Abstract
Polarized growth in the budding yeast Saccharomyces cerevisiae depends upon the asymmetric localization and enrichment of polarity and secretion factors at the membrane prior to budding. We examined how these factors (i.e., Cdc42, Sec4, and Sro7) reach the bud site and found that their respective mRNAs localize to the tip of the incipient bud prior to nuclear division. Asymmetric mRNA localization depends upon factors that facilitate ASH1 mRNA localization (e.g., the 3′ untranslated region, She proteins 1 to 5, Puf6, actin cytoskeleton, and a physical association with She2). mRNA placement precedes protein enrichment and subsequent bud emergence, implying that mRNA localization contributes to polarization. Correspondingly, mRNAs encoding proteins which are not asymmetrically distributed (i.e., Snc1, Mso1, Tub1, Pex3, and Oxa1) are not polarized. Finally, mutations which affect cortical endoplasmic reticulum (ER) entry and anchoring in the bud (myo4Δ, sec3Δ, and srp101) also affect asymmetric mRNA localization. Bud-localized mRNAs, including ASH1, were found to cofractionate with ER microsomes in a She2- and Sec3-dependent manner; thus, asymmetric mRNA transport and cortical ER inheritance are connected processes in yeast.
The establishment and maintenance of cell polarity in eukaryotes require coordination of the cell cycle, actin and microtubule cytoskeletons, and the secretory pathway to regulate growth along a given axis (9, 19, 38, 49). Asymmetric cell growth is necessary for a variety of cellular processes, including division (3, 6, 34), axon extension (29), motility (30, 51), and body morphogenesis (3, 24, 66). Polarization necessitates a role for polarity factors that are capable of regulating the cell cycle, cytoskeleton, and the secretory pathway but that are also responsive to extracellular signals (i.e., chemotactic signals, pheromones, and cell-cell contacts) in order to promote directed growth. Small GTPases of the Rho family (i.e., Cdc42 and Rho) have a conserved role in evolution as being master regulators of polarity (31, 49). In the budding yeast, Saccharomyces cerevisiae, polarized growth processes (e.g., budding and mating) require Cdc42 activation, which is necessary for polarization of the actin cytoskeleton (13, 31, 49). During budding, the commitment to polarize occurs during the G1 phase of the cell-cycle and leads to the formation of a cap of activated Cdc42 at the presumptive bud site. This asymmetric clustering of Cdc42 is sufficient to induce actin polymerization and subsequent polarization (32, 53, 61). Spatial regulation of the Cdc42 cap formation is aided by the existence of landmark proteins (or anchors) which enhance selection of a given bud site, usually adjacent to the previous bud scar in haploid cells (13, 54). However, regulation of the local Cdc42 concentration is sufficient to induce and maintain polarization (32, 53, 61). In G1-arrested unbudded cells, as well as cells lacking either landmarks or polarized actin, Cdc42 can become polarized and induce directed growth (32, 61). Recent work also suggests a role for endocytosis and protein recycling in maintenance of the Cdc42 cap in newly forming buds (33). In addition to Cdc42, other polarity and secretion factors (POLs) concentrate at the presumptive bud site and incipient bud, including components of the exocyst (e.g., Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84), Rho and Sec4 GTPases, Sec1, Myo2, and others (9, 27, 34, 50). Maintenance of their local concentrations may involve endocytosis, as suggested for v-SNARE recycling within the bud (59), although many of these proteins are soluble and have no direct means to target to the endocytic pathway.
Other studies suggest a role for mRNA localization as a means of regulating the local concentration of proteins. For example, the asymmetric localization of ASH1 mRNA to the bud restricts the localization of Ash1 protein in order to regulate mating type switching in yeast (42, 57). In higher eukaryotes, mRNA localization regulates embryonic patterning in Drosophila, as well as the subcellular localization of axonal proteins in neurons and β-actin in motile cells (15, 38, 66). ASH1 mRNA trafficking in yeast depends upon the SHE1/MYO4 and SHE2-5 genes, an intact secretory pathway, and in part upon its 3′ untranslated region (UTR). SHE1/MYO4 encodes a type V myosin that mediates ASH1 mRNA transport along actin filaments (35). SHE2 encodes an mRNA binding protein, and SHE3 encodes the adaptor that connects She2 to She1/Myo4 (7, 44, 58). SHE4 encodes a protein involved in actin-dependent endocytosis (62), and SHE5 encodes a formin ortholog involved in actin regulation (21). In our studies, we found that ASH1 mRNA localization depends upon the integrity of the secretory pathway, suggesting a connection between the processes of mRNA transport and exocytosis (1).
We postulated that the local enrichment of POLs in the incipient bud might be due to targeted mRNA delivery to the presumptive bud site and subsequent local translation. By examining the localization of POL mRNAs and proteins in situ and in vivo, we found that mRNA localization precedes local protein enrichment and subsequent bud emergence. mRNAs encoding bud-localized POLs (e.g., CDC42, SEC4, and SRO7) localize asymmetrically to the tip of the emerging bud in a manner dependent upon their 3′ UTRs, She1-5, Puf6, Myo2, and She2-binding. Bud enrichment of a nonanchored POL, Sro7, was mRNA transport dependent while that of membrane-anchored factors (e.g., Cdc42 and Sec4) was mRNA transport independent. We propose that mRNA transport plays an important role in the bud enrichment of proteins that lack membrane-anchoring signals and that asymmetric POL mRNA localization may facilitate polarization.
As Myo4 and She3 are required for both asymmetric mRNA transport and the retention of cortical endoplasmic reticulum (ER) in the early bud (ER inheritance) (20, 63, 64), we examined whether these processes are connected. We found that POL mRNA localization to the tip of the emerging bud correlated directly with the presence of the cortical ER. Correspondingly, a mutation in SEC3, which encodes an exocyst component involved in exocytosis (27) and ER inheritance (64), blocked asymmetric mRNA localization. Finally, cell fractionation studies demonstrated the cofractionation of ASH1 and bud-localized POL mRNAs with ER microsomes in a She2- and Sec3-dependent manner. Thus, asymmetric mRNA localization and ER inheritance are connected. This may ensure that membrane proteins encoded by bud-localized mRNAs can undergo translocation upon translation, as well as to spatially restrict the translation of soluble proteins.
MATERIALS AND METHODS
Media, yeast, cell culture, and imaging.
Yeast cells were typically grown to mid-log phase in standard synthetic or rich (yeast extract, peptone, and dextrose) growth medium containing 2% glucose as a carbon source. For the galactose-inducible RFP-SRO7 construct, wild-type (WT) cells were grown in raffinose-containing synthetic medium prior to the shifting of cells to medium containing galactose (3.5%). For the galactose-inducible integrated GFP-SRO7, cells were shifted to galactose-containing medium for 6 to 14 h. Strains are listed in Table 1. Cells transformed with plasmids for fluorescent protein expression were grown on selective medium. For the induction of MS2 fused to green fluorescent protein (MS2-GFP), cells were switched to medium lacking methionine for 1.5 h at 26°C, with the exception of myo2-66 cells, which were induced for 2.5 h at either 26°C or 37°C. Cells were taken directly for live cell imaging by fluorescence or confocal microscopy. To obtain quantitative data on the localization of mRNA and red fluorescent protein (RFP) in each strain examined, cells with small visible buds (i.e., cells in S and early G2/M phase) were scored for the localization of GFP (mRNA) and RFP (protein) signals. The following wavelengths were used: for GFP, excitation at 480 nm and emission at 530 nm; for rhodamine or monomeric RFP, excitation at 545 nm and emission at 560 to 580 nm. Cells having mRNA granules in both the tip of the daughter cell, as well as in the mother cell, were scored as having bud tip-localized mRNA.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1a | MATaade2 can1 his3 leu2 lys2 trp1 ura3 | J. Hirsch |
| W303-1b | MATα ade2 can1 his3 leu2 lys2 trp1 ura3 | J. Hirsch |
| NY1002 | MATα his3 leu2 trp1 ura3 myo2−66 | P. Novick |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| myo4Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 myo4Δ::kanMX | Euroscarf |
| puf6Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 puf6Δ::kanMX | Euroscarf |
| she2Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 she2Δ::kanMX | Euroscarf |
| she3Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 she3Δ::kanMX | Euroscarf |
| she4Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 she4Δ::kanMX | Euroscarf |
| she5Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 she5Δ::kanMX | Euroscarf |
| NY2450 | MATahis3Δ200 leu2-3,112 ura3-5 sec3Δ::kanMX4 | P. Novick |
| RGY1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 she2Δ::kanMX sro7::his5+::GAL1-GFP-SRO7 | This study |
| RGY2 | MATa/α ade2/ADE2 can1/CAN1 his3/his3Δ1 leu2/leu2Δ0 lys2/LYS2 MET15/met15Δ0 ura3/ura3Δ0 SHE2/she2Δ::kanMX SRO7/sro7::his5+::GAL1-GFP-SRO7 | This study |
| WPY154 | MATα ura3-52 his3Δ200 leu2Δ1 srp101-47 | W. Prinz |
Plasmids.
Plasmids encoding an ASH1 gene fragment with MS2 viral coat protein binding sites in its 3′ UTR (pIIIA/ASH1-UTR) and a gene fusion of the MS2 coat protein and GFP (pCP-GFP) (4) were generously provided by Kerry Bloom (University of North Carolina, NC). A plasmid, pSL1180, which encodes 12 loops of the viral MS2 coat protein binding site was generously provided by Robert Singer (Albert Einstein College of Medicine, NY). A multicopy plasmid expressing Sec63-RFP, pSM1960, was generously provided by S. Michaelis (John Hopkins, Baltimore, MD). pAD6 is a multicopy expression vector containing the LEU2 marker and encodes the myc epitope downstream of the ADH1 promoter. Plasmids used in this study are listed in Table 2. An integration construct, pFA6a-His3MX6-PGAL1-GFP (43), was used along with a specific oligonucleotide pair to SRO7 in order to generate a GAL1-GFP-SRO7 integration at the SRO7 locus. Integration was verified by PCR analysis.
TABLE 2.
Plasmids created for this study
| Plasmid namea | Gene | Backbone | Sitesb | Type | Selectable marker | Source |
|---|---|---|---|---|---|---|
| pAD54-SRO7 | SRO7 | pAD54 | SalI-SmaI | 2μm | LEU2 | This study |
| pAD54-RFP-SRO7 | RFPc | pAD54-SRO7 | SalI-SalI | 2μm | LEU2 | This study |
| pAD54-SRO7-MS2 | MS2d | pAD54-SRO7 | SmaI | 2μm | LEU2 | This study |
| pAD54-SRO7-3′UTR | SRO7-3′-UTR (+500 bp)e | pAD54 | SalI-SmaI | 2μm | LEU2 | This study |
| pAD54-RFP-SRO7-3′ UTR | RFPc | pAD54-SRO7-3′ UTR | SalI-SalI | 2μm | LEU2 | This study |
| pAD54-MS2-SRO7-3′ UTR | MS2d | pAD54-SRO7-3′ UTR | [HindIII] | 2μm | LEU2 | This study |
| pAD54-RFP-SRO7-MS2-3′ UTR | MS2-3′-UTRSRO7 (+500 bp)e | pAD54-RFP-SRO7 | SmaI-SacI | 2μm | LEU2 | This study |
| pGAL-RFP-SRO7-MS2-3′ UTR | RFP-SRO7-MS2-3′-UTRf | pYES2 | HindIII | 2μm | URA3 | This study |
| pAD54-SEC4 | SEC4 | pAD54 | SalI-SacI | 2μm | LEU2 | This study |
| pAD54-RFP-SEC4 | RFPc | pAD54-SEC4 | SalI-SalI | 2μm | LEU2 | This study |
| pAD54-SEC4-MS2 | MS2d | pAD54-SEC4 | SmaI | 2μm | LEU2 | This study |
| pAD54-SEC4-3′ UTRsg | 3-UTRSEC4 (+136 bp)e | pAD54-SEC4 | SalI-Sma1 | 2μm | LEU2 | This study |
| pAD54-RFP-SEC4-3′ UTRsg | RFPc | pAD54-Sec4-3′ UTRs | SalI-Sal1 | 2μm | LEU2 | This study |
| pAD54-MS2-SEC4-3′ UTRsg | MS2d | pAD54-SEC4-3′ UTRs | [SalI] | 2μm | LEU2 | This study |
| pAD54-RFP-SEC4-MS2-3′ UTR | MS2-3′-UTRSEC4 (+358 bp)e | pAD54-RFP-SEC4 | [SacI] | 2μm | LEU2 | This study |
| pAD54-RFP-SEC4AA-MS2-3′ UTR | SEC4A214/A215 | pAD54-RFP-SEC4-MS2-3′ UTR | 2μm | LEU2 | This study | |
| pAD54-CDC42 | CDC42 | pAD54 | SalI-SmaI | 2μm | LEU2 | This study |
| pAD54-RFP-CDC42 | RFPc | pAD54-CDC42 | SalI-SalI | 2μm | LEU2 | This study |
| pAD54-CDC42-MS2 | MS2d | pAD54-CDC42 | SmaI | 2μm | LEU2 | This study |
| pAD54-RFP-CDC42-MS2-3′ UTR | MS2-3′-UTRCDC42 (+435 bp)e | pAD54-RFP-CDC42-3′ UTR | SmaI-SacI | 2μm | LEU2 | This study |
| pAD54-SNC1-MS2 | MS2d | pAD54-RFP-cSNC1 | [SacI] | 2μm | LEU2 | This study |
| pAD54-RFP-SNC1-MS2-3′ UTR | MS2-3′-UTRSNC1 (+285 bp)e | pAD54-RFP-cSNC1 | [SacI] | 2μm | LEU2 | This study |
| pAD54-RFP-MS2 | MS2d | pADH54-RFP | SmaI | 2μm | LEU2 | This study |
| pAD54-PEX3 | PEX3 | pAD54 | SalI-SmaI | 2μm | LEU2 | This study |
| pAD54-PEX3-MS2-3′ UTR | MS2-3′-UTRPEX3 (+474 bp)e | pAD54-PEX3 | SacI | 2μm | LEU2 | This study |
| pAD4Δ-OXA1h | OXA1 | pAD4Δ | SalI-SmaI | 2μm | LEU2 | This study |
| pAD4Δ-OXA1-RFPh | RFP (without ATG) | pAD4Δ-OXA1 | SmaI-SacI | 2μm | LEU2 | This study |
| pAD4Δ-OXA1-RFP-MS2-3′ UTRh | MS2-3′-UTROXA3 (+500 bp)e | pAD4Δ-OXA1-RFP | SacI | 2μm | LEU2 | This study |
| pMET-RFP-MSO1-MS2-3′ UTR-MSO1h | RFP-MSO1-MS2-3′-UTRMSO1 (+447 bp)e | pUG36i | SmaI-SpeIk | CEN | URA3 | This study |
| pAD6-myc-SHE2-3′ UTRshort | SHE2-3′-UTR (+40 bp)e | pAD6 | SalI-SacI | 2μm | LEU2 | This study |
| pCP-GFP | MS2-GFP | CEN | HIS3 | K. Bloom | ||
| pIIIA/ASH1-3′ UTR | 3′-UTRASH1 | 2μm | URA3 | K. Bloom | ||
| pSL-MS2-12X | MS2d (12 loops) | R. Singer | ||||
| pSL-MS2-3′ UTR-SRO7 | 3′-UTRSRO7 (500 bp) | pSL-MS2-12 | SacI-BglII | This study | ||
| pSL-MS2-3′ UTR-SEC4 | 3′-UTRSEC4 (358 bp) | pSL-MS2-12 | SacI-[BglII] | This study | ||
| pSL-MS2-3′ UTR-CDC42 | 3′-UTRCDC42 (435 bp) | pSL-MS2-12 | SacI-BglII | This study | ||
| pSL-MS2-3′ UTR-OXA1 | 3′-UTROXA1 (500 bp) | pSL-MS2-12Sacj | BglII-SacI | This study | ||
| pSL-MS2-3′ UTR-PEX3 | 3′-UTRPEX3 (474 bp) | pSL-MS2-12Sacj | BglII-SacI | This study | ||
| pSL-MS2-3′ UTR-MSO1 | 3′-UTRMSO1 (447 bp) | pSL-MS2-12 | BglII-SacI | This study |
POL of interest is in boldface.
