Abstract
Antiretroviral treatment failure is associated with the emergence of resistant human immunodeficiency virus type 1 (HIV-1) populations which often express altered replicative capacity (RC). The resistance and RC of clinical HIV-1 strains, however, are generally assayed using activated peripheral blood mononuclear cells (PBMC) or tumor cell lines. Because of their high proliferation rate and concurrent high deoxynucleoside triphosphate (dNTP) content, both resistance and RC alterations might be misestimated in these cell systems. We have evaluated the resistance of HIV-1 clones expressing a variety of RT resistance mutations in primary human macrophages using a single cycle system. Our experiments indicate that d4T, ddI, and 3TC are more potent in macrophages than in HeLa-derived P4 tumor cells. Mutant viruses bearing thymidine analogue mutations (TAMs) or the K65R mutation had similar resistance levels in the two cell types. Strikingly, however, the M184V mutant, although fully resistant to 3TC in P4 cells, maintained some susceptibility to 3TC in macrophages from 8 of 11 donors. Using the same system, we found that the impact of resistance mutations on HIV RC was minimal in activated PBMC and in P4 cells. In contrast, mutant viruses exhibited strongly impaired RC relative to the wild type (WT) in macrophages, with the following RC order: WT > two TAMs > four TAMs = M184V > K65R. In undifferentiated monocytes, WT virus replication could be detected in three of six donors, but replication of all mutant viruses remained undetectable. Altogether, our results confirm that nucleoside reverse transcriptase inhibitors (NRTIs) are powerful antiviral agents in differentiated macrophages, reveal that HIV resistance to some NRTIs may be less efficient in these cells, and indicate that resistance-associated loss of RC is more pronounced in macrophages than in high-dNTP content cell systems.
Nucleoside analogues, also referred to as nucleoside reverse transcriptase inhibitors (NRTIs) are one of the main drug classes used in the treatment of human immunodeficiency virus type 1 (HIV-1) infection. After phosphorylation into their active triphosphate form by cellular kinases, these compounds, which lack a 3′-hydroxyl group, inhibit reverse transcription through termination of viral DNA synthesis. HIV-1 resistance to NRTIs is achieved via two distinct and generally exclusive mechanisms: (i) decreased incorporation of the triphosphorylated NRTI or (ii) excision of the triphosphorylated NRTI and primer unblocking. Decreased NRTI incorporation is mediated by mutations such as the M184V substitution, which induces high-level resistance to β-l-2′,3′-dideoxy-3′-thiacytidine (3TC or lamivudine) (24, 35, 61, 64), as well as other mutations such as K74V, K65R, or the Q151M complex. These mutations promote resistance through impairment of the correct positioning of triphosphate NRTIs within the RT active site (19, 67). In contrast, thymidine analogue mutations (TAMs), such as the M41L, the D67N, the K70R, the L210W, the T215F/Y, and the K219Q substitutions, which are essentially selected for in vivo by thymidine analogues (20, 37, 51, 57), induce resistance to most nucleoside analogues by favoring excision of the incorporated NRTI from the terminated viral DNA strand. This excision process occurs through transfer of the chain-terminating residue to an ATP or pyrophosphate nucleotide acceptor by HIV-1 RT. ATP-mediated removal of thymidine analogues such as 3′-azido-3′-deoxythymidine (AZT or zidovudine) or 2′,3′-didehydro-2′,3′-dideoxythymidine (d4T or stavudine), as well as of other NRTIs such as ddATP (the active form of 2′,3′-dideoxyinosine [ddI or didanosine]) and 2′,3′-dideoxycytidine (ddC or zalcitabine), is increased in RTs carrying TAMs (40-43, 46, 48).
Antiviral activity of nucleoside analogues and viral resistance to these compounds is highly dependent on the metabolism of nucleoside and nucleotides in HIV target cells. First, the extent of dideoxynucleoside triphosphate (ddNTP)-mediated chain termination has been found to be dependent upon the state of cellular proliferation and upon both dNTP and NRTI triphosphate pools, based on experiments with different cell types or on in vitro reverse transcription assays (8, 11, 25). Second, primer unblocking by TAMs is known to be affected by intracellular dNTP concentrations. Indeed, in vitro, while primer rescue of AZT-terminated DNA by TAMs is highly efficient regardless of dNTP concentrations, pyrophosphorolysis of d4T is strongly inhibited by high concentrations of the next complementary dNTP (50% inhibitory concentration [IC50] ≈ 0.5 μM) (40-42). It is assumed that higher concentrations of dNTPs favor translocation of the chain-terminated primer within the HIV RT primer grip, thereby reducing the chances that ATP or pyrophosphate react with the phosphodiester bond that links the terminating NRTI to the upstream nucleotide. Such differences in susceptibility to excision are generally explained by the fact that, unlike d4T, AZT has a large 3′ radical that precludes the formation and further translocation of a stable complex between the chain-terminated primer-template and RT (IC50 for dNTP-mediated AZT excision of >100 μM) (41) and is therefore not affected by dNTP concentrations.
In spite of the potential impact of cell division and DNA turnover on antiviral activity of NRTIs and on HIV resistance to these compounds, HIV-1 drug susceptibility is usually measured in mitogen-activated peripheral blood mononuclear cells (PBMC) or in tumor cell lines. Similarly, the replicative capacity (RC) of resistant viruses, which has been shown to be highly affected by cellular concentrations of dNTPs, is also measured in cells where DNA turnover and dNTP content are markedly higher than in most primary human cells (3, 8, 25, 65). This is particularly true in quiescent lymphocytes, in which dNTP concentrations are 6- to 20-fold lower than in activated lymphocytes (3, 8, 25), and in cells of the monocyte/macrophage lineage, which also display low dNTP content and do not undergo significant cell division cycles after differentiation (65). Together with CD4+ T lymphocytes, monocytes/macrophages are essential target cells for HIV-1 and are believed to play a crucial role in HIV infection and pathogenesis. Cells of the monocyte/macrophage lineage are thought to be involved in spreading of the virus throughout the body because they are widely distributed in all body tissues and compartments, including the brain, where they constitute the majority of cells infected by HIV-1 (32, 38, 39). Moreover, because of their long half-life and relative resistance to the cytopathogenic effect of the HIV, they may act as a viral reservoir that may play an important part in transmission of HIV to CD4+ T lymphocytes (1, 26, 28, 33, 38, 39, 49, 50, 62).
Although most NRTIs, non-NRTIs, and protease inhibitors have been shown to have potent antiviral activity in monocytes/macrophages (2, 4, 7, 27, 52, 54), little is known about resistance and resistance associated changes in viral RC in this cellular compartment. To our knowledge, the resistance of viruses carrying TAMs or other NRTI-associated resistance mutations in the low dNTP cellular context of monocytes/macrophages has not been addressed. However, the high ddNTP/dNTP ratio in macrophages may have a direct impact on the outcome of phenotypic assays in that it may affect not only the antiviral activity of NRTIs on the wild-type (WT) virus, as already reported by Aquaro et al. for some NRTIs (1, 2), but also resistance due to mutations that act through decreased incorporation (M184V and K65R). The low endogenous dNTP content of these cells might unmask the loss of processivity of viral RT due to resistance mutations (14) and have an effect on the excision of incorporated NRTIs with a small 3′ radical, such as d4T or ddI.
Here we studied resistance of HIV-1 mutant viruses harboring TAMs and other NRTI resistance mutations to AZT, d4T, ddI, 3TC, and TDF in primary human monocyte-derived macrophages (MDMs). Our results show that overall resistance phenotypes of TAM-bearing viruses and of the K65R mutant virus are similar in primary human MDMs and in a HeLa-derived tumor cell line. Interestingly, however, the M184V mutant virus exhibited a markedly lower change in 3TC IC50 relative to the WT virus in MDMs from 8 of 11 donors than in the HeLa-derived tumor cell line, due essentially to a higher 3TC-triphosphate (3TCTP)/dCTP ratio in MDMs. We also compared the RCs of these mutant viruses in undifferentiated monocytes, in MDMs, in phytohemagglutinin (PHA)-stimulated PBMC, and in the HeLa-derived tumor cell line. We report that the RC of most RT mutants is significantly lower in MDMs than in stimulated PBMC or tumor cells.
MATERIALS AND METHODS
Nucleoside analogues.
