Abstract
A transcription corepressor, MAT1-mediated transcriptional repressor (MMTR), was found in mouse embryonic stem cell lines. MMTR orthologs (DMAP1) are found in a wide variety of life forms from yeasts to humans. MMTR down-regulation in differentiating mouse embryonic stem cells in vitro resulted in activation of many unrelated genes, suggesting its role as a general transcriptional repressor. In luciferase reporter assays, the transcriptional repression activity resided at amino acids 221 to 468. Histone deacetylase 1 (HDAC1) interacts with MMTR both in vitro and in vivo and also interacts with MMTR in the nucleus. Interestingly, MMTR activity was only partially rescued by competition with dominant-negative HDAC1(H141A) or by treatment with an HDAC inhibitor, trichostatin A (TSA). To identify the protein responsible for HDAC1-independent MMTR activity, we performed a yeast two-hybrid screen with the full-length MMTR coding sequence as bait and found MAT1. MAT1 is an assembly/targeting factor for cyclin-dependent kinase-activating kinase which constitutes a subcomplex of TFIIH. The coiled-coil domain in the middle of MAT1 was confirmed to interact with the C-terminal half of MMTR, and the MMTR-mediated transcriptional repression activity was completely restored by MAT1 in the presence of TSA. Moreover, intact MMTR was required to inhibit phosphorylation of the C-terminal domain in the RNA polymerase II largest subunit by TFIIH kinase in vitro. Taken together, these data strongly suggest that MMTR is part of the basic cellular machinery for a wide range of transcriptional regulation via interaction with TFIIH and HDAC.
Cell type-specific gene expression is regulated by the combinatorial actions of general transcriptional machinery, tissue-specific DNA binding factors, and transcriptional coregulators (23, 26). In previous studies, by screening the cDNA library of mouse embryonic stem cells (mESCs), we identified genes that may play important roles in early development and differentiation (22). Embryonic stem cells are pluripotent cells with self-renewing capabilities and broad differentiation plasticity (9, 27). The capacity of embryonic stem cells to differentiate in vitro in a specifically controlled fashion provides unique opportunities for experimental analysis of how genes are regulated when cells commit to a specific lineage of differentiation during early embryogenesis (34). There are a number of genes that play important roles in regulating temporal and spatial embryonic development and tissue differentiation (3, 34). On the assumption that many of these genes are differentially expressed in embryonic stem cells during differentiation, we made an attempt to identify them and found several interesting novel cDNA clones with these characteristics. Among them, we have found that HBMG010 (GenBank accession no. AF438610) is expressed very weakly in undifferentiated mESCs but is increased and maintained at a high level of expression during differentiation (22). HBMG010 has repressor activity by directly interacting with menage a trois 1 (MAT1), a component of the general transcription factor TFIIH complex (see below). Thereafter, we named this clone MAT1-mediated transcriptional repressor (MMTR).
While we were characterizing MMTR, the same clone was independently isolated by another group via its interaction with DNA methyltransferase 1 (DNMT1) in humans and was given a different name (DNA methyltransferase 1-associated protein 1 [DMAP1]) (38). DMAP1 is implicated in gene regulation through modification of chromatin. DMAP1 associates with DNMT1 and histone deacetylase 2 (HDAC2) during the late S phase to mediate transcriptional repression. DMAP1 also binds to the transcriptional corepressor TSG101 (38). Recently, Doyon et al. (6) reported that DMAP1 was involved in the NuA4 histone acetyltransferase multisubunit complex, which is responsible for acetylation of histone H4 and H2A N-terminal tails in yeast and plays primary roles in transcription, cellular response to DNA damage, and cell cycle control. DMAP1 acts as a corepressor and enhances the repression activities of interacting partner proteins. For example, DMAP1 interacts with RNA polymerase II subunit 5-mediating protein (RMP), facilitating the exclusive nuclear localization of RMP and increasing its corepressor activity in a dose-dependent manner (5). It has been reported that the transcriptional activity of DMAP1 is inhibited by RGS6, a member of a subfamily of mammalian regulators of G protein signaling proteins which were discovered as negative regulators of heterotrimeric G protein signaling (25). Coexpression of DMAP1 with RGS6 promotes the nuclear migration of RGS6. DMAP1 was also shown to enhance Daxx-mediated repression of glucocorticoid receptor transcriptional activity, and Daxx protected protein degradation of DMAP1 in vivo (28). Despite these findings, the precise mechanism of DMAP1 in regulation of gene expression in mammalian cells remains to be further explored. In this study, we investigated the functional characteristics of DMAP1/MMTR.
MMTR is ubiquitously expressed in most mouse tissues and localized predominantly in the nucleus. We have found that MMTR has transcriptional repression activity without direct binding to DNA elements or specific transcription factors. The N-terminal part of MMTR interacts with HDAC1, and this interaction is shown to be partially responsible for MMTR's transcriptional repression activity. To further elucidate the functional mechanism of MMTR, a yeast two-hybrid screen was performed to isolate the MMTR-interacting protein(s). As a result, we identified menage a trois 1 (MAT1), a component of the general transcription factor TFIIH complex (see below), which is able to rescue the transcriptional repression activity of MMTR. Indeed, MMTR-mediated transcription repression activities of various human promoters were completely restored by MAT1 in the presence of the HDAC1 inhibitor trichostatin A (TSA). Interestingly, intact MMTR protein showed in vitro inhibition of the phosphorylation of the TFIIH kinase substrate, the C-terminal domain (CTD) of the RNA polymerase II (Pol II) largest subunit. Taken together, our data indicate that MMTR is a key component of RNA Pol II-mediated gene expression through its interaction with HDAC1 and the modulation of TFIIH kinase activity via MAT1.
MATERIALS AND METHODS
Cell culture, transfection, and antibodies.
293T (human embryonic kidney cell line) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (HyClone), 50 units/ml penicillin, and 50 μg/ml streptomycin (Gibco BRL). 293T cells were transfected using a conventional calcium phosphate transfection method (46). Antibodies used in this study, glutathione S-transferase (GST) and hemagglutinin (HA) antibodies (Zymed), Flag antibody (Sigma), and MAT1 and β-tubulin antibodies (Santa Cruz), were purchased from their respective suppliers. The MMTR antisera were produced by subcutaneous injection of recombinant protein (300 μg of protein per injection) into rabbits four times every 2 weeks. Immunized serum was obtained from blood collected from cardiac puncture. HDAC1 antibody was kindly donated by Sung Hee Baek (Seoul National University, Seoul, Korea).
Plasmids.
