Abstract
MEKK2 and MEK5 encode Phox/Bem1p (PB1) domains that heterodimerize with one another. MEKK2, MEK5, and extracellular signal-related kinase 5 (ERK5) form a ternary complex through interactions involving the MEKK2 and MEK5 PB1 domains and a 34-amino-acid C-terminal extension of the MEK5 PB1 domain. This C-terminal extension encodes an ERK5 docking site required for MEK5 activation of ERK5. The PB1 domains bind in a front-to-back arrangement, with a cluster of basic amino acids in the front of the MEKK2 PB1 domain binding to the back-end acidic clusters of the MEK5 PB1 domain. The C-terminal moiety, including the acidic cluster of the MEKK2 PB1 domain, is not required for MEK5 binding and binds MKK7. Quiescent MEKK2 preferentially binds MEK5, and MEKK2 activation results in ERK5 activation. Activated MEKK2 binds and activates MKK7, leading to JNK activation. The findings define how the MEKK2 and MEK5 PB1 domains are uniquely used for differential binding of two mitogen-activated protein kinase kinases, MEK5 and MKK7, for the coordinated control of ERK5 and c-Jun N-terminal kinase activation.
MEKK2 and MEKK3 are unique among the 20 or more mitogen-activated protein kinase (MAPK) kinase kinases (MKKKs) in that they encode the Phox and Bem1p domains referred to as the PB1 domain (18). We have previously shown that the MEKK2 and MEKK3 PB1 domains bind the PB1 domain of MEK5, the MAPK kinase (MKK) that phosphorylates and activates extracellular signal-related kinase 5 (ERK5) (17). PB1 domains are used to form heterodimers between proteins such as MEKK2 (or MEKK3) and MEK5, using a β-grasp topology in a front-to-back orientation of the two PB1 domains (11, 13, 35). This ubiquitin-like grasp domain involves a cluster of basic residues in the front of one PB1 domain that bind clusters of acidic amino acids in the back of a second PB1 domain. Alignment of mouse and human PB1 domains demonstrates a significant conservation of front-end basic and rear-end acidic residues in this β-grasp topology. The back-end acidic cluster in MEKK2 and MEKK3 is divergent from typical PB1 domains that use this sequence to bind the front-end basic cluster of a second PB1 domain (16, 18). Divergence of the back-end acidic cluster of MEKK2 and MEKK3 PB1 domains relative to many other PB1 domain sequences suggests a unique function for this region of the PB1 domain in MEKK2 and MEKK3.
Residues 98 to 131 immediately C terminal to the MEK5 PB1 domain are required for ERK5 binding. In addition, amino acids encoded within residues 18 to 25 at the front end of the N-terminal region of the MEK5 PB1 domain are also required for ERK5 binding. Thus, the MEK5 PB1 domain has evolved to be a scaffold or platform to organize a MEKK2 (or MEKK3)-MEK5-ERK5 signaling module (18). The MEK5 PB1 domain organization of such a signaling module ensures the rapid activation of ERK5 upon activation of either MEKK2 or MEKK3 (18, 25).
Typically, docking interactions among MAPKs and other proteins are achieved via conserved sequences in the MAPKs (c-Jun N-terminal kinase [JNK], ERK1/2, p38, ERK5) that are selective for the specific upstream MKKs and substrates (2, 12, 26). For all MAPKs a cluster of negatively charged amino acids is encoded C-terminal to the kinase domain sequence and referred to as the common docking (CD) domain (23, 28). The CD domain is used to bind specific MKKs, MAPK phosphatases, substrates, and scaffold proteins. In addition, the ED (Glu-Asp) domain and docking groove function as conserved motifs that also contribute to the interactions of MAPKs with other proteins (4, 29). Docking site sequences in MKKs, MAPK substrates, MAPK phosphatases, and MAPK scaffolds share a conserved motif of R/K-X4-ØA-X-ØB (where ØA and ØB are Leu, Ile, or Val hydrophobic residues) (1, 26). The basic residues in this docking site bind the acidic residues of the MAPK CD domain (28), and the hydrophobic residues reside in a docking groove that engages the ØA-X-ØB hydrophobic motif of the docking site (4). These docking site interactions provide the specificity for MAPK interactions and play a role in the activation of the kinase.
Specific motifs have also been defined for the control of interactions between specific MKKKs and MKKs. A docking site termed domain for versatile docking (DVD) is found in several MKKs, including MEK1, MKK3/6, and MKK4/7 (27). The DVD site is encoded in specific MKKs near the extreme C terminus of the MKK and was shown to be involved in binding MKKKs, including MEKK1, MEKK4 (MTK1), ASK1, TAO2, TAK1, and Raf-1. It appears that the N lobe within the kinase domain of the MKKK binds the MKK DVD site. No such interaction has been shown for MEKK2/3 and MEK5.
Herein, we demonstrate the unique function of the MEK5 PB1 domain in organizing signaling functions controlled by MEKK2. We demonstrate an ERK5 docking site within the C-terminal extension (residues 98 to 131) of the MEK5 PB1 domain. The MEK5 sequence 115RNIHGLKV122 conforms to the consensus docking site motif defined for other MKKs (1, 26); it is required for ERK5 activation and does not interact with ERK1, JNK, or p38. In addition, we demonstrate that the nonconserved C-terminal acidic region of MEKK2, but not that of MEKK3, can bind MKK7, which activates JNK (31). The ability of the MEKK2 PB1 domain to bind both MEK5 and MKK7 allows MEKK2 to coordinately activate the ERK5 and JNK pathways. Our studies define the properties of the MEKK2 and MEK5 PB1 domains for the organization of kinase complexes for activation of two MAPK pathways, ERK5 and JNK, in response to stimuli that activate MEKK2.
MATERIALS AND METHODS
Cell culture and reagents.
MEKK2−/− mouse embryonic fibroblasts (MEFs) were isolated as described previously and grown in Iscove's modified Dulbecco's medium (Life Technologies, Inc.) with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 7% CO2. HEK293 cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum. A mouse monoclonal antibody (MAb) for MEKK2 was generated against recombinant MEKK2. The MAb (12CA5) against the hemagglutinin (HA) epitope was from Roche Molecular Biochemicals. Anti-FLAG (M5) and -ERK5 Abs and anti-Xpress Ab were from Sigma and Invitrogen, respectively. The mouse MAb for MEK5 was purchased from BD PharMingen. Anti-phospho-p38, -phospho-JNK, -phospho-ERK1/2, -phospho-ERK5, -p38, -JNK, -ERK2, -MKK1, -MKK4, and -MKK7 Abs were obtained from Cell Signaling. Anti-MKK3 Ab was from Santa Cruz Biotechnology.
DNA constructs.
All glutathione S-transferase (GST) fusions were constructed in pGEX 5X-1 using the cDNA fragments amplified by a PCR. The indicated specific residues were replaced by alanine (Ala) using PCR-based mutagenesis, and coding amino acids are described in the figure legends. For retroviral infections, MEKK2 constructs were prepared in pMSCVpuro vector, and Phoenix cells were used as packaging. For transient expression in HEK293 cells, HA-MEKK2 and -MEKK3 and FLAG-MEK5 were prepared in pCMV5 vector, and FLAG-MKK7 and Xpress-ERK5 were prepared in pcDNA3 vector. A constitutively active MEK5 mutant (311S315T-DD MEK5) was generated by replacement of Ser and Thr (amino acids [aa] 311 and 315) with Asp.
Pulldown assays with GST fusion proteins.
Different GST fusion proteins were coupled to glutathione beads. Total cell lysates were prepared from HEK293 cells transiently transfected with DNA constructs using a solubilizing buffer as described below. GST fusion proteins on glutathione beads were incubated with 1 mg HEK293 cell lysate at 4°C for 2 h. The beads were washed five times with solubilizing buffer, and the protein complex isolated on the beads was subjected to SDS-PAGE and immunoblotting.
Immunoprecipitation and immunoblotting analysis.
Cells were washed three times with ice-cold phosphate-buffered saline and lysed with solubilizing buffer (1% NP-40, 10 mM Tris [pH 7.5], 150 mM NaCl, 0.4 mM EDTA, 10 mM NaF, 2 mM Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml α1-antitrypsin, and 1 mM phenylmethylsulfonyl fluoride), and cleared supernatants were retained for further processing. Lysates were subjected to immunoblotting or immunoprecipitation with anti-FLAG, -MEKK2, or -MKK7 Ab. For immunoprecipitation, immune complexes were collected with protein G Sepharose beads, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. After blocking, membranes were blotted with the indicated Abs and visualized using the Supersignal West Pico detection system (Pierce).