AZT, d4T, and ddI were purchased from Sigma. 3TC was a generous gift from Glaxo Smith Kline, and TDF was kindly donated by Gilead.
Cells.
LTR-LacZ HeLa-derived CD4+ P4 tumor cells (18) were cultured in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U of penicillin/ml, 100 μg of streptomycin/ml (all from Gibco-BRL), and 500 μg of Geneticin/ml to select for the stably transfected LTR-LacZ gene. PBMC were isolated from the buffy coats from healthy blood donors by Ficoll-Hypaque density gradient centrifugation (Pharmacia Biotech), washed twice in cold phosphate-buffered saline (PBS)-0.3 mM EDTA, and washed once in cold PBS alone. PBMC were resuspended in RPMI medium supplemented with 10% heat-inactivated FCS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at a density of 106 cells/ml; stimulated with 1 U of PHA (J2L Elitech)/ml for 72 h; and then cultured in RPMI medium containing 5 U of recombinant human interleukin-2 (Roche)/ml (referred to as PHA-stimulated PBMC).
Monocytes were isolated from PBMC by adherence to plastic for 1 h at 37°C in RPMI medium enriched with 100 U of penicillin/ml, 100 μg of streptomycin/ml, nonessential amino acids, 50 μg of β-mercaptoethanol/ml, and HEPES (all from Gibco-BRL) (hereafter referred to as MDM medium) and 2% heat-inactivated human AB serum (ABS; Institut Jacques Boy, France); nonadherent cells were washed off, and adherent cells were washed a further three times with PBS and then cultured overnight in MDM medium containing 10% heat-inactivated human ABS. The following morning, monocytes were harvested by incubation at 4°C for 1 h, vortexing, and gentle scraping. Cells were counted, seeded at a concentration of 105 cells/well in 96-well plates in MDM medium enriched with 10% heat-inactivated human ABS, and allowed to differentiate into macrophages for 6 to 8 days. MDM populations expressed >95.5% CD14, CD64, CD32, and CD16, whereas CD2 and CD3 contaminants remained at <2.5%, as assessed by flow cytometry.
Construction of plasmids.
HIV infectious molecular clones were derived from an NL4-3 backbone carrying a Luc (Firefly luciferase) reporter gene in the place of nef and a stop codon in the env gene (NL4-3LUC+ΔNefΔEnv, a generous gift from T. Dragic, Albert Einstein College of Medicine, Bronx, NY). Several resistance mutations were introduced into the RT sequence of the NL4-3LUC+ΔNefΔEnv plasmid. For viruses bearing two TAMs at positions 41 and 215 (M41L+T215Y), four TAMs at positions 41, 215, 67, and 210 (M41L+T215Y+D67N+L210W), or the M184V mutation, the SalI-SpeI fragment from previously described NL4-3XCS mutant vectors (14, 34) was ligated into the SalI-SpeI fragment of NL4-3LUC+ΔNefΔEnv. NL4-3XCS is a variant of NL4-3 that carries an XbaI site upstream of the protease coding region, a ClaI site at the junction between the protease and the RT, and a SnaBI site at the junction between the RT and the RNase H coding sequences. The K65R mutation was inserted by site-directed mutagenesis of a pBluescript II SK plasmid (Stratagene) carrying the ClaI-EcoRI fragment of HIV-1 NL4-3 (pSK-RTS plasmid). The K65R mutation was introduced by using the following primers: sense, 5′-CCATTTAGTACTGTCTTTTTCTCTTTATGGCAAATACTGG-3′; and antisense, 5′-CCAGTATTTGCCATAAAGAGAAAAAGACAGTACTAAATGG-3′. The correct introduction of the mutation was verified by direct sequencing. The ClaI-SnaBI fragment was cloned into NL4-3XCS, and the SalI-SpeI fragment of the NL4-3XCS-K65R plasmid was cloned back into the SalI-SpeI sites of pNL4-3LUC+ΔNefΔEnv. The intermediate cloning step into NL4-3XCS was necessary because pNL4-3LUC+ΔNefΔEnv bears two ClaI restriction sites: one at position 2562 (within the RT) and one within the Luc gene.
Preparation of viral stocks.
To produce pseudotyped viral stocks for infection of primary cells (monocytes, MDMs, and PBMC), HEK 293T cells (6 × 106 cells in T75 flasks) were cotransfected with 16 μg of the pNL4-3LUC+ΔNefΔEnv-derived mutant plasmids and 16 μg of a CMV-VSV-G expression vector by using the calcium phosphate precipitation method. For infection of P4 cells, viral stocks were produced by cotransfecting 16 μg of the pNL4-3LUC+ΔNefΔEnv-derived vectors and only 0.2 μg of the CMV-VSV-G expression vector, in order to avoid VSV-G-mediated cytotoxicity in these cells. Cells were washed 6 h posttransfection and 10 ml of fresh medium were added. The viral supernatant was harvested 24 h later and centrifuged, and aliquots were frozen at −80°C.
Resistance assays.
For experiments with MDMs, all drug and virus dilutions, as well as infection experiments, were carried out in MDM medium supplemented with 10% heat-inactivated FCS, in order to avoid human serum-induced particle inhibition (31). For experiments with P4 cells, Dulbecco modified Eagle medium enriched with 10% FCS was used.
In order to measure NRTI resistance of the various NL4-3LUC+ΔNefΔEnv-derived viruses in MDMs, 105 MDMs were pretreated with serial dilutions of AZT (0.0001 to 10 μM), d4T (0.0001 to 10 μM), ddI (0.0001 to 100 μM), 3TC (0.0001 to 100 μM), and TDF (0.0001 to 100 μM) for 16 h or for 4 h prior to infection or exposed to the drug at the time of infection and infected with the NL4-3LUC+ΔNefΔEnv-derived viruses (80 ng of p24/ml). Drugs were maintained throughout the culture.
For resistance experiments in P4 cells, 104 cells were plated in 96-well plates 24 h before infection and treated with serial dilutions of AZT (0.0005 to 50 μM), d4T (0.008 to 25 μM), ddI (0.0016 to 50 μM), 3TC (0.1 to 300 μM), and TDF (0.0001 to 100 μM) for 16 h, for 4 h prior to infection, or exposed to the drugs concomitantly to infection with 20 ng of p24/ml of the NL4-3LUC+ΔNefΔEnv-derived pseudotyped viruses. No drug-dependent cell toxicity could be detected by using an MTT toxicity assay, even with 100 μg of AZT, d4T, ddI, or TDF or with 300 μg of 3TC.
Input virus (as estimated by HIV p24 ELISA [Innogenetics]) was adjusted to the number of cells in the well in order to have equivalent amounts of p24 per cell.
Infection was monitored by quantifying the luciferase activity in cellular lysates 72 h postinfection for MDMs and 40 h postinfection for P4 cells. The percent infection in the presence of each NRTI with respect to infection in untreated cells was calculated, and the means were plotted against the NRTI concentration to calculate the IC50 for each drug by using Prism software. Resistance was expressed as the fold change in IC50, i.e., the ratio of the IC50 of the mutant clone to the IC50 of the WT virus.
RC assays.
Viral RC was assayed in P4 tumor cells, PHA-stimulated PBMC, monocytes, and MDMs. Cells were infected with serial dilutions of the NL4-3LUC+ΔNefΔEnv-derived viral clones carrying the previously described resistance mutations. Viral input was normalized between the different cell types to match the level of p24 used to infect 105 MDMs (i.e., if the highest virus concentration used to infect 105 MDMs and 105 monocytes was 400 ng/ml, then 40 ng of p24/ml was used to infect 104 P4 cells and 160 ng of p24/ml was used to infect 105 PBMC containing ca. 40% CD4+ T lymphocytes). The RC was determined by calculating the slope of the curve plotting luciferase activity against p24 input in its linear portion. The relative RC of the mutant viruses was expressed as a percentage of the WT.
Infection was monitored by quantifying the luciferase activity in cell lysates at 40 h postinfection in HeLa P4 cells and at 72 h postinfection in primary cells (PHA-stimulated PBMC, monocytes, and MDMs).
Quantification of intracellular dNTP pools.