To get the full-length cDNA of MMTR, the original 1.3-kb MMTR clone from the mESC cDNA library was employed as a probe to screen the mouse erythroid leukemia cell ZAP express (Stratagene) cDNA phage library. Resulting phages were plaque purified, and inserts were rescued as pBK-CMV-MMTR, which contains 1,543 nucleotides encoding a complete open reading frame with 468 amino acids. For the construction of the prokaryotic expression vector, a BamHI/SmaI fragment of the MMTR cDNA from pBK-CMV-MMTR was inserted into the BamHI/XhoI (gap filled by Klenow polymerase) site of the pGEX-3X vector (Amersham). A pCMV-HA-MMTR vector was constructed by subcloning the EcoRI/XhoI fragment of pBK-CMV-MMTR into the EcoRI/XhoI site of pCMV-HA (Clontech). MMTR expression vectors containing various deletions were obtained by PCR cloning strategies. The N-terminal half region of MMTR, encoding amino acids 1 to 220, was amplified with the MMTR 1F (EcoRI) and MMTR 1R (XhoI) primers (Table 1). A primer pair for the C-terminal half region of MMTR encoding amino acids 221 to 468 includes MMTR 2F (EcoRI) and MMTR 2R (XhoI) (Table 1). Each PCR product was cloned into the pGEM-T Easy vector (Promega). EcoRI/XhoI fragments of deletion mutants were subcloned into the EcoRI/XhoI site of the pCMV-HA vector. To generate the GAL4 binding domain-MMTR fusion protein expression vector (GAL4-MMTR), the XbaI fragment from pBK-CMV-MMTR was subcloned into the XbaI site of pcDNA1.1 (Invitrogen), which contains a GAL4 binding domain upstream of the multicloning site (pCMV-0). GAL4-MMTR deletion mutants were generated by combinatorial PCR procedures using a primer set for the N-terminal half region of MMTR 1F (NotI) and MMTR 1R (XhoI) and the C-terminal half region of MMTR 2F (NotI) and MMTR 2R (XhoI) (Table 1). Each PCR product was cloned into the pGEM-T easy vector. NotI fragments of each mutant were subcloned into the NotI site of the pCMV-0 vector to generate pGAL4-MMTR-N and pGAL4-MMTR-C. To generate pEGFP-MMTR, an EcoRI/SmaI fragment (blunted by Klenow polymerase) from pBK-CMV-MMTR was subcloned into the SmaI site of pEGFP-C1 (Clontech). Full-length MAT1 was PCR cloned from the cDNA library of K562 (a human erythroleukemia cell line) and subcloned into Flag-tagged pcDNA3. The primer set for full-length MAT1 includes MAT1 primer 1 (EcoRI) and MAT1 primer 2 (XbaI) (Table 1). The plasmid pcDNA3-Flag-MAT1-a (encoding amino acids 1 to 114) was PCR cloned from pcDNA3-Flag-MAT1 and subcloned into the site of EcoRI/XbaI in Flag-tagged pcDNA3. The primer pair for MAT1-a is MAT1 primer 1 (EcoRI) and MAT1 primer 3 (XbaI). The plasmid pcDNA3-Flag-MAT1-b (encoding amino acids 109 to 309) was generated by subcloning the EcoRI/BamHI (blunt) fragment from pB42AD-MAT1 into the EcoRI/XhoI (blunt) site of Flag-tagged pcDNA3. The plasmid pcDNA3-Flag-MAT1-c (encoding amino acids 109 to 175) was PCR cloned from pcDNA3-Flag-MAT1 and subcloned into the EcoRI/XhoI site of Flag-tagged pcDNA3. The primer set for MAT1-c is MAT1 primer 4 (EcoRI) and MAT1 primer 5 (XhoI). The plasmid pcDNA3-Flag-MAT1-d (encoding amino acids 179 to 309) was PCR cloned from pcDNA3-Flag-MAT1 and subcloned into the site of EcoRI/XbaI of Flag-tagged pcDNA3. The primer set for MAT1-d is MAT1 primer 6 (EcoRI) and MAT1 primer 2 (XbaI). The plasmid pCDNA3-Flag-HDAC1(H141A) was donated by Sung Hee Baek (Seoul National University, Seoul, Korea). The GST-CTD expression vector was provided by Robert P. Fisher (Memorial Sloan-Kettering Cancer Center, New York, NY). The plasmid pBJ5-HDAC1-Flag was kindly donated by Jongbok Yoon (Yonsei University, Seoul, Korea). The plasmid pLV-TH was provided by Didier Trono (University of Geneva, Geneva, Switzerland). p53, Rb, and c-Myc cDNAs were PCR cloned from the K562 cDNA library.
TABLE 1.
Oligonucleotides used in this studya
| Name | Sequence of oligonucleotide | Orientation | Positionb |
|---|---|---|---|
| MMTR 1F (EcoRI) | 5′-GAATTCTA ATGGCTACGGGC-3′ | Forward | +58 |
| MMTR 1R (XhoI) | 5′-CTCGAGAGCATCAAACACTGG-3′ | Reverse | +718 |
| MMTR 2F (EcoRI) | 5′-GAATTCTA GGGCATGAGAGACGG-3′ | Forward | +719 |
| MMTR 2R (XhoI) | 5′-CTCGAGTGG TTTCTTGGCTTTC-3′ | Reverse | +1462 |
| MMTR 1F (NotI) | 5′-GCGGCCGCATGGCTACGGGC-3′ | Forward | +58 |
| MMTR 2F (NotI) | 5′-GCGGCCGCGGGCATGAGAGACGG-3′ | Forward | +719 |
| MAT1 primer1 (EcoRI) | 5′-CTGAATTCATGGACGATCAGGG-3′ | Forward | +102 |
| MAT1 primer2 (XbaI) | 5′-TCTAGATTAACTGGGCTGCC-3′ | Reverse | +1029 |
| MAT1 primer3 (XbaI) | 5′-TCTAGAATCCACATTGTTGG-3′ | Reverse | +444 |
| MAT1 primer4 (EcoRI) | 5′-CTGAATTCACCAACAATGTGGA-3′ | Forward | +429 |
| MAT1 primer5 (XhoI) | 5′-CTCGAGTAGAATCTGCTGCAG-3′ | Reverse | +627 |
| MAT1 primer6 (EcoRI) | 5′-CTGAATTCAAGCAGGCTTTTTTA-3′ | Forward | +639 |
The specific restriction site-tagged sequences are underlined, with their names in parentheses.
Position relative to transcription start site.
Immunoprecipitation.
For immunoprecipitations, 1 mg of cell extract was precleared with 20 μl of protein A/G-Sepharose beads (Sigma) by rotation for 1 h at 4°C. After centrifugation, the supernatant was incubated with 4 μg of HA or Flag or HDAC1 or MMTR antibody on a rocking platform for 16 h at 4°C, and then 20 μl of the protein A/G-agarose was added, followed by incubation for an additional 3 h. The immune complex was washed three times with lysis buffer, and bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting.
Dual-luciferase assays.
Luciferase reporter constructs containing the pGL3-basic vector (Promega) containing the synthetic mouse CP2 binding elements (pGL3-CP2-syn) (2) or minimal promoters (Tert, Tcf, Cdc25A, cyclin E, HKII, and Ras) (41) were used in these experiments. To analyze the functional relationships between HDAC1 or MAT1 and MMTR, the pGL2-basic vector (Promega) containing upstream GAL4 binding sites (pGL2-GAL4) was cotransfected with GAL4-MMTR, GAL4-MMTR-N, pGAL4-MMTR-C, Flag-HDAC1, or Flag-HDAC1(H141A) expression vectors and the pRL-TK vectors (Promega) in the absence or presence of TSA (5 nM). The GAL4-CRTR (36) repression domain (RD) was used as a positive control. The 293T cells (5 × 104) were plated in 24-well dishes, and a total of 2 μg of DNA combining both reporter and an appropriate combination of expression vectors was transfected using the calcium phosphate method. At 36 h after transfection, cells were harvested in 50 μl of passive lysis buffer (Promega) and 20 μl of lysate was used to perform the luciferase assay on a Lumat LB9501 luminometer (Berthold). The dual-luciferase assay system (Promega) was employed according to the manufacturer's instructions, and firefly luciferase expression was normalized against Renilla luciferase. All transfections were repeated at least three times.