In vitro protein phosphatase and kinase assays.
As to protein phosphatase treatment, MEKK2 immunoprecipitate was incubated with λ protein phosphatase (New England Biolabs) in 50 μl of reaction buffer supplied by the manufacturer for 50 min at 30°C. For the in vitro kinase assay, MEKK2 immunoprecipitate was incubated with 50 μl of kinase buffer (0.05% NP-40, 20 mM HEPES [pH 7.5], 50 mM NaCl, 2.5 mM MgCl2, 20 mM β glycerophosphate, 0.1 mM Na3VO4, 2 mM dithiothreitol, and 0.1 mM ATP) for 50 min at 30°C. After the reaction, immunoprecipitates were washed three times with solubilizing buffer and set aside for further processing. In the case of the in vitro kinase assay with [γ-32P]ATP, the kinase buffer contained 5 μCi [γ-32P]ATP and 20 μM ATP instead of 0.1 mM ATP. The reaction was conducted at 30°C for 20 min. The kinase activity of MEKK2 was visualized by autoradiography.
TR-FRET assay.
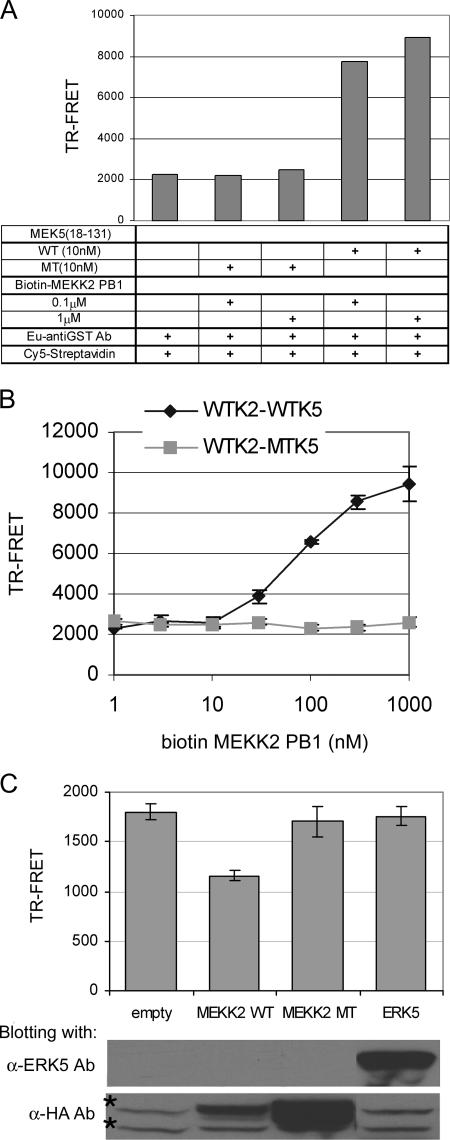
GST-MEK5 (aa 18 to 131; wild-type [WT] and mutant 64D65E-AA) was purified with glutathione-conjugated beads. A cDNA encoding hexahistidine-tagged MEKK2 PB1 was generated by PCR and introduced into the Avi-tag vector (Avidity). The Avi-tag sequence was inserted into the N-terminal end of the six-His MEKK2 PB1 coding region, which is specifically biotinylated using biotin protein ligase (BirA enzyme). The recombinant biotinylated protein was purified by Ni-NTA agarose. The indicated amount of biotinylated MEKK2 PB1 domain and 10 nM GST-MEK5 (aa 18 to 131) were mixed in a buffer (150 mM NaCl, 10 mM Tris [pH 8.0], 0.1% bovine serum albumin). For the competition assay, 10 nM GST-MEK5 PB1 and 100 nM biotinylated MEKK2 PB1 were incubated in a buffer in the presence of transfected cell lysates. After an hour at room temperature, Eu-TMT [europium chelate of terpyridine-bis(methylenamine) tetraacetic acid] anti-GST Ab (Perkin Elmer) and Cy5 streptavidin (Amersham) were added to the mixture and incubated for 30 min at room temperature. The final assay volume was 50 μl in the wells of a 96-well plate. The time-resolved fluorescence resonance energy transfer (TR-FRET) signal was measured using a PHERAstar plate reader (BMG labtech). The Eu donor was excited using a 340-nm excitation filter. The emission of Eu and Cy5 was measured with 615- and 665-nm emission filters, respectively. After an 80-μs postexcitation delay, 100 μs of integration time was used for the signal detection. The Z′ factor calculated under this assay condition was 0.75. Z′ was calculated according to the equation Z′ = 1 − 3 × (σp + σn)/|μp − μn|, where σp and σn are the standard deviations of the positive and negative controls, respectively, and μp and μn are average values for the positive and negative controls, respectively. The data shown in Fig. 2 are expressed as averages of the background-subtracted Cy5 emission signal.
FIG. 2.
TR-FRET assay of MEKK2 PB1-MEK5 PB1 interaction. (A) TR-FRET assay using 10 nM of GST-WT or 64D65E-AA (MT) MEK5 (aa 18 to 131) and the indicated amount of biotinylated MEKK2 PB1 domain. For TR-FRET measurement, 10 nM Eu anti-GST Ab and 10 nM Cy5 streptavidin were added into the mixture, and the TR-FRET signal was detected by use of a plate reader. (B) Titration of biotinylated MEKK2 PB1 domain. A TR-FRET assay was conducted with serially diluted biotin-MEKK2 PB1 domain and 10 nM GST-WT or -MT MEK5 (aa 18 to 131). Each data point is represented as the mean TR-FRET value ± standard deviation (error bars) for individual assays conducted in triplicate. (C) MEKK2 but not ERK5 interferes with TR-FRET between MEKK2 and MEK5. A TR-FRET assay using WT or MT MEK5 (aa 18 to 131) and MEKK2 PB1 domain was conducted in the presence of the transfected cell lysates. First, GST-MEK5 (aa 18 to 131) and the indicated cell lysates (200 μg protein of empty vector, HA-WT MEKK2, HA-47K-A MEKK2 (MEKK2 MT), or Xpress-ERK5 transfectant) were incubated in each well for 1 h. Subsequently, biotinylated MEKK2 PB1 domain was added into each well for 1 h. Each TR-FRET value was calculated by subtracting a background (TR-FRET of a combination of MT MEK5 [aa 18 to 131] and MEKK2 PB1 domain) from TR-FRET of WT MEK5-MEKK2. The data are represented as the mean TR-FRET value ± standard deviations (error bars) for individual assays conducted in triplicate. The expression of each molecule in the transfectant is shown in the bottom two panels. Nonspecific bands in the anti-HA Ab blot are marked with asterisks.
RESULTS
Identification of an ERK5 docking site in MEK5.
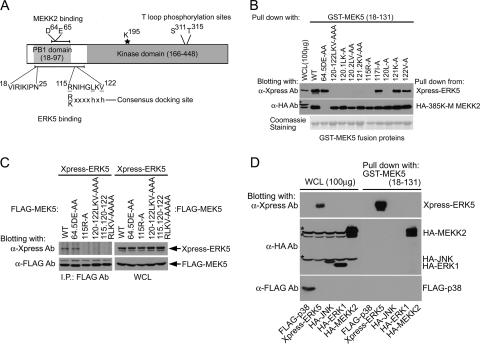
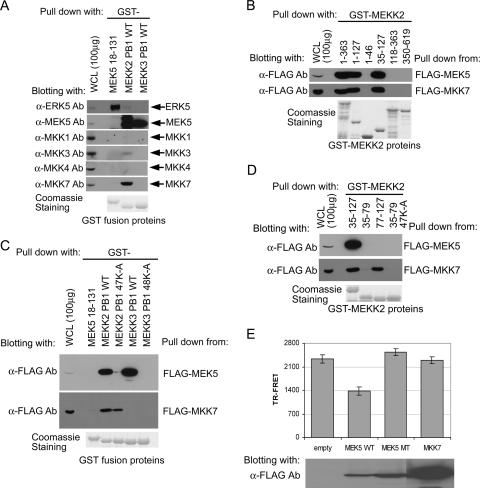
Fig. 1A shows a diagram of MEK5 domains involved in interacting with MEKK2 (or MEKK3) and ERK5. The MEK5 PB1 domain is encoded within residues 18 to 97. We (18) and another group (25) have shown that the acidic residues 64D and 65E are required for MEKK2 binding. Furthermore, we have shown that residues 18 to 25 within the MEK5 PB1 domain and a C-terminal extension of the PB1 domain within aa 98 to 131 were required for ERK5 binding. The MEK5 kinase domain is encoded within residues 166 to 448.