A total of 5 × 106 MDMs or 107 P4 cells were cultured in the absence of any drug or in the presence of 1, 0.1, 0.01, or 0.001 μM AZT, d4T, ddI, or 3TC for 6 h or for 24 h. Cells were then washed twice with 0.9% NaCl (pH 7.5), lysed with 500 μl of lysis buffer (0.05 M Tris-HCl [pH 7.4]-70% ethanol solution), and frozen at −80°C. Cell extracts were analyzed by liquid chromatography coupled to mass spectroscopy to quantify endogenous dNTP and NRTI-triphosphate concentrations in control and treated MDMs and P4 cells (12, 13, 29). Mean endogenous dNTP concentrations/106 cells were compared in both cell types (Fig. 1). In order to minimize differences inherent to donor cells in MDMs, the NRTI-triphosphate/paired endogenous dNTP concentration ratios were compared (see Fig. 5).
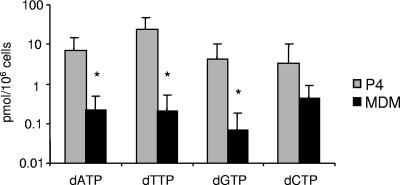
FIG. 1.
dNTP concentrations in MDMs and in HeLa P4 cells. Endogenous dNTPs were measured in MDM and in HeLa P4 tumor cells by high-pressure liquid chromatography. The results reported here represent the mean concentrations of seven independent experiments for HeLa P4 cells and of 15 independent donors for MDMs. The data are expressed as dNTP concentrations/106 cells. An asterisk indicates that these values were significantly different in MDMs than in P4 cells (P < 0.01).
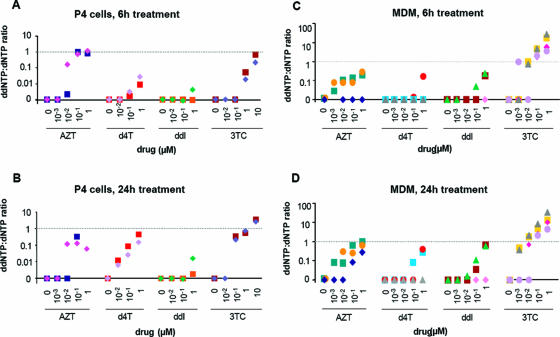
FIG. 5.
Triphosphate NRTIs in P4 cells and in MDMs. P4 cells (107) and MDMs (5 × 106) were exposed to serial dilutions (ranging from 0.001 to 1 μM) of AZT, d4T, ddI, and 3TC for 6 or 24 h. The cells were washed, and NRTI-triphosphates, as well as the corresponding dNTP were quantified in each cell lysate by high-pressure liquid chromatography. The NRTI-triphosphate/corresponding dNTP ratios cells were calculated in 106 P4 cells treated for 6 h (A) or 24 h (B) and in MDMs treated for 6 h (C) or 24 h (D). The NRTI-triphosphate/dNTP ratios are represented as single shape and color symbols on the graph: for P4 cells, each symbol represents an independent experiment; for MDMs, each symbol represents the NRTI-triphosphate/dNTP ratio for cells from a single donor.
Statistical analyses.
The IC50 values and fold changes in IC50 were compared by using a one-way analysis of variance (ANOVA) test, unless otherwise specified. A P value of <0.05 was considered to be significant. When groups differed significantly, a Bonferroni's multiple-comparison post-test was performed to make comparisons two by two. dNTP contents in MDMs and P4 cells were compared by using an unpaired t test.
RESULTS
Intracellular dNTP concentrations in MDMs.
Primary human macrophages have been reported to have lower dNTP levels than CD4+ T lymphocytes or tumor cells (23, 60). To confirm that this was indeed the case in the experimental system used here, we measured dNTP concentrations in MDMs from 15 healthy blood donors and in HeLa-derived P4 tumor cells by liquid chromatography coupled to mass spectroscopy. As shown in Fig. 1, mean dATP, dGTP, and dTTP concentrations were >100-fold lower in MDMs than in P4 cells, and dCTP concentrations were 15-fold lower in MDMs than in P4 cells. The differences in dATP, dGTP, and dTTP contents were statistically significant (P = 0.002 for dATP, P = 0.003 for dGTP, and P = 0.002 for dTTP). dCTP was the most abundant and dGTP was the most scarce nucleotide in the MDMs.
Susceptibility of WT NL4-3 to NRTIs in MDMs.
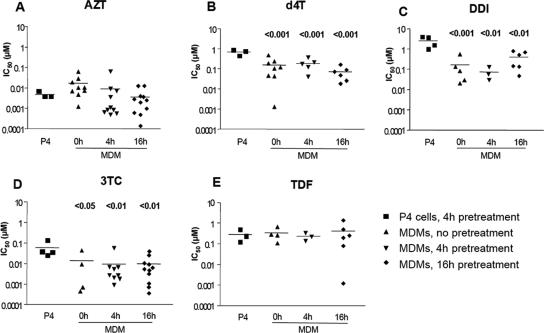
Although numerous studies (2, 4, 27, 52, 54) have reported on the activity of NRTIs in MDMs, we wanted to calibrate the activity of these compounds in the single-cycle, luciferase-based, VSV-pseudotype MDM infection assay system developed here. We compared the antiviral activity of five NRTIs—AZT, d4T, ddI, 3TC, and TDF—in MDMs and in P4 cells by using a WT NL4-3-derived, VSV-G-pseudotyped virus bearing a Luc reporter gene in the place of Nef. In these experiments, MDMs and P4 cells were precultured with the drugs for different time periods prior to infection: a long pretreatment (16 h), a short pretreatment (4 h), or no pretreatment, where cells were exposed to the drug at the time of infection (0 h). In P4 cells, the time of exposure of the cells to the drug prior to infection did not affect the IC50 values for any of the drugs tested. Therefore, further experiments with P4 cells in the present study were all carried out with a 4-h pretreatment. The mean IC50 values were 0.004 μM for AZT, 0.61 μM for d4T, 2.62 μM for ddI, 0.06 μM for 3TC, and 0.27 μM for TDF (Table 1).
TABLE 1.
IC50 values for five NRTIs in P4 cells and in MDMs infected with WT NL4-3-derived virus
| NRTI | Mean IC50 ± SEM (no. of expts)a in:
|
|||
|---|---|---|---|---|
| P4 cells (4-h pretreatment) | MDMs
|
|||
| No pretreatment | 4-h pretreatment | 16-h pretreatment | ||
| AZT | 0.004 ± 0.001 (3) | 0.015 ± 0.052 (9) | 0.007 ± 0.005 (10) | 0.003 ± 0.001 (11) |
| d4T | 0.61 ± 0.11 (3) | 0.14 ± 0.05‡ (8) | 0.17 ± 0.05‡ (5) | 0.07 ± 0.02‡ (6) |
| ddI | 2.62 ± 0.76 (4) | 1.70 ± 0.10‡ (5) | 0.07 ± 0.030† (3) | 0.36 ± 0.14† (6) |
| 3TC | 0.06 ± 0.038 (4) | 0.015 ± 0.011* (4) | 0.010 ± 0.006† (9) | 0.010 ± 0.004† (11) |
| TDF | 0.27 ± 0.10 (3) | 0.33 ± 0.12 (4) | 0.23 ± 0.06 (3) | 0.40 ± 0.20 (6) |
The mean IC50 values were calculated from at least three independent experiments. The number of independent experiments is indicated in parentheses.
, P < 0.05;
, P < 0.01;
, P < 0.001 (comparison of IC50 values in MDMs and in P4 cells).
In MDMs, IC50 values displayed important interdonor variability. The mean IC50 values measured in MDMs, however, were comparable to those measured in P4 cells for AZT and TDF, independently of the time of pretreatment (Fig. 2 and Table 1). For d4T, ddI, and 3TC, the mean IC50 values measured in MDMs were significantly lower than those measured in P4 cells (Fig. 2 and Table 1). For d4T, the mean IC50 value in MDMs was slightly lower in cells treated for 16 h before infection (0.065 μM) than in cells treated for 4 h (0.171 μM) or in cells treated at the time of infection (0.141 μM). With ddI, the mean IC50 values varied from 1.699 μM in cells treated at the time of infection to 0.074 μM in cells pretreated for 4 h and to 0.359 μM in MDMs pretreated for 16 h. For 3TC, the mean IC50 was fivefold lower in MDMs than in P4 cells (Fig. 2 and Table 1) in accordance with what has been previously described in a study performed in PBMC (58). Taken together, these data illustrate the impact of the cellular context on the antiviral activity of some NRTIs such as d4T, ddI, and 3TC. The data suggest that some NRTIs may be more potent RT inhibitors in MDMs than in cells with high dNTP concentrations, as previously reported (2, 4, 52, 54).