Immunocytochemistry.
Cells were seeded on 22-by-22-mm glass coverslips and grown to 50 to 60% confluence. After a wash with phosphate-buffered saline, cells were fixed in an ice-cold methanol-acetone mixture (1:1) for 5 min at room temperature (RT) and treated with blocking solution (3% bovine serum albumin [BSA] in phosphate-buffered saline) at RT for 1 h. The cells were incubated at RT for 1 h with HDAC1 antibodies. Bound antibodies were revealed by incubation for 1 h at RT with Cy5-conjugated anti-mouse immunoglobulin G (IgG) antibody (Jackson Immunoresearch). Nuclei were stained with Hoechst 33258. Subcellular localization of green or red fluorescence was detected by fluorescence microscopy (Olympus BX50).
GST pull-down assays.
GST fusion proteins were expressed in the Escherichia coli strain BL21(DE3) and purified by using glutathione-Sepharose 4B beads (Amersham) according to the manufacturer's protocols. GST or GST-MMTR protein immobilized on 200 μl of the beads was incubated on a rotary shaker for 90 min at 4°C with precleared 293T whole-cell extracts expressing Flag-HDAC1 or Flag-MAT1 protein. After three washes with lysis buffer, proteins were recovered by boiling them in 2× SDS-PAGE loading buffer and subjected to SDS-PAGE followed by Western blot analysis.
Kinase assays.
293T cells were cotransfected with Flag-CDK7, Flag-cyclin H, and Flag-MAT1 expression vectors using the calcium phosphate method. Cells were lysed in a lysis buffer (20 mM Tris-Cl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Roche]) at 36 h posttransfection and immunoprecipitated with 4 μg of Flag antibody. Finally, 25 μl of protein A/G-Sepharose bead-bound cdk-activating kinase (CAK) complexes were resuspended in 300 μl of lysis buffer. Integrity of the immune complex was confirmed by Western blotting for every purification step. Ten microliters of protein slurry and recombinant GST-CTD proteins (2.5 μg) were added to an in vitro kinase assay buffer (20 mM HEPES, pH 7.5, 10 mM magnesium chloride, 1 mM dithiothreitol, 10 μM ATP, and 5 μCi [γ-32P]ATP [Amersham]). The reaction mixture was incubated at 30°C for 30 min and quenched with loading buffer (50% [vol/vol] glycerol, 2% bromophenol blue, and 1% xylene cyanol). The kinase reaction mixture was electrophoresed on a 10% SDS-PAGE gel and wet transferred to a polyvinylidene difluoride membrane (Stratagene) according to the manufacturer's instructions. Western blotting was employed to confirm that an equal amount of CAK complex was used in each reaction. 32P-labeled protein was detected by autoradiography. To confirm the effect of MMTR on the phosphorylation of CTD, increasing amounts of purified GST, GST-MMTR, GST-MMTR-N, or GST-MMTR-C were added to reactions and the phosphorylation level of CTD was measured by autoradiography.
RESULTS
N-terminal half region of MMTR has transcriptional repression activity.
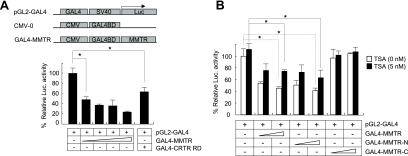
To dissect the mechanism underlying the transcriptional repression activity of MMTR, full-length MMTR was fused to the GAL4 DNA binding domain (GAL4-MMTR) and transiently transfected into 293T cells. The transcriptional repression activity of this construct was tested on a GAL4-responsive minimal luciferase reporter gene (pGL2-GAL4). The CRTR RD (amino acids 1 to 52) (20) was used as a positive control. Similar to results with GAL4-CRTR RD, GAL4-MMTR reduced the expression level of the reporter activity by 75% in a dose-dependent manner (Fig. 1A).
FIG. 1.
N-terminal half of MMTR has transcriptional repression activity. (A) GAL4-MMTR fusion protein-based luciferase (Luc.) reporter assays were performed with 293T cells. pGL2-GAL4 reporter vectors were transfected with increasing amounts of GAL4-MMTR expression vector, and reporter activity was measured at 36 h posttransfection. CRTR RD was used as a positive control. (B) pGL2-GAL4 reporter vectors were cotransfected into 293T cells with various combinations of GAL4-MMTR, GAL4-MMTR-N, and GAL4-MMTR-C expression vectors. Cells were treated with 5 nM TSA for 48 h after transfection. The cell extracts were subjected to luciferase assays. Results with a P value (*) of less than or equal to 0.05, which is considered statistically significant, are presented. Error bars indicate standard deviation from triplicate transfections.
To determine which domain of MMTR is responsible for transcriptional repression activity, we compared the activities of the N-terminal half region (amino acids 1 to 220) to the C-terminal half region (amino acids 221 to 468, containing a coiled-coil domain). These deletion forms of MMTR proteins were found to be expressed at similar levels as determined by Western blot analysis (data not shown; also, see Fig. 5 for an example). When these constructs were tested for their transcriptional activities in transiently transfected 293T cells along with the pGL2-GAL4 reporter construct, the N-terminal half region of MMTR showed strong repression activity comparable to that of full-length MMTR (Fig. 1B). Full-length MMTR and the N-terminal half region of MMTR reduced the luciferase activity to 50% of that of the control, while the carboxy-terminal half region of MMTR did not affect the luciferase activity. These data indicated that the repression activity resided in the N-terminal half region of MMTR.
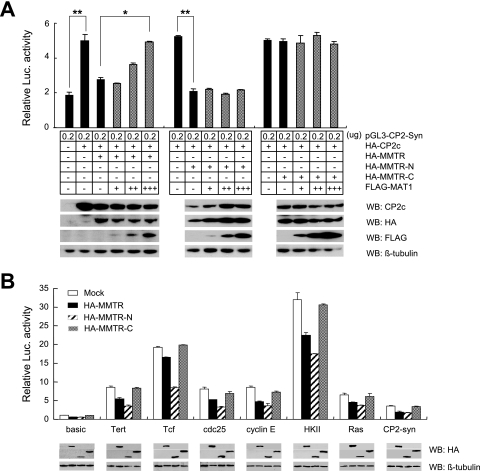
FIG. 5.
MAT1 inhibits MMTR-mediated transcriptional repression by interaction with C-terminal half region of MMTR. (A) The pGL3-CP2-syn reporter vector was transfected into 293T cells with various combinations of HA-CP2c, MMTR, MMTR-N, and MMTR-C and increasing amounts of MAT1 expression vectors. Cell extracts were assayed for luciferase (Luc.) activity at 36 h posttransfection. Same cell extracts were also subjected to SDS-PAGE and subsequently analyzed by Western blotting (WB) with CP2c, HA, Flag, or β-tubulin antibody. Results with a P value of less than or equal to 0.05 (*) or 0.01 (**) are presented. Error bars indicate standard deviation from triplicate transfections. (B) Luciferase activities of pGL3-luciferase reporter vectors containing minimal promoters of eight genes were analyzed in the absence or presence of intact HA-MMTR or deletion mutant (MMTR-N and MMTR-C) expression vectors. Luciferase activities were measured 36 h posttransfection. The expression level of each transfected MMTR polypeptide was also determined by Western blotting with HA antibody. Error bars indicate standard deviation from triplicate transfections.