FIG. 1.
Mapping of the ERK5 binding site in MEK5 PB1 C-terminal extension. (A) Schematic diagram of MEK5 functional domains and motifs. MEKK2 (aa 56 to 97) and ERK5 (aa 18 to 131) binding sites are indicated with bars. Crucial residues for the binding are highlighted with amino acid sequences. The identified ERK5 docking site is aligned with the consensus MAPK docking site sequence (where “h” is a hydrophobic residue [Leu, Val, or Ile]). (B) ERK5 and MEKK2 binding sites in MEK5 were analyzed using MEK5 mutants. GST fusion proteins of the indicated MEK5 fragments (aa 18 to 131) were used to analyze the binding to full-length Xpress-ERK5 and HA-kinase-inactive MEKK2 (385K-M). The adsorbents and 100 μg (10% of the lysates used for the pulldown assay) of each whole-cell lysate were analyzed with anti-Xpress (for ERK5) or anti-HA (for MEKK2) Ab blotting. A nonspecific band in the anti-HA Ab blot is marked with an asterisk. (Bottom) Expression of GST fusion proteins used in the assay. (C) Binding in cells of full-length ERK5 to MEK5 mutants. The indicated FLAG-tagged MEK5 mutants were coexpressed with Xpress-ERK5 and immunoprecipitated with anti-FLAG Ab. ERK5 binding was detected by anti-Xpress Ab blotting of the immunoprecipitates (left). The expression of each protein was determined by immunoblotting of whole-cell lysates (right). (D) Binding specificity of MEK5 docking site for MAPKs. GST-MEK5 (aa 18 to 131) fusion proteins were incubated with each lysate of FLAG-p38, Xpress-ERK5, HA-JNK, HA-ERK1, or HA-MEKK2-transfected HEK293 cells. Binding was detected by immunoblotting with anti-FLAG Ab (for p38), anti-Xpress Ab (for ERK5), or anti-HA Ab (for JNK, ERK1, and MEKK2) (right). One hundred micrograms of cell lysate was used to determine the expression of each protein (left). Nonspecific bands in the anti-HA blot are marked with asterisks.
Visual scanning of the MEK5 sequence encoded within residues 98 to 131 identified a putative consensus MAPK docking site (Fig. 1A). The sequence 115RNIGHLKV122 conforms to the MAPK consensus docking site R/Kxxxxhxh, where h is Leu, Val, or Ile (1, 26). The critical residues within this sequence were mutated to alanine in a GST fusion encoding MEK5 residues 18 to 131 and used in pulldown assays to determine the amino acid requirements for binding of ERK5. As a control, the 64D65E → AA GST-MEK5 (aa 18 to 131) protein was used in pulldown assays. Figure 1B shows that the mutation 115R → A or 120L → A resulted in loss of ERK5 binding. The mutation 117I → A or 125R → A (not shown) had no effect on ERK5 binding. The mutation 121K → A or 122V → A alone had no effect on the ERK5 binding in MEK5 (aa 18 to 131) pulldown assays, but the double mutation 121K122V → AA resulted in the loss of ERK5 binding. These findings indicate that 115R and 120L are critical for ERK5 binding. The valine at residue 122 itself is not essential for ERK5 binding, but disruption of the 120LKV122 sequence by mutation of 120L or dual mutation of 121K122V inhibits ERK5 pulldown with MEK5 (aa 18 to 131). These findings are in absolute agreement with 115RNIGHLKV122 being a required docking site for ERK5 binding. Importantly, each mutant protein with alanine substitutions in the ERK5 docking site retained binding to MEKK2 (Fig. 1B). As demonstrated previously (18), the mutation of 64D65E in the MEK5 PB1 domain caused loss of binding to MEKK2 but not to ERK5. These results demonstrate that each MEK5 (aa 18 to 131) peptide expressed as a GST fusion protein was folded properly to form a functional PB1 domain and that the loss of specific binding was due to alanine replacements of important amino acids required for ERK5 (115R and 120LKV122) or MEKK2 (64D and 65E) binding.
Figure 1C shows that in cells, residues 115R and 120LKV122 in the MEK5 full-length sequence are required for interaction with ERK5. Coimmunoprecipitations show that MEK5 and ERK5 form a complex in cells and that mutation of the critical docking site residues as defined in Fig. 1B disrupts the interaction of MEK5 and ERK5. Importantly, the MEK5 (aa 18 to 131) sequence that includes the 115RNIGHLKV122 docking site does not bind p38, JNK, or ERK1 (Fig. 1D). Cumulatively, these findings define an ERK5-specific docking site within the N-terminal region of MEK5 that, in combination with sequences within the PB1 domain, is used to specifically bind ERK5.
A TR-FRET assay was developed for the direct in vitro measurement of MEKK2 and MEK5 PB1 domain interactions (Fig. 2). Panel A shows the characteristics of the assay. A robust TR-FRET signal is observed with the interaction of WT MEKK2 PB1 domain and the MEK5 PB1 domain having the 34-aa C-terminal extension (MEK5 aa 18 to 131). The mutation 64D65E → AA in the acidic cluster in the back half of the MEK5 PB1 domain abolished measurable MEKK2-MEK5 PB1 domain interaction. The TR-FRET signal increases significantly with increasing concentrations of recombinant MEKK2 PB1 domain protein (Fig. 2B). Even at high concentrations of MEKK2 PB1 domain there is no TR-FRET signal with 64D65E-AA (MT) MEK5, demonstrating the absolute requirement of the MEK5 acidic cluster for the MEKK2/MEK5 PB1-PB1 domain interaction. The TR-FRET assay is highly reproducible with a Z′ value of >0.75 and is a robust in vitro assay to monitor PB1-PB1 domain interactions. Lysates from transfected HEK293 cells were then used to determine whether full-length expressed MEKK2 or ERK5 would interfere with MEKK2/MEK5 PB1-PB1 domain interactions. Figure 2C shows that WT MEKK2 but not 47K-A MEKK2, which does not bind MEK5 due to disruption of its front-end basic cluster of the PB1 domain, inhibits recombinant MEKK2/MEK5 PB1 domain interactions. Lysates from ERK5 expressing HEK293 cells have no effect on MEKK2/MEK5 PB1 domain interactions. This finding indicates that ERK5 does not interfere with MEKK2 binding to the back-end acidic cluster in the MEK5 PB1 domain and C-terminal extension (aa 18 to 131). The fact that ERK5 does not interfere with MEKK2 PB1 domain binding to the MEK5 PB1 domain is consistent with ERK5 binding to the docking domain encoded in the 115RNIGHLKV122 sequence in the C-terminal extension of the MEK5 PB1 domain and an N-terminal region of the MEK5 PB1 domain independent of the rear-end acidic cluster.
ERK5 activation requires the MEK5 docking site.
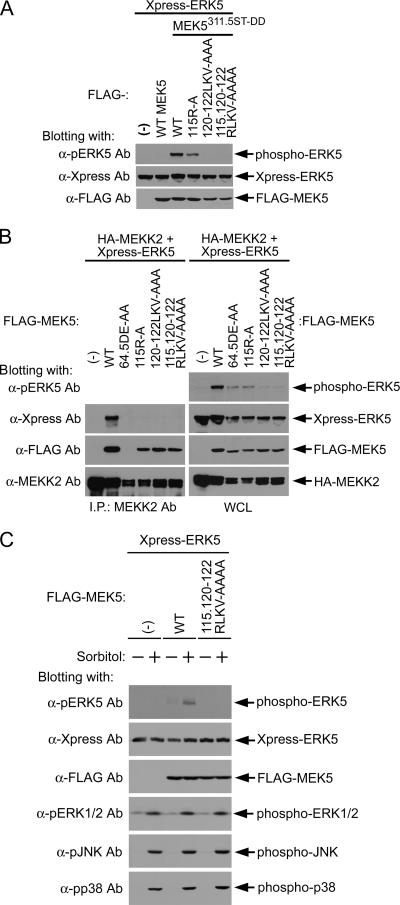
The mutation of 311S315T in the T loop of MEK5 to aspartates (311S315T-DD) results in constitutively activated MEK5 kinase activity. The expression of activated MEK5 activates ERK5 as determined by phospho-ERK5 Ab immunoblotting that measures the phosphorylated active state of ERK5 (Fig. 3A). Mutation of the 120LKV122 sequence to alanines resulted in the loss of ERK5 activation in response to constitutively active MEK5. The mutation of 115R to alanine resulted in a partial inhibition of ERK5 activation. This contrasts somewhat with the results shown in Fig. 1, where mutation of 115R caused a complete loss of ERK5 association as measured by a GST-MEK5 pulldown assay and MEK5 coimmunoprecipitation of ERK5. This finding suggests that the 115R → A MEK5 mutant protein has a diminished ability to interact with and activate ERK5 and that the change in interaction is sufficient to cause a loss of stringent binding but not sufficient to inhibit weak interactions in cells that give a partial activation of ERK5. In contrast, the 120LKV122 → AAA mutant protein is unable to activate ERK5, indicating that this 3-aa sequence within the docking site is essential for MEK5-ERK5 interaction and phosphorylation of ERK5.