Brackets indicate blunt-end ligation.
Without ATG and termination codons.
Indicates MS2 binding sites (12 copies).
Indicates length of 3′ UTR.
Amplified by PCR and subcloned.
Short 3′ UTR.
These constructs lack the HA epitope at the amino terminus.
GFP was removed and the XbaI site was destroyed by fill-in and self-ligation.
Indicates addition of a SacI site 5′ to the MS2 loop sequences by Pfu site-directed mutagenesis.
A [PaeI]/SpeI fragment containing the RFP-MSO1-MS2-3′-UTR cassette was created first in pAD54, amplified by PCR, transferred to pGEM-Teasy, and then cloned into these sites.
FISH and immunofluorescence.
mRNAs were detected by fluorescence in situ hybridization (FISH), as described previously (1). RNAs were synthesized using PCR-amplified DNA fragments of the appropriate genes as templates. The templates included the following: CDC42 (bp 1 to 600), RHO3 (1 to 696), YPT1 (1 to 621), SEC1 (1 to 648), SEC4 (1 to 530), EXO84 (1 to 400), SEC3 (1 to 480), TUB1 (1 to 550), SRO7 (1 to 400), SRO77 (1 to 480), SSO1 (1 to 280), and SNC1 (1 to 310). RNA probes were synthesized in sense and antisense orientations, using a digoxigenin RNA labeling kit (Roche, Mannheim, Germany). After hybridization, cells were washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and blocked in 1× phosphate-buffered saline containing 0.1% Triton X-100 and 10% horse serum. Cells were incubated with horseradish peroxidase-conjugated antidigoxigenin monoclonal antibodies (1:500) (Jackson ImmunoResearch, West Grove, PA) in blocking buffer for 2 h. After a washing step, cells were incubated with Cy5-conjugated anti-horseradish peroxidase antibodies (1:100) (Jackson ImmunoResearch) for 2 h at room temperature. Cells were then washed and mounted in 0.01 M Tris-Cl, pH 8.4, 90% glycerol, 1 mg/ml p-phenylenediamine, and 0.1 μg/ml propidium iodide (Sigma, St. Louis, MO).
Fluorescence imaging was performed using a Zeiss laser scanning confocal microscope. The following wavelengths were used: for rhodamine-phalloidin, excitation at 545 nm and emission at 560 to 580 nm; for Cy5, excitation at 650 nm and emission at 680 nm. Control experiments for in situ hybridization showed no observable background using the sense RNA probes. No signal was observed in the absence of primary antibodies. To obtain quantitative data on mRNA localization in each strain, between 80 to 120 cells with visible small buds (i.e., cells in G1/S and early G2/M phase) were scored for the localization of mRNA signals using FISH. The data are summarized in Fig. 1.
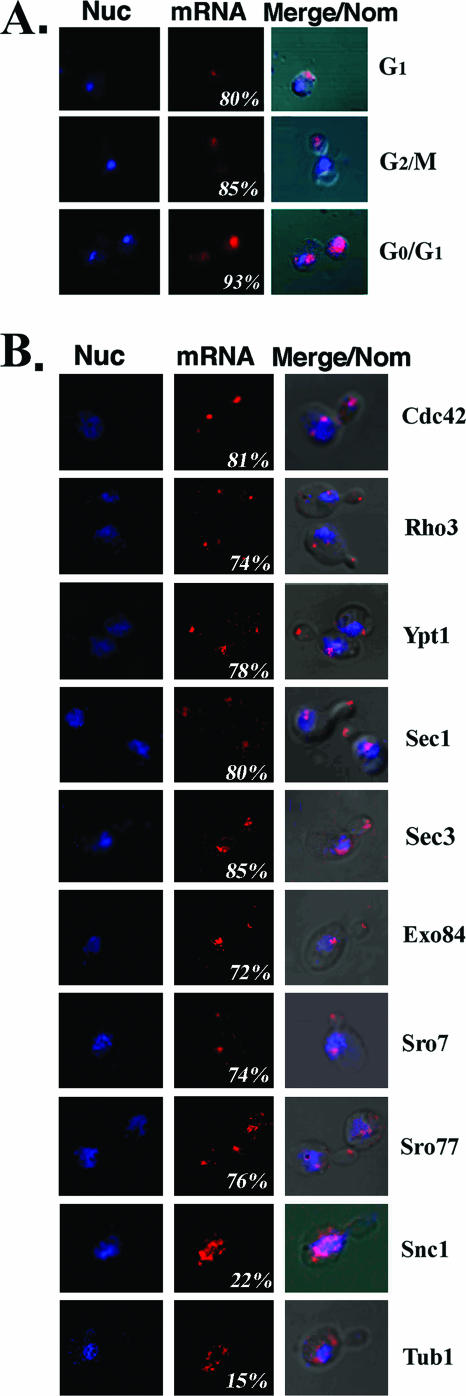
FIG. 1.
In situ localization of endogenous POL mRNAs. (A) Endogenous SEC4 mRNA localizes to the bud tip prior to nuclear division and segregation. WT yeast cells were grown on rich medium at 26°C prior to fixation and in situ hybridization with a specific digoxigenin-labeled RNA antisense probe for SEC4. The panels show nuclear staining performed with propidium iodide (Nuc), mRNA visualized using Cy5-conjugated antidigoxigenin antibodies (mRNA), and merging of the Nuc/mRNA windows with Nomarski-visualized cells (Merge/Nom). Numbers represent the percentage of small-budded cells having mRNA present in the bud prior to nuclear division. The phase of the cell cycle is listed, based upon the observed cellular morphology. (B) Endogenous mRNAs encoding bud-localized proteins also localize to the bud tip prior to nuclear division. WT yeast cells were treated as above and hybridized in situ with specific digoxigenin-labeled RNA antisense probes for different POL genes (as labeled). Representative small-budded (early G2/M) cells are shown.
Immunoprecipitation of She2 and mRNA detection by reverse transcription-PCR (RT-PCR).
WT yeast cells (W303-1a) containing either pAD6 or pADH-myc-She2 were grown on synthetic medium to an optical density at 600 nm (OD600) of 1, centrifuged at 3,000 × g for 5 min at 4°C, and resuspended in 100 OD600 units per ml in lysis buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, 2 mM MgCl2, 100 U/ml RNasin [Promega], 0.1% NP-40, 1 mM dithiothreitol, 100 μM vanadyl complex, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 0.5 μg/ml leupeptin, and 0.01 μg/ml benzamidine). Cells (at a density of 100 OD600 units) were broken by vortexing with glass beads (0.5 mm) at 4°C and centrifuged at 4,000 × g for 10 min at 4°C. For immunoprecipitation (IP) and Western analysis, 500 μg of protein of the supernatant diluted in lysis buffer (final volume, 0.5 ml) was subjected to IP with 1 μg of anti-myc antibody (Roche) and detection with the same antibody (1:1,000 dilution) following immunoblotting. For IP and RNA extraction experiments, 200 μl of lysate (representing 20 OD600 units of cells) was incubated with 5 μg of anti-myc antibody for 2 h at 4°C with rotation, after which 50 μl of protein A-G agarose slurry was added and incubated for an additional 2 h at 4°C. The agarose beads were then washed (three times) with wash buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, 2 mM MgCl2, and 0.05% NP-40), and protein-RNA complexes were eluted by incubation in 200 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 0.1% sodium dodecyl sulftate [SDS], in diethyl pyrocarbonate-treated water) at 65°C for 10 min. Total RNA was purified from the eluate after DNase I treatment using a Masterpure Yeast RNA purification kit (Epicenter, Madison, WI), according to the manufacturers instructions. After precipitation with added glycogen (Roche), the total RNA was resuspended in 30 μl of diethyl pyrocarbonate-treated water, and 1- to 3-μl aliquots were taken for reverse transcription (RT) using the Moloney murine leukemia virus RT RNase H(−) (Promega), according to the manufacturer's instructions. Total RNA was purified from the lysate in a similar fashion, and 1 μg of RNA was taken for RT. For the detection of specific transcripts, 40 ng of reverse-transcribed RNA was amplified by PCR (30 cycles) with the same oligonucleotide pairs (50 pg of each oligonucleotide per reaction) used to create the DNA templates necessary to generate RNA probes for FISH (see “FISH and immunofluorescence” above). For ASH1 and SNC1, primers corresponding to 143 bp (bp 1703 to 1767 +79 bp of the 3′ UTR) and 285 bp (3′ UTR) fragments were used instead, respectively. Control reactions (i.e., PCR amplification of total RNA without RT, of immunoprecipitated RNA without RT, and of the PCR mix without template [either RNA or first-strand cDNA]) were performed in parallel. Products from the PCRs were run on 1% agarose gels and documented.
Cell fractionation and mRNA detection by RT-PCR.
Yeast cells were grown at 30°C in selective medium (500 ml) to and OD600 of 0.5 to 0.6. Cycloheximide was added to a final concentration of 100 μg/ml, and cultures were grown for another 15 min at 30°C. Cells were harvested and resuspended in 10 ml of ice-cold SK buffer (0.1 M KPO4 [pH 7.5], 1.2 M sorbitol, cycloheximide 100 μg/ml). After a 5-min incubation on ice, cells were centrifuged for 3 min at 3,000 rpm, mechanically disrupted in liquid nitrogen using a mortar and pestle, and resuspended in 1 ml of ice-cold lysis buffer (10 mM Tris-HCl [pH 7.5], 0.25 M sucrose, 30 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml cycloheximide, 200 U/ml RNasin [Promega], and a standard protease inhibitor mix). Crude lysates were obtained by centrifugation for 5 min at 3,000 rpm. Lysates (900 μl each) were then equilibrated with 2.5 M sucrose containing 150 mM KCl, 10 mM MgCl2, and 10 mM Tris-HCl (pH 7.5) to a final concentration of 2.1 M sucrose (final volume, 6.25 ml) and placed in the bottom of a 17-ml ultracentrifuge tube. Next, a discontinuous sucrose gradient was formed by adding consecutive layers of 2.05 M (6.25-ml volume) and 1.3 M sucrose (2-ml volume) containing 150 mM KCl, 10 mM MgCl2, and 10 mM Tris-HCl (pH 7.5). Gradients were centrifuged at 25,000 rpm (90,000 × g) in an SW32 rotor (Beckman) at 40°C for 5 h. Next, fractions were collected, and total RNA from the membrane (interface between 1.3 M and 2.05 M sucrose layers) and the cytoplasmic (2.1 M sucrose layer) fractions was isolated using a MasterPure Yeast RNA Purification Kit, including DNase I treatment, according to the manufacturer's recommendations. Reverse transcription was performed using Moloney murine leukemia virus RT RNase H(−) (Promega) under conditions suggested by the manufacturer. For each mRNA to be detected, the RT reaction was performed for the same quantity of total RNA (2.5 μg). cDNA used for PCR amplification was tested using serial dilutions in order to avoid reaching a plateau phase during PCR. Specific oligonucleotide pairs were used to amplify genes of interest. For each mRNA, the same quantity of cDNA from the RT reaction was taken from the ER and cytosolic fractions for PCR. Control reactions (i.e., PCR amplification of total RNA without RT, PCR amplification of RNA from ER and cytosolic fractions without RT, or PCR without template) were performed in parallel and, unless mentioned, yielded no products after PCR. Samples of the PCRs were electrophoresed on 1% agarose gels and documented.