MMTR activity is partially mediated by direct interaction with HDAC1.
Recent reports have shown that MMTR/DMAP1 interacts with HDAC2 and represses transcription partially through direct interaction with HDAC2 (38). In this study, we tested whether MMTR can associate with another corepressor, HDAC1, which belongs to the same class as HDAC2 (14). To determine whether MMTR and HDAC1 act cooperatively or independently, we performed the GAL4 fusion protein-based luciferase assays and tested the effect of TSA, a potent inhibitor of HDAC, on MMTR activity (Fig. 1B). Interestingly, TSA treatment could only partially rescue the repression activity of MMTR, from 50% up to 70%. This result suggests that transcriptional repression by MMTR is in part mediated through HDAC1-mediated chromatin remodeling and also via an HDAC-1-independent mechanism(s).
To analyze the effect of HDAC1 on the transcriptional repression activity of MMTR, we performed GAL4 fusion protein-based luciferase assays. 293T cells were cotransfected with the GAL4-MMTR and Flag-HDAC1 or dominant-negative form of HDAC1 [Flag-HDAC1(H141A)] expression vectors (17), and the relative luciferase activities of the pGL2-GAL4 reporter were examined. MMTR repressed the luciferase activity additively with HDAC1 (Fig. 2A, lane 6). When the dominant-negative form of HDAC1 [Flag-HDAC1(H141A)] was introduced, it reduced the repression activity of HDAC1 but did not completely remove the repression (Fig. 2A, compare lanes 7 and 8 to lane 2). These data indicate that repression activity of MMTR possibly involves a direct interaction with HDAC1.
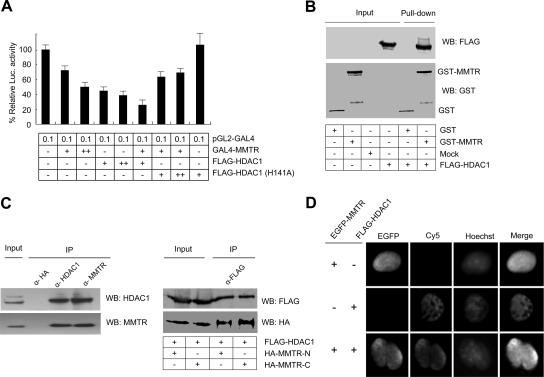
FIG. 2.
MMTR recruits HDAC1 for transcriptional repression activity. (A) pGL2-GAL4 reporter vector was transfected into 293T cells with various combinations of GAL4-MMTR, Flag-HDAC1, and Flag-HDAC1(H141A) expression vectors. Cell extracts were assayed for luciferase (Luc.) activity at 36 h posttransfection. Error bars indicate standard deviation from triplicate transfections. (B) Recombinant GST or GST-MMTR bound to glutathione-Sepharose beads was incubated with extracts from 293T cells expressing Flag-HDAC1. Bound material was subjected to SDS-PAGE, and Western blot analysis was performed using Flag antibody. (C, left panel) 293T cells were transfected with Flag-MMTR or Flag-HDAC1 expression vectors. Cell lysates were immunoprecipitated with an HDAC1 or MMTR antibody, and the precipitates were subjected to SDS-PAGE followed by Western blotting with MMTR or HDAC1 antibody. (C, right panel) 293T cells were cotransfected with HA-MMTR-N or HA-MMTR-C and Flag-HDAC1 expression vectors. Cell lysates were immunoprecipitated with Flag antibody and the precipitates were subjected to SDS-PAGE followed by Western blotting with Flag or HA antibody. (D) 293T cells were cotransfected with EGFP-MMTR and Flag-HDAC1 expression vectors. The HDAC1 proteins were visualized by indirect immunofluorescence using primary anti-rabbit HDAC1 antibody and Cy5-conjugated anti-mouse IgG. Nuclei were visualized by using Hoechst 33258.
MMTR recruits HDAC1 for transcriptional repression activity.
To determine whether HDAC1 is able to interact with MMTR in vitro, GST pull-down assays were performed. GST-MMTR fusion proteins were expressed in E. coli, purified using glutathione-Sepharose beads, and incubated with the 293T cell extracts overexpressing Flag-HDAC1. After extensive washing, the proteins that bound to glutathione-Sepharose beads were separated by SDS-PAGE and detected by Western blotting. Flag-HDAC1 was efficiently retained by GST-MMTR (Fig. 2B). We also confirmed that MMTR directly interacts with HDAC1 in vivo by using coimmunoprecipitation assays (Fig. 2C). The Flag-HDAC1 expression vector was transiently transfected into 293T cells with the Flag-MMTR expression vector. Cell extracts were immunoprecipitated with HDAC1 or MMTR antibody, respectively, and precipitates were analyzed by Western blotting. HA antibody was used as a negative control for immunoprecipitation. The result showed that HDAC1 was coprecipitated with MMTR antibody (Fig. 2C, left) and vice versa.
To analyze the domain responsible for the interaction between MMTR and HDAC1, the Flag-HDAC1 expression vector was cotransfected with the HA-MMTR-N or HA-MMTR-C expression vector. HDAC1 interacts with both the N- and C-terminal half regions of MMTR (Fig. 2C, right). Intracellular colocalization of MMTR and HDAC1 observed by immunocytochemistry further confirmed the in vivo interaction between the two proteins (Fig. 2D). 293T cells were cotransfected with enhanced green fluorescent protein (EGFP)-MMTR and Flag-HDAC1 expression vectors, and the MMTR protein was observed 36 h posttransfection using green fluorescent protein fluorescence. The HDAC1 protein was visualized by indirect immunofluorescence using primary anti-rabbit HDAC1 antibody and Cy5-conjugated anti-mouse IgG. Colocalization of EGFP-MMTR and Flag-HDAC1 in the nuclei of transfected 293T cells was observed. These data strongly suggest that MMTR and HDAC1 interact both in vitro and in vivo and that treatment with TSA or cotransfection with dominant-negative HDAC1 could not fully inhibit the repression activity of MMTR. In support of this hypothesis, we sought to identify MMTR-interacting proteins in addition to HDAC1 that contribute to the transcriptional repression activity of MMTR.
MAT1 is an interacting partner of MMTR.
To identify the postulated partner proteins for MMTR, we performed a yeast two-hybrid screen of a human brain cDNA library using MMTR cDNA as bait. MAT1, a component of the general transcription factor complex TFIIH, was identified as one of five positive clones (data not shown). To confirm the interaction of MMTR and MAT1 in vivo, coimmunoprecipitation assays were performed. When 293T cell extracts expressing HA-MMTR were immunoprecipitated with HA antibody, MAT1 was coimmunoprecipitated with HA-MMTR (Fig. 3A, upper panel). To map the regions of MMTR responsible for interaction with MAT1, the full-length MAT1 coding sequence was cotransfected with the HA-MMTR-N or HA-MMTR-C expression vector. Only the C-terminal half region of MMTR (HA-MMTR-C) was coimmunoprecipitated with Flag-MAT1 (Fig. 3A, lower panel). These results indicate that MMTR interacts with MAT1 via its C terminus (amino acids 221 to 468). Because the most highly conserved region in MMTR among species from yeast to animals is between amino acids 229 and 247 (see Fig. S2 in the supplemental material), this 19-amino-acid sequence in MMTR could be a putative binding motif for MAT1.