FIG. 2.
IC50 values for five NRTIs in P4 cells and in MDMs infected with WT NL4-3. HeLa P4 cells (104 cells in 96-well microtiter plates) were pretreated with different drugs for 4 h and then infected with 8 ng of WT NL4-3ΔenvΔNef-Luc+/VSV-G virus p24/ml. MDMs (105 cells in 96-well microtiter plates) were pretreated with the same NRTIs at the time of infection (0 h) or for 4 or 16 h prior to infection with 80 ng/ml p24 of WT NL4-3ΔenvΔNef-Luc+/VSV-G-pseudotyped virus. Infection was monitored by quantifying the luciferase activity 40 h postinfection in P4 cells and 72 h postinfection in MDMs. The experiments presented here were performed with MDMs from at least three different independent donors tested in independent experiments and represent the means of quadruplicate cultures for each donor. Each point represents the mean of quadruplicate cultures of cells from a single donor. Graphs report the IC50 values measured for AZT (A), d4T (B), ddI (C), 3TC (D), and TDF (E). The differences in mean IC50 measured for each drug in HeLa P4 tumor cells and in MDMs pretreated with the NRTIs for different times were compared by using a one-way ANOVA test, followed by a Bonferroni's multiple-comparison post-test comparing the p4 IC50 to each of the IC50s measured in MDMs for each pretreatment length when differences between groups were significant. When significant, the P values are reported on the graph.
Resistance to NRTIs in MDMs.
We next studied resistance in MDMs of HIV-1 RT mutants carrying resistance mutations representative of three main resistance pathways. TAMs, which emerge in vivo as a result of insufficient HIV suppression by antiretroviral regimens that include AZT or d4T, are known to confer different degrees of resistance to most nucleoside and nucleotide analogues. The viruses tested here carried either two TAMs (M41L and T215Y) or four TAMs (M41L+T215Y+L210W+D67N). Of note, although mutation D67N is most often found in association with mutations T215F and K219Q or K219E, it is also frequently associated with the mutations present on viruses tested here. The typical 3TC resistance mutation M184V was also used for experiments on resistance to 3TC, and the K65R substitution was used for experiments on resistance to TDF. As with WT NL4-3, preliminary analyses revealed that the time of pretreatment did not affect the IC50 of the NRTIs in P4 cells infected with mutant viruses. Therefore, in all P4 experiments, cells were pretreated with the NRTI for 4 h before infection. In MDMs, however, resistance assays were performed with different pretreatment times. Again, important interdonor variability was observed with MDMs, probably due to intrinsic differences between donors regarding the state of differentiation and activation of MDMs. In most cases, this variability decreased with longer preexposure of the cells to the drug before infection.
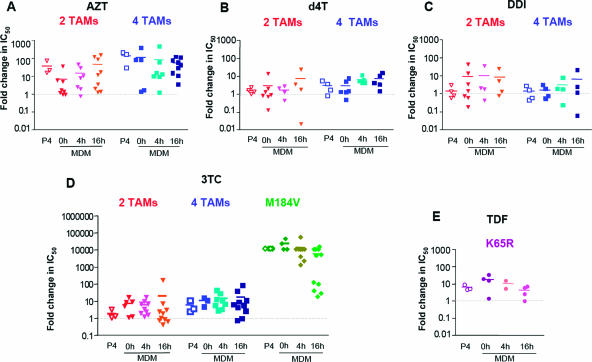
Overall, resistance profiles and fold changes in IC50 for nucleoside analogues were similar in P4 cells and in MDMs infected with the two-TAM (M41L+T215Y) and four-TAM (M41L+T215Y+D67N+L210W) mutants. TAM-mediated resistance to AZT was comparable in both cell types: for the two-TAM virus, the fold change in IC50 ranged from 8- to 54-fold in MDMs and was 43-fold in P4 cells; for the four-TAM virus, the fold change in IC50 ranged from 64- to 150-fold in MDMs and was 164-fold in P4 cells (Fig. 3A and Table 2). For d4T and ddI, TAM-mediated resistance in MDMs was very low, with fold changes in IC50 ranging from 1.65- to 7.13-fold for d4T and from 1.5- to 8.9-fold for ddI with both TAM-bearing mutant viruses, depending on the time of pretreatment (Fig. 3 and Table 2). Although resistance of the four-TAM virus to d4T seemed to slightly increase with a 16-h pretreatment prior to infection (Fig. 3 and Table 2), probably due to the phosphorylation kinetics of this drug in MDMs, the fold changes in IC50 in the two cell types were not significantly different. Likewise, the fold change in IC50 for ddI was slightly higher in MDMs (ranging from 7.3- to 8.9-fold) than in P4 cells (1.3-fold) for the two-TAM virus, but the difference between these figures did not reach statistical significance. The mean fold changes in IC50 for TDF in MDMs infected with the K65R mutant virus were slightly modified by the time of pretreatment, ranging from 12.3-fold when MDMs were exposed to the drug at the time of infection to 3.1-fold in cells pretreated for 16 h (Fig. 3 and Table 2). As with the clones carrying TAMs, these values were similar to the mean fold change in IC50 measured in P4 cells infected with the K65R mutant (4.6 μM) (Fig. 3 and Table 2). Taken together, our data show that viruses bearing TAMs or the K65R substitution expressed similar levels of resistance to AZT, d4T, ddI, and TDF in P4 cells and primary human macrophages.
FIG. 3.
Fold changes in IC50 for five NRTIs in P4 cells and in MDMs infected with NL4-3-derived RT mutant viruses. P4 cells (104 cells in 96-well microtiter plates) were exposed to different drugs for 4 h and then infected with 8 ng/ml p24 of the WT or mutant NL4-3ΔenvΔNef-Luc+/VSV-G-pseudotyped viruses. MDMs (105 cells in 96-well microtiter plates) were exposed to the same drugs at the time of infection (0 h) or for 4 or 16 h prior to infection with 80 ng/ml p24 of the WT or mutant NL4-3ΔenvΔNef-Luc+/VSV-G-pseudotyped viruses. Infection was monitored by quantifying the luciferase activity at 40 h postinfection in P4 cells and at 72 h postinfection in MDMs. The experiments presented here were performed with MDMs from at least four different independent donors tested in independent experiments and represent the means of quadruplicate wells for each donor. Each point represents the mean of quadruplicate cultures of cells from a single donor. Graphs report the fold change in IC50 for viruses harboring mutations with respect to WT NL4-3 for AZT (A), d4T (B), ddI (C), 3TC (D), and TDF (E). The differences in mean fold changes in IC50 measured for each drug in P4 cells and in MDMs pretreated with the NRTIs for different times were compared by a one-way ANOVA test.
TABLE 2.
Fold changes in IC50 values for five NRTIs in P4 cells and in MDMs infected with mutant NL4-3-derived viruses carrying two TAMs, four TAMs, or the M184V or K65R substitution
| NRTI | Mutations or substitution | Fold change ± SEM (no. of expts)a
|
|||
|---|---|---|---|---|---|
| P4 cells (4-h pretreatment) | MDMs
|
||||
| No pretreatment | 4-h pretreatment | 16-h pretreatment | |||
| AZT | Two TAMs | 43.48 ± 21.07 (3) | 8.43 ± 5.3 (8) | 17.91 ± 8.11 (5) | 54.62 ± 24.3 (7) |
| Four TAMs | 164.02 ± 66.15 (3) | 134.05 ± 85.3 (5) | 150.80 ± 77 (7) | 64.97 ± 18.17 (9) | |
| d4T | Two TAMs | 1.50 ± 0.26 (3) | 2.79 ± 1.80 (6) | 1.65 ± 0.5 (4) | 7.13 ± 5.3 (4) |
| Four TAMs | 2.89 ± 1.01 (3) | 2.84 ± 1.13 (6) | 6.25 ± 1.31 (5) | 6.81 ± 2.46 (5) | |
| ddI | Two TAMs | 1.29 ± 0.49 (3) | 8.22 ± 5.36 (7) | 8.97 ± 7.62 (4) | 7.31 ± 5.12 (4) |
| Four TAMs | 1.28 ± 0.50 (3) | 1.5 ± 0.56 (4) | 2.79 ± 1.57 (4) | 5.78 ± 4.65 (4) | |
| 3TC | Two TAMs | 1.54 ± 0.55 (3) | 9.04 ± 3.32 (6) | 13.10 ± 3.60 (9) | 16.90 ± 15.12 (10) |
| Four TAMs | 4.96 ± 2.36 (3) | 5.06 ± 1.55 (3) | 11.86 ± 3.9 (8) | 13.79 ± 6.651 (10) | |
| M184V | >10,000 (3) | >10,000 (4) | >10,000 (9) | 2,989 ± 1,789 (11) | |
| TDF | K65R | 4.63 ± 0.95 (3) | 12.36 ± 4.58 (4) | 7.42 ± 3.51 (2) | 3.09 ± 1.02 (4) |
The mean fold changes in IC50 relative to the reference virus NL4-3 were calculated from at least three independent experiments. The number of independent experiments is indicated in parentheses.
