Abstract
The gonadotropins luteinizing hormone (LH) and follicle-stimulating hormone (FSH) are produced in the embryonic pituitary in response to delivery of the hypothalamic gonadotropin releasing hormone (GnRH). GnRH has a pivotal role in reestablishing gonadotropin levels at puberty in primates, and for many species with extended reproductive cycles, these are reinitiated in response to central nervous system-induced GnRH release. Thus, a clear role is evident for GnRH in overcoming repression of these genes. Although the mechanisms through which GnRH actively stimulates LH and FSH β-subunit (FSHβ) gene transcription have been described in some detail, there is currently no information on how GnRH overcomes repression in order to terminate reproductively inactive stages. We show here that GnRH overcomes histone deacetylase (HDAC)-mediated repression of the gonadotropin β-subunit genes in immature gonadotropes. The repressive factors associated with each of these genes comprise distinct sets of HDACs and corepressors which allow for differentially regulated derepression of these two genes, produced in the same cell by the same regulatory hormone. We find that GnRH activation of calcium/calmodulin-dependent protein kinase I (CaMKI) plays a crucial role in the derepression of the FSHβ gene involving phosphorylation of several class IIa HDACs associated with both the FSHβ and Nur77 genes, and we propose a model for the mechanisms involved. In contrast, derepression of the LH β-subunit gene is not CaMK dependent. This demonstration of HDAC-mediated repression of these genes could explain the temporal shut-down of reproductive function at certain periods of the life cycle, which can easily be reversed by the actions of the hypothalamic regulatory hormone.
In the mouse embryo, the pituitary gonadotropes become fully differentiated between embryonic day (E)11.5 when the common α-subunit is first expressed, and E16.5 when the luteinizing hormone β-subunit (LHβ) gene is first detected, while expression of the follicle-stimulating hormone β-subunit (FSHβ) gene appears on the following day. The expression of these genes is facilitated by unique cell-specific groups of transcription factors which are expressed in response to local signals, and most are present by E9 to 13.5 (6, 23, 50). Expression of these transcription factors (e.g., SF-1, Egr-1, and Pitx-1 for LH and SF-1, AP-1, Lhx3, Ptx1, and Ptx2 for FSH) has been shown to be sufficient to induce activity of the transiently transfected LHβ and FSHβ promoters in reporter gene assays in heterologous cells (9, 20, 27, 41, 55, 56, 61, 67). However, LHβ and FSHβ gene expression in the developing gonadotrope appears only following the migration of the gonadotropin releasing hormone (GnRH) neurons in the hypothalamus and GnRH delivery to the pituitary, which starts around E16. This indicates a role for GnRH in initiating gonadotropin gene expression, which might be distinct from its stimulating increases in gonadotropin gene expression in mature gonadotropes, in which the gonadotropin genes are already expressed at a basal level.
Gonadotropin expression continues to increase in response to GnRH during gestation (18, 43). After birth, however, the levels of LH and FSH vary in different species: in mice, levels of LH and FSH increase to day 12, corresponding with increases in GnRH levels and its pulsatile secretion as the hypothalamus matures during the first 2 weeks postparturition (43). In primates, gonadotropin synthesis is repressed soon after birth as a result of insufficient release of GnRH from the hypothalamus, and it is reversed at puberty when GnRH synthesis and release are restored (18, 47). Subsequently, many mammals and nonmammalian species have reproductive seasons or extended cycles, which are reinitiated in response to central nervous system-induced GnRH synthesis and release. Thus, a clear role is evident for GnRH in not only stimulating gonadotropin gene expression above basal levels but also overcoming a repressed state of inactivity of these genes. Although the mechanisms through which GnRH actively stimulates LHβ and FSHβ gene transcription have been described in some detail, there is currently no information on how GnRH overcomes repression in order to terminate reproductively inactive stages.
Reversible suppression of gene expression is often achieved through the actions of DNA-associated factors, such as nuclear receptors, some of which recruit corepressors and chromatin-modifying enzymes. These repressors act collectively to block the binding sites of activators, to prevent interaction with the general transcription machinery, and/or to compact the chromatin, making it less accessible to DNA-binding activators. The best-studied chromatin modification leading to chromatin compaction is histone deacetylation, in which histone deacetylases (HDACs) remove acetyl groups from the lysine residues of core histone N-terminal tails through a reaction that is easily reversed by histone acetyltransferases. The absence of acetyl groups on these lysines restores the positive charge to the histones and thus strengthens their interaction with the DNA, increasing chromatin compaction and making the chromatin less accessible to the activators. The modification of these lysines may also provide specific binding surfaces for the recruitment of additional proteins (30, 54).
Mammalian HDACs are grouped into three classes based on their sequence similarity to Saccharomyces cerevisiae histone deacetylases. Class I includes HDACs 1, 2, 3, 8, and 11 and class II includes HDACs 4, 5, 6, 7, 9, and 10, while the third class of HDACs is the conserved NAD-dependent Sir2 family of deacetylases. Although the class I HDACs are mostly nuclear, class II HDACs can be found in the nucleus and/or cytoplasm, with the subclass IIa HDACs actively shuttling between the two cellular localizations. Notably, HDAC6 has a rather different function as it is primarily found in the cytoplasm, where it deacetylates α-tubulin, thereby regulating microtubule-dependent cell motility (8, 29, 57, 64).
In comparison to the class I HDACs, class II HDACs are much larger proteins and contain several domains for interacting with diverse proteins, such as other HDACs, transcription factors, and corepressors. HDACs are usually found associated with the chromatin in multiprotein complexes together with corepressors that lack their own enzymatic activity but help recruit the HDACs and/or stabilize the complex (13, 24). Two such corepressors, the nuclear receptor corepressor (NCoR) and silencing mediator of retinoic and thyroid hormone receptors (SMRT), were first described as corepressors for the unliganded thyroid hormone and retinoic acid receptors but also serve as common corepressors for other DNA-bound factors (13). NCoR contains three distinct repression domains in its N terminus which allow interaction with several HDACs and also with another common corepressor, mSin3A. Sin3 is a large multidomain protein that can form a scaffold upon which the rest of the complex assembles. It also interacts directly with HDACs 1 and 2, several class IIa HDACs, and various DNA-bound factors (13, 17).
Regulation of the activity of repressive complexes may be directed at any or several of its components. In the case of nuclear receptors, the binding of a ligand induces conformational change of the receptor so that its interaction with the HDAC-containing repressive complex is lost, and histone acetyltransferases are recruited instead (13, 26, 48). The activity of HDACs can also be regulated: for example, phosphorylation of several class IIa HDACs by calcium/calmodulin-dependent protein kinases (CaMKs) or possibly protein kinase D allows their interaction with 14-3-3 proteins, leading to their subsequent nuclear export (16, 22, 32, 37, 39, 44, 58). In contrast, extracellular signal-regulated kinase 1 and 2 was reported to phosphorylate HDAC4, leading to its nuclear accumulation, while the activity of HDAC3 is increased following its interaction with protein phosphatase 4 (68, 69).
In an attempt to explore the ways in which gonadotropin gene expression is repressed, we revealed initially that the expression of both LHβ and FSHβ genes is repressed in the αT3-1 murine gonadotrope cells by HDAC activity and that GnRH is able to overcome this repression. These cells were produced by targeted oncogenesis and are thought to represent a partially differentiated gonadotrope around E11.5; they do not express the gonadotropin β-subunit genes, although the transcription factors known to activate them are present (1, 14, 46). We hypothesized that to derepress these genes by disrupting the HDAC-containing repressive complexes, GnRH must employ a distinct mechanism that likely differs from the activation of LHβ and FSHβ gene transcription above basal activity. The aim of this study was thus to characterize these repressive complexes and to determine the mechanisms through which GnRH is able to overcome their effects.
MATERIALS AND METHODS
Cell culture and transfections.
The immature murine gonadotrope αT3-1 cells were cultured in minimum essential medium, supplemented with 10% fetal calf serum, 10 mM HEPES, 0.1 mM minimum essential medium nonessential amino acids, 1 mM sodium pyruvate solution, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco-Invitrogen). During the course of the study, at least three different batches of cells from different sources were used in order to confirm the consistency of the results. The more mature gonadotrope LβT2 cells, which express both subunit genes (LHβ quite abundantly and FSHβ at a much lower level) and represent gonadotropes at approximately E16.5, were cultured in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose with 10% certified fetal calf serum, 10 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco-Invitrogen). Cells were cultured in six-well plates unless otherwise stated. All cells were maintained at 37°C in 5% CO2. Transfections in both cell types were carried out at 50 to 60% confluence as described previously (40) using FuGene6 (Roche Diagnostics, Basel, Switzerland) or according to the method of Luo et al. (35) using GenePORTER 2 (Gene Therapy Systems, San Diego, CA) transfection reagents. As appropriate, cells were treated with GnRH (d-Ala6-LH-releasing hormone [LHRH], where indicated; otherwise, 10 to 100 nM buserelin for 1 to 24 h) and/or trichostatin A (TSA; 20 to 200 ng/ml for 24 h, added at the time of GnRH), cyclosporine A (20 μM, added 1 h before the GnRH), or KN-93 (10 μM, added 30 min before the GnRH) (all from Sigma-Aldrich, St. Louis, MO); all were added at 0.1% of the culture volume.