FIG. 3.
Requirement of MEK5 PB1 domain for ERK5 activation. (A) FLAG-WT MEK5 or constitutively active MEK5 (311S315T-DD) (MEK5311.5ST-DD) having the indicated mutations were coexpressed with Xpress-ERK5. Whole-cell lysates for each condition were blotted sequentially with Abs for phospho-ERK5 to measure ERK5 activation, Xpress to measure ERK5 protein, and FLAG to measure MEK5 protein. (B) MEKK2-MEK5-ERK5 ternary complex composite and ERK5 activation. FLAG-tagged MEK5 mutant protein, Xpress-tagged ERK5, and HA-tagged MEKK2 were expressed in HEK293 cells. HA-MEKK2 was immunoprecipitated (I.P.) from the cell lysates with anti-MEKK2 Ab. The HA-MEKK2 immunocomplexes (left) and whole-cell lysate (WCL; right) were analyzed with anti-Xpress (for ERK5), anti-FLAG (for MEK5), and anti-MEKK2 Abs. Additionally, whole-cell lysates were blotted with an anti-phospho-ERK5 Ab that measures activated ERK5. (C) A docking site mutation in MEK5 abrogates stimulus-dependent activation of ERK5. FLAG-MEK5 or mutant FLAG-MEK5 together with Xpress-ERK5 was expressed in HEK293 cells. Twenty-four hours after transfection, the cells were deprived of serum for 6 h and then stimulated with 100 mM sorbitol for 20 min. Cell lysates were blotted with anti-phospho-ERK5, -ERK1/2, -JNK, and -p38 Abs. Anti-Xpress and -FLAG Abs were used to show the expression of Xpress-ERK5 and FLAG-MEK5, respectively.
Testing was then performed to determine whether MEKK2-mediated activation of ERK5 requires the MEK5 docking site for ERK5 activation (Fig. 3B). A significant fraction of MEKK2, when expressed by transient transfection, is activated, resulting in the activation of the MEK5-ERK5 pathway. The mutation 64D65E → AA in the PB1 domain of MEK5 caused a loss of coimmunoprecipitation with MEKK2 (Fig. 3B, left) but not with ERK5 (Fig. 1C). The 64D65E → AA MEK5 mutant protein is markedly inhibited in its ability to activate ERK5 (Fig. 3B, top right). Disruption of the MEK5-ERK5 interaction by mutation of the MEK5 docking site abolishes the binding of MEK5 to ERK5 but not to MEKK2, leading to the disruption of MEKK2 activation of ERK5. Similar to the results shown in Fig. 3A, the mutation 115R → A caused a marked but not complete inhibition of ERK5 activation in response to MEKK2, whereas the mutation 120LKV122 → AAA causes a complete loss of MEKK2-mediated activation of ERK5. The results in Fig. 3B also show that MEKK2, MEK5, and ERK5 form a ternary complex (left) where ERK5 can be coimmunoprecipitated with MEKK2 because of the interaction of both MEKK2 and ERK5 with MEK5. Figure 3C shows that the 115R120LKV122 → AAAA MEK5 mutant protein was also unable to activate ERK5 in response to sorbitol stimulation. In contrast, p38, JNK, and ERK1/2 activation in response to sorbitol was similar to that in WT cells. Thus, disruption of the MEK5 docking site-ERK5 interaction inhibits MEKK2 activation of ERK5 but not of other MAPK pathways.
MEK5 interaction sites in ERK5.
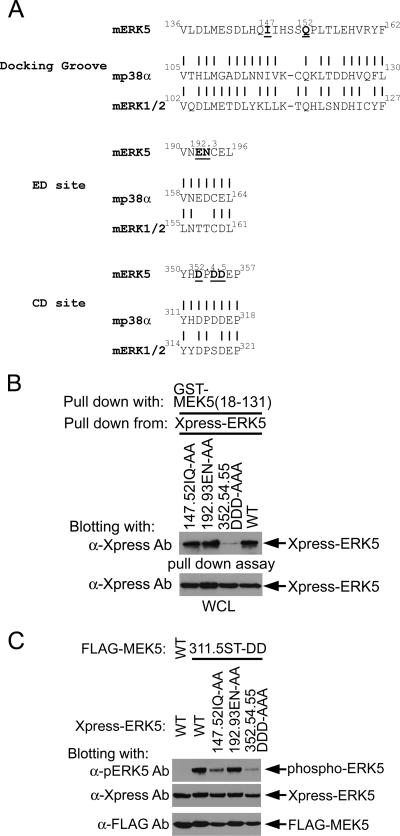
Figure 4A shows the homology between the predicted docking groove, ED, and CD sites for ERK5 compared to that of p38α and ERK1/2 (4, 23, 28, 29). Residues predicted to be critical for each of these sites are highlighted in the ERK5 sequence and were mutated in ERK5 to alanines. Figure 4B shows that the mutation of 147I and 152Q in the docking groove and 192E and 193N in the ED site did not measurably affect GST-MEK5 (aa 18 to 131) pulldown of ERK5. In contrast, the mutation of 352D, 354D, and 355D to alanines in the CD site inhibited interaction of GST-MEK5 (aa 18 to 131) with ERK5 as measured in pulldown assays.
FIG. 4.
Requirement of ERK5 CD site for MEK5 binding and MEK5-dependent ERK5 activation. (A) Sequence alignment of MAPK kinase binding sites in MAPKs. Based on the reports of docking motifs in p38 and ERK1/2, the docking groove, ED site, and CD site in ERK5 are predicted. ERK5 Ala mutation sites used in this study are shown in boldface and underlined. (B) GST-MEK5 pulldown assay using ERK5 docking domain mutants. Xpress-tagged WT and the indicated mutants of ERK5 were expressed in HEK293 cells. These cell lysates (WCL) were provided for the GST-MEK5 (aa 18 to 131) binding assay. ERK5 in the absorbents was detected using anti-Xpress Ab. (Bottom) Equal expression of ERK5 proteins in HEK293 cells. (C) Loss of ERK5 activation in ERK5 common docking domain mutant. WT or constitutively active MEK5 (311.5ST-DD) was coexpressed with WT or the indicated docking domain mutants ERK5 in HEK293 cells. The whole-cell lysates were sequentially blotted with anti-phospho-ERK5, anti-Xpress (for ERK5), and anti-FLAG (for MEK5).
Figure 4C shows the effect of docking groove, ED, and CD site mutations on ERK5 activation by constitutively active MEK5. The 352DPDD355 → APAA mutation in the CD site markedly inhibited ERK5 activation. Mutation of 147I and 152Q in the docking groove partially inhibited ERK5 activation even though these mutations had no effect on the ability of GST-MEK5 (aa 18 to 131) to pull down ERK5 (Fig. 4B). This finding indicates that 147I and 152Q in the docking groove are important for ERK5 activation and that 352D, 354D, and 355D are essential for MEK5 activation of ERK5. The ERK5 CD domain for MEK5 binding that we have identified is the same site recently characterized as a CD site for ERK5 binding to p90 RSK (21).
A rear-end acidic cluster in the MEKK2 PB1 domain binds MKK7.