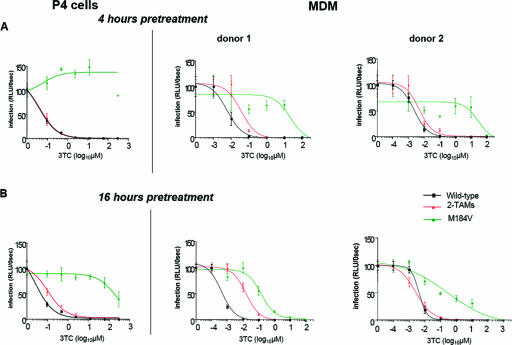
In contrast to TAM- or K65R-mediated resistance to AZT, ddI, d4T, and TDF, the pattern of resistance to 3TC promoted by the M184V mutant appeared to differ in MDMs and in P4 cells. In P4 cells infected with the M184V mutant, the 3TC IC50 could never be calculated, since the inhibition measured at the highest 3TC concentration never exceeded 50% (Fig. 4). In MDMs from the majority of donors, however, the IC50 for 3TC could be readily measured in spite of the M184V mutation. In MDMs, this effect appeared to be strongly influenced by the batch of MDMs studied and by the time of pretreatment with 3TC. Two examples of the impact of pretreatment times on 3TC inhibition curves in MDMs are shown on Fig. 4. When MDMs were exposed to 3TC at the time of infection or 4 h before infection, the IC50 could be measured in 2 of 4 and in 3 of 10 experiments, respectively, and ranged from 9 to 120 μM. When MDMs from the same donors underwent a 16-h 3TC pretreatment, M184V-mediated resistance decreased, and the IC50 could be measured in cells from 8 of 11 donors. In these conditions, values for 3TC in MDMs infected with the M184V mutant ranged from 0.15 to 207 μM, with a mean of 39 μM. The difference in IC50 values measured in P4 cells and in 16-h-pretreated MDMs was statistically significant (P < 0.05). This result strongly suggests that in MDMs 3TC may retain some residual antiviral activity in spite of the M184V mutation. Likely because of the lower IC50 of 3TC on WT virus in MDMs, however, no significant differences were seen between MDMs and P4 cells and between MDMs pretreated by 3TC for different times when we considered the fold changes in IC50 relative to WT NL4-3.
FIG. 4.
Example of 3TC dose-response curves for WT, two-TAM (2-TAM), and M184V viruses. P4 cells and MDMs were pretreated with serial dilutions of 3TC for 4 h (A) or 16 h (B) and then infected with WT NL4-3ΔenvΔNef-Luc+/VSV-G (black) or with the two-TAM mutant (red) or with the M184V mutant (green). Infection was quantified by measuring the luciferase activity in cell lysates at 40 h postinfection for P4 cells and at 72 h postinfection for MDMs. Two representative experiments of the six in which 3TC maintained some antiviral activity despite the M184V mutation are shown here. The data represent the means of quadruplicate wells. Altogether, 11 independent experiments were performed with MDMs from different donors.
Quantification of triphosphorylated nucleoside analogues in MDMs.
In order to shed some light on the resistance profiles of viruses carrying resistance mutations in MDMs and in particular to determine whether residual 3TC activity in spite of the M184V substitution in MDMs could be related to differences in the metabolism of 3TCTP or of dCTP in these cells, triphosphorylated nucleoside analogues were quantified by liquid chromatography coupled to mass spectroscopy in both MDMs and P4 cells treated for 6 h (Fig. 5A and C) or 24 h (Fig. 5B and D) with concentrations of drugs that spanned across the range of measured IC50 values for the WT virus. The results were expressed as the ratio of the NRTI triphosphate to that of its natural endogenous counterpart (ddNTP/dNTP ratio). With the exception of AZT in P4 cells, the AZTTP/dTTP, d4T-TP:dTTP and ddATP:dATP ratios were either undetectable or inferior to 1 in both MDMs and P4 cells over the whole range of tested drug concentrations (Fig. 5). The absolute values for AZTTP, d4TTP and ddATP in primary human macrophages were also either indiscernible or barely detectable, even at 0.1 and 1 μM input drug concentrations, which are much higher than those that proved to have potent antiviral activity (Table 1). The ddATP/dATP ratio was the lowest in both cell types. In marked contrast, 3TCTP/dCTP ratios were the highest of all ddNTP/dNTP ratios in MDMs, in spite of the fact that endogenous dCTP concentrations were higher than the other dNTPs (Fig. 1). In MDMs, the 3TCTP/dCTP ratio was already ≥1 when cells were exposed to concentrations of 3TC as low as 0.01 μM for 24 h (Fig. 5D), a concentration 10- to 100-fold lower than for the other NRTIs. Of note, the 3TCTP/dCTP ratio was comparable to the other ddNTP/dNTP ratios in P4 cells (Fig. 5A and C). In MDMs from four different donors, the 3TCTP/dCTP ratios ranged from 4.4 to 33.3 with a mean of 15.3, contrasting with a ratio ranging 2.6 to 3.6 with a mean of 3.1 in P4 cells for cells exposed to 1 μM input 3TC for 24 h (Fig. 5D). Given the limited amounts of MDMs that could be obtained from each individual donor and the large cell numbers required for the measurement of ddNTP and dNTP concentrations, quantification of triphosphorylated nucleoside analogues and phenotypic resistance assays could not be conducted in parallel using MDMs from the same batches. Consequently, correlations between the 3TCTP/dCTP ratios and IC50 values for 3TC in MDMs infected with the M184V mutant virus could not be formally established. Nonetheless, these results indicate that the potent antiviral activity of 3TC in macrophages might be due to a particularly favorable metabolism of 3TC triphosphate in MDMs, leading to higher 3TCTP/dCTP ratios and permitting partial activity of 3TC in these cells even in the presence of the M184V substitution.
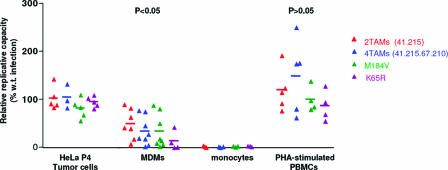
RC of RT mutants in MDMs.
Previous reports (9, 10, 14, 45) have revealed that the RC of viruses carrying the M184V resistance mutation or TAMs is impaired in conditions of low dNTP concentrations. We thus performed RC assays in different primary cell systems, using the same mutant viruses described above. The RC, as measured in a single cycle of replication, was evaluated in P4 cells, PHA-stimulated PBMC, undifferentiated monocytes, and differentiated MDMs. We observed that RCs of mutant viruses were similar to that of the WT virus in P4 cells and in PHA-stimulated PBMC (Fig. 6). In MDMs, however, the RCs of all mutant viruses were strongly impaired compared to WT (Fig. 6). Despite quantitative differences in loss of RC, a similar trend was observed in MDMs from all donors. Mean relative RC (RC of the mutant expressed as a percentage of WT virus RC) were 49.6% for the virus carrying two TAMs, 34.6% for the virus carrying four TAMs, 34.4% for the M184V mutant, and 14.3% for the K65R mutant (Fig. 6). These figures were significantly different from those measured in P4 tumor cells (P < 0.05 for the two-TAM virus and for the M184V mutant and P < 0.001 for the K65R mutant) and in PHA-stimulated PBMC (P < 0.01 for all viruses) as assessed using a one-way ANOVA test followed by a Bonferroni's paired post-test. The relative RCs did not differ between P4 cells and PHA-stimulated PBMC.
FIG. 6.