Reporter gene assays were carried out using 600 bp of the proximal murine FSHβ gene promoter fused to the firefly luciferase gene as described previously (41). Firefly luciferase values were normalized to those of Renilla luciferase which was cotransfected as an internal control. Experiments were carried out on at least three separate occasions, and representative results are shown.
Plasmid constructs.
For construction of small interfering RNA (siRNA)-encoding plasmids, the 19-nucleotide gene-specific sequences targeting each gene, which were repeated in reverse orientation with a 9-nucleotide spacer and with BglII- and HindII-compatible overhangs (according to instructions of Oligoengine, Seattle, WA) were as follows (corresponding NCBI accession numbers are given in parentheses): HDAC1 (NM_008228), CCAGAACACTAACGAGTAC; HDAC 2 (NM_008229), GAAGACCCGGACAAAAGAA; HDAC3 (NM[010411]), TGCTGAACCATGCACCCAG; HDAC4 (taken from XM_358028, in which the boldface C corresponds to a T in later entries e.g., NM_207225), ATGCGAGTGCATCCGCGGA; HDAC5 (NM_010412), GCAAGCATTCTACAACGAT; HDAC6 (AF006603), CCAGGACGATCTCCAAGAT; HDAC7 (AF207749), TTCAACTTTAGGCCCTCGG; HDAC8 (BC061257), GGAGGCTATAACCTTGCCA; HDAC9 (NM024124), CCACACATCACTGGATCAA; HDAC10 (NM_199198), CTTCTCCACTCCACTGCCA; HDAC11 (BC016208), TCATTGATCTCGATGCCCA; mSin3A (NM_011378), GTACTGGTGCAACTGGTGG; NCoR, AGGAAGAGTGTTCCTGATT; SMRT, TGACTACATCACCTCGCAG (NCoR and SMRT targets were taken from Zakaria et al. [67]). The siRNA construct targeting SMRT was also cotransfected with an equal amount of an additional construct containing the sequence CCCATAGAATCAAAGCACC. All siRNA oligonucleotides were checked by a BLAST search against the mouse genome to minimize any possibility that they might cross-react with other transcripts and to confirm that there is no sequence similarity in the sequences chosen to make these constructs with any of the other HDACs.
After constructs were annealed to their complementary sequences, the double-stranded DNA fragments were inserted into the BglII/HindIII-digested pSUPER vector (Oligoengine, Seattle, WA). All constructs were verified by sequencing, and their successful knockdown of target proteins was verified by Western analysis. In controls, the empty pSUPER vector was transfected, while a similar construct targeting green fluorescent protein (GFP) (35) was transfected to confirm that there was no nonspecific effect on the individual HDAC expression levels. All constructs were transfected at 2 to 4 μg of DNA per six-well plate.
Expression vectors for mouse Nur77, MEF2A, and MEF2D were created by inserting the coding sequences, amplified from LβT2 cell cDNA, into the pCS2 plasmid vector. The dominant negative Nur77 (dnNur77) was created by amplifying Nur77 without its activation domain (a deletion of the N terminus upstream of residue 152) as previously described (4). The expression vector for constitutively active calcineurin was created by amplification of its catalytic subunit A without the calmodulin-binding and autoinhibitory domains. The S354A and S354E mutant Nur77 constructs were a gift from Lester Lau (University of Illinois at Chicago). The pEGFP-HDAC4 (where EGFP is enhanced GFP), pEGFP-HDAC5, and pEGFP-HDAC6 constructs and the pEGFP-HDAC4(S246A S467A S632A) construct were a gift from Xiang-Jiao Yang (McGill University, Canada). The pEGFP-HDAC5(S259A S498A) construct was a gift from Eric Olson (University of Texas Southwestern Medical Center). The pYFP-HDAC7 (where YFP is yellow fluorescent protein) construct was a gift from Hung-Ying Kao (Case Western Reserve University). The dominant negative CaMKI (dnCaMKI) construct was a gift from Tom Soderling (Oregon Health Science University). The expression vectors were transfected at 1 to 1.5 μg per six-well plate.
RNA extraction, reverse transcriptase PCR, and real-time PCR.
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA), and the total RNA (5 μg) was reverse transcribed using either Superscript II (Invitrogen) or Moloney murine leukemia virus (Promega, Madison, WI) reverse transcriptase and oligo(dT) primers (5 mM; New England Biolabs, Beverly, MA). PCR amplification of LHβ or FSHβ transcripts was carried out using various sets of primers, some of which spanned introns, as indicated in the figure legends. Nur77 and calcineurin catalytic subunits were amplified using primers targeting 380 bp of the Nur77 coding sequence or the full-length 1.5-kb calcineurin catalytic subunit, respectively. Control reactions were carried out in which the reverse transcriptase was omitted to ensure that the amplicon did not result from genomic DNA contamination. Internal controls comprised amplification of the mouse 60s ribosomal protein (60sRP) or β-actin. Quantitative real-time PCR was carried out essentially as described previously (35) using primers to produce 120-bp amplicons from the β-subunit cDNAs and a 100-bp amplicon for 60sRP as an internal control. The comparative cycle threshold method was used to compare mRNA levels in the various samples to those in untreated samples after their normalization to the internal control. All samples were assayed in duplicate.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (35). Briefly, proteins and DNA were cross-linked using 1.5% formaldehyde for 15 min at 37°C and arrested using 1 M glycine. The cells were washed twice with phosphate-buffered saline (PBS) before being collected in 1 ml of PBS with protease inhibitors. The cells were then pelleted before resuspension in lysis buffer and were incubated on ice for 15 min before sonication to yield genomic DNA fragments between 500 bp and 1 kb. Cell debris was then precipitated (13,000 rpm for 10 min at 4°C), and the supernatant was transferred to fresh tubes. A 20-μl aliquot was removed as input, and the remainder was diluted and precleared with protein A-Sepharose beads before overnight incubation with primary antisera targeting the following: HDACs 1, 3, 4, 5, 6, and 7 (Cell Signaling Technology); HDACs 2 and 11 (Abcam); HDAC9 (Southwestern Medical Center, Center for Biomedical Inventions); HDAC10 (Novus Biologicals); mSin3A (Upstate Biotechnology); and HDAC8, SMRT, Nur77, pNur77, and MEF2 (Santa Cruz Biotechnology). For the controls, antiserum was omitted or replaced with normal rabbit serum. The antibody-bound complexes were captured with protein A-Sepharose beads and washed in a series of increasing salt concentrations. The complexes were then eluted, and the cross-links were reversed. Proteinase K treatment was carried out before phenol-chloroform extraction of the DNA. Regions of the proximal promoters of the LHβ or FSHβ genes were amplified from input and ChIP samples, using the following primers: 5′-GTGAAGCCCACCCACCACGC-3′ and 5′-CCTTGGGCACCTGGCTTTAT-3′ to amplify the LHβ proximal promoter from 579 to 13 bp upstream of the transcriptional start site; 5′-CACAGCCCATAGGAACAAGA-3′ and 5′-CCAAAGCAGTCTAAATGCCA-3′ to amplify the FSHβ proximal promoter from 436 to 137 bp from the transcriptional start site. All ChIP experiments were carried out on at least three separate occasions, and representative results are shown.
Western blot analysis.
Western blot analysis was carried out as previously described (35) using the same antisera as for ChIP analyses or antisera targeting the following: HDAC9 (Abcam), pCaMKI, pCaMKII, pCaMKIV or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology); or pHDAC4 (S632) and pHDAC5 (S498) (Signal Antibody Technology, San Francisco, CA).
Fluorescence microscopy.
The αT3-1 cells (1 × 105) were plated on 22-mm ethanol-washed grade 1 coverslips in six-well plates and transfected 24 h later with 500 ng of DNA. After 24 to 48 h, GnRH (10 nM) was added 2 to 8 h prior to fixing of both treated and untreated cells. The medium was aspirated, and cells were washed once with Hanks balanced salt solution before being fixed for 15 min at room temperature with 4% paraformaldehyde (Sigma). Thereafter, cells were washed twice with PBS and subsequently stained at 37°C with DAPI (4′,6′-diamidino-2-phenylindole) for 15 min. The cells were washed once more with PBS and rinsed with distilled water before the coverslips were mounted on glass slides using an antifading solution (PBS containing 15% polyvinyl alcohol, 33% glycerol, and 0.1% sodium azide). The preparations were viewed under an Olympus IX81 confocal microscope (FV500 confocal system). The localization of the HDACs was noted in an average of 60 cells for each construct transfected at every time point, and representative pictures are shown.
RESULTS
Gonadotropin β-subunit genes are repressed by HDACs in immature gonadotropes, and GnRH can overcome this repression.