To detect proteins by immunoblotting, 25 μg of protein from the total crude lysate or from the ER and cytosolic fractions was subjected to SDS-polyacrylamide gel electrophoresis on 8% acrylamide gels. Immunoblots were blocked in 5% nonfat milk in phosphate-buffered saline and incubated with a monoclonal anti-GFP antibody (1:1,000; Roche) to detect GFP-Sec63 or anti-phosphoglycerate kinase antibody diluted 1:1,000 (gift of Z. Elazar, Weizmann Institute of Science).
RESULTS
Many POL mRNAs localize to the bud prior to nuclear division.
As most POL proteins are enriched in the bud, we examined the localization of their mRNAs during cell division by FISH with specific digoxigenin-labeled antisense probes (Fig. 1). First, we examined the localization of endogenous mRNA encoding Sec4 (Fig. 1A), a bud-localized GTPase that mediates secretory vesicle fusion with the plasma membrane, in WT cells. FISH labeling of endogenous SEC4 mRNA yielded single punctate structures located near the cell surface prior to budding (G1 phase). These structures were distinct from the nucleus, which was revealed by propidium iodide staining (Fig. 1A). In cells undergoing division (early G2/M phase), nearly all SEC4 mRNA was found in the bud even though nuclear division and segregation had yet to occur. This pattern was observed in 85% of the cells examined. Cells in late G2/M also had SEC4 mRNA located in the nucleus of the mother cell. This may represent a maternal pool of SEC4 mRNA yet to be exported. After mitosis, SEC4 mRNA was observed in both mother and daughter cells.
As SEC4 mRNA is asymmetrically distributed to the bud prior to nuclear division, similar to ASH1 mRNA, we examined the localization of other POL mRNAs by FISH (Fig. 1B). Endogenous mRNAs encoding the Cdc42 and Rho3 GTPases (31); the Sec1, Sro7, and Sro77 SNARE regulators (23, 25); the Sec3 and Exo84 exocyst components (27); and Ypt1, a GTPase involved in ER-Golgi transport, localized at least in part to the buds of G2/M phase cells prior to nuclear division. This pattern was discerned in ≥70% of cells for each mRNA examined (Fig. 1B), indicating that these mRNAs were also exported from mother cells. Asymmetric POL mRNA localization correlates with the bud-specific pattern of protein localization seen during cell division.
However, not all mRNAs encoding factors involved in the cytoskeletal regulation or membrane transport show bud localization prior to nuclear division (Fig. 1B). For example, cells probed for the mRNAs encoding tubulin (TUB1) or the Snc1 exocytic v-SNARE (SNC1) showed a low percentage of G2/M phase cells having bud-localized mRNA (15% and 22%, respectively). As neither Snc1 nor Tub1 is specifically bud localized during cell division (10, 47), this could be reflected by the labeling of their mRNAs primarily in mother cells.
Localization of CDC42, SEC4, and SRO7 mRNA and protein in vivo.
In situ experiments (Fig. 1) imply that asymmetric POL mRNA localization may correlate with protein localization to the bud. Thus, we examined the localization of both mRNA and translated protein in vivo using mRNAs encoding monomeric RFP fused with Sec4, Cdc42, and Sro7. These fusions encode the hemagglutinin epitope upstream of RFP and bear 12 MS2 viral coat protein binding sites downstream of the termination codon but upstream to the 3′ UTR (Fig. 2A). Addition of the MS2 binding sites allows for visualization of mRNA transcripts in yeast expressing MS2-GFP (4, 5). The addition of RFP allows for visualization of the protein generated from the transcript. Protein expression from these dual-detection constructs (used in the experiments shown in Fig. 2, 3,5, 6, and 8 and the data of Table 6; see also movies S2 and S3 in the supplemental material) was verified by Western analysis, and functionality was tested by complementation in their respective conditional mutants (i.e., sec4-8, cdc42-6, and sro7Δ sro77Δ cells) (data not shown). A set of single-detection constructs expressing either functional RFP-POL protein fusions (for protein localization) or POL genes bearing MS2 binding sites (for mRNA localization) was also created and used in other experiments (Fig. 3 and Tables 3 and 4). Exogenous mRNA and protein levels expressed from these constructs were measured by RT-PCR and Western analysis, respectively, and typically yielded levels ∼10-fold over endogenous expression (data not shown).
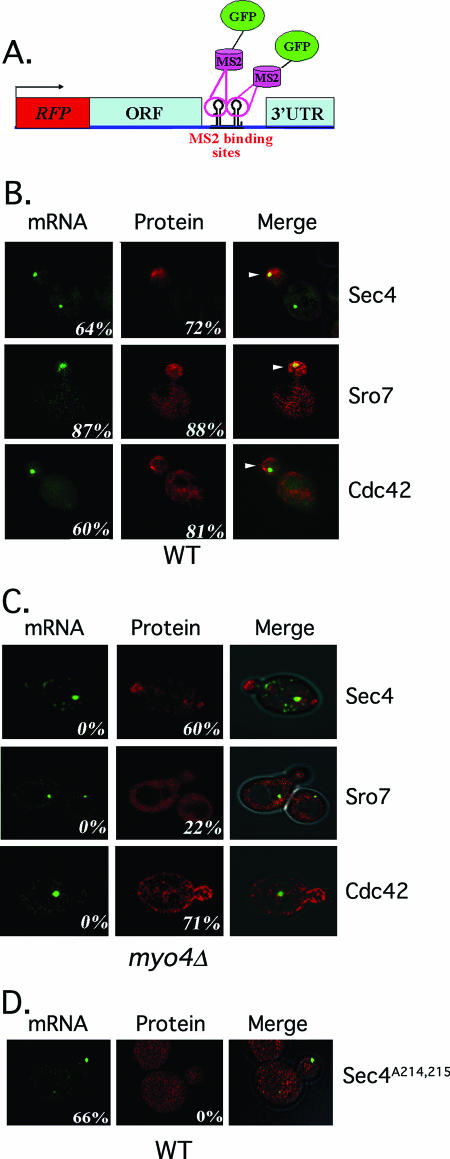
FIG. 2.
POL mRNA and protein localize to the bud tip. (A) A schematic of the expression construct used for dual mRNA and protein detection. This construct expresses monomeric RFP fused to the amino terminus of a given POL gene (ORF, representing CDC42, SEC4, or SRO7). A hemagglutinin epitope tag is present upstream of RFP (not shown). Downstream to the termination codon and upstream of the 3′ UTR are 12 MS2 coat protein binding sites. Full-length 3′ UTRs (CDC42, +435 bp; SEC4, +358 bp; SRO7, +500 bp) were placed downstream of the MS2 binding sites. Gene expression is under control of the ADH1 promoter. (B) POL mRNAs localize to thebud tip. WT yeast cells expressing MS2-GFP and the above RFP-CDC42, RFP-SEC4, or RFP-SRO7 expression constructs were examined by confocal fluorescence microscopy. GFP fluorescence (mRNA) and RFP-POL protein fluorescence (protein) are shown, and images are merged in the last row. Numbers indicate the percentage of cells bearing mRNA or protein at the bud tip. (C) POL mRNAs mislocalize to the mother in myo4Δ cells. myo4Δ yeast cells expressing MS2-GFP and the above CDC42, SEC4, or SRO7 expression constructs were examined by confocal microscopy. Numbers indicate the percentage of cells bearing mRNA or protein at the bud tip. (D) Sec4 lacking its geranylgeranylation site mislocalizes to the cytoplasm. WT yeast cells expressing MS2-GFP and RFP-SEC4A214,215, which lacks the C-terminal residues necessary for anchor attachment, were examined by confocal microscopy. Numbers indicate the percentage of cells bearing mRNA or protein at the bud tip.
FIG. 3.
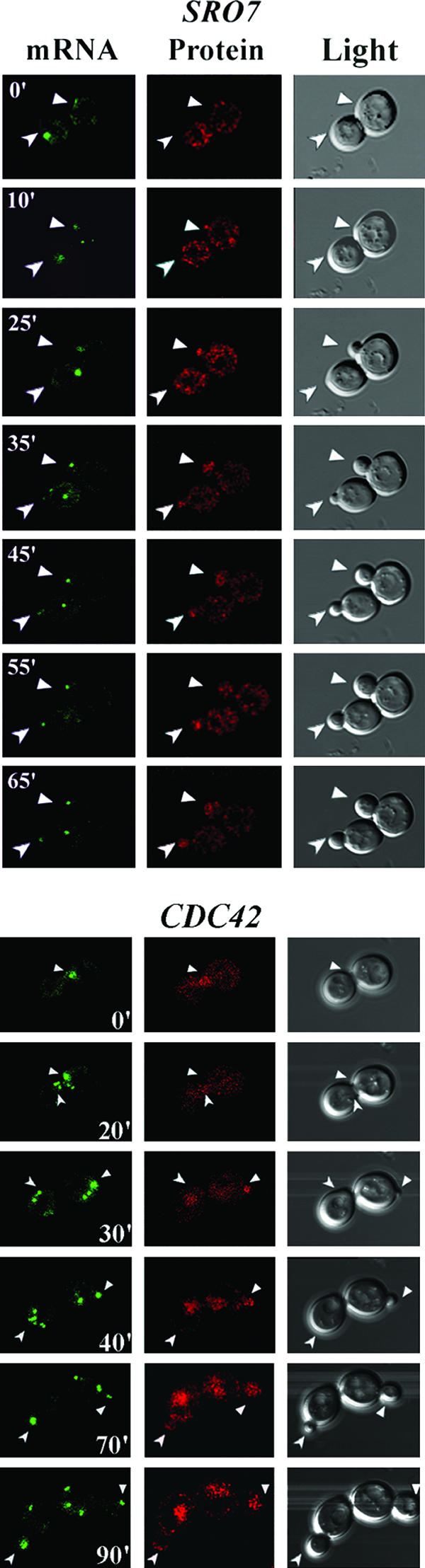
POL mRNA localization precedes POL protein enrichment and bud emergence. WT yeast cells expressing MS2-GFP and either the SRO7 or CDC42 mRNA and protein detection constructs were examined by time-lapse confocal microscopy. GFP fluorescence (mRNA), RFP-POL protein fluorescence (protein), and differential interference contrast microscopy (light) are shown. Numbers indicate time in minutes. Arrows in the respective windows indicate the localization of mRNA, protein, or the selected bud site(s). Full-length movies of the time-lapse experiments for SRO7 (Movie S2) and CDC42 (Movie S3) are available in the supplemental material.
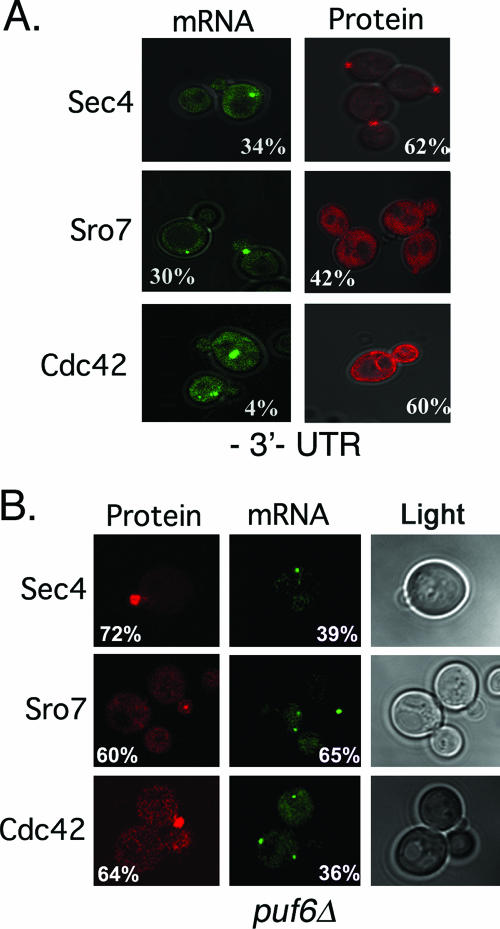
FIG. 5.
POL mRNAs partly mislocalize in the absence of their 3′ UTRs and in puf6Δ cells. (A) POL mRNAs mislocalize in the absence of their 3′ UTRs. WT yeast cells expressing MS2-GFP and either the RFP-SEC4, RFP-CDC42, or RFP-SRO7 mRNA and protein detection constructs lacking their 3′ UTRs were examined by confocal microscopy. GFP fluorescence (mRNA) and RFP-POL fluorescence (protein) are shown. Numbers indicate the percentages of cells showing mRNA or protein localization at the bud tip. (B) POL mRNAs are mislocalized in puf6Δ cells. WT and puf6Δ yeast cells expressing MS2-GFP and the RFP-SEC4, RFP-CDC42, or RFP-SRO7 dual-detection constructs were examined by confocal microscopy. GFP fluorescence (mRNA) and RFP-POL protein fluorescence (protein) are shown. Numbers indicate the percentages of cells showing mRNA or protein localization at the bud tip.