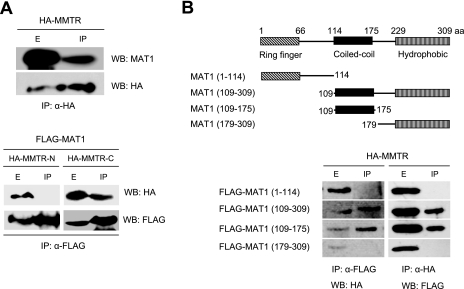
FIG. 3.
MMTR protein interacts with MAT1. (A, upper panel) 293T cells were transfected with HA-MMTR expression vectors, and the cell lysates were immunoprecipitated (IP) with HA antibody (α-HA) at 36 h posttransfection. The precipitates were subjected to SDS-PAGE, followed by Western blotting (WB) with an HA antibody or MAT1 antibody for detection of endogenous MAT1. (A, lower panel) 293T cells were cotransfected with HA-MMTR-N or HA-MMTR-C and Flag-MAT1 expression vectors. Cell lysates were immunoprecipitated with Flag antibody (α-FLAG), and the precipitates were subjected to SDS-PAGE followed by Western blotting with Flag or HA antibody. (B, upper panel) Schematic representation of MAT1 deletion mutants used in this experiment. (B, lower panel) 293T cells were cotransfected with each Flag-MAT1 deletion mutant and full-length HA-MMTR expression vectors. Cell lysates were immunoprecipitated with HA or Flag antibody, and precipitates were subjected to SDS-PAGE followed by Western blotting with Flag or HA antibody.
MAT1 is composed of three functional regions (23). The N-terminal ring finger region is required for transcription activation and RNA Pol II phosphorylation. The coiled-coil region of the middle is involved in XPD/XPB binding. The C-terminal hydrophobic region is required for cyclin-dependent kinase (cdk) activation and cdk/cyclin binding. To identify the region of MAT1 required for interaction with MMTR, Flag-MAT1 deletion mutants were transiently expressed along with HA-MMTR. Only deletion mutants of MAT1 containing the coiled-coil region [Flag-MAT1(109-308) and Flag-MAT1(109-175)] could interact with HA-MMTR (Fig. 3B). These results indicate that the coiled-coil regions of both MAT1 and MMTR are responsible for the interaction between the two proteins.
Transcriptional repression activity of MMTR is composed of two independent mechanisms involving HDAC1 and MAT1.
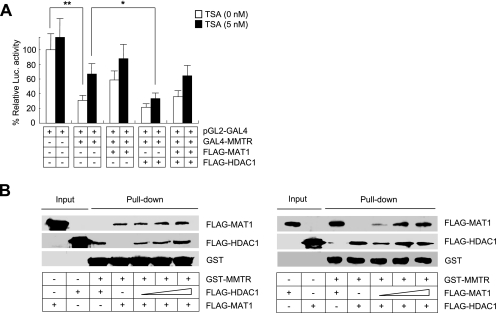
Identification of MAT1 provided an interesting link between MMTR and the general transcription machinery, TFIIH. Also, it explains the observation that TSA treatment could only partially rescue the transcriptional repression effects of MMTR (Fig. 1B). Based on the hypothesis that MMTR activity is connected with MAT1, we examined the effects of MAT1 on the transcriptional repression activity of MMTR. To achieve this goal, we tested to see if the MAT1-associated pathway can completely restore the MMTR effect. The pGL2-GAL4 luciferase vector and the GAL4-MMTR expression vector were cotransfected with various combinations of Flag-MAT1 or Flag-HDAC1 expression vectors into 293T cells in the presence or absence of TSA. The GAL4-MMTR-mediated repression activity of the reporter vector was almost rescued by overexpression of MAT1 in the presence of TSA (Fig. 4A). These results demonstrate that MMTR represses transcriptional activity by a combined mechanism that involves both HDAC1 and MAT1.
FIG. 4.
MAT1 inhibits transcriptional repression activity of MMTR. (A) pGL2-GAL4 reporter vectors were cotransfected with GAL4-MMTR and Flag-MAT1 expression vectors into 293T cells in the presence or absence of TSA. Cell lysates were assayed for luciferase (Luc.) activity at 36 h posttransfection. Results with a P value of less than or equal to 0.05 and 0.01 (* or **, respectively), which is considered statistically significant, were presented. Error bars indicate standard deviation from triplicate transfections. (B) Recombinant GST-MMTR bound to glutathione-Sepharose beads was incubated with extracts from 293T cells expressing Flag-MAT1 or Flag-HDAC1. Bound material was subjected to SDS-PAGE and subsequently analyzed by Western blotting with Flag antibody. Increasing amounts of cell lysates from 293T cells transfected with Flag-HDAC1 (left panel) or Flag-MAT1 (right panel) were used for the GST pull-down assay.
Having confirmed that MMTR associates with both HDAC1 (Fig. 2) and MAT1 (Fig. 3), we examined whether the binding of either HDAC1 or MAT1 to MMTR affects the other's binding to MMTR. As evidenced by GST pull-down assays with GST-MMTR, increasing the level of HDAC1 (left panel) or MAT1 (right panel) expression did not affect the binding of the other to MMTR (Fig. 4B). Taken together, these data indicate that the transcriptional repression activity of MMTR is composed of two independent mechanisms involving HDAC1 and MAT1.
The intact MMTR protein is required for MAT1-mediated transcriptional repression of MMTR.
Based on the hypothesis that MMTR represses CAK activity in the TFIIH complex by interaction with MAT1, we examined the effects of MAT1 on the transcriptional repression activity of MMTR. To achieve this goal, we employed a luciferase assay vector under the control of the synthetic eukaryotic promoter, CP2-Syn (20). The CP2-Syn reporter, which contains four copies of the CP2c binding elements upstream of the luciferase gene, was cotransfected into 293T cells with HA-CP2c and various combinations of the HA-MMTR, HA-MMTR-N, HA-MMTR-C, and Flag-MAT1 expression vectors. At 36 h posttransfection, the dual-luciferase assay was performed with cell extracts. Overexpression of CP2c increased the reporter activity about 2.5-fold compared to that of the control, whereas the cotransfection of MMTR clearly repressed CP2c-mediated luciferase activity in comparison with the reporter activity of the cells transfected with the CP2c expression vector alone (Fig. 5A). Interestingly, coexpression of increasing amounts of the MAT1 construct relieved this repression in a dose-dependent manner (Fig. 5A, third bar versus fourth, fifth, and sixth bars). Since the N-terminal half region of MMTR alone was sufficient to repress transactivation of the reporter but could not interact with MAT1 (Fig. 3), we tested the effect of MAT1 on repression activity of HA-MMTR-N. HA-MMTR-N repressed CP2c-mediated activation of the reporter, but MAT1 could not activate MMTR-N-mediated repression activity (Fig. 5A, 8th bar versus 9th, 10th and 11th bars). On the other hand, the C-terminal half region of MMTR (HA-MMTR-C), which could bind to MAT1 but was devoid of repression activity, did not affect CP2-mediated reporter transactivation, nor could MAT1 affect reporter activity (Fig. 5A, 13th bar versus 14th, 15th, and 16th bars). These results demonstrate that the N-terminal half region of MMTR has the transcriptional repression activity and MAT1 inhibits the MMTR-mediated transcriptional repression activity by interaction with the C-terminal half region of MMTR.