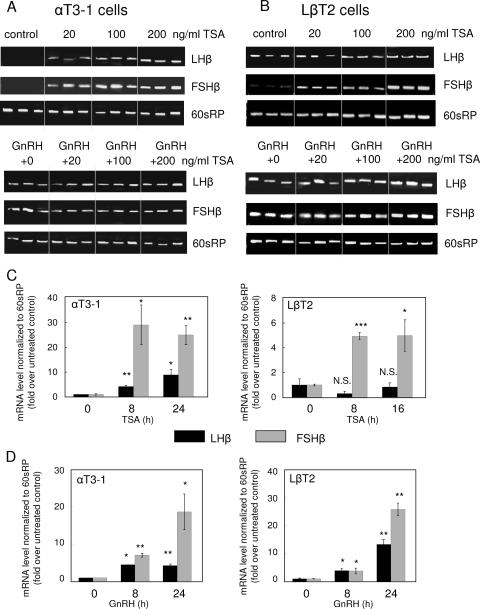
Immature gonadotrope αT3-1 cells, which do not normally express either the LHβ or the FSHβ gene, were exposed to the nonspecific inhibitor of class I and class II HDACs, TSA (at 0, 20 or 200 ng/ml), and/or to GnRH (100 nM d-Ala6-LHRH) for 24 h. Reverse transcription-PCR (RT-PCR) over 25 cycles failed to detect either transcript in untreated cells, while either TSA or GnRH was sufficient to facilitate expression of both genes, with TSA having an apparent dose-related effect. The combined treatment did not increase levels beyond those of the GnRH treatment alone (Fig. 1A).
FIG. 1.
LHβ and FSHβ gene expression is repressed in immature gonadotropes by HDACs, and this is overcome by GnRH. RT-PCR analysis of LHβ and FSHβ mRNA levels was carried out in immature αT3-1 (A) and mature LβT2 (B) gonadotrope cells following a 24-h exposure to TSA (20, 100, or 200 ng/ml) and/or GnRH (d-Ala6-LHRH; 100 nM) in three repeats for each treatment. Total RNA was isolated from the cells, and cDNA was synthesized and used as template for PCR. Control reactions were carried out in which the reverse transcriptase was omitted to ensure that the amplicon did not result from genomic DNA contaminations (not shown). The primers were designed to amplify fragments of the LHβ and FSHβ cDNAs, while amplification of a fragment of the mouse 60sRP cDNA is shown as a control. Total RNA was extracted from αT3-1 or LβT2 cells treated with TSA (C) or GnRH (D), and quantitative real-time PCR was performed. Changes in cycle threshold values were normalized to those of 60sRP and compared to the levels in untreated controls. Values are means ± standard errors of the means (n = 3). *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
The mature gonadotrope LβT2 cells express the LHβ quite abundantly but only very low levels of FSHβ. TSA treatment in these cells did not alter the LHβ transcript levels but increased those of FSHβ in a dose-related manner. GnRH treatment also led to an increase in the FSHβ mRNA levels while the combined treatment failed to increase these levels any further (Fig. 1B).
These findings were confirmed using quantitative real-time PCR, which showed an increase in both transcript levels in αT3-1 cells following 8 or 24 h of treatment with TSA (100 ng/ml), but only the FSHβ transcript responded to TSA in the LβT2 cells (Fig. 1C). Both transcripts were elevated in both cell lines following 8 or 24 h of GnRH treatment (Fig. 1D). This indicates that both genes are repressed by HDACs in the αT3-1 cells but only the FSHβ gene is repressed by HDACs in the LβT2 cells and that GnRH is able to overcome this repression.
Distinct sets of HDACs repress the LHβ and FSHβ genes.
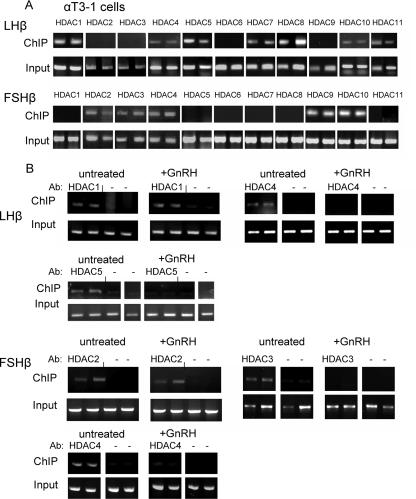
In order to understand more about the mechanism of repression of these genes in the immature cells, we carried out ChIP analysis to identify the HDACs that are associated with each of the β-subunit gene promoters. HDACs 1, 4, 5, 7, 8, 10, and 11 were found associated with the LHβ gene promoter, whereas HDACs 2, 3, 4, 9, and 10 were found associated with the FSHβ promoter (Fig. 2A). These experiments were repeated using antisera against the class I and some of the class II HDACs that associate with these genes, following GnRH treatment for 6 h, to see whether they were released. This time frame was chosen because the derepression of the LHβ and FSHβ gene expression was usually apparent following an 8-h exposure to GnRH. This revealed dissociation of HDACs 3, 4, and 5 from the LHβ and/or FSHβ promoters following the GnRH treatment, while HDACs 1 or 2 were still associated (Fig. 2B).
FIG. 2.
Distinct sets of HDACs are associated with LHβ and FSHβ genes in the immature gonadotropes. (A) ChIP analysis was carried out in duplicate to determine which HDACs are associated with the LHβ and FSHβ gene promoters in immature αT3-1 cells; an aliquot from the same cells before precipitation was designated as the input sample. PCR amplified a 567-bp or 300-bp region of the LHβ or FSHβ proximal promoters, respectively. (B) The ChIP analysis was repeated as relevant for HDACs 1 to 5 on LHβ (upper panels) or FSHβ (lower panels) gene promoters, following a 6-h GnRH exposure; the treatments were carried out in duplicate. Ab, antibody.
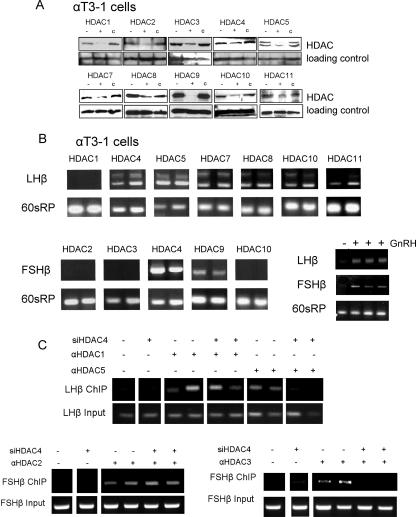
To elucidate the role of these HDACs in regulating the expression of each of the gonadotropin β-subunit genes, we knocked down their expression levels using an siRNA approach. The ability of the constructs to reduce the level of the respective protein was confirmed by Western analysis (Fig. 3A). Knockdown of any of the associated HDACs, except for HDAC1, was sufficient to allow LHβ gene expression. Notably, two bands were detected in these experiments, which were sequenced and confirmed to represent spliced and unspliced LHβ transcripts; the use of template that had not undergone reverse transcription confirmed that this unspliced form was not a result of genomic contamination but, rather, of incomplete splicing. Expression of the FSHβ gene occurred only after knock-down of HDAC4 or -9 (Fig. 3B).
FIG. 3.
Knockdown of the associated HDACs reveals their crucial roles in the repression. (A) Transfection with an siRNA construct (2 to 4 μg) targeting the HDACs found associated with the respective genes was employed to knock down the HDAC expression levels in αT3-1 cells, and the efficiency was verified by Western analysis. The left lane (−) shows the protein in nontransfected cells, the middle lane (+) is following transfection with the specific siRNA, and in the right lane (c) the sample is from cells transfected with a control siRNA sequence. Also shown in the lower box in each row is the loading control, GAPDH or polymerase II. (B) Total RNA was isolated 48 h after transfection with the various siRNA HDAC constructs, and RT-PCR analysis of LHβ and FSHβ mRNA levels was carried out. The primers designed to amplify the LHβ transcript targeted two different exons, while primers targeting FSHβ and 60sRP were as described in the legend of in Fig. 1. Also shown are untreated (first lane) or GnRH-treated (100 nM for 24 h; next three lanes) controls. (C) ChIP was carried out to test the association of HDACs 1 and 5 (for LHβ) or 2 and 3 (for FSHβ) following siRNA-mediated knockdown of HDAC4. All transfections were carried out in duplicate 48 h before harvest. siHDAC4, HDAC4-targeting siRNA construct; α, antisera against.
Given that HDAC4 was found associated with both promoters and appeared absolutely required for their repression, we assessed its role in the integrity of the complex through its siRNA-mediated knockdown followed by ChIP analysis to test the association of HDACs 1, 2, 3, or 5 with the relevant promoters. This indicated that HDAC4 has a role in the recruitment of HDAC3 or -5 but not HDAC1 or -2 to the respective promoters (Fig. 3C).
Corepressors that repress the FSHβ gene are identified.