FIG. 6.
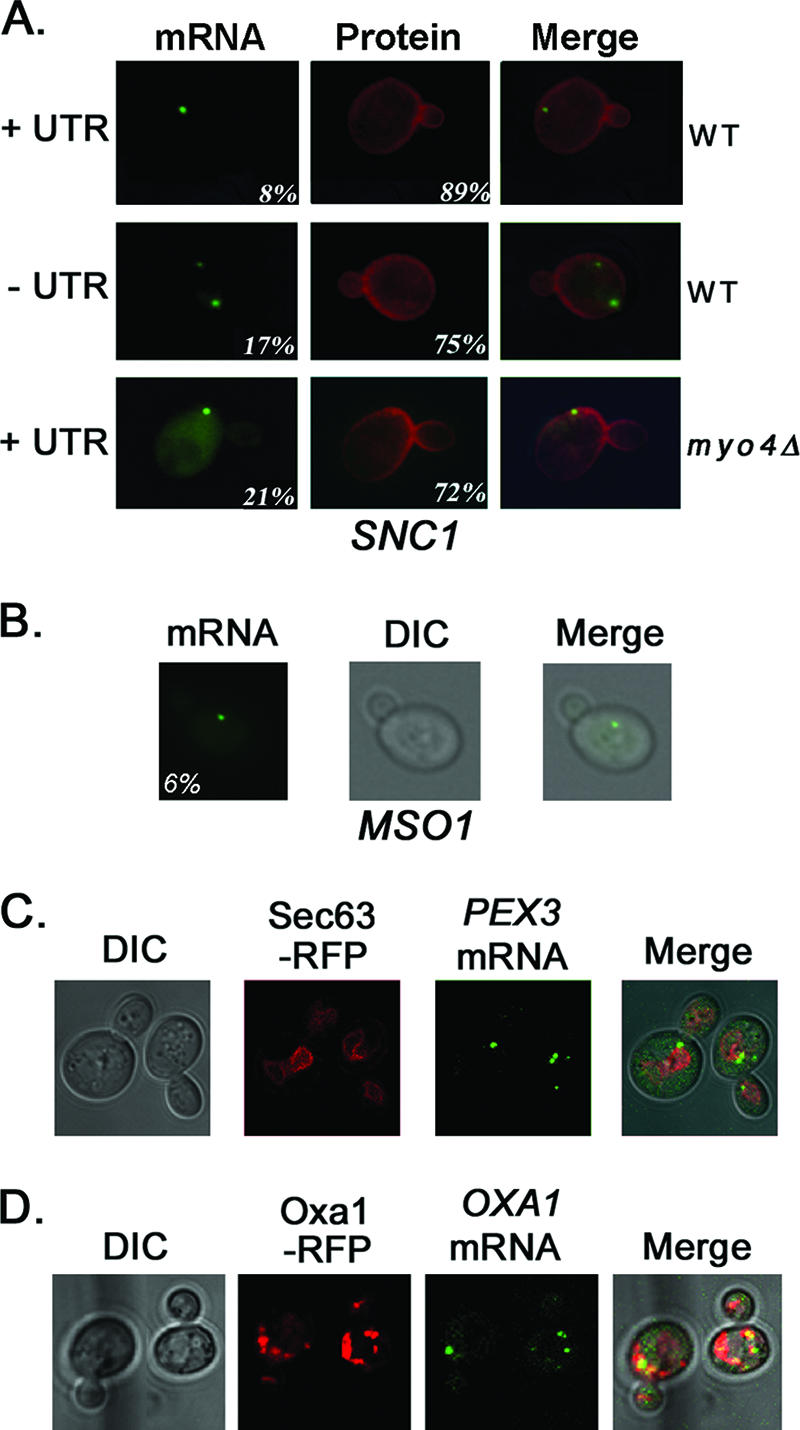
SNC1, MSO1, PEX3, and OXA1 mRNAs are not polarized in yeast. (A) SNC1 mRNA localizes to mother cells and not to the bud tip. WT and myo4Δ yeast cells expressing MS2-GFP and RFP-SNC1 bearing MS2 sites (dual-detection constructs) were examined by fluorescence microscopy. RFP-SNC1 bearing its 3′ UTR (+UTR) or lacking its 3′ UTR (−UTR) was examined in WT cells. In myo4Δ yeast, RFP-SNC1 bearing the 3′ UTR was examined. MS2-GFP fluorescence (mRNA) and RFP-Snc1 fluorescence (protein) are shown. Numbers indicate the percentages of cells showing either mRNA localization to the bud tip or protein localization to the plasma membrane of both mother and bud. The numbers of cells counted for mRNA and protein were as follows, respectively: 129 and 166 for WT cells with the UTR; 105 and 97 WT cells lacking the UTR; and 121 and 117 for myo4Δ cells with the UTR. (B) MSO1 mRNA localizes to mother cells. WT cells expressing MSO1 bearing MS2 binding sites and its native 3′ UTR and MS2-GFP were examined by fluorescence microscopy. Numbers indicate the percentages of cells showing mRNA localization to the bud tip. (C) PEX3 is not polarized and colocalizes with Sec63-RFP-labeled membranes. WT cells expressing PEX3 bearing MS2 binding sites upstream of its native 3′ UTR, SEC63-RFP, and MS2-GFP were examined by confocal microscopy. (D) OXA1 mRNA is not polarized and colocalizes with Oxa1-RFP. WT cells expressing OXA1-RFP bearing MS2 binding sites upstream of its native 3′ UTR and MS2-GFP were examined by confocal microscopy.
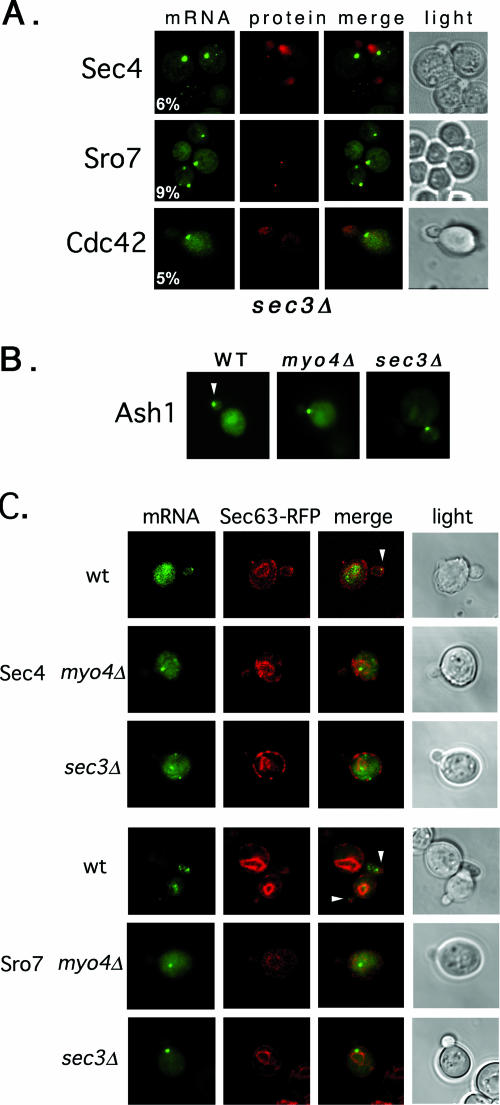
FIG. 8.
SEC3 is required for ER inheritance and mRNA localization. (A) POL mRNA localization is Sec3 dependent. sec3Δ yeast cells expressing RFP-SEC4, RFP-CDC42, or RFP-SRO7 gene fusions bearing MS2 binding sites in their 3′ UTRs (dual-detection constructs) and MS2-GFP were examined by confocal microscopy. (B) ASH1 mRNA localization is Sec3 dependent. WT, myo4Δ, and sec3Δ yeast cells expressing an ASH1 gene fragment bearing MS2 binding sites prior to the ASH1 3′ UTR and MS2-GFP were examined by confocal microscopy. Only MS2-GFP fluorescence is shown. Arrowheads indicate localization of the mRNA granule to the bud tip. (C) POL mRNA localization correlates with ER localization. WT, sec3Δ, and myo4Δ yeast cells expressing either SEC4 or SRO7 bearing MS2 binding sites upstream of their 3′ UTRs (single-detection constructs), as well as MS2-GFP and RFP-SEC63 from plasmids, were examined by confocal microscopy. Arrowheads indicate localization of the mRNA granule to the bud tip.
TABLE 6.
Percent localization of mRNA and protein in small-budded cells
| Gene and strain | Temp (°C) | % Localization
|
No. of cells scored
|
||||
|---|---|---|---|---|---|---|---|
| Bud tip
|
Mother
|
||||||
| mRNA | Protein | mRNA | Protein | mRNA | Protein | ||
| SEC4 +3′ UTR WT | 26 | 76 | 92 | 24 | 8 | 220 | 160 |
| myo2-66 | 26 | 84 | 95 | 16 | 5 | 68 | 96 |
| myo2-66 | 37 | 6 | 12 | 94 | 88 | 95 | 89 |
| puf6Δ | 26 | 39 | 72 | 61 | 28 | 159 | 165 |
| sec3Δa | 26 | 6 | 94 | 141 | |||
| srp101a | 26 | 79 | 21 | 58 | |||
| srp101a | 37 | 4 | 96 | 69 | |||
| SEC4 −3′ UTR WT | 26 | 34 | 62 | 66 | 38 | 115 | 140 |
| SRO7 +3′-UTR WT | 26 | 84 | 71 | 16 | 29 | 169 | 110 |
| puf6Δ | 26 | 65 | 60 | 35 | 40 | 173 | /44 |
| sec3Δa | 26 | 9 | 91 | 82 | |||
| srp101a | 26 | 94 | 6 | 48 | |||
| srp101a | 37 | 42 | 58 | 57 | |||
| SRO7 −3′ UTR WT | 26 | 30 | 42 | 70 | 58 | 130 | 110 |
| CDC42 +3′ UTR WT | 26 | 64 | 78 | 36 | 22 | 184 | 96 |
| puf6Δ | 26 | 36 | 64 | 64 | 36 | 140 | 105 |
| sec3Δa | 26 | 5 | 95 | 100 | |||
| CDC42 −3′ UTR WT | 26 | 4 | 60 | 96 | 40 | 89 | 96 |
| ASH1 WTa | 26 | 100 | 0 | 56 | |||
| myo4Δa | 26 | 3 | 97 | 36 | |||
| sec3Δa | 26 | 16 | 84 | 52 | |||
Only mRNA localization was determined.
TABLE 3.
Percent localization of SEC4 mRNA and protein in small-budded cells
| Strain | Temp (°C) | Localization of SEC4
|
No. of cells scored
|
||||
|---|---|---|---|---|---|---|---|
| Bud tipa
|
Mother
|
mRNA | Protein | ||||
| mRNA | Protein | mRNA | Protein | ||||
| WT | 26 | 88 | 98 | 12 | 2 | 148 | 99 |
| 37 | 76 | 89 | 24 | 11 | 95 | 120 | |
| myo4Δ | 26 | 0 | 60 | 100 | 40 | 100 | 122 |
| she2Δ | 26 | 0 | 66 | 100 | 34 | 112 | 145 |
| she3Δ | 26 | 0 | 69 | 100 | 31 | 150 | 113 |
| she4Δ | 26 | 0 | 73 | 100 | 27 | 89 | 130 |
| she5Δ | 26 | 0 | 56 | 100 | 44 | 118 | 95 |
This scoring includes cells having mRNA granules present in both mother and tip of the daughter cell.
TABLE 4.
Percent localization of SRO7 mRNA and protein in small-budded cells
| Strain | Temp (°C) | % Localization of SR07
|
No. of cells scored
|
||||
|---|---|---|---|---|---|---|---|
| Bud tipa
|
Mother
|
mRNA | Protein | ||||
| mRNA | Protein | mRNA | Protein | ||||
| WT | 26 | 72 | 84 | 28 | 16 | 120 | 116 |
| 37 | 80 | 76 | 20 | 24 | 78 | 90 | |
| myo4Δ | 26 | 0 | 35 | 100 | 65 | 128 | 88 |
| she2Δ | 26 | 0 | 33 | 100 | 67 | 100 | 94 |
| she3Δ | 26 | 12 | 45 | 88 | 55 | 123 | 10 |
| she4Δ | 26 | 2 | 34 | 98 | 66 | 90 | 83 |
| she5Δ | 26 | 0 | 26 | 100 | 74 | 75 | 96 |
| WT (1 h)b | 26 | 71 | 29 | 89 | |||
| WT (6 h)c | 26 | 68 | 79 | 32 | 21 | 150 | 142 |
This scoring includes cells having mRNA granules present in both the mother and the tip of daughter cell.
WT (GAL-RFP-SRO7) after 1 h of induction on galactose-containing medium. No RFP fluorescence was detected.
WT (GAL-RFP-SRO7) after 6 h of induction on galactose-containing medium.