To confirm whether this MMTR-mediated transcriptional inhibition is a general mechanism maintained during important biological processes, such as differentiation and development, we assessed the effect of MMTR knock-down on transcription in a mESC line. As shown in Fig. S4 in the supplemental material, reduced expression of MMTR by antisense MMTR generally caused elevated expression of randomly chosen genes, suggesting that MMTR is implicated in a wide range of transcriptional repression. Data in Fig. 5B further confirm this finding by use of reporter assays with 293T cells. The reporter vectors used in the experiments contain promoters of cell cycle-related genes, such as cyclin E, cdc25, and Tcf genes, oncogenes (Tert, HK, and ras genes), and the ubiquitous CP2c binding elements of the mouse α-globin gene promoter (CP2-Syn). We found that the N-terminal half region of MMTR effectively decreased luciferase reporter activity in most of the promoters tested, but the C-terminal half region did not show any significant inhibitory effect (Fig. 5B), supporting the hypothesis that MMTR is involved in a wide range of transcriptional repression via its N-terminal half region. Interestingly, we have found that the N-terminal half region of MMTR could more effectively down-regulate transcription from cellular promoters than full-length MMTR. This discrepancy might be explained by a sequential binding model. Full-length MMTR can either bind to the TFIIH complex via interaction between the C-terminal half region and MAT1 in the TFIIH complex or to HDAC1 by the N-terminal half region of MMTR. The full-length MMTR molecules which happen to bind first to MAT1 via their C-terminal regions of MMTR cannot repress transcription unless their N-terminal regions bind HDAC1. Data in Fig. 5B suggest that full-length MMTR which first binds to MAT1 might have decreased affinity toward HDAC1 and the recruitment of HDAC1 to the promoter region might be a necessary requirement for initiation of transcriptional repression. In comparison, the full-length MMTR molecules which bound first to HDAC1 via their N-terminal regions can be recruited to the MAT1-containing complex. Therefore, the fraction of full-length MMTR which bound first to MAT1 cannot initiate transcriptional repression, so they would, rather, decrease overall transcriptional repression activity by full-length MMTR populations. For full understanding of this model, further investigation is warranted.
MMTR inhibits TFIIH kinase-mediated CTD phosphorylation.
Since MAT1 is a member of the general transcription factor complex TFIIH, which is involved in transcription initiation and elongation, it can be postulated that interaction of MMTR with MAT1 inhibits the transcriptional activity of TFIIH by a regulatory function of the TFIIH complex. Therefore, we tested whether MMTR could regulate the activity of RNA Pol II-dependent transcription by inhibiting the function of the MAT1-containing CAK complex. The most widely studied substrate of CAK is the CTD of the largest subunit of RNA Pol II (24). The mammalian RNA Pol II CTD consists of 52 repeats of a consensus heptapeptide, YSPTSPS, and is efficiently phosphorylated in vitro by CAK activity on the serine at position 5 (13, 42, 44).
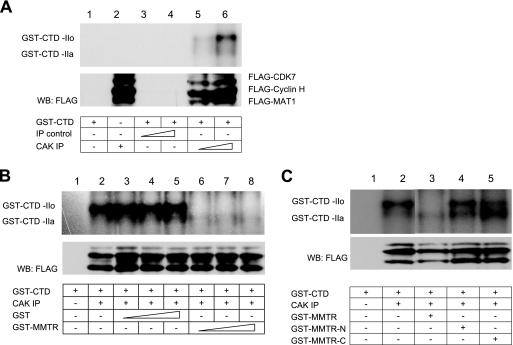
To determine whether CAK activity is modulated by MMTR, we purified GST-CTD fusion protein in E. coli and examined its phosphorylation by TFIIH kinase activity. For preparation of the active CAK complex, Flag-CDK7, Flag-cyclin H, and Flag-MAT1 expression vectors were cotransfected into 293T cells and immunoprecipitated with HA or Flag antibody. Recombinant GST-CTD was incubated with either the immunoprecipitation control (HA antibody) or CAK immunoprecipitation (Flag antibody) complex and [γ-32P]ATP in the reaction buffer. Kinase activities were analyzed by SDS-PAGE and subsequent autoradiography. Hyperphosphorylation (GST-CTD-IIo) and hypophosphorylation (GST-CTD-IIa) of GST-CTD were detected in a CAK immunoprecipitation complex dose-dependent manner (Fig. 6A). The result indicated that GST-CTD is specifically phosphorylated by the CAK immunoprecipitation complex.
FIG. 6.
MMTR inhibits CAK-mediated RNA polymerase II CTD phosphorylation. (A) 293T cells were cotransfected with Flag-CDK7, Flag-cyclin H, and Flag-MAT1 expression vectors. Cell extracts were immunoprecipitated with an HA or Flag antibody, and each precipitate was incubated with kinase reaction buffer with recombinant GST-CTD as a substrate. Phosphorylation of GST-CTD was visualized by SDS-PAGE, followed by autoradiography. (B) Kinase assays with GST-CTD were performed with increasing amounts of recombinant GST or GST-MMTR, respectively. (C) Kinase assays with CAK on GST-CTD comparing inhibition activities of MMTR N-terminal and C-terminal half regions.
To confirm the inhibitory effect of MMTR on CAK activity in vitro, various amounts of purified GST-MMTR were added to the kinase reaction. Exogenously added GST-MMTR effectively inhibited the phosphorylation of GST-CTD, while GST had no effect on the phosphorylation of GST-CTD (Fig. 6B). These results indicate that MMTR is required to down-regulate CAK activity in the TFIIH complex.
To determine the domain responsible for CAK activity, the GST-MMTR, GST-MMTR-N, or GST-MMTR-C protein was added to the kinase reaction. GST-MMTR significantly inhibited the phosphorylation of GST-CTD, but both GST-MMTR-N and GST-MMTR-C failed to show inhibitory effects (Fig. 6C). These results indicate that intact MMTR is required to down-regulate CAK activity in the TFIIH complex.
DISCUSSION
In this study, we provide the first direct evidence that MMTR is a potent corepressor functioning as a component of the basic cellular machinery that regulates gene expression in general. Since MMTR has no recognizable DNA-binding domain or DNA binding protein interaction domain (see Fig. S1 in the supplemental material), we propose here that MMTR functions as a corepressor. Corepressors harbor an intrinsic transcription-silencing ability and actively repress transcription but do not bind DNA directly. They are recruited by transcription factors bound to the regulatory regions of target genes and contribute to the silencing ability of transcriptional silencers or repress the transcriptional activity of gene activators (19, 33). Indeed, MMTR did not bind to the promoters of a set of genes containing binding sites for several transcription factors and also did not bind to the specific transcription factor CP2c protein (data not shown). The elucidation of an intranuclear MMTR-interacting transcription factor warrants further investigation. Similarly, an atypical orphan nuclear receptor, small heterodimer partner, lacks a conventional DNA binding domain and represses the transcriptional activity of various nuclear receptors (21). BCL-6-interacting corepressor (BCoR) is also known to function as a corepressor (16). The corepressor function of BCoR is also modulated by interaction with specific class I and II HDACs. Therefore, MMTR, like small heterodimer partner and BCoR, functions as a corepressor without direct binding to DNA elements.