The atypical sequence of the rear-end acidic cluster of the MEKK2 PB1 domain led us to search for potential binding partners other than MEK5. Since MEKK2 regulates the JNK pathway in addition to ERK5 (5, 7, 8, 20), we screened for interaction of the MEKK2 PB1 domain with other MKKs (Fig. 5). Immunoblotting showed that HEK293 cells express endogenous MKK1, MKK3, MKK4 and MKK7, and MEK5. Figure 5A shows that the MEKK2 PB1 domain binds endogenous MEK5 as expected. The PB1 domain from MEKK3 also binds endogenous MEK5. In addition to MEK5, the MEKK2 PB1 domain binds endogenous MKK7 but not MKK1, MKK3, or MKK4. Although the MEKK3 PB1 domain binds MEK5, it does not bind MKK7; thus, the MEKK2 PB1 domain, but not the MEKK3 PB1 domain, selectively binds MKK7. Used as a control, the MEK5 PB1 domain and its C-terminal extension were shown to bind ERK5 but failed to bind any of the MKKs tested. Pulldown assays showed that the back-end acidic cluster region of the MEKK2 PB1 domain is required for binding MKK7 (Fig. 5D). Neither the front-end basic cluster of the MEKK2 PB1 domain nor the kinase domain of MEKK2 binds MKK7 (Fig. 5B and C). Deletion of the front-end basic region or a 47K → A mutation in the MEKK2 PB1 domain abolishes MEK5 binding but has little or no effect on MKK7 binding. Only the acidic cluster back-end region (aa 77 to 127) of the MEKK2 PB1 domain is required for MKK7 binding (Fig. 5D). Finally, we used the TR-FRET in vitro MEKK2/MEK5 PB1 domain binding assay to show that MKK7, although it binds to the MEKK2 PB1 domain, does not compete with MEK5 for binding to the MEKK2 PB1 domain (Fig. 5E). Cumulatively, the findings shown in Fig. 5 demonstrate that the front-end basic region and rear-end acidic region of the MEKK2 PB1 domain selectively bind MEK5 and MKK7, respectively.
FIG. 5.
Role of MEKK2 PB1 domain nonconserved acidic cluster in MKK7 binding. (A) Selective binding of MEKK2 PB1 domain to MKK7 as well as MEK5. Plain HEK293 cell lysate (10 mg/sample) was provided for a pulldown assay with GST-fused MEKK2 (aa 35 to 127) and MEKK3 (aa 36 to 128) PB1 domains and MEK5 PB1 domain plus the C-terminal region (aa 18 to 131). The proteins bound were analyzed with the indicated anti-MAPK kinase or anti-ERK5 Ab. The expression of each protein (whole-cell lysate [WCL]) is shown in the left lane. (Bottom) Coomassie staining of GST fusion protein used for the assay. (B) MEKK2 PB1 domain is required for MKK7 binding. The indicated GST-fused MEKK2 fragments were used for a pulldown assay with 1 mg FLAG-MKK7 or -MEK5 transfected cell lysates. The adsorbed MKK7 and MEK5 proteins were detected with anti-FLAG Ab. (C) A conserved basic residue in MEKK2 PB1 domain is not involved in MKK7 binding. A pulldown assay was conducted with GST-fused MEKK2 (WT or 47K-A) and MEKK3 (WT or 48K-A) PB1 domains from FLAG-MEK5 or -MKK7 transfected cell lysates. The MKK7 and MEK5 proteins were detected with anti-FLAG Ab. GST-MEK5 (aa 18 to 131) was used as a negative control for the assay. (D) A nonconserved acidic cluster in the MEKK2 PB1 domain is required for MKK7 binding. FLAG-MKK7 or -MEK5 was pulled down from the transfected cells with GST-fused MEKK2 PB1 domain (aa 35 to 127) and the first or latter half of the PB1 domain (aa 35 to 79 and 77 to 127). The bound proteins were detected with anti-FLAG Ab. (E) MEK5, but not MKK7, interferes with MEK5 in MEKK2 binding. A TR-FRET assay of MEK5 (aa 18 to 131) and MEKK2 PB1 domain was conducted in the presence of the transfected cell lysates. A mixture of biotinylated MEKK2 PB1 domain and transfected cell lysates (empty vector, FLAG-WT, or -MT [64D65E-AA] MEK5 and FLAG-MKK7) was incubated in a well, and then GST-WT MEK5 (aa 18 to 131) was added to each well. The subsequent TR-FRET measurement and evaluation of the TR-FRET value were the same as described in the legend to Fig. 2.
MEKK2-MKK7 interaction does not require the MKK7 DVD site.
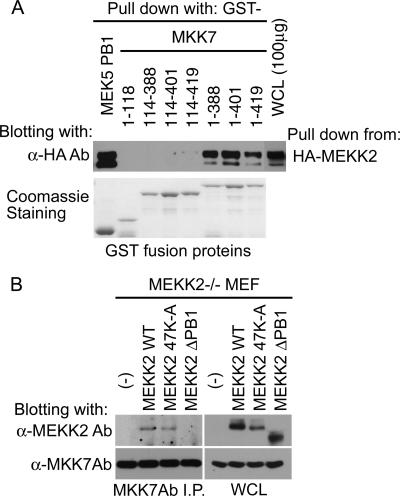
Recently, Takekawa et al. (27) reported a conserved docking site, termed DVD, in several MKKs that was shown to be necessary for binding specific MKKKs. MEK5, MEKK2, and MEKK3 were absent from the analysis. The DVD is a short sequence of approximately 20 aa which is C-terminal to the kinase domain of MKKs. To test the requirement of the MKK7 DVD site for MEKK2 binding, different GST-MKK7 fusion proteins covering the MKK7 peptide sequence were used for pulldown assays with full-length MEKK2 (Fig. 6A). There was no difference in MEKK2 binding to full-length (aa 1 to 419) MKK7 versus C-terminal MKK7 deletions with the DVD site (aa 1 to 401) or without the DVD site (aa 1 to 388) encoded in the fusion protein. These findings indicate that the DVD site in MKK7 is not required for MEKK2 binding. Interestingly, both the MKK7 N-terminal regulatory region (aa 1 to 118) and the kinase domain (aa 114 to 419) appear to be necessary for a functional interaction with MEKK2 that is clearly independent of the MKK7 DVD site.
FIG. 6.
Lack of requirement for the DVD site and N lobe in MEKK2-MKK7 binding. (A) MEKK2 binding site in MKK7. HA-MEKK2 was pulled down from the transfected cell lysates with the indicated GST-fused MKK7 fragments or MEK5 PB1 domain. The expression of HA-MEKK2 in whole-cell lysate (WCL) is presented in the right panel. MEKK2 in the precipitates was detected by anti-HA Ab blotting. (B) The requirement of MEKK2 PB1 domain for MKK7 binding in cells. MEKK2−/− MEF cells stably expressing empty vector, WT, 47K-A, or ΔPB1 MEKK2 were prepared using retroviral infection and puromycin selection. MKK7 in each growing transfectant was immunoprecipitated (I.P.) with anti-MKK7 Ab, and the immunoprecipitate was sequentially blotted with anti-MEKK2 and -MKK7 Abs (left). (Right) Expression of MEKK2 and MKK7 in the whole-cell lysates.
Further analysis of the requirements for interaction between MEKK2 and MKK7 were performed with added-back WT and mutant forms of MEKK2 in MEKK2−/− MEFs, which provided a null background of WT MEKK2 (8). Using retroviral infection, WT, 47K-A, or PB1 domain truncation mutant (ΔPB1) MEKK2 was stably expressed in the the MEKK2−/− MEFs. Assays were then performed to test the ability of immunoprecipitations of endogenous MKK7 to coimmunoprecipitate WT and mutant forms of MEKK2. Both WT and 47K-A MEKK2, but not ΔPB1 MEKK2, were detected in MKK7 immunoprecipitates (Fig. 6B), confirming that the PB1 domain was required for MKK7 binding. MKK7 binding of MEKK2 is independent of the basic residue 47K that is required for MEK5 binding.
Functional role of MEKK2 PB1 domain in JNK and ERK5 activation.