All three POL mRNAs gave MS2-GFP fluorescence in the form of RNA granules (gRNA) that localized to the tip of the emerging bud in the majority of small-budded cells (S/early G2 phase). These granules were similar to those seen for ASH1 mRNA using MS2-GFP detection (1, 4, 5). MS2-GFP granules localized to the bud tips in the majority of small-budded WT cells at 26°C (RFP-SEC4, 64% of cells; RFP-CDC42, 60%; RFP-SRO7, 87%) (Fig. 2B). Similar results were obtained with mRNAs lacking the RFP gene (Tables 3 and 4), though the percentage of cells having SEC4 mRNA at the bud tip was higher (88% of cells at 26°C) (Table 3). Bud-localized MS2-GFP granules were observed at the mother-bud junction only in large-budded cells (i.e., postmitosis) during the reversal of polarity. MS2-GFP granules could sometimes be detected late in the cell cycle in mother cells (Fig. 2B, Sec4), which may represent maternal mRNA being readied for the next division. Overall, the localization of exogenously expressed POL mRNAs in vivo (Fig. 2B and Tables 3 and 4) was similar to that seen with endogenous mRNAs in situ (Fig. 1).
As POL mRNAs localize to the tip of the emerging bud, we evaluated POL protein localization concomitantly by RFP fluorescence (Fig. 2B). In all cases there was a strong correlation between placement of the MS2-GFP granule and the local intensity of RFP fluorescence. For example, RFP-Sec4 localized almost exclusively to the bud (bud enrichment), as shown for native and GFP-tagged Sec4 (8, 60). This was observed in 72% of cells at 26°C and correlated with the presence of the MS2-GFP granule (Fig. 2B). As a control, we examined localization of an mRNA encoding RFP and the MS2 sites alone. RFP fluorescence was evenly dispersed in these cells, and only a small percentage of cells had MS2-GFP granules near the bud tip (e.g., 18%; number of cells, 150). Thus, the RFP sequence does not contribute significantly to mRNA targeting.
RFP-Cdc42 protein, like RFP-Sec4, concentrated in newly forming buds in 81% of WT cells at 26°C but was present on other structures including the plasma and vacuolar membranes (Fig. 2B). This is consistent with previous Cdc42 localization studies (53). RFP-Sro7 protein was found to be bud enriched in 88% of cells at 26°C (Fig. 2B), which differs from earlier studies that showed nonpolarized or cytoplasmic Sro7 localization (37, 39). However, those studies localized exogenously expressed epitope-tagged SRO7 that lacked its 3′ UTR. To determine the localization of endogenous Sro7, we created a galactose-inducible GFP-SRO7 integration at the SRO7 locus and examined localization upon induction with galactose (see Fig. S1 in the supplemental material). We found GFP-Sro7 concentrated at the bud tip in SHE2/she2Δ (∼WT) cells. Thus, Sro7 protein mislocalizes in the absence of its 3′ UTR. Finally, we note that values for both RFP-Sec4 and RFP-Sro7 localization were basically similar with or without the MS2 binding sites (Tables 3 and 4) (bud enrichment of RFP-Sec4 and RFP-Sro7 was observed in 98% and 84% of WT cells, respectively), although we cannot exclude the possibility that the MS2 sites and/or MS2-GFP binding has minor effects upon protein localization.
Overall, POL protein localization correlated well with POL mRNA localization in vivo. Although the dual mRNA-protein detection constructs employ strong constitutive promoters, both RFP-Sec4 and RFP-Cdc42 localization was similar to that previously described. Moreover, RFP-Sro7 bearing its 3′ UTR localized in a manner more reflective of its exocytic function. This is supported by localization studies employing GFP-Sro7 expression from the SRO7 locus (see Fig. S1 in the supplemental material) as well as exogenous RFP-SRO7 mRNA and protein expression from a galactose-inducible promoter (Table 4). Using the latter, we found no significant difference with results obtained using the constitutive ADH1 promoter (Table 4). Together, these results suggest that mRNA translation occurs in the bud during polarization and leads to the local enrichment of POL proteins.
mRNA placement precedes bud emergence and polarized growth.
To determine whether mRNA placement and bud emergence correlate, we examined the localization of RFP-SRO7 and RFP-CDC42 mRNA and protein in WT cells using a dual-detection system and time-lapse microscopy (Fig. 3; see Movies S2 and S3 in the supplemental material). Both RFP-SRO7 and RFP-CDC42 mRNAs localized to sites where protein enrichment was later detected and, importantly, where bud emergence occurred. This correlation between mRNA localization and bud emergence was observed in all cells examined (n = 50) and occurred within minutes of the granule settling at a specific site in unbudded cells (e.g., early G1). For example, note gRNA placement in the upper cell of RFP-SRO7-expressing cells at 0, 10, and 25 min and in the lower cell at 10, 25, and 35 min and the subsequent localization of RFP-Sro7 and bud emergence (Fig. 3). We note that these granules showed mobility until attachment at the presumptive bud site, as observed for ASH1 gRNA (4, 5). Once anchored, the granules basically remained at the tip of the newly forming bud until mitosis, when they migrated to the next site of polarized growth. This movement correlates with the reversal of polarity that occurs during mitosis. We identified three sizes of fluorescent RNA granules, as revealed by MS2-GFP labeling: a granule of 0.3 to 0.5 μm that is close to, or inside, the nucleus of the mother late during budding; a small granule of 0.1 to 0.2 μm that is exported from the mother to the tip of the emerging bud; and a third granule of 0.3 to 0.5 μm present at the bud tip which likely represents the cumulative delivery of small granules. We note that these fluorescent granules do not measure the actual sizes of gRNA.
POL mRNA localization is She protein dependent.
mRNA localization necessitates the formation of ribonucleoprotein complexes between RNA binding proteins and cis-acting signals present in the mRNA. Ribonucleoproteins form in the nucleus and are exported to the cytoplasm prior to mRNA translation. Previous studies demonstrated that ASH1 mRNA transport and localization are dependent upon a complex formed between She1/Myo4, She3, and She2, with She2 binding directly to ASH1 mRNA (7, 35, 42, 44, 57).
We examined whether mutations in the SHE genes affect POL mRNA and protein localization. We initially determined the localization of RFP-SEC4, RFP-CDC42, and RFP-SRO7 mRNA and protein using a dual-detection system in a myo4Δ/she1Δ mutant (Fig. 2C). However, no MS2-GFP granules localized to the tip of the emergent bud in any cells (0% bud tip localization) (Fig. 2C) but were retained within mother cells (Tables 3 and 4 include data regarding SRO7 and SEC4 mRNA lacking RFP in myo4Δ cells). Thus, POL mRNAs are mislocalized in myo4Δ/she1Δ cells. Concomitantly, we examined the localization of RFP-Sec4, RFP-Cdc42, and RFP-Sro7 protein. Interestingly, RFP-Sec4 was enriched in the buds of most myo4Δ cells (Fig. 2C), as in WT cells (60% versus 72% of cells showed bud enrichment, respectively) (Fig. 2B) although some RFP fluorescence was observed in the cytoplasm. Similar results were obtained for RFP-Cdc42, in which 71% of myo4Δ cells showed bud enrichment (Fig. 2C) versus 81% of WT cells (Fig. 2B). In contrast, RFP-Sro7 was not enriched in the buds of myo4Δ cells (22% of cells showed bud enrichment versus 88% of WT cells) (Fig. 2B and C) and localized to the cytoplasm, as seen previously (37, 39). Results obtained for both SEC4 and SRO7 mRNA and protein using the single-detection constructs (Tables 3 and 4) were similar to those obtained using dual-detection constructs (Fig. 2C), though there was a reduction in RFP-Sec4 bud enrichment in myo4Δ cells (Table 3). Nevertheless, She1/Myo4 appears necessary for the asymmetric localization of POL mRNAs to the bud, as shown previously for ASH1 (42, 57), as well as for the proper distribution of Sro7 protein. That the majority of Sec4 and Cdc42 reaches the bud implies that these membrane-anchored proteins target correctly even in the absence of mRNA transport. This may result from SEC4 and CDC42 translation in the mother cell and transport into the bud via the secretory pathway, as shown for Sec4 (50). However, the localization of proteins lacking a membrane-anchoring signal, like Sro7 and Ash1, are adversely affected in the absence of MYO4.
To verify that membrane-anchoring confers the WT localization of RFP-Sec4 in myo4Δ cells and has no bearing upon SEC4 mRNA localization, we mutated the geranylgeranylation site involved in lipid anchor attachment. We examined the localization of RFP-SEC4A214,215 mRNA and protein in WT cells and found that anchor removal caused the loss of RFP-Sec4 enrichment in the bud and led to cytoplasmic localization (Fig. 2D). In contrast, removal of the geranylgeranylation site had no detectable effect upon mRNA localization (Fig. 2D). Thus, proteins lacking membrane-targeting signals mislocalize even when the mRNA is properly localized.
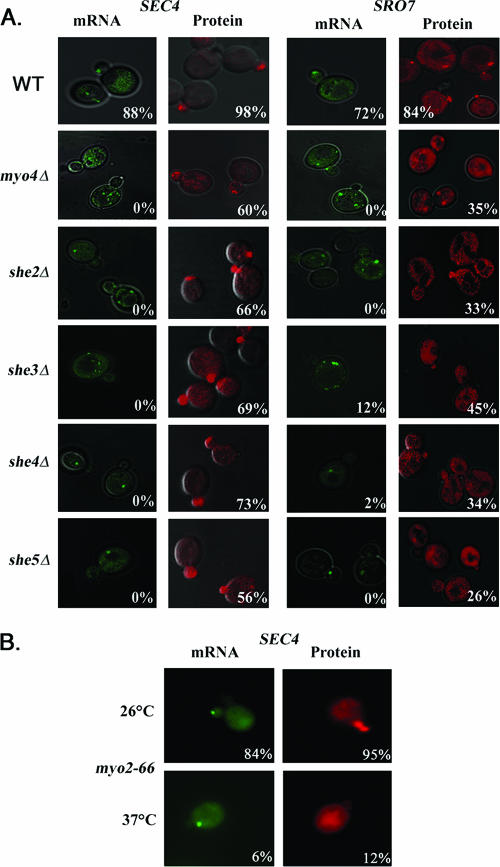
Next, POL mRNA and protein localization was examined in the other sheΔ mutants using single-detection expression constructs (Fig. 4A and Tables 3 and 4). SEC4 mRNA possessing a shortened 3′ UTR (+136 bp, as used in experiments in Table 3 and Fig. 4) was employed and was as effective (if not more so) as SEC4 mRNA with its entire 3′ UTR (+358 bp, as used in Fig. 2B and C) in localizing to the tip of the emergent bud in WT yeast at 26°C (e.g., 88% bud localization with the short 3′ UTR [Table 3] versus 64% with the long 3′ UTR [Fig. 2B]). However, SEC4 and SRO7 mRNAs were mislocalized in the she2-5Δ mutants (Fig. 4A and Tables 3 and 4), and few cells had MS2-GFP granules at the bud tip. In most cases the granules were fragmented and dispersed within mother cells (Fig. 4A), as seen for ASH1 mRNA in the sheΔ mutants (4, 5). Formation of the large gRNA particles was greatly compromised in all sheΔ mutants (Table 5), being reduced from 85 to 90% in WT cells to 10 to 20% in most cells, with the exception of she3Δ cells (Table 5). Thus, gRNA assembly is adversely affected in absence of She proteins. We examined the stability of these mRNAs indirectly using RT-PCR but observed no difference in the amounts obtained from total RNA derived either from WT or sheΔ cells (data not shown).
FIG. 4.
POL mRNAs are mislocalized in sheΔ mutants and in temperature-shifted myo2-66 cells. (A) POL mRNAs are mislocalized in sheΔ cells. To localize mRNA, sheΔ yeast cells were transformed with plasmids expressing MS2-GFP and either an SEC4 or SRO7 single-detection construct bearing MS2 binding sites upstream of the 3′ UTR (+136 bp and +500 bp, respectively). To localize protein, sheΔ yeast cells were transformed in parallel with plasmids expressing RFP fusions with either SEC4 (with the SEC4 3′ UTR +136 bp) or SRO7 (with the SRO7 3′ UTR +500 bp). Cells were grown at 26°C prior to scoring for mRNA localization to the bud tip (mRNA) or RFP protein enrichment (protein) in the bud (see also Tables 3 and 4). (B) SEC4 mRNA is mislocalized in temperature-shifted myo2-66 cells. To localize mRNA, myo2-66 yeast cells were transformed with plasmids expressing MS2-GFP and a SEC4 single detection construct bearing MS2 binding sites upstream of the 3′ UTR (+136 bp). To localize protein, myo2-66 yeast cells were transformed in parallel with plasmids expressing an RFP fusion with SEC4 (3′ UTR +136 bp). Cultures were grown and maintained at 26°C or shifted to 37°C for 2.5 h prior to scoring as above.
TABLE 5.
Formation of gRNA granules in small-budded cells
| Strain | gRNA formation (% of cells)
|
No. of cells scored
|
||
|---|---|---|---|---|
| SEC4 | SRO7 | SEC4 | SRO7 | |
| WT | 86 | 90 | 89 | 120 |
| myo4Δ | 14 | 19 | 100 | 88 |
| she2Δ | 13 | 15 | 115 | 144 |
| she3Δ | 46 | 59 | 97 | 110 |
| she4Δ | 20 | 20 | 84 | 100 |
| she5Δ | 24 | 26 | 120 | 149 |
Bud enrichment of RFP-Sec4 protein (expressed from single-detection constructs) was observed in the she2-5Δ mutants and remained unchanged from WT cells (Fig. 4A and Table 3). In contrast, the percentage of cells having RFP-Sro7 enriched in the bud declined significantly (e.g., 33%, 45%, 34%, and 26% of she2Δ, she3Δ, she4Δ, and she5Δ cells, respectively, versus 84% of WT cells at 26°C) (Fig. 4A and Table 4). Thus, RFP-Sro7 bud enrichment depends upon She-dependent SRO7 mRNA transport. This is supported by endogenous Sro7 localization studies, in which GFP-Sro7 was found to be mislocalized in she2Δ cells (see Fig. S1 in the supplemental material). No difference in the level of GFP-Sro7 protein was observed in she2Δ cells in comparison to SHE2+ cells (data not shown).
mRNA localization depends upon an intact secretory pathway and cytoskeleton.