RCs of viruses carrying NRTI-associated resistance mutations in P4 cells, PBMC, MDMs, and monocytes. P4 tumor cells, PHA-stimulated PBMC, monocytes, and MDMs were infected with serial dilutions of the WT (black) or of viruses bearing two-TAMs (red), four-TAMs (blue), or the M184V (green) or K65R (purple) substitutions. Infection was monitored by measuring the luciferase activity in cell lysates. Luciferase activity was plotted against p24 input, and the slope of the resulting curve in its linear portion was considered to quantify RC. The relative RC was defined to be the RC of the mutant virus as a percentage of that of the WT. Relative RCs of mutant viruses in HeLa P4 cells, in MDMs, and in PHA-stimulated PBMC are shown. The significance of the differences in relative RCs in the three cell types was compared by using a one-way ANOVA test, followed, when groups were significantly different, by a Bonferroni's multiple-comparison post-test. The corresponding P values are indicated.
This phenomenon was even more striking in undifferentiated monocytes. When undifferentiated monocytes were infected with the same pseudotyped mutant clones, no infection could be detected in three of six experiments performed with cells from six different donors. In the remaining three experiments, only replication of the WT virus could be detected (Fig. 6). Replication of the WT virus was between 200- and 1,500-fold lower in monocytes than in the corresponding differentiated MDMs at the highest virus input tested (400 ng/ml in monocytes and MDMs). As for mutant clones, replication was never detected in monocytes (Fig. 6).
Overall, our results indicate that resistance mutations might not affect RC in actively dividing cells such as P4 tumor cells or PHA-stimulated PBMC. In cells with low DNA turnover and low dNTP concentrations, however, such as MDMs and even moreso monocytes, resistance mutations may have a strong impact on viral RC, reflecting that in these cells the mutation-induced loss of processivity of the viral RT is amplified by low dNTP pools.
DISCUSSION
Although often referred to as a model of HIV-1 replication in a low-dNTP context, primary human macrophages have not been studied to date for their capacity to influence HIV resistance to nucleoside analogues and RC. HIV resistance phenotype and fitness are usually measured in transformed cell lines or in PHA-stimulated PBMC, i.e., in cells characterized by high DNA turnover and high dNTP content. These cells, however, are very different from cellular targets for HIV-1 in vivo. The study presented here was designed to examine (i) the specific antiviral activity of nucleoside analogues in MDMs compared to tumor cells, using WT HIV-1; (ii) the phenotypic effect of known NRTI-resistance mutations in MDMs compared to tumor cells; and (iii) the impact of these mutations on HIV-1 RC in MDMs compared to stimulated PBMC and to tumor cell lines.
We report here that the specific antiviral activity of nucleoside analogues can markedly differ in tumor cells and in macrophages, according to the nucleoside analogue studied. With WT HIV-1, d4T, ddI, and 3TC were clearly more active in MDMs than in P4 cells, whereas AZT and TDF had similar antiviral potencies in both cell types. These results differ slightly from those of previous reports, which found that all nucleoside analogues tested, including AZT, ddC, and ddI, were more potent in MDMs than in PBMC, owing to higher intracellular ddNTP/dNTP ratios in MDMs (2, 4, 52, 53). Although we could not directly explain why AZT did not display higher antiviral activity in MDMs, we hypothesize that this difference could relate to the different experimental system used here. Although most previous studies relied on productive infection of MDMs over several days of propagation, we used a single-cycle infection system. We believe that this experimental approach may be more reliable for a quantitative assessment of drug susceptibility, in view of a more synchronous infection, a wider dynamic range of infectivity output values permitted by the luciferase system, and a more accurate control of drug concentrations in the cultures over a short period of incubation. In addition, the present study did not directly examine whether the differences in IC50 observed for some nucleoside analogues are a consequence of lower HIV RC in MDMs. Although this remains a possibility, we believe that it would not be consistent with the fact that differences in IC50 were seen with some analogues but not with others.
In contrast to studies examining the intrinsic antiviral activity of nucleoside analogues on wild-type HIV-1, there had been no previous reports on HIV resistance to nucleoside analogues in MDMs. Using HIV-1 clones expressing a variety of RT resistance mutations, the present study provides two principal findings. First, we observed that resistance to nucleoside analogues conferred by TAMs did not significantly differ in MDMs and in HeLa-derived tumor cells. This observation is particularly noteworthy regarding resistance to drugs such as d4T, for which it has been proposed that resistance could markedly differ according to the cellular dNTP content. In vitro, after termination of DNA synthesis by d4TTP, but not by AZTTP, the rate of primer unblocking mediated by TAMs has been indeed found to be strongly modulated by the concentration of the next dNTP (40, 41). This phenomenon is explained by a dNTP-mediated increase in translocation of the d4T-terminated primer in the RT primer grip, contrasting with the slow and dNTP-independent rate of translocation of the AZT-terminated primer. Correspondingly, using phenotypic assays based on cell systems with a high dNTP content, TAMs promote high-level resistance to AZT but only minimal resistance to d4T, ddI, 3TC, and tenofovir, in spite of manifest treatment failure in vivo. It has therefore been assumed, but not demonstrated, that the apparent inability of phenotypic assays to measure significant resistance to d4T or ddI was related to the high proliferation rate of the cells used in such systems, in which efficient TAM-mediated resistance would be blunted by high dNTP concentrations. Our data, however, do not support this hypothesis: using primary human macrophages with a dNTP content 100-fold lower than tumor cells, the d4T and ddI IC50s measured for viruses carrying TAMs remain much lower than the IC50 measured for AZT.
Second, we observed that resistance to 3TC mediated by the M184V mutation was, with a number of MDM donors, remarkably lower than what is usually measured for this mutation in tumor cell-based assays (30, 36, 59). When MDMs were exposed to 3TC shortly before infection with the M184V mutant, the IC50 value of 3TC could be measured in a limited number of MDM batches. When MDMs were treated for 16 h before infection, however, the IC50 values were often markedly lower than those measured in both P4 cells and MDMs pretreated for a short period (Fig. 3D and 4). This finding is in line with those of Quan et al. in cell-free RT assays and in PBMC (58) and with those of Aquaro et al. (5), who found that the emergence of 3TC resistance in culture through acquisition of the M184V mutation is significantly slower and more difficult in MDMs than in other cell types. Although this latter finding could be related to an intrinsic lower rate of viral replication in MDMs, it may pertain to the fact that in these cells the selective advantage conferred by the M184V mutation, even in the presence of 3TC, is smaller than in other cell types. Consistent with our resistance results, measurement of intracellular 3TCTP and dCTP content in MDMs revealed that the 3TCTP/dCTP ratios were markedly higher than for other nucleoside analogues and also markedly higher in macrophages than in tumor cells (Fig. 5). Resistance to 3TC induced by the M184V substitution is the consequence of reduced incorporation of the analogue in the nascent chain of viral DNA. Indeed, this mechanism of resistance, which involves a direct competition between the analogue and its natural endogenous counterpart, is more likely to be affected by increases in the 3TCTP/dCTP ratio than would be resistance mediated by primer unblocking. In view of these results, we hypothesize that in vivo, where HIV primary target cells are far less actively dividing than mitogen-stimulated PBMC or tumor cells, 3TC may retain some residual antiviral activity in spite of the M184V mutation. Although few trials were specifically designed to test the hypothesis of a clinical benefit drawn from selectively maintaining the M184V substitution through continued use of 3TC despite the emergence of the mutation, several clinical observations support this hypothesis (47, 56). Interruption of nucleoside analogues, including 3TC, in patients with multiple drug resistance and stable plasma viral load results in a significant increase in viremia (21). In the same type of patients, treatment maintenance with 3TC alone results in a better virological and clinical outcome than complete treatment interruption (17), again suggesting that some residual benefit is retained in the prescription of 3TC in spite of genotypic resistance to this drug.