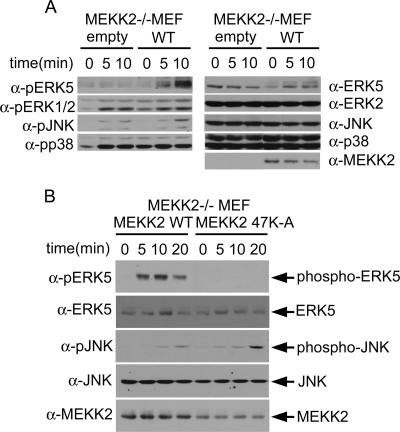
We have shown previously, using MEKK2−/− cells, that MEKK2 can stimulate both ERK5 and JNK activity (5, 8). Others have also shown that MEKK2 regulates JNK activity (6). We thus examined ERK5, ERK1/2, JNK, and p38 MAPK activity with or without adding MEKK2 back in MEKK2−/− MEFs. Hyperosmotic stress induced by sorbitol addition to the medium is a strong activator of both the ERK5 and JNK pathways. Cells stably expressing null or WT MEKK2 were stimulated with sorbitol, and MAPK activity was measured by using phosphospecific Abs that measure the activated state of specific MAPKs (Fig. 7A, left). In the absence of MEKK2 expression, sorbitol does not stimulate ERK5 activity. MEKK2 expression in the MEKK2−/− MEFs reconstituted a strong ERK5 activation in response to sorbitol. JNK activation in response to sorbitol is modest in MEKK2−/− and is augmented by adding back MEKK2 expression. The presence or absence of MEKK2 expression had little effect on ERK1/2 or p38 activation in response to sorbitol. Thus, MEKK2 regulates both ERK5 and JNK but not ERK1/2 or p38 in the MEF response to sorbitol.
FIG. 7.
Functional role of MEKK2 PB1 domain in JNK and ERK5 activation. (A) MEKK2 regulates both JNK and ERK5 activation. MEKK2−/− MEF cells stably expressing empty vector or WT-MEKK2 which had been serum starved for 8 h were treated with 100 mM sorbitol for the indicated time periods. The activation of ERK5, ERK1/2, JNK, and p38 was measured with anti-phospho-specific Ab for each molecule (left). The total protein expression of each molecule was shown on the right. (B) The 47K-A MEKK2 mutant abrogates the activation of ERK5 but enhances JNK activation. After serum starvation, MEKK2−/− MEF cells stably expressing WT or 47K-A MEKK2 were treated with 100 mM sorbitol for the indicated time periods. The cells collected were subjected to immunoblotting with anti-phospho-ERK5 and -JNK Abs. Total ERK5 or JNK expression was used to confirm the loading of equal amounts. MEKK2 expression is shown in the bottom panel.
The ability to add MEKK2 back to MEKK2−/− MEFs to reconstitute the ERK5 and augment the JNK response to sorbitol provided the assay to prove that the 47K→A MEKK2 mutant inhibited ERK5 but not JNK activation. Stable adding back of 47K→A MEKK2 in MEFs indeed augmented JNK activation but failed to reconstitute ERK5 activation (Fig. 7B). Thus, the MEKK2 PB1 domain is required for MEKK2 regulation of both ERK5 and JNK activation, with the basic region in the front end of the MEKK2 PB1 domain being essential for activation of ERK5 and the rear-end acidic region being essential for activation of JNK.
Quiescent MEKK2 preferentially binds MEK5, and activated MEKK2 binds MKK7.
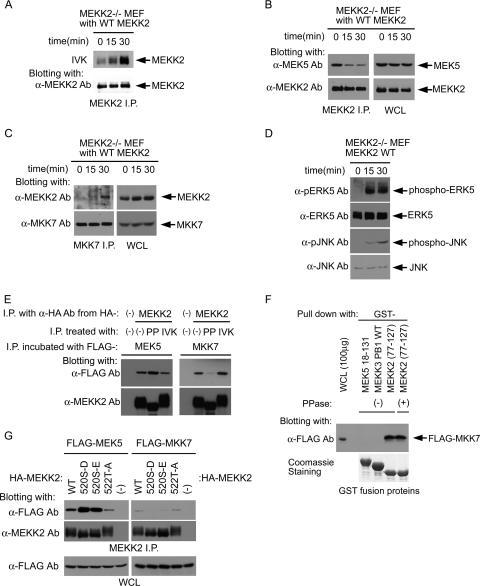
Figure 8A shows that sorbitol stimulation of MEFs activates MEKK2 kinase activity as measured by autophosphorylation in immunoprecipitates from sorbitol-stimulated cells. Coimmunoprecipitation of MEKK2 and endogenous MEK5 shows that the two kinases form a complex in unstimulated cells (Fig. 8B). Sorbitol stimulation diminishes the amount of MEK5 that coimmunoprecipitates with MEKK2, consistent with MEK5 dissociation being induced by MEKK2 activation of MEK5, which we have demonstrated using live-cell FRET analysis of MEKK2-MEK5 interactions (18). In contrast, MKK7 binding to MEKK2 increases over time following sorbitol stimulation (Fig. 8C). Figure 8D shows that ERK5 activation is faster than JNK activation in the MEF response to sorbitol. Thus, the kinetics of ERK5 and JNK activation in response to sorbitol stimulation of MEFs is consistent with MEK5-ERK5 being in a complex with MEKK2, as we have demonstrated, and MKK7 binding to MEKK2 and activation of JNK correlates temporally with the dissociation of MEK5 from the MEKK2 complex.
FIG. 8.
MEKK2 activity reciprocally regulates MEK5 and MKK7 binding. (A) MEKK2 activation upon sorbitol stimulation. After serum starvation, MEKK2−/− MEF cells stably expressing WT-MEKK2 were treated with 100 mM sorbitol for the indicated time periods. MEKK2 was immunoprecipitated (I.P.) with anti-MEKK2 Ab. An in vitro kinase reaction (IVK) was conducted with the immunoprecipitates. (Top) MEKK2 kinase activity was measured by its autophosphorylation. (Bottom) Anti-MEKK2 Ab immunoblotting of the upper panel. (B) MEK5 binding is decreased upon MEKK2 activation. MEKK2 immunoprecipitates were prepared as described for panel A. (Left) The immunoprecipitates were analyzed with anti-MEK5 and -MEKK2 Abs sequentially. (Right) Expression of MEK5 and MEKK2 in whole-cell lysates (WCL). (C) MKK7 binding is induced by MEKK2 activation. The cells were prepared and treated as described above. MKK7 was immunoprecipitated with anti-MKK7 Ab. The absorbents were analyzed with anti-MEKK2 and -MKK7 Abs. (D) Kinetics of ERK5 and JNK activation with respect to the stimulation. A portion of the total cell lysates used for A were subjected to anti-phospho-ERK5 or -JNK Ab blotting. The expression of each molecule is shown as anti-ERK5 or -JNK Ab blotting. (E) Reciprocal binding of MEK5 and MKK7 to MEKK2 in vitro. MEKK2 was immunoprecipitated from the cells transiently expressing HA-MEKK2 or empty vector with anti-HA Ab and protein G Sepharose beads. MEKK2 on the beads was treated without or with either protein phosphatase (PP) or IVK. After extensive washing, the beads were incubated with the lysates from the cells transiently expressing FLAG-MEK5 (left) or -MKK7 (right). (Top) MEK5 or MKK7 in MEKK2 immunoprecipitates was analyzed with anti-FLAG Ab. (Bottom) Amount of MEKK2 on the beads. The phosphorylation status of MEKK2 was reflected by the mobility difference as shown on a gel. (F) Phosphatase treatment of the MEKK2 (aa 77 to 127) peptide has no effect on its ability to bind MKK7. The indicated GST fusion proteins on glutathione beads were treated with or without protein phosphatase and incubated with the lysates from cells transiently expressing FLAG-MKK7. Absorbent was analyzed by blotting with anti-FLAG Ab. (Bottom) Coomassie staining of GST fusion proteins used in the assay. The activity of protein phosphatase used for this experiment was confirmed by the mobility shift of MEKK2 characteristic of its dephosphorylation (data not shown). (G) Effect of MEKK2 T-loop activation site mutation on MEK5 and MKK7 binding to MEKK2. HA-WT-MEKK2 and one of its mutants, FLAG-MEK5 or FLAG-MKK7, were individually expressed in HEK293 cells. Cell lysates (1 mg/each) expressing WT or mutant MEKK2 and either MEK5 or MKK7 were mixed and incubated for 2 h at 4°C. MEKK2 was immunoprecipitated by use of anti-MEKK2 Ab. The immunoprecipitates were blotted using anti-FLAG and anti-MEKK2 Abs.