Mutations in the actin cytoskeleton or secretory pathway result in the mislocalization of ASH1 mRNA (1, 42, 57). We examined SEC4 mRNA localization in cells bearing a temperature-sensitive mutation in the type V myosin, Myo2, which confers both Sec4 and secretory vesicle transport along actin filaments (50). SEC4 mRNA mislocalized within 1 h of the shift to restrictive temperatures (37°C) (data not shown) and only 6% of small-budded cells had mRNA anchored to the bud tip after a 2.5-h shift (Fig. 4B and Table 6). Thus, the requirements for both POL and ASH1 mRNA transport appear identical and require an intact secretory pathway or actin cytoskeleton to mediate mRNA localization. Protein localization was also strongly affected, as 88% of small-budded cells lacked RFP-Sec4 enrichment in the bud after 2.5 h at 37°C (Fig. 4B and Table 6). Thus, active secretion is necessary to localize both Sec4 mRNA and protein, probably because of its general effect on actin polarization (1, 22, 48, 61).
Role of the 3′ UTR in POL mRNA and protein localization.
As the 3′ UTR of ASH1 participates in asymmetric ASH1 mRNA localization and the requirements for POL mRNA localization are identical to those for ASH1 (Fig. 2C and 4 and Tables 3 and 4), we examined whether removal of the 3′ UTR affects POL mRNA and protein localization (Fig. 5A and Table 6). Removal of the 3′ UTRs from RFP-SEC4, RFP-CDC42, or RFP-SRO7 resulted in a significant loss in mRNA localization to the tip of the emerging bud (34%, 4%, and 30% bud tip localization with a deletion of the 3′ UTRs versus 76%, 64%, and 84% with the 3′ UTRs, respectively) (Fig. 5A and Table 6). Thus, the 3′ UTR is required for asymmetric POL mRNA localization, suggesting that it contains cis-acting motifs necessary for mRNA trafficking. As limited bud tip localization was observed for the SEC4 and SRO7 mRNAs without their 3′ UTRs, it suggests that the coding regions may also have cis-acting signals, as shown earlier for ASH1 (14). Indeed, compared to known consensus sequences (45), we found She2 binding motifs in both the coding region (bp 1822 to 1853) and 3′ UTR (bp +333 to 385) of SRO7.
The enrichment of RFP-Sec4 and RFP-Cdc42 in the bud was only partly affected upon removal of the 3′ UTR (62% and 60% of cells showed enrichment with a deletion of the 3′ UTR versus 98% and 78% with the 3′ UTR, respectively) (Fig. 5A and Table 6), due to the presence of their membrane-targeting domains. However, the number of cells showing RFP-Sro7 enrichment in the bud declined significantly (42% without the 3′ UTR versus 71% with the 3′ UTR) (Fig. 5A and Table 6). We note that the latter value was somewhat lower than that seen in other experiments (Fig. 2B, 88 %; Table 4, 84%).
Role of Puf6 in POL mRNA and protein localization.
ASH1 mRNA localization and translation are regulated by another RNA binding protein, Puf6 (26). As Puf6 binds to the 3′ UTR alone, unlike She2 (45), it may have a specific role in asymmetric mRNA localization. We examined POL mRNA and protein localization in puf6Δ cells (Fig. 5B and Table 6) and found a reduction in the number of cells having bud-localized RFP-SEC4 and RFP-CDC42 mRNA to about half of WT values (to 39% and 36%, respectively). In contrast, the deletion of PUF6 had less effect upon RFP-SRO7 mRNA localization (60% bud tip localization), as seen previously with ASH1 mRNA (26). Moreover, RFP-Sro7 protein localization was largely unaffected, showing 60% bud enrichment in puf6Δ cells (Fig. 5B and Table 6), compared to 71% in control WT cells (Table 6). Thus, Puf6 may play a more significant role in the localization of mRNAs encoding the membrane-anchored small GTPases.
SNC1 and MSO1 mRNAs do not localize to the bud tip.
Asymmetric POL mRNA trafficking results in the enrichment of POL proteins in the bud and polarized growth. Yet not all POL mRNAs and proteins are bud enriched. For example, SNC1 mRNA localized predominantly to the mother cell early in cell division (Fig. 1B), and Snc1 protein decorates the plasma membrane of both mother and bud (41, 47). To verify this, we examined the localization of RFP-SNC1 mRNA and protein in vivo (Fig. 6A). RFP-SNC1 mRNA localized predominantly to mother cells and not to the small buds of either WT or myo4Δ cells, and removal of the SNC1 3′ UTR had no effect upon mRNA localization. RFP-Snc1 protein localized to the plasma membrane of both mother and bud in both WT and myo4Δ cells (Fig. 6A), as seen previously.
Next, we examined the in vivo localization of MSO1 mRNA, which encodes a Sec1-interacting factor that also localizes to the plasma membrane of both mother and bud (11). Similar to SNC1 mRNA, MSO1 mRNA localized to mother cells (Fig. 6B). These results suggest that the symmetric distribution of Snc1 and Mso1 arises from the translation of mother-localized transcripts early during cell division and delivery to the cell surface via the secretory pathway.
PEX3 and OXA1 mRNAs do not localize to the bud tip.
To verify that mRNAs encoding nonasymmetrically distributed proteins are not polarized, we examined the localization of PEX3 and OXA1 mRNA in vivo. PEX3 encodes a peroxisome assembly protein that translocates first to the ER (28). We expressed PEX3 mRNA bearing MS2 binding sites in WT cells (Fig. 6C) and found that nearly all cells were labeled with multiple fluorescent granules. These localized principally to mother cells but also to large buds (i.e., late in the cell cycle). Moreover, PEX3 mRNA granules colocalized with membranes labeled by Sec63-RFP, an ER marker, suggesting that this mRNA is in contact with the ER. Next, we examined the localization of OXA1 mRNA, which localizes to mitochondria in a manner dependent upon its 3′ UTR (16). We expressed OXA1-RFP bearing MS2 binding sites in WT cells and also observed multiple fluorescent granules in mother cells and large buds (Fig. 6D). These granules colocalized with structures labeled by Oxa1-RFP (Fig. 6D), which are likely to be mitochondria. These results suggest that not all mRNAs are polarized in yeast and that our in vivo labeling strategy can monitor mRNA localization to organelles.
She2 binds to polarized POL mRNAs.
As certain POL mRNAs are polarized in a She-dependent manner, we examined whether they bind to She2, like ASH1 mRNA. We performed IP with myc-tagged She2 (Fig. 7A) and examined the immunoprecipitates for the presence of mRNAs encoding Ash1 and various POL proteins (Fig. 7B and C). After isolation of total RNA from the precipitates, DNase treatment, and RT-PCR with specific oligonucleotides, we found that the ASH1, SRO7, SRO77, SEC4, CDC42, EXO84, SEC1, SEC3, and RHO3 mRNAs were all present in precipitates from cells expressing myc-She2 but not in precipitates from control cells expressing the tagging vector alone (Fig. 7B and C). Control PCRs proved negative for these mRNAs (Fig. 7B and C). Thus, mRNAs encoding bud-localized proteins bind to She2 and are likely to be transported together with ASH1 mRNA. In contrast, mRNAs encoding TUB1 and SNC1 were not detected in immunoprecipitates from myc-She2-expressing cells, implying that their localization to the mother cell is independent of She2.
FIG. 7.
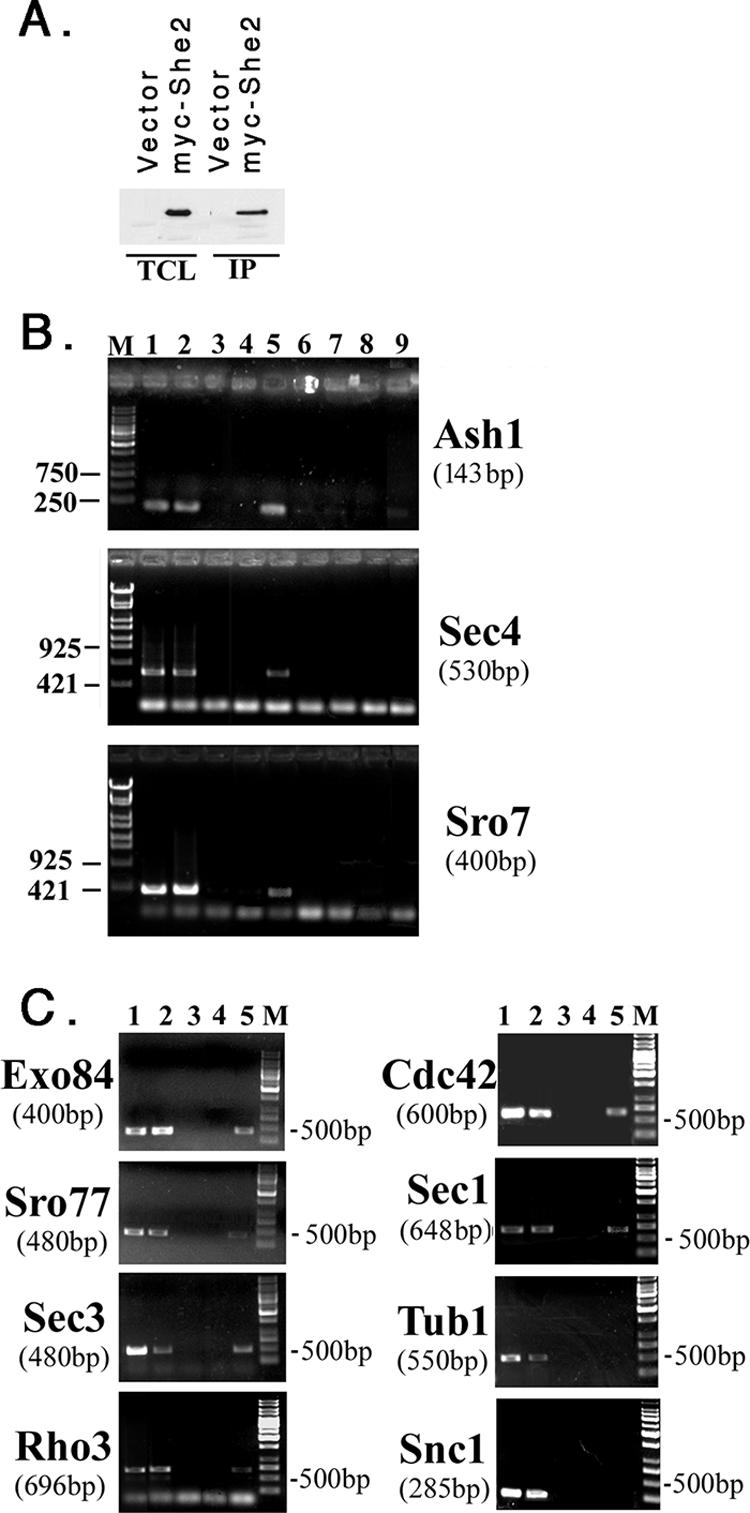
She2 binds to ASH1 and POL mRNAs. (A) IP of She2. WT yeast cells expressing myc-tagged She2 (myc-She2) or a control vector (vector) were lysed, subjected to IP with anti-myc antibodies, and detected by immunoblotting with anti-myc antibodies (dilution of 1:1,000). TCL, total cell lysate (50 μg of protein). (B) ASH1, SEC4, and SRO7 mRNAs coimmunoprecipitate with myc-She2. WT yeast cells expressing myc-She2 or a control vector were lysed and subjected to IP, RNA extraction, and RT-PCR with specific oligonucleotide pairs to ASH1, SEC4, or SRO7. Samples were electrophoresed on 1% agarose gels. Lane M, molecular mass markers (bp); lane 1, RNA derived from control lysate with RT-PCR; lane 2, RNA derived from myc-She2 lysate with RT-PCR; lane 3, No template with RT-PCR; lane 4, RNA derived from control IP with RT-PCR; lane 5, RNA derived from myc-She2 IP with RT-PCR; lane 6, RNA derived from control lysate without RT-PCR; lane 7, RNA derived from myc-She2 lysate without RT-PCR; lane 8, RNA derived from control IP with RT-PCR; and lane 9, RNA derived from myc-She2 IP with RT-PCR. (C) Other mRNAs coimmunoprecipitate with myc-She2, except SNC1 and TUB1. The experiment is the same as described in panel B, except that specific oligonucleotide pairs were used in the PCR to detect other POL mRNAs (i.e., EXO84, SRO77, SEC3, and SNC1) as well as TUB1. The same pairs were used for generating templates for FISH probes (see Materials and Methods), except for SNC1. Lane M, mass markers (bp); lane 1, RNA derived from control lysate with RT-PCR; lane 2, RNA derived from myc-She2 lysate with RT-PCR; lane 3, no template with RT-PCR; lane 4, RNA derived from control IP with RT-PCR; and lane 5, RNA derived from myc-She2 IP with RT-PCR.
SEC3 is required for the bud tip localization of POL and ASH1 mRNAs.