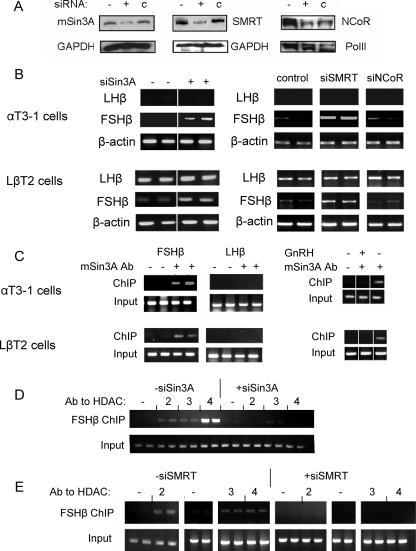
In order to characterize further the repressive complexes at each of these gene promoters, we evaluated the effect of siRNA-mediated knockdown of mSin3A, SMRT, or NCoR; the degree of knockdown was assessed by Western analysis (Fig. 4A). Transfection of the siRNA vector to reduce mSin3A expression was sufficient to restore expression of the FSHβ gene in both cell lines, while the LHβ transcript remained undetected in the αT3-1 cells, and its levels were unaffected in LβT2 cells. Similarly, transfection of the siRNA constructs targeting SMRT had a clear stimulatory effect on the FSHβ transcript levels in both αT3-1 and LβT2 cells while apparently not affecting the levels of the LHβ transcript. The siRNA construct targeting NCoR appeared to have no effect on the levels of either transcript in both cell lines (Fig. 4B).
FIG. 4.
Corepressors that repress the FSHβ gene in both cell types are identified. siRNA constructs (2 to 4 μg) targeting the corepressors mSin3A, SMRT, or NCoR were transfected into αT3-1 and LβT2 cells. (A) Western analysis was utilized to confirm the degree of knockdown as described in the legend of Fig. 3A, except that the control siRNA for NCoR was against SMRT. (B) RT-PCR was carried out to assess the effects of these knockdowns on LHβ and FSHβ mRNA levels in both cell lines as described in the legend of Fig. 1, with β-actin as an internal control. All transfections were carried out in duplicate. (C) ChIP analysis was carried out in both cell lines, using antisera to mSin3A in untreated or GnRH-treated cells (100 nM for 6 h), as indicated. DNA fragments were amplified as described in the legend of Fig. 2B from the precipitate and input samples. ChIP analysis was also carried out in αT3-1 cells after transfection of the siRNA construct to knock down mSin3A (D) or SMRT (E) expression and test the association of HDACs 2, 3, and 4 with the FSHβ gene, as described in the legend of Fig. 2. Ab, antibody; siSin3A, Sin3A-targeting siRNA construct; siSMRT, SMRT-targeting siRNA construct.
ChIP analysis was carried out to determine whether these corepressors associate directly with the FSHβ gene promoter. We found mSin3A associated with the FSHβ promoter in both cell types, but this association was lost following treatment of the cells with GnRH (100 nM for 6 h). No association of mSin3A with the LHβ gene promoter was detected (Fig. 4C). SMRT appeared to be associated only with the FSHβ promoter in untreated cells but was found associated with both promoters following GnRH treatment (data not shown).
In order to understand the role of these cofactors in the FSHβ-repressive complexes, ChIP was carried out to test the association of HDACs 2, 3, or 4 after siRNA-mediated knockdown of mSin3A or SMRT. The mSin3A knockdown abolished association of HDACs 2 and 4 and reduced that of HDAC3 to levels that were barely detectable (Fig. 4D), while the knock-down of SMRT led to a loss of association of all three HDACs (Fig. 4E). Coimmunoprecipitation experiments indicated that mSin3A and SMRT do not coprecipitate in these cells (not shown). However, both mSin3A and SMRT coprecipitate with HDAC3, while mSin3A also coprecipitates with HDACs 1, 2, or 4 (not shown).
GnRH causes the nuclear export of HDACs 4, 5, and 7.
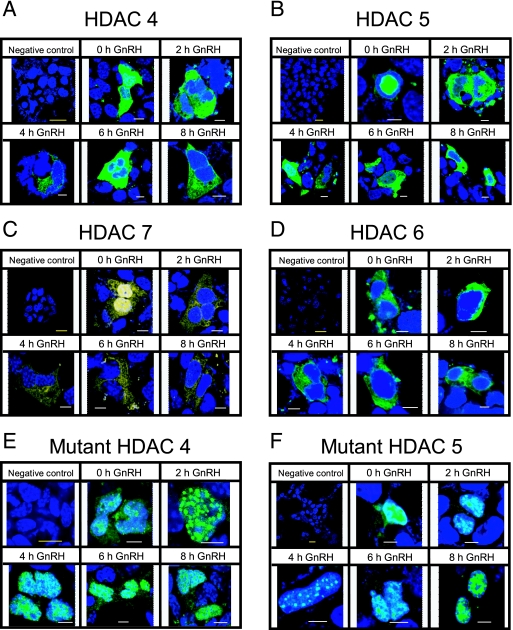
Knowing that GnRH activates a number of phosphorylation pathways, some of which might target class IIa HDACs and alter their cellular localization, we examined the localization of four class II HDACs following GnRH treatment in the immature gonadotrope cells. EGFP-tagged HDAC 4, 5, or 6 or YFP-tagged HDAC7 was transfected into the cells, and after 24 to 48 h the cells were treated with GnRH (10 nM) for 0 to 8 h. HDAC4 was found initially distributed throughout the cell (in 86% of the cells in which it was expressed), but after 4 to 8 h of treatment, it was exclusively cytoplasmic in the majority of expressing cells (59 to 63%) (Fig. 5A). HDAC5 was predominantly exclusively nuclear in untreated cells (80%) but already after a 2-h treatment was exclusively cytoplasmic in most cells (82%) (Fig. 5B). HDAC7 was initially pan-cellular in most cells (92%) but was found exclusively in the cytoplasm in the majority of cells (67 to 73%) after 4 to 8 h of treatment (Fig. 5C). HDAC6, which served as a control, was expressed only in the cytoplasm, where it remained following the treatment (Fig. 5D).
FIG. 5.
GnRH stimulates nuclear export of HDACs 4, 5, and 7. Expression vectors (500 ng) encoding fluorescently tagged class IIa HDACs 4, 5, and 7 (A to C) or HDAC6 (D) were transfected into αT3-1 cells which, at 24 to 48 h after transfection, were then exposed to GnRH (10 nM for 0 to 8 h). (E and F) Alternatively, vectors encoding mutant HDAC4 or -5 in which the serines (in HDAC4, residues S246, S467, and S632; in HDAC5, residues S259 and S498) were mutated to alanines, were transfected, and cells were similarly treated. Cells were then stained with DAPI; micrographs are representative of the localization of the HDACs in an average of 60 cells that were noted at each time point. Also shown is the negative control in which none of the HDAC constructs was transfected. Yellow scale bars (in negative controls), 10 μm; white scale bars, 5 μm.
To test whether the nuclear export might involve GnRH-induced phosphorylation of the 14-3-3 recognition sites, we transfected mutant forms of EGFP-tagged HDAC4 or HDAC5 in which these serines (in HDAC4, S246, S467 and S632; in HDAC5, S259 and S498) were mutated to alanines. The mutant HDAC4 was initially nuclear or pan-cellular and remained so in 99% of the cells following the GnRH treatment (Fig. 5E). The mutant HDAC5 was initially located only in the nucleus and remained exclusively nuclear in the majority (77%) of the cells after 8 h of treatment (Fig. 5F). The observed speckled localization of HDACs 4 and 5 has previously been reported when these HDACs are found primarily or exclusively in the nucleus. This pattern of localization is attributed to recruitment of HDACs 4 and 5 by SMRT (also seen in speckles following transfection or detection of the endogenous protein) and is thought to relate to a class of nuclear structures in which SMRT is found and where it recruits HDACs 4 and 5 (58, 62, 64).
GnRH-mediated derepression of the FSHβ but not the LHβ gene involves activation of CaMKI and its phosphorylation of HDACs 4 and 5.
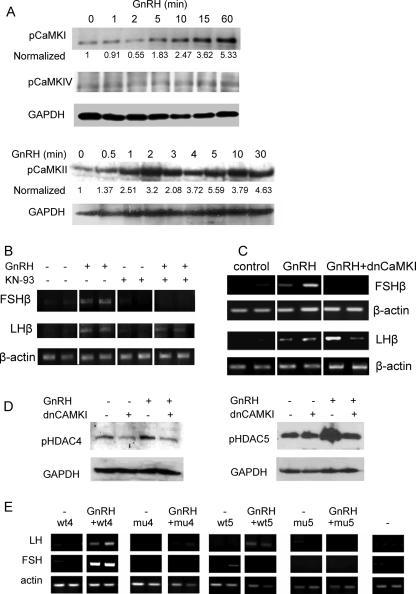
Our results indicate that the complexes repressing expression of these genes in the immature gonadotropes include several class IIa HDACs which move out of the nucleus following GnRH treatment, possibly as a result of their phosphorylation. Previous studies have indicated that CaMKI and CaMKIV can phosphorylate these HDACs, though neither has previously been linked to GnRH signaling; we therefore sought to determine which of these enzymes is activated by GnRH by examining changes in the levels of phosphorylated CaMKs (pCaMKs). After GnRH treatment, levels of both pCaMKI and pCaMKII were increased but with different kinetics: pCaMKI showed a gradual increase through the first hour of treatment, at which point it was at a considerably higher level than in the untreated controls. pCaMKII, on the other hand, showed a rapid response within the first minute, but the levels then remained similarly elevated for the duration examined. The pCaMKIV was barely detectable, and its levels were apparently unaltered by the GnRH treatment (Fig. 6A).