To demonstrate that MEK5 can bind the inactive state of MEKK2 and that MKK7 preferentially binds the phosphorylated, activated state of MEKK2, we treated MEKK2 immunoprecipitates with or without λ protein phosphatase or Mg2+-ATP (Fig. 8E). The λ phosphatase dephosphorylates MEKK2 and returns it to a basal quiescent state, while addition of Mg2+-ATP results in further phosphorylation and activation of MEKK2. Activated, phosphorylated MEKK2 is shifted upward in SDS gels, which is characteristic of the phosphorylation-dependent mobility of many proteins (Fig. 8A and E) (7). Treatment with λ phosphatase reverses the phosphorylation, resulting in a faster-migrating MEKK2 protein (Fig. 8E). The addition of Mg2+-ATP to immunoprecipitates resulted in phosphorylation and an upward shift of MEKK2 on gels. Following the treatment of immunoprecipitates with or without λ phosphatase or the addition of Mg2+-ATP for in vitro kinase phosphorylation, the pellets were washed extensively and analyzed for the binding of MEK5 or MKK7. The presence of MEK5 is greatest in the λ phosphatase-treated immunoprecipitates, consistent with MEK5 being able to bind tightly to inactive MEKK2, although there is MEK5 binding in control and Mg2+-ATP-treated samples. This result is most simply interpreted as indicating that MEK5 preferentially binds inactive MEKK2 over activated MEKK2. Most strikingly and to the point, however, is the absence of MKK7 in λ phosphatase-treated immunoprecipitates, showing that MEK5 and MKK7 have converse binding preferences for active and inactive MEKK2 (Fig. 8E, right). Furthermore, the MEKK2 fragment from residues 77 to 127 efficiently binds MKK7 (Fig. 5D), and phosphatase treatment of this peptide has no effect on MKK7 binding (Fig. 8F), indicating that kinase activity independent of MEKK2 PB1 domain phosphorylation regulates MEK5 and MKK7 binding. Figure 8G shows the mutation of the T-loop phosphorylation site in MEKK2 (520S) to either an aspartate or glutamate, which renders MEKK2 an inactive kinase (37). Inactivation of MEKK2 kinase activity significantly augments MEK5 binding and attenuates MKK7 binding. Mutation of an adjacent threonine (522T) in the MEKK2 T loop has little effect on MEK5 or MKK7 binding to MEKK2. Whereas MEK5 preferentially binds to the inactive state of MEKK2, it is unequivocal that MKK7 does not bind to inactive MEKK2 and preferentially binds to the activated state of MEKK2. The results indicate that phosphorylation of the MEKK2 T-loop residues required for MEKK2 kinase activation influences the binding of MEK5 and MKK7 at the distant PB1 domain near the N terminus of the kinase.
DISCUSSION
PB1 domains have been classified into A-, B-, or AB-type PB1 domains depending on their mechanisms of interaction with other PB1 domains (16, 35). A-type PB1 domains use the acidic cluster, B-type PB1 domains use the basic cluster, and AB-type PB1 domains use both the basic and acidic clusters to bind other PB1 domains (35). According to this classification MEKK2 and MEKK3 have B-type domains and MEK5 has A-type PB1 domains. Thus, MEKK2 (or MEKK3) and MEK5 form AB-type PB1 domain heterodimers. This leaves the potential for the acidic cluster in the MEKK2 and MEKK3 PB1 domains and the basic cluster of the MEK5 PB1 domain to interact with other proteins.
The present study shows a noncanonical function for the back-end acidic cluster of the MEKK2 PB1 domain. This region of the MEKK2 PB1 domain was shown to bind MKK7, and this interaction appears to be necessary for MEKK2 activation of the JNK pathway. MEKK2 and MEKK3 have 76% homology in their PB1 domain sequences, and the back-end acidic cluster region is even more conserved, with an 81% sequence homology. Despite the high sequence homology in MEKK2 and MEKK3 PB1 domains, only MEKK2 binds MKK7. Screening for interactions of MEKK2, MEKK3, and MEK5 PB1 domains with PB1 domains from p62, Par6, and protein kinase C ζ has shown the MEK5 PB1 domain to bind the adaptor protein p62. The p62 PB1 domain was previously shown to bind protein kinase C ζ (10, 35) as well as MEK5 (13, 19), but we have not shown the p62-MEK5 interaction to be physiological in cells. So far we have not found an MKK other than MEK5 that binds MEKK3. Similar to our finding that PB1 domains have noncanonical functions in addition to binding other PB1 domains, it was recently reported that p62 binds ERK1/2 (22). It was found that the basic region of the p62 PB1 domain binds and sequesters ERK1/2 from activation by MKK1/2. The targeted knockout of p62 expression resulted in increased ERK1/2 activity. It remains unclear what other potential function the MEKK3 PB1 domain rear-end acidic cluster may have in terms of protein-protein interactions. Based on the p62 results of Rodriguez et al. (22), it is possible that MEKK3 is binding a protein other than a MKK with the acidic cluster region in its PB1 domain. Obviously, the inference is that PB1 domains may play a much greater role in forming protein complexes than PB1 hetero- or homodimers.
The ERK5 docking site 115RNIHGLKV122 in MEK5 conforms to a consensus docking site motif (R/Kxxxxhxh) by sequence alignment. It was shown experimentally that a ternary complex of MEKK2, MEK5, and ERK5 is formed in cells (Fig. 3B) (18). It should be noted that others have previously reported that MEKK2 and ERK5 share a common binding site in the MEK5 PB1 domain (25), but we are unable to see these interactions and cannot explain the data difference at this time. In the present report, we have shown that the MEKK2 PB1 domain binds MKK7 for JNK activation. Thus, the MEKK2 and MEK5 PB1 domains have developed noncanonical functions to organize different signaling complexes for the MEKK2-dependent coordinated activation of two MAPK pathways. Defining the noncanonical function of PB1 domains in organizing the coordinated regulation of two MAPK pathways illustrates for the first time how such domain architecture can be used in MKKK-MKK interactions to build specificity into the control of MAPK signaling.
It needs to be noted that the function of the MEKK2 (and MEKK3) and MEK5 PB1 domains is quite different from that of the recently described DVD site (27). MKKs have a DVD site that binds different MKKKs. The DVD site, located next to the C terminus of the kinase domain, basically interacts with the N lobe of the MKKK kinase domain. The N lobe functions primarily to catalyze substrate phosphorylation, whereas the C lobe of the kinase domain is involved in the control of kinase activity. Thus, the MKKK N-lobe interaction with the MKK DVD site seems more related to substrate recognition by MKKKs, whereas the PB1 domain of MEKK2 and MEK5 functions to form a ternary complex including ERK5, independently of the kinase domain recognition of substrate. MEKK2 and MEKK3 appear unique among MKKKs in using PB1 domains for organizing MKKK-MKK-MAPK signaling complexes. The previously reported (6) MKK7 interaction with the kinase domain of MEKK2 most likely occurs via the MKK7 DVD and the N lobe of the MEKK2 kinase domain that we have shown is not required for MEKK2-dependent activation of JNK (6). This interaction is most likely secondary to the binding of MKK7 to the MEKK2 PB1 domain acidic cluster region. We have shown that MEKK2-MEK5 and MEKK2-MKK7 complexes involve PB1 domains of MEKK2 independent of N-lobe interactions with a DVD, which are weaker and less-specific interactions than those formed by the PB1 domain of MEKK2. In this sense, MEKK2 and MEK5 recognition independent of the kinase domain of either is similar to that of yeast Pbs2p, a MKK that binds Ste11 or Ssk2/Ssk22 MKKKs. Pbs2p binding of Ssk2/Ssk22 occurs within its N-terminal regulatory domain, independently of the Pbs2p kinase domain (30).
An interesting question arises as to why MEKK2, MEKK3, and MEK5 utilize PB1 domains for the formation of a ternary signaling complex. Other MAPK signaling complexes frequently utilize scaffold proteins for organizing MAPK signaling complexes. Examples include KSR (kinase suppressor of Ras) for Raf-MKK1/2-ERK1/2 activation in response to Ras.GTP (15) and JNK-interacting proteins for MLK-MKK4/7-JNK activation (34). These scaffold proteins function in controlling the spatiotemporal dynamics of MAPK signaling in response to different stimuli and generally regulate a single MAPK pathway: ERK1/2, JNK, or p38. To date, neither MEK5 nor ERK5 has been discovered to bind to any scaffold protein for the control of ERK5 activation. MEKK3 has been shown to bind the scaffold protein OSM/CCM2 independent of the MEKK3 PB1 domain (32). The OSM (osmosensing scaffold for MEKK3)/CCM2 (cerebral caverous malformation 2) scaffold also binds MKK3, selectively engineering the activation of p38 and not ERK5 in response to different stimuli. Thus, PB1 domains can selectively organize MEKK2 (and MEKK3) and MEK5 in complexes for ERK5 regulation, while a scaffold protein like OSM/CCM2 that binds an MKKK like MEKK3 and an MKK like MKK3 would be used for preferential MEKK3 regulation of p38 activation. An analogy would be Ste5 organizing the pheromone response by scaffolding the Ste11, Ste7, and Fus3 proteins and Pbs2p organizing the osmotic stress response via the Ssk2/Ssk22-Pbs2p-Hog1 signaling complex (24). In addition, as known for Ste11, different MKKKs, such as MEKK3, are able to selectively control one or more MAPK pathways by the scaffold assembly of different MKKs and MAPKs.