In addition to their requirement in ASH1 mRNA localization, Myo4 and She3 are necessary for the retention of cortical ER in the early bud (20). Interestingly, components of the exocyst (e.g., Sec3 and Sec5) also affect ER inheritance (52, 63, 64) and ASH1 mRNA localization (1). Because of this correlation, we examined whether POL mRNA localization is affected in sec3Δ cells (Fig. 8). We found that RFP-SEC4, RFP-CDC42, and RFP-SRO7 mRNAs were highly mislocalized in sec3Δ yeast (6%, 5%, and 9% bud tip localization, respectively) (Fig. 8A and Table 6). However, only RFP-Sro7 localization was diffuse, while RFP-Sec4 and RFP-Cdc42 remained bud enriched (Fig. 8A), presumably due to delivery to the bud via the secretory pathway. We examined the localization of ASH1 mRNA in sec3Δ cells and found that it was also mislocalized (16% bud tip localization) (Fig. 8B and Table 6). Thus, Sec3 is necessary for the polarized localization of ASH1 and POL mRNAs during budding (Fig. 8A and Table 6), like Myo4 (Fig. 2C and 4; Tables 3 and 4), She2, and She3 (Fig. 4 and Tables 3 and 4). This effect was independent of changes in the actin cytoskeleton, as labeling with rhodamine-conjugated phalloidin was normal in sec3Δ cells (64; also data not shown).
Srp101 in POL mRNA trafficking.
SRP101 encodes a component of the signal recognition particle receptor that affects maintenance and structure of the cortical ER (46). Thus, we examined POL mRNA trafficking in temperature-sensitive srp101-47 cells (Table 6). We found that RFP-SEC4 mRNA was mislocalized (4% bud tip localization) after a shift to 37°C but not at 26°C. Localization of RFP-SRO7 mRNA was affected to a lesser extent (42% bud tip localization at 37°C). Coupled with the results obtained with mutants defective in cortical ER inheritance (i.e., myo4Δ, she3Δ, and sec3Δ), it would appear that all mutations that regulate ER transport and bud retention also affect mRNA trafficking. This implies that cortical ER inheritance and mRNA localization are connected.
POL mRNA localization correlates with the cortical ER.
Estrada et al. (20) demonstrated that Myo4 and She3 cofractionate with the ER. Yet as IST2 mRNA localization was unaffected in a mutant in cortical ER inheritance (aux1Δ), they suggested that ER inheritance and mRNA localization are separate processes (20). However, bud enrichment of IST2 mRNA and protein occurs in a Myo2- and secretory pathway-independent fashion (36), which differs from the secretion-dependent localization of Sec4, Cdc42, and other membrane-anchored POLs (i.e., Snc1) (41, 48, 67). Thus, we examined whether POL mRNAs associate with the ER using fluorescent protein labeling and ER fractionation experiments. First, we followed the colocalization of Sec63-RFP with SEC4 or SRO7 mRNAs bearing MS2 binding sites in myo4Δ, sec3Δ, and control WT cells (Fig. 8C). We found that Sec63-RFP labeling of the bud was readily evident in small-budded (i.e., S/early G2) WT cells, less evident in myo4Δ cells, and hardly seen in sec3Δ cells (Fig. 8C). Values for the percentages of cells showing Sec63-RFP in the bud tip in WT, myo4Δ, and sec3Δ strains were 100 (57 cells), 86 (59 cells; the extent of labeling in the bud, however, was much less than that of WT cells), and 26% (84 cells), respectively. The reduction in cortical ER present in the bud, seen in both myo4Δ and sec3Δ cells, implies less efficient ER inheritance (52, 64). Upon MS2-GFP expression, granules corresponding to both SEC4 and SRO7 mRNAs were present at the tips of emerging buds in small-budded WT cells and in close proximity to the ER (Fig. 8C). In contrast, these granules mislocalized to mother cells in myo4Δ and sec3Δ yeast, as shown above (Fig. 2C, 4, and 8A) but remained more or less associated with the perinuclear ER (Fig. 8B). Thus, mRNA localization to the early bud correlates with ER inheritance.
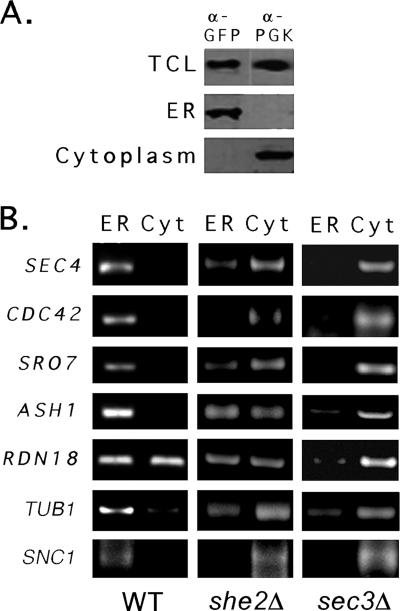
ASH1 and POL mRNAs are associated with the ER fraction.
Given the connection between asymmetric POL mRNA localization and cortical ER trafficking, we examined whether mRNAs are physically connected to the ER (Fig. 9). Yeast cells were lysed under conditions that preserve mRNA, and gradient density centrifugation was performed to obtain separate ER membrane and cytosolic fractions. The ER microsomal fraction was verified using antibodies to detect Sec63-GFP, while the cytosolic fraction was verified using antibodies to phosphoglycerate kinase (Fig. 9A). Next, ASH1 and other mRNAs were detected in the fractions by semiquantitative RT-PCR (Fig. 9B). Importantly, mRNAs encoding Sec4, Cdc42, Sro7, and Snc1 were specifically enriched in the ER fraction, along with ASH1 mRNA (Fig. 9B), which was shown to be enriched on the ER (18). mRNAs encoding RDN18, a ribosomal rRNA, and TUB1 were found in both the ER and cytosolic fractions (Fig. 9B). The cofractionation of RDN18 was expected, given its translation on both free and ER-anchored polysomes; and although SNC1 mRNA is not asymmetrically localized, it probably resides on the ER to allow for translocation upon translation. That all polarized mRNAs associate with the ER suggests that one mechanism might mediate both cortical ER inheritance and mRNA localization to the bud.
FIG. 9.
POL mRNAs are enriched in the ER fraction of WT cells but not in she2Δ and sec3Δ cells. (A) Fractionation of lysates into ER microsome and cytosolic fractions. WT, she2Δ, and sec3Δ yeast cells expressing Sec63-GFP were lysed and subjected to sucrose density gradient centrifugation (see Materials and Methods). Aliquots from the total crude lysate (TCL) and different fractions obtained through centrifugation were subjected to SDS-polyacrylamide gel electrophoresis and detected in blots with antibodies against an ER marker, Sec63 (anti [α]-GFP), or a cytosolic marker, phosphoglycerate kinase (anti-PGK). Data from WT cells are shown; no differences were observed with the mutants. (B) mRNAs encoding ASH1 and POLs are enriched in the ER microsome fraction in WT but not she2Δ or sec3Δ cells. Total RNA isolated from the fractions obtained using density gradient centrifugation was subjected to DNase I treatment and RT-PCR with oligonucleotide pairs specific to ASH1, SEC4, CDC42, SRO7, RDN18, TUB1, and SNC1. Samples were electrophoresed on agarose gels and documented by ethidium bromide labeling.
To demonstrate the involvement of either She2 or Sec3 in the association of mRNAs with the ER, we fractionated she2Δ and sec3Δ cells to obtain microsomal and cytosolic fractions and examined them for mRNA by semiquantitative RT-PCR (Fig. 9B). In contrast to WT cells, most mRNAs were enriched in the cytosolic fraction from she2Δ cells. This effect was even more dramatic in sec3Δ cells, in which little mRNA association with the ER remained (Fig. 9B). These results suggest that Sec3 plays a significant role in mRNA anchoring to the ER.
DISCUSSION
Polarized growth involves both the activation of Cdc42 and an increase in the local concentration of Cdc42 and other POLs at the presumptive bud site (53, 61, 67). Several models have been put forth to explain how Cdc42 becomes polarized. One model suggests that landmark proteins associated with previous bud sites recruit Cdc42 to drive rearrangement of the actin cytoskeleton and exocytic apparatus (54). Another proposes that a septin barrier surrounding the Cdc42 cap prevents the diffusion of cortical proteins and leads to their enrichment (2). A third model proposes that actin-dependent endocytosis within the bud recycles POLs back to the site where polarity was established and maintains polarization (33, 59). Yet none of these models is mutually exclusive, and polarization may employ all three to concentrate POLs at the bud site.
mRNA localization plays a developmental role in Drosophila and Caenorhabditis elegans by effectively altering the local concentration of proteins in different cell types to control morphogenesis (15, 38, 66). By employing in situ and in vivo labeling, we found that many POL mRNAs are trafficked to the tip of the incipient bud in yeast, like ASH1 mRNA (Fig. 1 to 3; Tables 3, 4, and 6). Asymmetric POL mRNA placement preceded both POL protein enrichment and subsequent bud emergence, as evidenced by time-lapse microscopy (Fig. 3; see Movies S2 and S3 in the supplemental material). Thus, mRNA recruitment to the presumptive bud site may contribute to polarization, in addition to the mechanisms described above.
Asymmetrically localized POL mRNAs utilize the same SHE gene-dependent mechanism (Fig. 2C and 4A and Tables 3 and 4; see Fig. S1 in the supplemental material) involved in ASH1 mRNA delivery to the bud and even bind to She2 (Fig. 7). At the physiological level, She-dependent mRNA localization modulated the local concentration of Sro7 (Fig. 2C and 4 and Table 4; see Fig. S1 in the supplemental material), a POL (25) that has no membrane anchor. In contrast, POLs with membrane anchors, like Sec4 or Cdc42, reached the bud tip in a She-independent fashion (Fig. 2C and 4A; Table 3). Thus, defects in mRNA transport affect the localization of only a subset of POL proteins. Given that essential polarity and exocytosis factors (i.e., Cdc42 and Sec4) utilize lipid anchors (as protein targeting sequences) and the secretory pathway to reach the presumptive bud site (48, 61, 67) (Fig. 2D and 4B), this may explain why the SHE genes are dispensable for growth. Membrane anchoring may also allow for protein enrichment at the bud site via endocytosis and recycling (33, 59).
Sro7 may typify factors that lack intrinsic membrane anchors (e.g., Sec1 and Sro77) and, therefore, employ mRNA localization to maintain higher concentrations at the site of exocytosis. These factors interact physically with other components of the export machinery (i.e., t-SNAREs) to localize to the site of secretion (23). Local concentrations of exocyst components may be regulated in a similar fashion, as suggested by the She2 binding and polarized distribution of the SEC3 and EXO84 mRNAs (Fig. 1B and 7). Thus, we propose that the intracellular targeting and localization of POL mRNA are not merely a redundant pathway to ensure protein localization but a means to facilitate polarization. To test this more directly, we deleted SHE2 in either sro77Δ or sro7Δ sro77Δ cells and examined them for defects in growth, particularly at low temperatures (15 to 18°C), where sro7Δ sro77Δ cells are inhibited (37, 39). However, we found no enhancement or inhibition in the growth of either sro77Δ she2Δ (wherein Sro7 function is necessary for cold-resistant growth) or sro7Δ sro77Δ she2Δ cells in comparison to sro77Δ or sro7Δ sro77Δ cells, respectively, at any temperature (data not shown). We also examined whether the deletion of SHE2 in cdc42-6 or rho3-V51 cells might confer synthetic growth defects due to the mislocalization of CDC42 or RHO3 mRNA in the mutant backgrounds. However, no additional defects in growth were observed in rho3-V51 she2Δ or cdc42-6 she2Δ cells at any temperature (14 to 37°C) (data not shown). This implies that defects in the functioning of these small GTPases are not further enhanced by changes in their local translation, presumably as the mutant proteins are targeted correctly via the secretory pathway. Thus, POL mRNA trafficking does not fulfill an essential role during budding, although we presume it is likely to affect the kinetics of polarization under certain circumstances. Interestingly, Sro7 orthologs from Drosophila and mice associate with Fmr1, an mRNA targeting protein, to form a complex involved in neural development (68). Thus, Sro7 may play a direct role in mRNA trafficking.
An earlier study demonstrated the enrichment of 22 other mRNAs in the buds of exponentially growing cells; however, these transcripts showed no specificity in terms of function, nor do they have a role in cell polarization (56). Thus, they may constitute mRNAs necessary for general metabolic functions that allow the bud to become autonomous for protein synthesis prior to nuclear division. If so, a large number of mRNAs may become bud enriched late in the cell cycle. This might explain why the POL mRNAs shown here were not identified in the earlier screen if they are polarized only during one phase of the cell cycle (i.e., S phase). Yet not all mRNAs are asymmetrically localized. For example, SNC1 and MSO1 mRNA localize to mother cells early in the cell cycle and only reach the bud after nuclear division occurs (Fig. 1B, 6A, and 6B). This might reflect mRNA trafficking on ER peripheral to the nucleus. TUB1 mRNA is also not asymmetrically distributed (Fig. 1B), and all three proteins (Tub1, Snc1, and Mso1) have a symmetric pattern of localization (10, 11, 41, 47). Unlike asymmetrically localized mRNAs, TUB1 and SNC1 mRNA did not associate with She2 (Fig. 7), implying that their localization is She independent. Other mRNAs, such as PEX3 and OXA1, were also not asymmetrically localized but were found mainly in mother cells and closely associated with the ER membranes and mitochondria, respectively (Fig. 6C and D). Yet TUB1 mRNA is present on the ER along with other mRNAs (Fig. 9). This exemplifies a known phenomenon whereby mRNAs encoding soluble proteins are associated with membrane-bound polysomes (40). Thus, whether an mRNA is asymmetrically localized or not, it is likely to be associated with ER membranes. In the case of polarized mRNAs, however, we suggest that they are specifically delivered along with cortical ER (Fig. 8C and 9; Table 6).