Most resistance mutations in HIV-1 have been found to be associated with some loss in RC. Because reduction in RT function resulting from such mutations could be partially compensated for by high concentrations of dNTPs, we have compared the RCs of different RT mutants in MDMs, in mitogen-stimulated PBMC, and in HeLa-derived P4 cells. We observed that all mutants displayed reduced RCs relative to WT virus in MDMs, whereas no significant differences were seen in P4 cells or in mitogen-stimulated PBMC. The relative RCs in MDMs were as follows: two TAMs > four TAMs = M184V > K65R (Fig. 6). Viruses bearing the K65R mutation were clearly more strongly impaired in their RCs than previously described in other cell systems, with a mean RC of 14.3% of the WT. Thus, using cells with a remarkably low dNTP content, mutations that did not appear to reduce HIV-1 RC in cells with high DNA turnover were found to have a strong impact on viral RC. This phenomenon confirms the findings of a number of different previous studies, which documented the effect of a low-dNTP environment on reduced HIV replication or reduced RT processivity, mostly in the context of the M184V mutation (9, 10, 14, 15, 45, 55, 63, 66). Although an early study by Caliendo et al. (16) documented that a four-TAM mutant displayed increased processivity in vitro under limited dNTP levels, our results confirm recent findings by Bouchonnet et al. (14) that reduced efficiency of DNA synthesis by mutant RT in the presence of low dNTP concentrations, which may involve enzymatic properties other than enzyme processivity, is not restricted to the M184V mutation. In particular, the impact of many combinations of TAMs on HIV RC is seen both here in MDMs and in the low-dNTP-content cells after treatment with hydroxyurea (14). It could also explain reduced RCs in PBMC, in which the dNTP content is generally below that seen in tumor cells. Of note, as previously reported by some authors (22, 44, 67) but contradicted by others (6), our results emphasize that among the different mutation profiles studied, the replicative impact of mutation K65R may be stronger than that of other RT mutations, provided that it is studied in cells with a low-dNTP environment.
In patients failing antiretroviral therapy, plasma viral load often appears to stabilize to levels below pretherapy levels, whereas CD4 counts also stabilize to levels above pretherapy counts, despite viral genotypes and phenotypes indicating complete resistance to the current therapeutic regimen. Such a profile has been attributed either to decreased viral RC or to residual activity of the drugs. Using MDMs as a surrogate model for HIV replication in primary cells, which we hypothesize to be more relevant to HIV replication in vivo than cell systems commonly used to monitor resistance phenotype and RC, we found that both mechanisms could be involved in the clinical profiles described above. The low dNTP content in MDMs enhances the replicative defect of HIV variants carrying RT mutations. In parallel, the high ddNTP/dNTP ratio, particularly striking in MDMs in the case of 3TC, explains partly why some nucleoside analogues appear to retain residual activity in vivo. Although differentiated macrophages are clearly not the main source of replicating virus in HIV-infected individuals, our results strongly reemphasize the fact that culture systems based on artificially stimulated primary cells or on tumor cell lines may, in some instances, be poorly reflective of the actual expression of HIV resistance to nucleoside analogues and of resistance-associated loss of RC in vivo.
Acknowledgments
This study was supported by the Agence Nationale de Recherche sur le Sida, by European Union grant QLK2-CT-2002-001311, and by grants from Bristol-Myers Squibb and Glaxo SmithKline.
We thank Anne Claire Brehin for participating in MDM purification for intracellular dNTP quantifications and Allan Hance for helpful discussions.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Aquaro, S., P. Bagnarelli, T. Guenci, A. De Luca, M. Clementi, E. Balestra, R. Calio, and C. F. Perno. 2002. Long-term survival and virus production in human primary macrophages infected by human immunodeficiency virus. J. Med. Virol. 68:479-488. [DOI] [PubMed] [Google Scholar]
- 2.Aquaro, S., E. Balestra, S. Panti, A. Cenci, F. Serra, M. Francesconi, D. Abdelahad, R. Calio, and C. F. Perno. 1998. Correlation between HIV-inhibiting drug activity in human macrophages and clinical outcome. Clin. Ter. 149:37-41. (In Italian.) [PubMed] [Google Scholar]
- 3.Aquaro, S., R. Calio, J. Balzarini, M. C. Bellocchi, E. Garaci, and C. F. Perno. 2002. Macrophages and HIV infection: therapeutic approaches toward this strategic virus reservoir. Antivir. Res. 55:209-225. [DOI] [PubMed] [Google Scholar]
- 4.Aquaro, S., C. F. Perno, E. Balestra, J. Balzarini, A. Cenci, M. Francesconi, S. Panti, F. Serra, N. Villani, and R. Calio. 1997. Inhibition of replication of HIV in primary monocyte/macrophages by different antiviral drugs and comparative efficacy in lymphocytes. J. Leukoc. Biol. 62:138-143. [DOI] [PubMed] [Google Scholar]
- 5.Aquaro, S., V. Svicher, F. Ceccherini-Silberstein, A. Cenci, F. Marcuccilli, S. Giannella, L. Marcon, R. Calio, J. Balzarini, and C. F. Perno. 2005. Limited development and progression of resistance of HIV-1 to the nucleoside analogue reverse transcriptase inhibitor lamivudine in human primary macrophages. J. Antimicrob. Chemother. 55:872-878. [DOI] [PubMed] [Google Scholar]
- 6.Arion, D., G. Borkow, Z. Gu, M. A. Wainberg, and M. A. Parniak. 1996. The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem. 271:19860-19864. [DOI] [PubMed] [Google Scholar]
- 7.Arner, E. S., A. Valentin, and S. Eriksson. 1992. Thymidine and 3′-azido-3′-deoxythymidine metabolism in human peripheral blood lymphocytes and monocyte-derived macrophages. A study of both anabolic and catabolic pathways. J. Biol. Chem. 267:10968-10975. [PubMed] [Google Scholar]
- 8.Arts, E. J., J. P. Marois, Z. Gu, S. F. Le Grice, and M. A. Wainberg. 1996. Effects of 3′-deoxynucleoside 5′-triphosphate concentrations on chain termination by nucleoside analogs during human immunodeficiency virus type 1 reverse transcription of minus-strand strong-stop DNA. J. Virol. 70:712-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 11.Balzarini, J., R. Pauwels, M. Baba, P. Herdewijn, E. de Clercq, S. Broder, and D. G. Johns. 1988. The in vitro and in vivo anti-retrovirus activity, and intracellular metabolism of 3′-azido-2′,3′-dideoxythymidine and 2′,3′-dideoxycytidine are highly dependent on the cell species. Biochem. Pharmacol. 37:897-903. [DOI] [PubMed] [Google Scholar]
- 12.Becher, F., A. Pruvost, C. Goujard, C. Guerreiro, J. F. Delfraissy, J. Grassi, and H. Benech. 2002. Improved method for the simultaneous determination of d4T, 3TC and ddI intracellular phosphorylated anabolites in human peripheral-blood mononuclear cells using high-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 16:555-565. [DOI] [PubMed] [Google Scholar]
- 13.Becher, F., D. Schlemmer, A. Pruvost, M. C. Nevers, C. Goujard, S. Jorajuria, C. Guerreiro, T. Brossette, L. Lebeau, C. Creminon, J. Grassi, and H. Benech. 2002. Development of a direct assay for measuring intracellular AZT triphosphate in humans peripheral blood mononuclear cells. Anal. Chem. 74:4220-4227. [DOI] [PubMed] [Google Scholar]
- 14.Bouchonnet, F., E. Dam, F. Mammano, V. de Soultrait, G. Hennere, H. Benech, F. Clavel, and A. J. Hance. 2005. Quantification of the effects on viral DNA synthesis of reverse transcriptase mutations conferring human immunodeficiency virus type 1 resistance to nucleoside analogues. J. Virol. 79:812-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer, P. L., and S. H. Hughes. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castagna, A., A. Danise, S. Menzo, L. Galli, N. Gianotti, E. Carini, E. Boeri, A. Galli, M. Cernuschi, H. Hasson, M. Clementi, and A. Lazzarin. 2006. Lamivudine monotherapy in HIV-1-infected patients harbouring a lamivudine-resistant virus: a randomized pilot study (E-184V study). AIDS 20:795-803. [DOI] [PubMed] [Google Scholar]
- 18.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription: a termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 19.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 20.Coakley, E. P., J. M. Gillis, and S. M. Hammer. 2000. Phenotypic and genotypic resistance patterns of HIV-1 isolates derived from individuals treated with didanosine and stavudine. AIDS 14:F9-F15. [DOI] [PubMed] [Google Scholar]