FIG. 6.
GnRH derepression of the FSHβ gene involves CaMKI. (A) Western analysis was carried out to detect phosphorylated forms of CaMKI, -II, and -IV in αT3-1 cells after exposure for 0 to 60 min to GnRH using antisera as marked; GAPDH is shown as a loading control, to which CaMK levels were normalized, and these values are expressed as a ratio to the levels in untreated cells. (B) αT3-1 cells were exposed to GnRH (10 nM for 8 h) and/or the CaMK inhibitor KN-93 (10 μM; added 30 min before the GnRH) before RT-PCR analysis of FSHβ and LHβ mRNA levels. (C) Similarly, the dnCaMKI expression vector (1.5 μg) was transfected into cells 16 h before treatment with GnRH (10 nM for 24 h), and the effect on GnRH derepression of both genes was assessed by RT-PCR in the same way. Transfections and treatments were carried out in duplicate. (D) Levels of phosphorylated HDAC4 and -5 (pHDAC4 and -5) were assessed by Western analysis in control cells and cells treated with GnRH (2 h), with or without transfection (24 h before) of the dnCaMKI construct. (E) Wild-type (wt) or mutant (mu) HDAC4 and -5 constructs (as described in the legend of Fig. 5) were overexpressed in the presence or absence of GnRH, and their effects on the derepression of LHβ and FSHβ were assessed by RT-PCR, as described in the legend of Fig. 3. Transfections and treatments were carried out in duplicate.
The role of CaMKs in the derepression of LHβ and FSHβ genes by GnRH was tested, initially using a general CaMK inhibitor, KN-93. The addition of KN-93 abolished the effect of GnRH on the FSHβ gene while not affecting the GnRH induction of LHβ transcription (Fig. 6B). In order to ascertain whether this was through actions of CaMKI or CaMKII, we used dnCaMKI, which was transfected prior to GnRH treatment, and the effects on the mRNA transcripts were measured by RT-PCR. The dnCaMKI abolished the GnRH-stimulated increase in FSHβ mRNA levels while not altering the effect of GnRH on the levels of LHβ (Fig. 6c).
In order to confirm the connection between GnRH activation of CaMKI and the phosphorylation of the class II HDACs, we carried out Western analysis to measure the changes in levels of phosphorylated HDAC4 and HDAC5 after GnRH treatment, with or without transfection of the dnCaMKI construct. The levels of both phosphorylated HDACs were clearly increased following GnRH treatment, and this was abated with transfection of the dnCaMKI construct (Fig. 6D).
We reasoned that overexpression of the class II HDACs would likely allow their replacement of the endogenous HDACs on the gonadotropin gene promoters and that they would be subject to similar posttranslational regulation. We therefore transfected wild-type or mutant HDAC4 or HDAC5 to assess the importance of the CaMK phosphorylation sites in the derepression by GnRH. Notably, GnRH was able to derepress the LHβ or FSHβ gene following transfection of the wild-type HDAC4, but its effect was abolished following transfection of the mutant HDAC4. Similarly, the mutant, but not the wild-type, HDAC5 prevented the GnRH effect on the LHβ gene, while interestingly overexpression of either form of HDAC5 had a repressive effect on GnRH-induced derepression of the FSHβ gene (Fig. 6E).
GnRH regulation of Nur77 plays a role in derepression of the FSHβ gene in the immature gonadotropes.
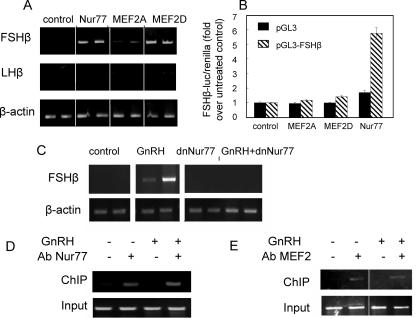
In order to try to elucidate the DNA-bound transcription factor(s) responsible for recruiting the repressive complexes to the LHβ and FSHβ gene promoters, we investigated the effects of various transcription factors on activating expression of these genes. We found that overexpression of Nur77 or MEF2D markedly increased the transcript levels of the FSHβ gene without affecting those of LHβ, while a marginal effect on the FSHβ gene was also seen following overexpression of MEF2A (Fig. 7A). We tested the effects of overexpression of these transcription factors on the promoter activity of FSHβ in transient transfection assays and found that Nur77 stimulated its activity, while MEF2A or -D had no effect (Fig. 7B). We confirmed a role for Nur77 in mediating the GnRH derepressive effect on FSHβ gene expression by transfecting dnNur77, which expresses the transcription factor without its activation domain, and abolished the effects of GnRH (Fig. 7C). However, ChIP assays showed that Nur77 and MEF2 (the antisera recognizes both MEF2A and MEF2D) were associated with the FSHβ gene promoter in αT3-1 cells both before and after GnRH treatment (Fig. 7D and E).
FIG. 7.
Nur77 induces expression of the FSHβ gene in the immature gonadotropes and plays a role in the GnRH derepressive effect. (A) Nur77, MEF2A, or MEF2D expression vectors (1 μg) were transfected into αT3-1 cells, and the effects on FSHβ and LHβ mRNA levels were assessed 40 h later by RT-PCR, as described in the legend of Fig. 1. (B) The effects of the same expression vectors (100 ng) on the transiently transfected FSHβ gene promoter fused to a firefly luciferase reporter gene (200 ng) were also tested. Firefly luciferase values were normalized to those of Renilla luciferase, and these values are expressed as a ratio to levels in untreated cells. The empty promoterless vector (pGL3) was also transfected as a control. Values are means ± standard errors of the means (n = 4 to 6). (C) The role of Nur77 in the GnRH effect on the FSHβ gene was tested using a dnNur77 which was overexpressed, and cells were then treated with GnRH before RT-PCR to detect FSHβ mRNA; β-actin was used as a control. Transfections and treatments were carried out in duplicate. ChIP analysis was carried out using antisera to Nur77 (D) or MEF2 (recognizes MEF2A, -C, and -D) (E) to test association of these proteins with the FSHβ gene promoter in untreated and GnRH-treated (10 nM for 2 h) αT3-1 cells, as described in the legend of Fig. 2A. Ab, antibody.
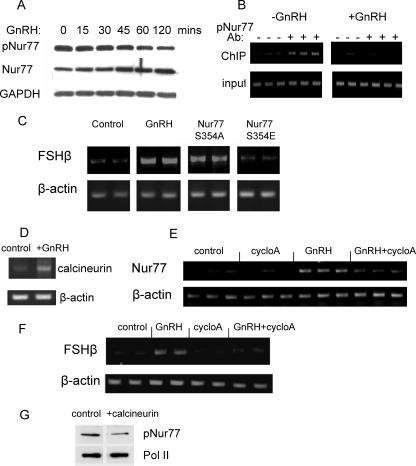
We postulated that the role of Nur77 in mediating the GnRH derepressive effect on FSHβ gene expression might be through its posttranslational modification, and therefore we looked at its phosphorylation status following GnRH treatment. The levels of S354-phosphorylated Nur77 (pNur77) decreased in response to the treatment, while the total levels of Nur77 increased, resulting in a clear drop in the ratio of pNur77 to unphosphorylated Nur77 (Fig. 8A). In order to confirm whether the Nur77 at the FSHβ gene promoter is indeed phosphorylated and whether this changes after GnRH treatment, ChIP was carried out using the pNur77 antiserum. pNur77 clearly was present before treatment and was undetectable following the GnRH exposure (Fig. 8B). In order to understand the impact of this phosphorylation, we overexpressed two Nur77 mutants, with a change of serine 354 to alanine (S354A) or to glutamic acid (S354E), and tested their effects on derepressing the FSHβ gene. Only the S354A mutant was able to induce FSHβ gene expression (Fig. 8C).
FIG. 8.
Calcineurin mediates the GnRH effect on Nur77. (A) Western analysis was carried out on αT3-1 cells following treatment for 0 120 min with GnRH using antiserum that recognizes the S354 phosphorylated Nur77 (pNur77), the total Nur77, or GAPDH as a control. (B) The pNur77 antisera was also used for ChIP analysis to test its association with the FSHβ gene promoter in untreated and GnRH-treated cells (10 nM for 2 h), carried out in triplicate. (C) The implication of phosphorylation in the regulation of FSHβ transcription was tested by overexpressing Nur77 with residue S354 mutated to an alanine or a glutamic acid before RT-PCR to assess changes in the FSHβ mRNA levels. (D) The levels of the calcineurin catalytic subunit mRNA in untreated or GnRH-treated αT3-1 cells were assessed by RT-PCR. (E and F) αT3-1 cells were treated with cyclosporine A 1 h before addition of GnRH (100 nM) for 1 h (E) or 8 h (F), and the effects on mRNA levels of Nur77 (E) or FSHβ (F) were assessed by RT-PCR, as described before. (G) The effects of calcineurin overexpression on pNur77 levels were tested in αT3-1 cells and assessed by Western analysis as described for panel A, using polymerase II (Pol II) as a loading control. cycloA, cyclosporine A; Ab, antibody.
