] [Google Scholar]
- 12.Gu, H., Y. R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155-1164. [DOI] [PubMed] [Google Scholar]
- 13.Halleck, M. S., J. J. Lawler, S. Blackshaw, L. Gao, P. Nagarajan, C. Hacker, S. Pyle, J. T. Newman, Y. Nakanishi, H. Ando, D. Weinstock, P. Williamson, and R. A. Schlegel. 1999. Differential expression of putative transbilayer amphipath transporters. Physiol. Genomics 1:139-150. [DOI] [PubMed] [Google Scholar]
- 14.He, W., F. J.-P. Miao, D. C.-H. Lin, R. T. Schwandner, Z. Wang, J. Gao, J.-L. Chen, H. Tian, and L. Ling. 2004. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature 429:188-193. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. V. Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer-Fender, S., C. Costanzi, and J. R. Pehrson. 2000. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 258:254-260. [DOI] [PubMed] [Google Scholar]
- 17.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 24:1911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. K., J. J. Fillmore, Y. Chen, C. Yu, I. K. Moore, M. Pypaert, E. P. Lutz, Y. Kako, W. Velez-Carrasco, I. J. Goldberg, J. L. Breslow, and G. I. Shulman. 2001. Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc. Natl. Acad. Sci. USA 98:7522-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraegen, E. W., P. W. Clark, A. B. Jenkins, E. A. Daley, D. J. Chisholm, and L. H. Storlien. 1991. Development of muscle insulin resistance after liver insulin resistance in high-fat-fed rats. Diabetes 40:1397-1403. [DOI] [PubMed] [Google Scholar]
- 20.Laping, N. J., B. A. Olson, and Y. Zhu. 2001. Identification of a novel nuclear guanosine triphosphate-binding protein differentially expressed in renal disease. J. Am. Soc. Nephrol. 12:883-890. [DOI] [PubMed] [Google Scholar]
- 21.Marahrens, Y., B. Panning, J. Dausman, W. Strauss, and R. Jaenisch. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11:156-166. [DOI] [PubMed] [Google Scholar]
- 22.Mountford, P., B. Zevnik, A. Duwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ntambi, J. M. 1999. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40:1549-1558. [PubMed] [Google Scholar]
- 24.Pehrson, J. R. 2004. Core histone variants, p. 181-204. In J. Zlatanova and S. H. Leuba (ed.), Chromatin structure and dynamics: state of the art, vol. 29. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 25.Pehrson, J. R., C. Costanzi, and C. Dharia. 1997. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 65:107-113. [DOI] [PubMed] [Google Scholar]
- 26.Pehrson, J. R., and V. A. Fried. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257:1398-1400. [DOI] [PubMed] [Google Scholar]
- 27.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of macroH2A subtypes and domains. Nucleic Acids Res. 26:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohl, J., A. Ring, U. Korkmaz, R. Ehehalt, and W. Stremmel. 2005. FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Mol. Biol. Cell 16:24-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preiss-Landl, K., R. Zimmermann, G. Hammerle, and R. Zechner. 2002. Lipoprotein lipase: the regulation of tissue specific expression and its role in lipid and energy metabolism. Curr. Opin. Lipidol. 13:471-481. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen, T. P., T. Huang, M. A. Mastrangelo, J. Loring, B. Panning, and R. Jaenisch. 1999. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 27:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schussler, G. C. 2000. The thyroxine-binding proteins. Thyroid 10:141-149. [DOI] [PubMed] [Google Scholar]
- 32.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh, S., M. Y. Tsai, Q. Xu, X. M. Mu, H. Lardy, K. E. Huang, H. Lin, S. D. Yeh, S. Altuwaijri, X. Zhou, L. Xing, B. F. Boyce, M. C. Hung, S. Zhang, L. Gan, and C. Chang. 2002. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci. USA 99:13498-13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaragonza-Hermans, N., and J. P. Felber. 1971. Studies of the metabolic effect induced in the rat by a high fat diet. II. Disposal of orally administered (14C)-glucose. Horm. Metab. Res. 4:25-30. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, J. S., L. Wang, H. Huang, M. Nelson, and D. I. Smith. 2001. Keratin 23 (K23), a novel acidic keratin, is highly induced by histone deacetylase inhibitors during differentiation of pancreatic cancer cells. Genes Chromosomes Cancer 30:123-135. [PubMed] [Google Scholar]
- 36.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]
- 37.Zhu, Q., G. W. Anderson, G. T. Mucha, E. J. Parks, J. K. Metkowski, and C. N. Mariash. 2005. The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology 146:3343-3350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.