- 21.Deeks, S. G., R. Hoh, T. B. Neilands, T. Liegler, F. Aweeka, C. J. Petropoulos, R. M. Grant, and J. N. Martin. 2005. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J. Infect. Dis. 192:1537-1544. [DOI] [PubMed] [Google Scholar]
- 22.Deval, J., K. L. White, M. D. Miller, N. T. Parkin, J. Courcambeck, P. Halfon, B. Selmi, J. Boretto, and B. Canard. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279:509-516. [DOI] [PubMed] [Google Scholar]
- 23.Diamond, T. L., M. Roshal, V. K. Jamburuthugoda, H. M. Reynolds, A. R. Merriam, K. Y. Lee, M. Balakrishnan, R. A. Bambara, V. Planelles, S. Dewhurst, and B. Kim. 2004. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279:51545-51553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, Q., Z. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, W. Y., T. Shirasaka, D. G. Johns, S. Broder, and H. Mitsuya. 1993. Differential phosphorylation of azidothymidine, dideoxycytidine, and dideoxyinosine in resting and activated peripheral blood mononuclear cells. J. Clin. Investig. 91:2326-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garaci, E., M. C. Caroleo, L. Aloe, S. Aquaro, M. Piacentini, N. Costa, A. Amendola, A. Micera, R. Calio, C. F. Perno, and R. Levi-Montalcini. 1999. Nerve growth factor is an autocrine factor essential for the survival of macrophages infected with HIV. Proc. Natl. Acad. Sci. USA 96:14013-14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geleziunas, R., A. E., F. Boulerice, H. Goldman, and M. A. Wainberg. 1993. Effect of 3′-azido-3′-deoxythymidine on human immunodeficiency virus type 1replication in human fetal brain macrophages. Antimicrob. Agents Chemother. 37:1305-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, D. S. Burke, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennere, G., F. Becher, A. Pruvost, C. Goujard, J. Grassi, and H. Benech. 2003. Liquid chromatography-tandem mass spectrometry assays for intracellular deoxyribonucleotide triphosphate competitors of nucleoside antiretrovirals. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 789:273-281. [DOI] [PubMed] [Google Scholar]
- 30.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higashikawa, F., and L. Chang. 2001. Kinetic analyses of stability of simple and complex retroviral vectors. Virology 280:124-131. [DOI] [PubMed] [Google Scholar]
- 32.Koenig, S., H. E. Gendelman, J. M. Orenstein, M. C. Dal Canto, G. H. Pezeshkpour, M. Yungbluth, F. Janotta, A. Aksamit, M. A. Martin, and A. S. Fauci. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089-1093. [DOI] [PubMed] [Google Scholar]
- 33.Lambotte, O., Y. Taoufik, M. G. de Goer, C. Wallon, C. Goujard, and J. F. Delfraissy. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114-119. [DOI] [PubMed] [Google Scholar]
- 34.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margot, N. A., E. Isaacson, I. McGowan, A. K. Cheng, R. T. Schooley, and M. D. Miller. 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16:1227-1235. [DOI] [PubMed] [Google Scholar]
- 36.Masquelier, B., E. Race, C. Tamalet, D. Descamps, J. Izopet, C. Buffet-Janvresse, A. Ruffault, A. S. Mohammed, J. Cottalorda, A. Schmuck, V. Calvez, E. Dam, H. Fleury, and F. Brun-Vezinet. 2001. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrob. Agents Chemother. 45:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxeiner, H. G., W. Keulen, R. Schuurman, M. Bijen, L. de Graaf, A. van Wijk, N. Back, M. W. Kline, C. A. Boucher, and M. Nijhuis. 2002. Selection of zidovudine resistance mutations and escape of human immunodeficiency virus type 1 from antiretroviral pressure in stavudine-treated pediatric patients. J. Infect. Dis. 185:1070-1076. [DOI] [PubMed] [Google Scholar]
- 38.McElrath, M. J., J. E. Pruett, and Z. A. Cohn. 1989. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc. Natl. Acad. Sci. USA 86:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meltzer, M. S., M. Nakamura, B. D. Hansen, J. A. Turpin, D. C. Kalter, and H. E. Gendelman. 1990. Macrophages as susceptible targets for HIV infection, persistent viral reservoirs in tissue, and key immunoregulatory cells that control levels of virus replication and extent of disease. AIDS Res. Hum. Retrovir. 6:967-971. [DOI] [PubMed] [Google Scholar]
- 40.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, M. D. 2004. K65R, TAMs, and tenofovir. AIDS Rev. 6:22-33. [PubMed] [Google Scholar]
- 45.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 46.Miller, V., and B. A. Larder. 2001. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir. Ther. 6(Suppl. 3):25-44. [PubMed] [Google Scholar]
- 47.Miller, V., T. Stark, A. E. Loeliger, and J. M. Lange. 2002. The impact of the M184V substitution in HIV-1 reverse transcriptase on treatment response. HIV Med. 3:135-145. [DOI] [PubMed] [Google Scholar]
- 48.Naeger, L. K., N. A. Margot, and M. D. Miller. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orenstein, J. M., C. Fox, and S. M. Wahl. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857-1861. [DOI] [PubMed] [Google Scholar]
- 50.Orenstein, J. M., M. S. Meltzer, T. Phipps, and H. E. Gendelman. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 62:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pellegrin, I., J. Izopet, J. Reynes, M. Denayrolles, B. Montes, J. L. Pellegrin, P. Massip, J. Puel, H. Fleury, and M. Segondy. 1999. Emergence of zidovudine and multidrug-resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. STADI Group. AIDS 13:1705-1709. [DOI] [PubMed] [Google Scholar]
- 52.Perno, C. F., E. Balestra, M. Francesconi, D. Abdelahad, R. Calio, J. Balzarini, and S. Aquaro. 2004. Antiviral profile of HIV inhibitors in macrophages: implications for therapy. Curr. Top. Med. Chem. 4:1009-1015. [DOI] [PubMed] [Google Scholar]
- 53.Perno, C. F., R. Yarchoan, J. Balzarini, A. Bergamini, G. Milanese, R. Pauwels, E. De Clercq, G. Rocchi, and R. Calio. 1992. Different pattern of activity of inhibitors of the human immunodeficiency virus in lymphocytes and monocyte/macrophages. Antivir. Res. 17:289-304. [DOI] [PubMed] [Google Scholar]
- 54.Perno, C. F., R. Yarchoan, D. A. Cooney, N. R. Hartman, S. Gartner, M. Popovic, Z. Hao, T. L. Gerrard, Y. A. Wilson, D. G. Johns, et al. 1988. Inhibition of human immunodeficiency virus (HIV-1/HTLV-IIIBa-L) replication in fresh and cultured human peripheral blood monocytes/macrophages by azidothymidine and related 2′,3′-dideoxynucleosides. J. Exp. Med. 168:1111-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrella, M., M. Oliveira, D. Moisi, M. Detorio, B. G. Brenner, and M. A. Wainberg. 2004. Differential maintenance of the M184V substitution in the reverse transcriptase of human immunodeficiency virus type 1 by various nucleoside antiretroviral agents in tissue culture. Antimicrob. Agents Chemother. 48:4189-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrella, M., and M. A. Wainberg. 2002. Might the M184V substitution in HIV-1 RT confer clinical benefit? AIDS Rev. 4:224-232. [PubMed] [Google Scholar]
- 57.Picard, V., E. Angelini, A. Maillard, E. Race, F. Clavel, G. Chene, F. Ferchal, and J. M. Molina. 2001. Comparison of genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 isolates from patients treated with stavudine and didanosine or zidovudine and lamivudine. J. Infect. Dis. 184:781-784. [DOI] [PubMed] [Google Scholar]
- 58.Quan, Y., B. G. Brenner, M. Oliveira, and M. A. Wainberg. 2003. Lamivudine can exert a modest antiviral effect against human immunodeficiency virus type 1 containing the M184V mutation. Antimicrob. Agents Chemother. 47:747-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quan, Y., Z. Gu, X. Li, Z. Li, C. D. Morrow, and M. A. Wainberg. 1996. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (−)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J. Virol. 70:5642-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy, B., C. Beuneu, P. Roux, H. Buc, G. Lemaire, and M. Lepoivre. 1999. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal. Biochem. 269:403-409. [DOI] [PubMed] [Google Scholar]
- 61.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 66.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti-Infect. Ther. 2:147-151. [DOI] [PubMed] [Google Scholar]
- 67.White, K. L., N. A. Margot, T. Wrin, C. J. Petropoulos, M. D. Miller, and L. K. Naeger. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46:3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]