We have shown that GnRH induces expression of calcineurin (Fig. 8D; also our unpublished data), with an increase in protein levels already after 2 h (not shown), and GnRH has previously been reported to stimulate expression of Nur77 (63). We thus went on to test whether the effect of GnRH on the FSHβ and Nur77 genes is through calcineurin and found that the stimulation of transcription of both genes was virtually abolished following treatment with the calcineurin inhibitor, cyclosporine A (Fig. 8E and F). Given the ability of GnRH to dephosphorylate Nur77, we further tested whether calcineurin might also be able to activate Nur77 in this way and found a marked drop in pNur77 levels following calcineurin overexpression (Fig. 8G).
DISCUSSION
In the current study we demonstrate a role for HDACs in repressing the gonadotropin β-subunit genes in the immature gonadotropes, and we show that this is overcome by the hypothalamic releasing hormone, GnRH. Our work was carried out in immortalized cell lines, which are thought to represent embryonic gonadotrope precursor cells and have provided a useful model for us to address the role of HDAC-mediated repression of the gonadotropin genes while revealing a novel role for GnRH in their derepression. Although we generally administered GnRH continuously, we have found similar induction of gene expression in pulsatile experiments, which might represent a more physiologically relevant model but are more limiting in the number of repeats that can be performed. Importantly, however, this study focuses on the phenomenon of derepression, which has not previously been studied at the cellular level, and so reports on the importance of pulsatile administration in increasing LH and FSH synthesis and release may, at least initially, be less relevant (10). Further study will clearly be required in the animal model to confirm the physiological relevance of our findings and to address the limitations of using cell lines, but our results do indicate that HDAC-mediated repression of gonadotropin gene expression could provide a temporal mechanism for shutdown of the reproductive function at certain periods of the life cycle, which can be reversed by the hypothalamic regulatory hormone.
Partial characterization of the repressive factors associated with each of these genes has revealed that they include distinct sets of HDACs and corepressors. The LHβ gene promoter appears associated with a particularly large number of HDACs which, based on the effects of the siRNA-mediated knockdown, do not have redundant functions. Although the requirement for so many different proteins with apparently similar enzymatic activities remains puzzling, it is supported by evidence of severe phenotypes resulting from mutations in genes encoding individual HDACs (64). The corepressor SMRT does not appear to repress this gene, and its knockdown did not allow LHβ gene expression, contrasting strongly with the effect of knockdown of almost any of the HDACs on expression of this gene; it was also not found associated with the gene in untreated cells.
The FSHβ gene promoter is associated with two class I and three class II HDACs, together with mSin3A and SMRT which likely act as scaffolds. mSin3A has previously been shown to interact with HDAC1/2 and SMRT, as well as a number of other mSin3A-associated proteins (17, 24, 52), and our studies confirmed that mSin3A and HDAC2 do coprecipitate in these cells. SMRT reportedly associates either with mSin3A-HDAC1/2 complexes or with HDAC3 in Sin3-independent complexes which lack HDAC1/2 (21, 24, 28). We found that HDAC3 coprecipitated with both mSin3A and SMRT, but mSin3A and SMRT did not coprecipitate, while HDAC4 appears crucial in the recruitment of HDAC3 but not HDAC2. This suggests that there may be two distinct corepressor complexes on the FSHβ gene promoter, one with mSin3A-HDAC2 and the other with SMRT-HDAC3, while HDAC4 is likely found in both complexes and may form a key link in the recruitment of the HDAC3-containing complex, as has been shown previously (8). However, the knockdown of either mSin3A or SMRT resulted in the virtual loss of the associated HDACs and was sufficient to restore FSHβ gene expression, indicating a clear functional interaction. This might well involve the recruitment of additional histone-modifying enzymes to this locus: mSin3A has been reported to associate with the Swi/Snf chromatin-remodeling complex and also to recruit DNA and histone methylases to the DNA (52).
In contrast to mSin3A, which clearly dissociated from the FSHβ gene promoter following GnRH treatment, SMRT remained associated and was similarly found at the LHβ gene promoter following GnRH treatment. This is not totally surprising because SMRT and/or the related NCoR are often found associated with actively transcribed genes, and ChIP carried out at frequent intervals has suggested mechanisms of their cycling on and off certain promoters during the transcriptional process (12, 25, 45). As our ChIP assay was carried out only at one time point, we do not know whether the association seen after GnRH exposure is transitory. The association of HDACs 1 or 2 from the respective promoters appeared similarly unaffected by the GnRH treatment. There is, however, mounting evidence that class I HDACs may play a role in ongoing transcriptional activation of certain genes, and in yeast the deacetylation of specific lysines was found to be positively correlated with transcription (12, 25, 31, 42, 59). Notably, knockdown of either HDAC1 or -2 failed to facilitate the respective gene expression, lending weight to the possibility of the essential role of these HDACs in gene activation. Given that HDAC2 is likely recruited to the repressed FSHβ gene specifically through mSin3A, we envisage that after GnRH treatment, HDAC2 can be recruited to the gene through a different mechanism, possibly involving an altered SMRT complex which no longer contains HDAC3. SMRT might also recruit HDAC1 to the activated LHβ gene promoter through a similar mechanism that requires further investigation.
Our results showing that GnRH causes dissociation of the HDACs 3, 4, and 5 from the respective genes and that the knock-down of the relevant class IIa HDACs is sufficient to derepress the gene expression indicate a crucial role for the class II HDACs, even though their independent deacetylase enzymatic activity has not been proven in vitro (11). These class IIa HDACs more likely have a crucial role in formation and/or stabilization of the complex, which is supported by previous suggestions that they do not function in an enzymatic capacity but rather recruit additional enzymes via the corepressors (8, 11, 57). Furthermore, these class IIa HDACs are much more regulated than the class I HDACs; this occurs at the level of their expression, cellular localization, and protein-protein interactions, and they can be regulated, for example, by kinases, calcium influx, ubiquitylation, or sumoylation (8, 57, 64). This ability to respond to common intracellular signals, while also playing a key role in the stability of the complex, would make them ideal candidates for mediating regulation of the function of the catalytic class I HDAC-containing repressive complex.
We found that HDACs 4, 5, and 7 translocate into the cytoplasm following GnRH exposure by means of a mechanism that, at least for HDACs 4 and 5, requires the serines which are targeted by CaMKs and recognized by 14-3-3 proteins (38, 58). The actions of the various CaMKs have previously been shown to overlap, and specificity is thought to relate to their patterns of expression, spatial localization, and activation (5, 51). Notably, only CaMKI and -II appear to be activated by GnRH treatment; this effect on CaMKII has been reported previously (19). The fact that the GnRH effect on the FSHβ gene was blocked by KN-93 treatment and by the dnCaMKI indicates that GnRH-induced CaMKI is likely responsible for the GnRH-induced translocation of the HDACs, causing disruption of the repressive complex. This was confirmed by the finding that the dnCaMKI reduces the GnRH-stimulated increase in levels of phosphorylated HDACs 4 and 5. The overexpression of the mutant HDAC4 confirmed that the phosphorylation is required for derepressing both genes, while the mutant HDAC5 similarly prevented derepression of the LHβ gene. Inhibition of the GnRH derepressive effect on the FSHβ gene following overexpression of the wild-type or the mutant HDAC5 was surprising but might be due to effects on Nur77 gene expression, which HDAC5 also regulates, involving stimulation of MEF sumoylation which inhibits transcription (15).
Neither CaMK inhibitor blocked the GnRH effect on the LHβ gene, indicating that CaMKI-induced phosphorylation of HDACs is not required for derepression of the LHβ gene. Instead, GnRH likely directs its derepression effect on the LHβ gene downstream of CaMK activation. We can thus hypothesize that GnRH acts at a different level to remove the repressive complex from the LHβ gene, presumably through the DNA-bound transcription factor(s) or at an associated corepressor, in a CaMK-independent manner. Identification of this as yet elusive repressor will enable us to examine how it is recruited to the LHβ gene and how its activity is regulated and will allow for a greater understanding of the repression of the LHβ gene.
The ability of GnRH to derepress the FSHβ gene is also dependent on its ability to activate Nur77. Nur77 has previously been reported to be dramatically up-regulated by GnRH in gonadotropes (28, 63), and its expression in T cells is regulated by MEF2D, whose overexpression we also found stimulated FSHβ gene expression. MEF2D can act as a repressor or activator of Nur77, depending on whether it interacts with class IIa HDACs or with coactivators (34, 66). As a repressor, MEF2 complexes with the corepressor Cabin1 which recruits mSin3A, HDAC1/2, and HDAC4 (66). In the event of calcium signaling, MEF2 activity is reversed as a result of activated calmodulin, which displaces Cabin1 and competes with MEF2D for HDAC4 binding, leaving MEF2D free to interact with the coactivator p300. In addition, calmodulin-activated calcineurin causes the translocation of nuclear factor of activated T cells (NFAT) which complexes with MEF2D on the Nur77 promoter, thereby facilitating recruitment of p300 (2, 65, 66). We found MEF2 associated also with the FSHβ gene in its repressed and active states, while both calmodulin and calcineurin are activated by GnRH (reference 49 and this study). Given that calcineurin appears to have a crucial role in GnRH-induced transcription of both the Nur77 and FSHβ genes, we consider this a probable mechanism for GnRH-stimulated MEF2 activation of both genes.