The critical role of ERK5 signaling for vascular development in embryos and vascular maintenance in the adult appears to be a critical function of ERK5 signaling (9). Although MEKK2 knockouts have normal vasculature the knockout of MEKK3 (36), MEK5 (33), and the transcription factor MEF2C (14) mimics ERK5 deletion in vascular development. Targeting MEKK2/3 PB1-MEK5 PB1 domain interactions might provide a very specific disruption of ERK5 activation to modulate vascularization in different diseases. PB1 domain interactions or the ERK5-MEK5 docking site interaction is independent of the ATP binding sites of kinases and might prove to be tractable targets much like the JNK binding domain inhibitory peptide for JNK inhibition (3).
Acknowledgments
This work was supported by NIH grants GM68820 and GM30324.
We thank Nancy Johnson for assistance with graphics.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, L. 2006. Mechanisms of MAPK signalling specificity. Biochem. Soc. Trans. 34:837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogoyevitch, M. A. 2005. Therapeutic promise of JNK ATP-noncompetitive inhibitors. Trends Mol. Med. 11:232-239. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. I., B. E. Xu, R. Akella, M. H. Cobb, and E. J. Goldsmith. 2002. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9:1241-1249. [DOI] [PubMed] [Google Scholar]
- 5.Chayama, K., P. J. Papst, T. P. Garrington, J. C. Pratt, T. Ishizuka, S. Webb, S. Ganiatsas, L. I. Zon, W. Sun, G. L. Johnson, and E. W. Gelfand. 2001. Role of MEKK2-MEK5 in the regulation of TNF-alpha gene expression and MEKK2-MKK7 in the activation of c-Jun N-terminal kinase in mast cells. Proc. Natl. Acad. Sci. USA 98:4599-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J., J. Yang, Y. Xia, M. Karin, and B. Su. 2000. Synergistic interaction of MEK kinase 2, c-Jun N-terminal kinase (JNK) kinase 2, and JNK1 results in efficient and specific JNK1 activation. Mol. Cell. Biol. 20:2334-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, J., L. Yu, D. Zhang, Q. Huang, D. Spencer, and B. Su. 2005. Dimerization through the catalytic domain is essential for MEKK2 activation. J. Biol. Chem. 280:13477-13482. [DOI] [PubMed] [Google Scholar]
- 8.Garrington, T. P., T. Ishizuka, P. J. Papst, K. Chayama, S. Webb, T. Yujiri, W. Sun, S. Sather, D. M. Russell, S. B. Gibson, G. Keller, E. W. Gelfand, and G. L. Johnson. 2000. MEKK2 gene disruption causes loss of cytokine production in response to IgE and c-Kit ligand stimulation of ES cell-derived mast cells. EMBO J. 19:5387-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi, M., and J. D. Lee. 2004. Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J. Mol. Med. 82:800-808. [DOI] [PubMed] [Google Scholar]
- 10.Hirano, Y., S. Yoshinaga, K. Ogura, M. Yokochi, Y. Noda, H. Sumimoto, and F. Inagaki. 2004. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J. Biol. Chem. 279:31883-31890. [DOI] [PubMed] [Google Scholar]
- 11.Hirano, Y., S. Yoshinaga, R. Takeya, N. N. Suzuki, M. Horiuchi, M. Kohjima, H. Sumimoto, and F. Inagaki. 2005. Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J. Biol. Chem. 280:9653-9661. [DOI] [PubMed] [Google Scholar]
- 12.Holland, P. M., and J. A. Cooper. 1999. Protein modification: docking sites for kinases. Curr. Biol. 9:R329-R331. [DOI] [PubMed] [Google Scholar]
- 13.Lamark, T., M. Perander, H. Outzen, K. Kristiansen, A. Overvatn, E. Michaelsen, G. Bjorkoy, and T. Johansen. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568-34581. [DOI] [PubMed] [Google Scholar]
- 14.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison, D. K. 2001. KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci. 114:1609-1612. [DOI] [PubMed] [Google Scholar]
- 16.Moscat, J., M. T. Diaz-Meco, A. Albert, and S. Campuzano. 2006. Cell signaling and function organized by PB1 domain interactions. Mol. Cell 23:631-640. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, K., and G. L. Johnson. 2003. PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J. Biol. Chem. 278:36989-36992. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura, K., M. T. Uhlik, N. L. Johnson, K. M. Hahn, and G. L. Johnson. 2006. PB1 domain-dependent signaling complex is required for ERK5 activation. Mol. Cell. Biol. 26:2065-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda, Y., M. Kohjima, T. Izaki, K. Ota, S. Yoshinaga, F. Inagaki, T. Ito, and H. Sumimoto. 2003. Molecular recognition in dimerization between PB1 domains. J. Biol. Chem. 278:43516-43524. [DOI] [PubMed] [Google Scholar]
- 20.Pearson, G. W., S. Earnest, and M. H. Cobb. 2006. Cyclic AMP selectively uncouples mitogen-activated protein kinase cascades from activating signals. Mol. Cell. Biol. 26:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranganathan, A., G. W. Pearson, C. A. Chrestensen, T. W. Sturgill, and M. H. Cobb. 2006. The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch. Biochem. Biophys. 449:8-16. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, A., A. Duran, M. Selloum, M. F. Champy, F. J. Diez-Guerra, J. M. Flores, M. Serrano, J. Auwerx, M. T. Diaz-Meco, and J. Moscat. 2006. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3:211-222. [DOI] [PubMed] [Google Scholar]
- 23.Rubinfeld, H., T. Hanoch, and R. Seger. 1999. Identification of a cytoplasmic-retention sequence in ERK2. J. Biol. Chem. 274:30349-30352. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz, M. A., and H. D. Madhani. 2004. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38:725-748. [DOI] [PubMed] [Google Scholar]
- 25.Seyfried, J., X. Wang, G. Kharebava, and C. Tournier. 2005. A novel mitogen-activated protein kinase docking site in the N terminus of MEK5α organizes the components of the extracellular signal-regulated kinase 5 signaling pathway. Mol. Cell. Biol. 25:9820-9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharrocks, A. D., S. H. Yang, and A. Galanis. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25:448-453. [DOI] [PubMed] [Google Scholar]
- 27.Takekawa, M., K. Tatebayashi, and H. Saito. 2005. Conserved docking site is essential for activation of mammalian MAP kinase kinases by specific MAP kinase kinase kinases. Mol. Cell 18:295-306. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 29.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20:466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatebayashi, K., M. Takekawa, and H. Saito. 2003. A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J. 22:3624-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tournier, C., A. J. Whitmarsh, J. Cavanagh, T. Barrett, and R. J. Davis. 1999. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol. Cell. Biol. 19:1569-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlik, M. T., A. N. Abell, N. L. Johnson, W. Sun, B. D. Cuevas, K. E. Lobel-Rice, E. A. Horne, M. L. Dell'Acqua, and G. L. Johnson. 2003. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 5:1104-1110. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., A. J. Merritt, J. Seyfried, C. Guo, E. S. Papadakis, K. G. Finegan, M. Kayahara, J. Dixon, R. P. Boot-Handford, E. J. Cartwright, U. Mayer, and C. Tournier. 2005. Targeted deletion of mek5 causes early embryonic death and defects in the extracellular signal-regulated kinase 5/myocyte enhancer factor 2 cell survival pathway. Mol. Cell Biol. 25:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitmarsh, A. J. 2006. The JIP family of MAPK scaffold proteins. Biochem. Soc. Trans. 34:828-832. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, M. I., D. J. Gill, O. Perisic, M. T. Quinn, and R. L. Williams. 2003. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12:39-50. [DOI] [PubMed] [Google Scholar]
- 36.Yang, J., M. Boerm, M. McCarty, C. Bucana, I. J. Fidler, Y. Zhuang, and B. Su. 2000. Mekk3 is essential for early embryonic cardiovascular development. Nat. Genet. 24:309-313. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, D., V. Facchinetti, X. Wang, Q. Huang, J. Qin, and B. Su. 2006. Identification of MEKK2/3 serine phosphorylation site targeted by the Toll-like receptor and stress pathways. EMBO J. 25:97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]