Although general transcription factors and RNA polymerase allow only minimal transcriptional control, cells exert fine control over transcription via specific DNA binding transcription factors performing tissue- or stage-specific gene expression (40). One well-known mechanism by which several coregulators function involves attachment or removal of the acetyl group from N-terminal tails of histones through recruitment of histone acetyltransferases or HDACs, respectively (32, 47). HDACs exist in a complex with transcriptional repressor proteins and are implicated in transcriptional repression resulting in nucleosome remodeling (1, 30) or inactivation of transcription factors (18). Because the interaction of MMTR with HDAC1 was confirmed by in vivo and in vitro binding assays (Fig. 2), MMTR may act as a mediator that recruits the corepressor HDAC1 or HDAC1-containing corepressor complex to the basal transcription machinery. Similarly, it is also suggested that repression of transcription by the orphan nuclear receptor RVR/Rev-erbb and the corepressor N-CoR probably involves intimate contacts with the general transcriptional machinery (TFIIB) and the proteins (Sin3 and HDAC-1) which are involved in nucleosomal condensation (29). Nevertheless, neither the HDAC inhibitor TSA nor cotransfection with the dominant-negative form of HDAC1 could fully rescue the repression activity of MMTR in our experiments (Fig. 1 and 2). These results suggest that the transcription repression activity of MMTR is mediated by both HDAC1-dependent and -independent mechanisms. Similarly, LCoR (ligand-dependent corepressor) is a transcriptional corepressor with HDAC-dependent and -independent modes of action (10).
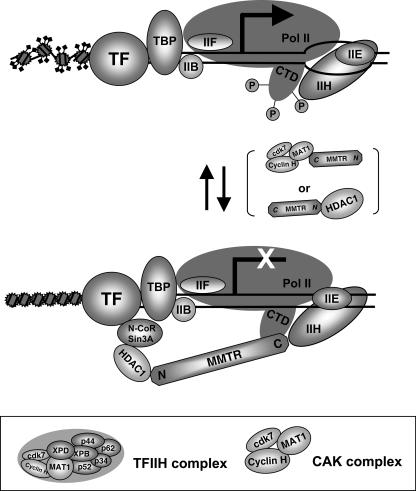
What other mechanisms might be involved in MMTR-mediated transcriptional repression? A substantial body of evidence indicates that nuclear hormone receptors and corepressors can interfere with the formation of a transcriptional preinitiation complex in vitro, apparently through direct inhibitory contacts with components of the general transcriptional machinery (11, 12). In this paper, we first report that the MMTR protein is capable of interaction with the general transcription factor TFIIH through binding to MAT1. MAT1 is a component of the CAK complex (cdk7, cyclin H, and MAT1), which is a subcomplex of TFIIH (35). TFIIH performs critical roles at initiation and postinitiation stages of transcription and in responses to DNA damage (43). Formation of an open promoter complex by RNA polymerase II appears to be mediated by TFIIH DNA helicase activity (8, 15). TFIIH also regulates the transition from transcription initiation to elongation, presumably mediated by phosphorylation of the C-terminal domain of RNA polymerase II by the CAK complex (4, 31). Recently, TFIIH was shown to promote the transition from very early elongation complexes to stable elongation complexes (7). In our results, MAT1 rescued reporter transcription activity which was decreased by MMTR overexpression (Fig. 4 and 5). Furthermore, MMTR was required for inhibition of phosphorylation of CTD in the RNA Pol II largest subunit by TFIIH kinase in vitro (Fig. 6). Therefore, we suggest that MMTR could also affect the functional activity of TFIIH via interaction with MAT1. As shown in Fig. 5B, initiation of transcriptional repression by MMTR first requires recruitment of HDAC1 via its N terminus. The C-terminal half region of MMTR cannot initiate transcriptional repression, though it can still bind MAT1 in transient-transfection assays. Once bound to HDAC1 via its N terminus, MMTR can then efficiently inhibit CTD phosphorylation. Data shown in Fig. 6C are also in accordance with this sequential binding model. To initiate full transcriptional repression activity, the intact MMTR molecule first binds HDAC1 via its N terminus and then is recruited to the TFIIH complex to inhibit CTD phosphorylation. Devoid of a functional N terminus which can bind HDAC1 to initiate transcriptional repression, the C-terminal half of MMTR cannot be efficiently recruited to the TFIIH complex to inhibit CTD phosphorylation. MAT1 also interacts with the XPB and XPD helicase subunits of TFIIH, which mediate the association of CAK with the core TFIIH (37, 39). When MMTR interacts with MAT1, a component of the CAK complex in TFIIH, the CTD of RNA Pol II is dephosphorylated and concomitantly HDAC1 is recruited to the basal transcription machinery. Both events may result in interference with the formation of the transcription initiation complex (Fig. 7).
FIG. 7.
Model of MMTR-mediated transcriptional repression. Proposed model of MMTR inhibition of transcription initiation complex formation by recruiting HDAC1 and/or dephosphorylating CTD via its interaction with MAT1, a component of the CAK complex (cdk7, cyclin H, and MAT1), which is a subcomplex of the general transcription machinery, TFIIH. In the model, initiation of transcriptional repression by MMTR first requires recruitment of HADC1 via its N terminus. The C-terminal half region of MMTR cannot initiate transcriptional repression, though it can still bind MAT1. Once bound to HDAC1 via its N terminus, MMTR can then efficiently inhibit CTD phosphorylation.
Since MMTR itself has transcriptional repression activity, MMTR may directly inhibit the activity of RNA Pol II-dependent transcription when it is recruited to the initiation complex via MAT1 of TFIIH. In addition, MMTR may serve as a mediator for a variety of factors regulating gene transcription. It has been recently reported that MMTR (DMAP1) facilitated the nuclear localization of RMP and the corepressor activity of RMP in a dose-dependent manner by interacting with the coiled-coil domain of RMP (5). RMP was reported to regulate transcription by competing with HBx to bind TFIIB and interacting with the RPB5 subunit of RNA Pol II as a corepressor of transcription regulators (24). RMP also regulates transcription through interaction with general transcription factors IIF, which assemble in the preinitiation complex and function in both transcription initiation and elongation (45). Inversely, it was also reported that MMTR transcriptional repressor activity was inhibited by RGS6 (25). RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity (25). Thus, our data strongly suggest that MMTR is part of the basic cellular machinery for a wide range of transcriptional regulation via interaction with TFIIH and HDAC.
Supplementary Material
Acknowledgments
We thank Sung Hee Baek (Seoul National University, Seoul, South Korea), Robert P. Fisher (Memorial Sloan-Kettering Cancer Center, New York, NY), Edward Seto (University of South Florida), Yang Shi (Harvard Medical School, Harvard, MA), Didier Trono (University of Geneva, Geneva, Switzerland), and Jongbok Yoon (Yonsei University, Seoul, South Korea) for their gift of plasmids or reagents. We also thank Yoon Shin Cho (Hanyang University, Seoul, South Korea), Sung-Hyun Kim (Hanyang University, Seoul, South Korea), Jianxin Bao (Washington University in St. Louis, School of Medicine), and Brian Watson (University of Washington) for their critical reading and comments on the manuscript.