We propose that She- and Sec3-dependent mRNA transport is connected with cortical ER transmittance to, and retention in, the incipient bud. This is because She1/Myo4, She3, Sec3, and Srp101 confer cortical ER inheritance (46, 52, 63, 64) and asymmetric POL mRNA trafficking (Fig. 2C, 4, and 8; Tables 3, 4, and 6). While a previous study discounted this connection (20), the tight correlation seen here between factors involved in cortical ER trafficking and POL mRNA localization suggests that mRNAs are transported on ER membranes to the emerging bud. Indeed, cell fractionation studies show the enrichment of POL and other mRNAs in ER fractions derived from WT cells (Fig. 9B). Moreover, both She2 and Sec3 are necessary for this enrichment (Fig. 9B), indicating that they help anchor mRNAs to the ER. Correspondingly, the deletion of SEC3, which yields a severe growth phenotype, has a more dramatic effect upon mRNA association than the deletion of SHE2. While the underlying mechanism is unknown, involvement of an exocyst component in both mRNA and ER transport could be important for connecting the processes of protein synthesis and secretion at the site of growth. For example, the cotransport of the ER and mRNA makes sense for membrane-anchored proteins, which require the ER as a platform for translation, translocation, and processing. As the mother-bud junction prevents the diffusion of plasma membrane proteins, ER inheritance without the cotrafficking of mRNA would be futile. Moreover, in the case of mRNAs encoding soluble proteins, it makes sense to localize translation in order to minimize protein diffusion away from the site of exocytosis.
Evidence from other systems supports our contention that mRNA localization is connected to the ER. For example, a Xenopus mRNA binding protein, Vera/Vg1RBP, is ER associated, and a number of mRNAs are linked to the ER in Xenopus oocytes (12, 17). Drosophila Trailer Hitch is a part of a large mRNA-protein complex that localizes to the ER and is required for the efficient secretion of Gurken and normal dorsal-ventral patterning in developing embryos (65). Finally, while this work was in progress, Schmid et al. (55) demonstrated by fluorescence microscopy that ASH1 mRNA is ER associated and that its localization to cortical ER is dependent upon She2 (55). In the absence of She2, ASH1 mRNA remains associated with nuclear ER and does not reach the bud tip (55). This may explain why some ASH1 mRNA remains associated with the ER fraction in she2Δ cells, as determined by RT-PCR (Fig. 9B).
This study shows an implicit relationship between mRNA localization, cortical ER inheritance, and subsequent polarized growth in yeast. Given current interest in the mechanisms governing the recruitment of POLs to the sites of secretion, it is critical to examine the contribution of local mRNA translation. Even more importantly, our results suggest that the 3′ UTRs of genes may be necessary for proper protein localization. Thus, proteins expressed from genes in which the 3′ UTR has been removed (e.g., in plasmid-based expression systems) or disconnected (e.g., in genome-wide carboxy-terminal GFP tagging studies) may be mislocalized. In conclusion, evidence from both simple and complex systems suggests that mRNA targeting is an evolutionarily conserved mechanism that may facilitate cell polarization.
Supplementary Material
Acknowledgments
We thank K. Bloom, Z. Elazar, S. Michaelis, P. Novick, and R. Singer for reagents and R. Shema for technical help.
This work was supported by grants to J.E.G. from the Minerva Foundation, Germany, and the Kahn Fund for Systems Biology, Weizmann Institute of Science, Israel. S.A. was supported in part by a postdoctoral fellowship from the Feinberg Graduate School. J.E.G. holds the Henry Kaplan Chair in Cancer Research.
Footnotes
Published ahead of print on 5 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aronov, S., and J. E. Gerst. 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J. Biol. Chem. 279:36962-36971. [DOI] [PubMed] [Google Scholar]
- 2.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. [DOI] [PubMed] [Google Scholar]
- 3.Baum, B. 2004. Animal development: crowd control. Curr. Biol. 14:R716—R718. [DOI] [PubMed] [Google Scholar]
- 4.Beach, D. L., E. D. Salmon, and K. Bloom. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569-578. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437-445. [DOI] [PubMed] [Google Scholar]
- 6.Betschinger, J., and J. A. Knoblich. 2004. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14:R674—R685. [DOI] [PubMed] [Google Scholar]
- 7.Bohl, F., C. Kruse, A. Frank, D. Ferring, and R. P. Jansen. 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19:5514-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, C., T. Hughes, M. Pypaert, and P. Novick. 2004. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167:889-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bretscher, A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke, D., P. Gasdaska, and L. Hartwell. 1989. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo-Flores, A., A. Weinberger, M. Robinson, and J. E. Gerst. 2005. Mso1 is a novel component of the yeast exocytic SNARE complex. J. Biol. Chem. 280:34033-34041. [DOI] [PubMed] [Google Scholar]
- 12.Chang, P., J. Torres, R. A. Lewis, K. L. Mowry, E. Houliston, and M. L. King. 2004. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol. Biol. Cell 15:4669-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chant, J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15:365-391. [DOI] [PubMed] [Google Scholar]
- 14.Chartrand, P., R. H. Singer, and R. M. Long. 2001. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 17:297-310. [DOI] [PubMed] [Google Scholar]
- 15.Condeelis, J., and R. H. Singer. 2005. How and why does beta-actin mRNA target? Biol. Cell 97:97-110. [DOI] [PubMed] [Google Scholar]
- 16.Corral-Debrinski, M., C. Blugeon, and C. Jacq. 2000. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20:7881-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshler, J. O., M. I. Highett, and B. J. Schnapp. 1997. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276:1128-1131. [DOI] [PubMed] [Google Scholar]
- 18.Diehn, M., M. B. Eisen, D. Botstein, and P. O. Brown. 2000. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 25:58-62. [DOI] [PubMed] [Google Scholar]
- 19.Drubin, D. G., and W. J. Nelson. 1996. Origins of cell polarity. Cell 84:335-344. [DOI] [PubMed] [Google Scholar]
- 20.Estrada, P., J. Kim, J. Coleman, L. Walker, B. Dunn, P. Takizawa, P. Novick, and S. Ferro-Novick. 2003. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evangelista, M., S. Zigmond, and C. Boone. 2003. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603-2611. [DOI] [PubMed] [Google Scholar]
- 22.Gao, X. D., S. Albert, S. E. Tcheperegine, C. G. Burd, D. Gallwitz, and E. Bi. 2003. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J. Cell Biol. 162:635-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerst, J. E. 2003. SNARE regulators: matchmakers and matchbreakers. Biochim. Biophys. Acta 1641:99-110. [DOI] [PubMed] [Google Scholar]
- 24.Gonsalvez, G. B., C. R. Urbinati, and R. M. Long. 2005. RNA localization in yeast: moving towards a mechanism. Biol. Cell 97:75-86. [DOI] [PubMed] [Google Scholar]
- 25.Grosshans, B. L., A. Andreeva, A. Gangar, S. Niessen, J. R. Yates, 3rd, P. Brennwald, and P. Novick. 2006. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J. Cell Biol. 172:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, W., Y. Deng, D. Zenklusen, and R. H. Singer. 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18:1452-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, W., M. Sacher, J. Barrowman, S. Ferro-Novick, and P. Novick. 2000. Protein complexes in transport vesicle targeting. Trends Cell Biol. 10:251-255. [DOI] [PubMed] [Google Scholar]
- 28.Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H. F. Tabak. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122:85-95. [DOI] [PubMed] [Google Scholar]
- 29.Horton, A. C., and M. D. Ehlers. 2003. Neuronal polarity and trafficking. Neuron 40:277-295. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz, R., and D. Webb. 2003. Cell migration. Curr. Biol. 13:R756—R759. [DOI] [PubMed] [Google Scholar]
- 31.Irazoqui, J. E., A. S. Gladfelter, and D. J. Lew. 2004. Cdc42p, GTP hydrolysis, and the cell's sense of direction. Cell Cycle 3:861-864. [PubMed] [Google Scholar]
- 32.Irazoqui, J. E., A. S. Gladfelter, and D. J. Lew. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062-1070. [DOI] [PubMed] [Google Scholar]
- 33.Irazoqui, J. E., A. S. Howell, C. L. Theesfeld, and D. J. Lew. 2005. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell 16:1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irazoqui, J. E., and D. J. Lew. 2004. Polarity establishment in yeast. J. Cell Sci. 117:2169-2171. [DOI] [PubMed] [Google Scholar]
- 35.Jansen, R. P., C. Dowzer, C. Michaelis, M. Galova, and K. Nasmyth. 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 84:687-697. [DOI] [PubMed] [Google Scholar]
- 36.Juschke, C., D. Ferring, R. P. Jansen, and M. Seedorf. 2004. A novel transport pathway for a yeast plasma membrane protein encoded by a localized mRNA. Curr. Biol. 14:406-411. [DOI] [PubMed] [Google Scholar]
- 37.Kagami, M., A. Toh-e, and Y. Matsui. 1998. Sro7p, a Saccharomyces cerevisiae counterpart of the tumor suppressor l(2)gl protein, is related to myosins in function. Genetics 149:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloc, M., N. R. Zearfoss, and L. D. Etkin. 2002. Mechanisms of subcellular mRNA localization. Cell 108:533-544. [DOI] [PubMed] [Google Scholar]
- 39.Lehman, K., G. Rossi, J. E. Adamo, and P. Brennwald. 1999. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146:125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerner, R. S., R. M. Seiser, T. Zheng, P. J. Lager, M. C. Reedy, J. D. Keene, and C. V. Nicchitta. 2003. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9:1123-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, M. J., B. J. Nichols, C. Prescianotto-Baschong, H. Riezman, and H. R. Pelham. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long, R. M., R. H. Singer, X. Meng, I. Gonzalez, K. Nasmyth, and R. P. Jansen. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277:383-387. [DOI] [PubMed] [Google Scholar]
- 43.Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 44.Munchow, S., C. Sauter, and R. P. Jansen. 1999. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112:1511-1518. [DOI] [PubMed] [Google Scholar]
- 45.Olivier, C., G. Poirier, P. Gendron, A. Boisgontier, F. Major, and P. Chartrand. 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 25:4752-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prinz, W. A., L. Grzyb, M. Veenhuis, J. A. Kahana, P. A. Silver, and T. A. Rapoport. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Protopopov, V., B. Govindan, P. Novick, and J. E. Gerst. 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74:855-861. [DOI] [PubMed] [Google Scholar]
- 48.Pruyne, D., L. Gao, E. Bi, and A. Bretscher. 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15:4971-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong, and A. Bretscher. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559-591. [DOI] [PubMed] [Google Scholar]
- 50.Pruyne, D. W., D. H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931-1945. [DOI] [PubMed] [Google Scholar]
- 51.Raftopoulou, M., and A. Hall. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23-32. [DOI] [PubMed] [Google Scholar]
- 52.Reinke, C. A., P. Kozik, and B. S. Glick. 2004. Golgi inheritance in small buds of Saccharomyces cerevisiae is linked to endoplasmic reticulum inheritance. Proc. Natl. Acad. Sci. USA 101:18018-18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schenkman, L. R., C. Caruso, N. Page, and J. R. Pringle. 2002. The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 156:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmid, M., A. Jaedicke, T. G. Du, and R. P. Jansen. 2006. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 16:1538-1543. [DOI] [PubMed] [Google Scholar]
- 56.Shepard, K. A., A. P. Gerber, A. Jambhekar, P. A. Takizawa, P. O. Brown, D. Herschlag, J. L. DeRisi, and R. D. Vale. 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 100:11429-11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takizawa, P. A., A. Sil, J. R. Swedlow, I. Herskowitz, and R. D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389:90-93. [DOI] [PubMed] [Google Scholar]
- 58.Takizawa, P. A., and R. D. Vale. 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 97:5273-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valdez-Taubas, J., and H. R. Pelham. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13:1636-1640. [DOI] [PubMed] [Google Scholar]
- 60.Walch Solimena, C., R. N. Collins, and P. J. Novick. 1997. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137:1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wedlich-Soldner, R., S. Altschuler, L. Wu, and R. Li. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299:1231-1235. [DOI] [PubMed] [Google Scholar]
- 62.Wendland, B., J. M. McCaffery, Q. Xiao, and S. D. Emr. 1996. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 135:1485-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiederkehr, A., J. O. De Craene, S. Ferro-Novick, and P. Novick. 2004. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 167:875-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiederkehr, A., Y. Du, M. Pypaert, S. Ferro-Novick, and P. Novick. 2003. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol. Biol. Cell 14:4770-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilhelm, J. E., M. Buszczak, and S. Sayles. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9:675-685. [DOI] [PubMed] [Google Scholar]
- 66.Wodarz, A. 2002. Establishing cell polarity in development. Nat. Cell Biol. 4:E39-44. [DOI] [PubMed] [Google Scholar]
- 67.Zajac, A., X. Sun, J. Zhang, and W. Guo. 2005. Cyclical regulation of the exocyst and cell polarity determinants for polarized cell growth. Mol. Biol. Cell 16:1500-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zarnescu, D. C., P. Jin, J. Betschinger, M. Nakamoto, Y. Wang, T. C. Dockendorff, Y. Feng, T. A. Jongens, J. C. Sisson, J. A. Knoblich, S. T. Warren, and K. Moses. 2005. Fragile X protein functions with lgl and the par complex in flies and mice. Dev. Cell 8:43-52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.