Less has been documented about the ability of Nur77 to act as a repressor of transcription, but it is reported to interact with SMRT, and this can be regulated by CaMKIV (53). Given that Nur77, which is present only at low levels in unstimulated immature gonadotrope cells, is also found at the FSHβ gene promoter before and after GnRH treatment, we considered that it, too, might convert to an activator after the hormonal treatment. The marked increase in the ratio of total Nur77 to pNur77, which occurred after GnRH exposure, indicated that dephosphorylation may activate Nur77, and this was corroborated by overexpression of Nur77 mutants showing that pNur77 is unable to derepress the FSHβ gene. This is reminiscent of the situation in YI adrenocortical cells in which adrenocorticotrope hormone was noted to stimulate Nur77 activity through its dephosphorylation at S354, although the phosphatase was not identified (33).
Previous reports have indicated that phosphorylation of Nur77 at S354, which is at the C-terminal end of the DNA binding domain, may lead to its reduced affinity for DNA (7), although in the present study it clearly was associated with the FSHβ gene promoter, possibly facilitated by protein-protein interactions with other DNA-bound factors. Notably, according to the gene context, Nur77 can bind the DNA as a monomer or a heterodimer together with other members of this family of transcription factors (e.g., Nurr1 and NOR1) or with retinoid X receptor (RXR) (60). Interaction with RXR involves the DNA binding domains of both proteins unless the RXR ligand is present, which causes a switch of the dimerization interface to the ligand binding domain (3). Clearly, interaction of Nur77 with coactivators and corepressors involves both N-terminal AF-1 and C-terminal AF-2 domains, and we can currently only hypothesize that phosphorylation at S354 might induce a conformational change to alter some of these interactions; this is currently an area of our ongoing study (3, 36, 53, 60).
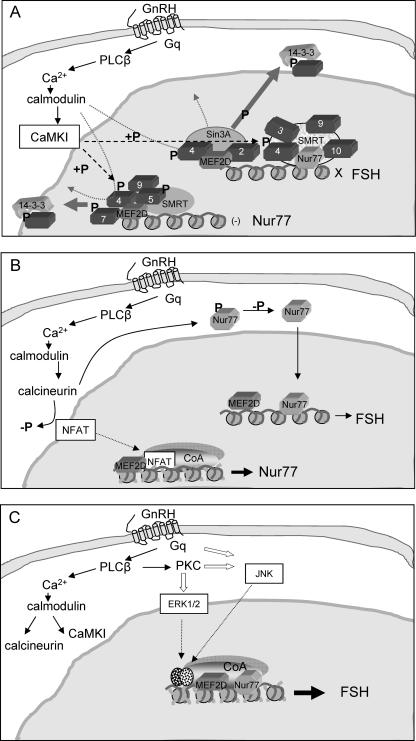
Taking these results together, we propose that GnRH derepresses the FSHβ gene in the immature gonadotrope by targeting both Nur77 and FSHβ genes, involving the following: (i) CaMKI phosphorylation of class IIa HDACs associated with both Nur77 and FSHβ gene promoters, causing disruption of the repressive complexes and their nuclear export, possibly involving also calmodulin competing with MEF2D for HDAC4 (65); (ii) GnRH activation of calcineurin, leading to the stimulation of Nur77 gene expression (presumably involving NFAT as has been shown in T cells [66]) and also likely dephosphorylation of Nur77; (iii) the dephosphorylated Nur77 then activates the FSHβ gene by facilitating the recruitment of coactivators and additional transcription factors recruited as a result of activation of the other GnRH signaling pathways (Fig. 9).
FIG. 9.
A model depicting the proposed mechanisms through which GnRH derepresses the FSHβ gene in immature gonadotropes. (A) GnRH binds to its G-protein-coupled receptor resulting in Ca2+ influx and activation of calmodulin. The calmodulin activates CaMKI, which phosphorylates class IIa HDACs at the FSHβ and Nur77 gene promoters, causing their dissociation from the complex and nuclear export, likely as a result of their binding 14-3-3 proteins. This may be helped by calmodulin's competing with MEF2D for binding HDAC4, both of which are associated with both promoters. (B) The activation of calmodulin by GnRH also leads to an increase in calcineurin levels, which stimulates Nur77 expression, likely as has been shown in T cells, through dephosphorylation of NFAT, which allows NFAT to translocate into the nucleus and activate Nur77 gene transcription. At the same time, calcineurin likely activates Nur77 through its dephosphorylation. (C) The increase in active nonphosphorylated Nur77 then activates the FSHβ gene by facilitating the recruitment of coactivators, possibly together with MEF2D and additional transcription factors (speckled ovals) recruited as a result of activation of other GnRH signaling pathways.
We have thus shown that the LHβ and FSHβ genes are repressed in immature gonadotropes by the actions of distinct repressive factors, which would allow for their differentially regulated derepression in the same cell by the same regulatory hormone. For the FSHβ gene, we are able to suggest the mechanisms through which GnRH overcomes this HDAC-mediated repression, involving activation of CaMKI and phosphorylation of class IIa HDACs associated with both FSHβ and Nur77 genes causing their nuclear export, and we propose a crucial role for GnRH-stimulated Nur77 activation and its dephosphorylation in facilitating FSHβ gene expression. In contrast, derepression of the LHβ gene is not CaMK dependent and likely occurs at the level of the DNA-bound repressor or corepressor; however, the exact mechanism of its derepression awaits further study.
Acknowledgments
We thank Hung-Ying Kao, Eric Olson, Xiang-Jiao Yang, Tom Soderling, and Lester Lau for their constructs and Pamella Mellon for the cells.
This work was supported by grants from the Academic Research Fund, National University of Singapore. S.L. is an A*STAR graduate scholar, M.K. is a recipient of a Singapore Millennium Foundation scholarship, and P.M. is a recipient of the Young Investigator Award, Office of Life Sciences, National University of Singapore.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Alarid, E. T., J. J. Windle, D. B. Whyte, and P. L. Mellon. 1996. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 122:3319-3329. [DOI] [PubMed] [Google Scholar]
- 2.Blaeser, F., N. Ho, R. Prywes, and T. A. Chatila. 2000. Ca2+-dependent gene expression mediated by MEF2 transcription factors J. Biol. Chem. 275:197-209. [DOI] [PubMed] [Google Scholar]
- 3.Cao, X. H., W. Liu, F. Lin, H. Li, S. K. Kolluri, B. Z. Lin, Y. H. Han, M. I. Dawson, and X. K. Zhang. 2004. Retinoid X receptor regulates Nur77/thyroid hormone receptor 3-dependent apoptosis by modulating its nuclear export and mitochondrial targeting. Mol. Cell. Biol. 24:9705-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro-Obregon, S., R. V. Rao, G. del Rio, S. F. Chen, K. S. Poksay, S. Rabizadeh, S. Vesce, X. K. Zhang, R. A. Swanson, and D. E. Bredesen. 2004. Alternative, nonapoptotic programmed cell death: mediation by arrestin 2, ERK2, and Nur77. J. Biol. Chem. 279:17543-17553. [DOI] [PubMed] [Google Scholar]
- 5.Corcoran, E. E., and A. R. Means. 2001. Defining Ca2+/calmodulin-dependent protein kinase cascades in transcriptional regulation. J. Biol. Chem. 276:2975-2978. [DOI] [PubMed] [Google Scholar]
- 6.Dasen, J. S., and M. G. Rosenfeld. 1999. Signaling mechanisms in pituitary morphogenesis and cell fate determination. Curr. Opin. Cell Biol. 11:669-677. [DOI] [PubMed] [Google Scholar]
- 7.Davis, I. J., T. G. Hazel, R.-H. Chen, J. Blenis, and L. F. Lau. 1997. Functional domains and the phosphorylation of the orphan receptor Nur77. Mol. Endocrinol. 7:953-964. [DOI] [PubMed] [Google Scholar]
- 8.de Ruijter, A. J. M., A. H. Van Gennip, H. N. Caron, S. Kemp, and A. B. P. Van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn, C., Q. L. Ou, J. Svaren, P. A. Crawford, and Y. Sadovsky. 1999. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J. Biol. Chem. 274:13870-13876. [DOI] [PubMed] [Google Scholar]
- 10.Ferris, H. A., and M. A. Shupnik. 2006. Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GnRH1. Biol. Reprod. 74:993-998. [DOI] [PubMed] [Google Scholar]
- 11.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 12.Gao, Z. G., P. Chiao, X. Zhang, X. H. Zhang, M. A. Lazar, E. Seto, H. A. Young, and J. P. Ye. 2005. Coactivators and corepressors of NF-κB in IκBα gene promoter. J. Biol. Chem. 280:21091-21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 14.Graham, K. E., K. D. Nusser, and M. J. Low. 1999. LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J. Endocrinol. 162:R1-5. [DOI] [PubMed] [Google Scholar]
- 15.Gregoire, S., and X. J. Yang. 2005. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25:2273-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozinger, C. M., and S. L. Schreiber. 2002. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9:3-16. [DOI] [PubMed] [Google Scholar]
- 18.Grumbach, M. M., and D. M. Styne. 2003. Puberty: ontogeny, neuroendocrinology, physiology, and disorders, p. 1115-1286. In P. R. Larsen, H. M. Kronenbergy, S. Melmed, and K. S. Polonsky (ed.), Williams textbook of endocrinology, 10th ed. W. B. Saunders, Philadelphia, PA.