This work was supported by the research fund of Hanyang University (HY-2004) and a grant (no. SC2010) from the Stem Cell Research Center of the 21st Century Frontier Research Program, funded by the Ministry of Science and Technology, Republic of Korea.
Footnotes
Published ahead of print on 19 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Chae, J. H., Y. H. Lee, and C. G. Kim. 1999. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem. Biophys. Res. Commun. 263:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Cripps, R. M., and E. N. Olson. 2002. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246:14-28. [DOI] [PubMed] [Google Scholar]
- 4.Dahmus, M. E. 1994. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 48:143-179. [DOI] [PubMed] [Google Scholar]
- 5.Delgermaa, L., N. Hayashi, D. Dorjsuren, T. Nomura, T. T. Thuy le, and S. Murakami. 2004. Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol. Cell. Biol. 24:8556-8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvir, A., R. C. Conaway, and J. W. Conaway. 1997. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl. Acad. Sci. USA 94:9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvir, A., K. P. Garrett, C. Chalut, J. M. Egly, J. W. Conaway, and R. C. Conaway. 1996. A role for ATP and TFIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J. Biol. Chem. 271:7245-7248. [DOI] [PubMed] [Google Scholar]
- 9.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, I., Y. Bastien, T. Wai, K. Nygard, R. Lin, O. Cormier, H. S. Lee, F. Eng, N. R. Bertos, N. Pelletier, S. Mader, V. K. Han, X. J. Yang, and J. H. White. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11:139-150. [DOI] [PubMed] [Google Scholar]
- 11.Fondell, J. D., F. Brunel, K. Hisatake, and R. G. Roeder. 1996. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol. Cell. Biol. 16:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fondell, J. D., A. L. Roy, and R. G. Roeder. 1993. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 7:1400-1410. [DOI] [PubMed] [Google Scholar]
- 13.Gebara, M. M., M. H. Sayre, and J. L. Corden. 1997. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J. Cell Biochem. 64:390-402. [PubMed] [Google Scholar]
- 14.Hassig, C. A., J. K. Tong, T. C. Fleischer, T. Owa, P. G. Grable, D. E. Ayer, and S. L. Schreiber. 1998. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 95:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holstege, F. C., P. C. van der Vliet, and H. T. Timmers. 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15:1666-1677. [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh, K. D., W. Fischle, E. Verdin, and V. J. Bardwell. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14:1810-1823. [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 19.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, H. C., J. H. Chae, Y. H. Lee, M. A. Park, J. H. Shin, S. H. Kim, S. K. Ye, Y. S. Cho, S. Fiering, and C. G. Kim. 2005. Erythroid cell-specific alpha-globin gene regulation by the CP2 transcription factor family. Mol. Cell. Biol. 25:6005-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. Y., K. Chu, H. J. Kim, H. A. Seong, K. C. Park, S. Sanyal, J. Takeda, H. Ha, M. Shong, M. J. Tsai, and H. S. Choi. 2004. Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix-loop-helix transcription factor BETA2/neuroD. Mol. Endocrinol. 18:776-790. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. J., J. H. Shin, J. Kim, S. H. Kim, J. H. Chae, E. J. Park, R. H. Seong, S. H. Hong, S. D. Park, S. Jeong, and C. G. Kim. 1999. Isolation of developmentally regulated novel genes based on sequence identity and gene expression pattern. Mol. Cells 9:207-218. [PubMed] [Google Scholar]
- 23.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Y., T. Nomura, J. Cheong, D. Dorjsuren, K. Iida, and S. Murakami. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 272:7132-7139. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Z., and R. A. Fisher. 2004. RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J. Biol. Chem. 279:14120-14128. [DOI] [PubMed] [Google Scholar]
- 26.Mannervik, M., Y. Nibu, H. Zhang, and M. Levine. 1999. Transcriptional coregulators in development. Science 284:606-609. [DOI] [PubMed] [Google Scholar]
- 27.Martin, G. R. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78:7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muromoto, R., K. Sugiyama, A. Takachi, S. Imoto, N. Sato, T. Yamamoto, K. Oritani, K. Shimoda, and T. Matsuda. 2004. Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J. Immunol. 172:2985-2993. [DOI] [PubMed] [Google Scholar]
- 29.Muscat, G. E., L. J. Burke, and M. Downes. 1998. The corepressor N-CoR and its variants RIP13a and RIP13Delta1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70. Nucleic Acids Res. 26:2899-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng, H. H., P. Jeppesen, and A. Bird. 2000. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 20:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien, T., S. Hardin, A. Greenleaf, and J. T. Lis. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75-77. [DOI] [PubMed] [Google Scholar]
- 32.Ordentlich, P., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 33.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 34.Prelle, K., N. Zink, and E. Wolf. 2002. Pluripotent stem cells—model of embryonic development, tool for gene targeting, and basis of cell therapy. Anat. Histol. Embryol. 31:169-186. [DOI] [PubMed] [Google Scholar]
- 35.Reardon, J. T., H. Ge, E. Gibbs, A. Sancar, J. Hurwitz, and Z. Q. Pan. 1996. Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH. Proc. Natl. Acad. Sci. USA 93:6482-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodda, S., S. Sharma, M. Scherer, G. Chapman, and P. Rathjen. 2001. CRTR-1, a developmentally regulated transcriptional repressor related to the CP2 family of transcription factors. J. Biol. Chem. 276:3324-3332. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol, M., I. Kolb-Cheynel, and J. M. Egly. 1997. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 16:1628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 39.Sandrock, B., and J. M. Egly. 2001. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J. Biol. Chem. 276:35328-35333. [DOI] [PubMed] [Google Scholar]
- 40.Sekimata, M., and Y. Homma. 2004. Regulation of Rb gene expression by an MBD2-interacting zinc finger protein MIZF during myogenic differentiation. Biochem. Biophys. Res. Commun. 325:653-659. [DOI] [PubMed] [Google Scholar]
- 41.Shin, J. H., J. K. Yi, Y. J. Lee, A. L. Kim, M. A. Park, S. H. Kim, H. Lee, and C. G. Kim. 2003. Development of artificial chimerical gene regulatory elements specific for cancer gene therapy. Oncol. Rep. 10:2063-2069. [PubMed] [Google Scholar]
- 42.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 43.Takagi, Y., C. A. Masuda, W. H. Chang, H. Komori, D. Wang, T. Hunter, C. A. Joazeiro, and R. D. Kornberg. 2005. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol. Cell 18:237-243. [DOI] [PubMed] [Google Scholar]
- 44.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 273:6769-6775. [DOI] [PubMed] [Google Scholar]
- 45.Wei, W., J. X. Gu, C. Q. Zhu, F. Y. Sun, D. Dorjsuren, Y. Lin, and S. Murakami. 2003. Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP). Cell Res. 13:111-120. [DOI] [PubMed] [Google Scholar]
- 46.Wigler, M., A. Pellicer, S. Silverstein, and R. Axel. 1978. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell 14:725-731. [DOI] [PubMed] [Google Scholar]
- 47.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.