- 19.Haisenleder, D. J., H. A. Ferris, and M. A. Shupnik. 2003. The calcium component of gonadotropin-releasing hormone-stimulated luteinizing hormone subunit gene transcription is mediated by calcium/calmodulin-dependent protein kinase type II. Endocrinology 144:2409-2416. [DOI] [PubMed] [Google Scholar]
- 20.Halvorson, L. M., M. Ito, J. L. Jameson, and W. W. Chin. 1998. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J. Biol. Chem. 273:14712-14720. [DOI] [PubMed] [Google Scholar]
- 21.Huang, E. Y., J. S. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh, Q. K., and T. A. McKinsey. 2006. Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism. Arch. Biochem. Biophys. 450:141-148. [DOI] [PubMed] [Google Scholar]
- 23.Ingraham, H. A., D. S. Lala, Y. Ikeda, X. R. Luo, W. H. Shen, M. W. Nachtigal, R. Abbud, J. H. Nilson, and K. L. Parker. 1994. The nuclear receptor steroidogenic factor-1 acts at multiple levels of the reproductive axis. Genes Dev. 8:2302-2312. [DOI] [PubMed] [Google Scholar]
- 24.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 25.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 26.Jones, P. L., and Y. B. Shi. 2003. N-CoR-HDAC corepressor complexes: Roles in transcriptional regulation by nuclear hormone receptors Curr. Topics Microbiol. Immunol. 274:237-268. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, J. S., C. C. Quirk, and J. H. Nilson. 2004. Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr. Rev. 25:521-542. [DOI] [PubMed] [Google Scholar]
- 28.Kakar, S. S., S. J. Winters, W. Zacharias, D. M. Miller, and S. Flynn. 2003. Identification of distinct gene expression profiles associated with treatment of LβT2 cells with gonadotropin-releasing hormone agonist using microarray analysis. Gene 308:67-77. [DOI] [PubMed] [Google Scholar]
- 29.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylase activity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 30.Kimura, A., M. Kazuko, and H. Masami. 2005. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J. Biochem. (Tokyo) 138:647-662. [DOI] [PubMed] [Google Scholar]
- 31.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 32.Li, X. F., S. Song, Y. Liu, S. H. Ko, and H. Y. Kao. 2004. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279:34201-34208. [DOI] [PubMed] [Google Scholar]
- 33.Li, Y. Z., and L. F. Lau. 1997. Adrenocorticotropic hormone regulates the activities of the orphan nuclear receptor Nur77 through modulation of phosphorylation. Endocrinology 138:4138-4146. [DOI] [PubMed] [Google Scholar]
- 34.Lu, J. R., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, M., M. Koh, J. Feng, Q. Wu, and P. Melamed. 2005. Cross talk in hormonally regulated gene transcription through induction of estrogen receptor ubiquitylation. Mol. Cell. Biol. 25:7386-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maira, M., C. Martens, E. Batsche, Y. Gauthier, and J. Drouin. 2003. Dimer-specifilic potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol. Cell. Biol. 23:763-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKinsey, T. A., C. L. Zhang, J. R. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melamed, P., M. Koh, P. Preklathan, L. Bei, and C. Hew. 2002. Multiple mechanisms for Pitx-1 transactivation of a luteinizing hormone β subunit gene. J. Biol. Chem. 277:26200-26307. [DOI] [PubMed] [Google Scholar]
- 41.Melamed, P., M. N. Abdul Kadir, A. Wijeweera, and S. Seah. 2006. Transcription of gonadotropin β subunit genes involves cross-talk between the transcription factors and co-regulators that mediate actions of the regulatory hormones. Mol. Cell Endocrinol. 252:167-183. [DOI] [PubMed] [Google Scholar]
- 42.Métivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 43.Ojeda, S. R., and M. K. Skinner. 2006. Puberty in the rat, p. 2061-2126. In J. D. Neill et al. (ed.), Knobil and Neill's physiology of reproduction, 3rd ed. Academic Press, New York, NY.
- 44.Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson, and E. Verdin. 2005. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280:13762-13770. [DOI] [PubMed] [Google Scholar]
- 45.Perissi, V., A. Aggarwal, C. K. Glass, D. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 46.Pernasetti, F., V. V. Vasilyev, S. B. Rosenberg, J. S. Bailey, H. J. Huang, W. L. Miller, and P. L. Mellon. 2001. Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology 142:2284-2295. [DOI] [PubMed] [Google Scholar]
- 47.Plant, T. M., V. L. Gay, G. R. Marshall, and M. Arslan. 1989. Puberty in monkeys is triggered by chemical stimulation of the hypothalamus. Proc. Natl. Acad. Sci. USA 86:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Privalsky, M. L. 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors Annu. Rev. Physiol. 66:315-360. [DOI] [PubMed] [Google Scholar]
- 49.Roberson, M. S., S. P. Bliss, J. J. Xie, A. M. Navratil, T. A. Farmerie, M. W. Wolfe, and C. M. Clay. 2005. Gonadotropin-releasing hormone induction of extracellular-signal regulated kinase is blocked by inhibition of calmodulin. Mol. Endocrinol. 19:2412-2423. [DOI] [PubMed] [Google Scholar]
- 50.Savage, J. J., B. C. Yaden, P. Kiratipranon, and S. J. Rhodes. 2003. Transcriptional control during mammalian anterior pituitary development. Gene 319:1-19. [DOI] [PubMed] [Google Scholar]
- 51.Schulman, H. 2004. Activity-dependent regulation of calcium/calmodulin-dependent protein kinase II localization. J. Neurosci. 24:8399-8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverstein, R. A., and K. Ekwall. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 53.Sohn, Y. C., E. Kwak, Y. J. Na, J. W. Lee, and S. K. Lee. 2001. Silencing mediator of retinoid and thyroid hormone receptors and activating signal cointegrator-2 as transcriptional coregulators of the orphan nuclear receptor Nur77. J. Biol. Chem. 276:43734-43739. [DOI] [PubMed] [Google Scholar]
- 54.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 55.Strahl, B. D., H. J. Huang, N. R. Pedersen, J. C. Wu, B. R. Ghosh, and W. L. Miller. 1997. Two proximal activating protein-1-binding sites are sufficient to stimulate transcription of the ovine follicle-stimulating hormone-beta gene. Endocrinology 138:2621-2631. [DOI] [PubMed] [Google Scholar]
- 56.Tremblay, J. J., and J. Drouin. 1999. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx and SF-1 to enhance luteinizing hormone β gene transcription. Mol. Cell. Biol. 19:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deactylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 58.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, A., S. K. Kurdistani, and M. Grunstein. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412-1414. [DOI] [PubMed] [Google Scholar]
- 60.Wansa, K. D. S. A., J. M. Harris, and G. E. O. Muscat. 2002. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J. Biol. Chem. 277:33001-33011. [DOI] [PubMed] [Google Scholar]
- 61.West, B. E., G. E. Parker, J. J. Savage, P. Kiratipranon, K. S. Toomey, L. R. Beach, S. C. Colvin, K. W. Sloop, and S. J. Rhodes. 2004. Regulation of the follicle-stimulating hormone beta gene by the LHX3 LIM-homeodomain transcription factor. Endocrinology 45:4866-4879. [DOI] [PubMed] [Google Scholar]
- 62.Wu, X. Y., H. Li, E. J. Park, and J. D. Chen. 2001. SMRTe inhibits MEF2C transcriptional activation by targeting HDAC4 and 5 to nuclear domains. J. Biol. Chem. 276:24177-24185. [DOI] [PubMed] [Google Scholar]
- 63.Wurmbach, E., T. Yuen, B. J. Ebersole, and S. C. Sealfon. 2001. Gonadotropin-releasing hormone receptor-coupled gene network. J. Biol. Chem. 276:47195-47201. [DOI] [PubMed] [Google Scholar]
- 64.Yang, X. J., and S. Grégoire. 2005. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25:2873-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youn, H. D., C. M. Grozinger, and J. O. Liu. 2000. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4 J. Biol. Chem. 275:22563-22567. [DOI] [PubMed] [Google Scholar]
- 66.Youn, H. D., T. A. Chatila, and J. O. Liu. 2000. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 19:4323-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zakaria, M. M., K. H. Jeong, C. Lacza, and U. B. Kaiser. 2002. Pituitary homeobox 1 activates the rat FSH beta (rFSH beta) gene through both direct and indirect interactions with the rFSH beta gene promoter. Mol. Endocrinol. 16:1840-1852. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, X. H., Y. Ozawa, H. Lee, Y. D. Wen, T. H. Tan, B. E. Wadzinski, and E. Seto. 2005. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 19:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, X. B., V. M. Richon, A. H. Wang, X. J. Yang, R. A. Rifkind, and P. A. Marks. 2000. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl. Acad. Sci. USA 97:14329-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]