Abstract
The epigenetic status of integrated adenovirus type 12 (Ad12) DNA in hamster cells cultivated for about 4 decades has been investigated. Cell line TR12, a fibroblastic revertant of the Ad12-transformed epitheloid hamster cell line T637 with 15 copies of integrated Ad12 DNA, carries one Ad12 DNA copy plus a 3.9-kbp fragment from a second copy. The cellular insertion site for the Ad12 integrate, identical in both cell lines, is a >5.2-kbp inverted DNA repeat. The Ad12 transgenome is packaged around nucleosomes. The cellular junction is more sensitive to micrococcal nuclease at Ad12-occupied sites than at unoccupied sites. Bisulfite sequencing reveals complete de novo methylation in most of the 1,634 CpGs of the integrated viral DNA, except for its termini. Isolated unmethylated CpGs extend over the entire Ad12 integrate. The fully methylated transgenome segments are characterized by promoter silencing and histone H3 and H4 hypoacetylation. Nevertheless, there is minimal transcriptional activity of the late viral genes controlled by the fully methylated major late promoter of Ad12 DNA.
A major part of mammalian genomes is made up of repetitive DNA, and viral retrotransposons comprise a substantial part of it. Little is known about the stability or numerical plasticity of the repetitive sequences. Repetitive DNA in mammalian genomes is heavily methylated (for recent reviews, see references 6, 29, and 47), possibly as a defense against the potential activity of foreign genes in an established genome (5, 50). The degree of methylation, particularly of the repetitive cellular DNA sequences, is subject to alterations in cells with integrated foreign DNA in the genome (13, 32). Transgenomes are frequently generated and exploited in experimental and applied molecular biology. Their structure and the effects of their insertion on the stability and functionality of the recipient genome require more detailed studies.
In a model system, we have investigated the stability as well as the methylation and transcriptional profiles of integrated adenoviral genomes in hamster cells. The transformed cell line T637 has been generated by infecting baby hamster kidney cell line BHK21 (40) with human adenovirus type 12 (Ad12) (41). The genomes of T637 cells carry about 15 copies of Ad12 DNA integrated at a single chromosomal site (18, 35, 39). Upon continuous cultivation of cell line T637, morphological revertants, from epitheloid to more fibroblastic, arose spontaneously (10). In one of these revertants, TR12, only one copy and a fragment of a second copy of Ad12 DNA persist stably (7).
The genomes of human adenoviruses are always chromosomally integrated and hypermethylated in Ad12-transformed cells and in Ad12-induced hamster tumor cells (26, 42, 43). Virion DNA or free intracellular adenoviral DNA is not methylated (11, 17). The integrated Ad12 genomes in the revertant cell line TR12 are more extensively methylated than those in the parental cell line, T637 (27). Hence, hypermethylation of transgenic DNA might be related to its stability in the recipient genome.
Cell lines T637 and TR12 offer the advantage of analyzing foreign DNA integrates of about 15 copies and 1 copy plus a 3.9-kbp fragment of Ad12 DNA, respectively, at the same chromosomal site and at the same time after insertion. By using the bisulfite genomic sequencing technique, we have determined the complete methylation map of a total of 1,634 CpG dinucleotides in the 34.1- plus 3.9-kbp stable Ad12 integrate in the TR12 revertant cell line. We have also compared this methylation pattern to that in selected Ad12 DNA segments in the original T637 cell line. DNA hypermethylation and absence of histone H3 and H4 acetylation as well as histone H3 lysine 9 trimethylation correlate with transcriptional inactivation. However, the hypermethylated state of the major late promoter (MLP) of Ad12 DNA still permits minimal residual activity of the late Ad12 structural genes in both cell lines. Conceivably, the transiently hemimethylated state following DNA replication in growing cells or even the completely methylated state of the MLP is compatible with low levels of transcription. The E1 genes of Ad12 are transcribed in cell line T637 and at a very low level in the revertant. The cellular DNA at the site of transgenome insertion exhibits increased sensitivity towards micrococcal nuclease digestion compared to the unoccupied insertion site. The viral transgenome itself is organized around nucleosomes.
MATERIALS AND METHODS
Cells and virus.
Propagation of cell lines T637 and TR12 and of Ad12 virions was described previously (15).
Standard methods of molecular biology were employed for Southern blotting (19, 37), DNA-DNA hybridization, DNA in vitro manipulations, transformation, DNA sequencing of integrated Ad12 DNA, and PCR amplification. For cloning and subcloning, the plasmid vectors pBS(+) (Stratagene, La Jolla, CA) and pGEM-T (Promega, Madison, WI) were used.
Characterization of the integration patterns of Ad12 DNA in T637 and TR12 cells and subcloning of Ad12 DNA fragments.
The viral-cellular and interviral DNA junctions of integrated viral genomes in T637 and TR12 cells were characterized (i) by hybridization of the restriction endonuclease-cleaved genomic DNA with α-32P-labeled specific DNA probes on Southern blots; (ii) by subcloning DNA fragments from the identified off-size bands that include junction sequences; and (iii) by PCR amplification, subcloning, and/or sequencing of junction fragments. Off-size viral DNA fragments do not correspond in length to authentic Ad12 DNA restriction fragments and are due to viral-cellular DNA junctions or rearranged viral DNA.
Quantitative reverse transcription-PCR of total RNA from cell line T637 or TR12.
Total RNA was extracted from T637 or TR12 cells that grew in exponential phase and had reached about 70% confluence. Genomic DNA was quantitatively removed by treating the total RNA samples twice with “on-column” DNase I (QIAGEN, Hilden, Germany) and subsequently with RQ1 DNase I (Promega, Madison, WI). For unidirectionally transcribed Ad12 regions (E1A, E1B, E4, β-actin), total RNA was reverse transcribed to cDNA using random hexamers. For regions known to be bidirectionally transcribed (hexon, penton, and endoprotease), cDNAs were generated using gene-specific primers. All quantitative PCRs (qPCRs) were performed in an Applied Biosystems 7500 real-time PCR system and by using SYBR-Green incorporation into double-stranded DNA for amplicon detection. Standard curves were generated with PCR products that were slightly longer than the qPCR amplicons. cDNA generated from 0.1 ng (β-actin) or 100 ng total RNA (Ad12 genes) was used in the qPCRs, which were all performed at least in duplicate. Amplification of possibly contaminating genomic DNA was excluded by running the qPCR with samples that had been generated by mock reverse transcription in the absence of reverse transcriptase. qPCR signals were not obtained under these conditions. The specificity of SYBR-Green incorporation was validated by generating melting curves and by separating the amplicons by electrophoresis in agarose gels. As an internal control, the amount of β-actin mRNA that was strongly transcribed in cell culture was determined.
Bisulfite protocol for the genomic sequencing technique.
A published procedure (3, 9) was slightly modified. DNAs from cell lines T637 and TR12 were isolated using standard procedures and were dialyzed for 3 days against several changes of Tris-EDTA (TE) buffer. Genomic DNA was sheared by 10 passes through a 23-gauge needle. In a single conversion reaction, 2 μg of sheared genomic DNA was bisulfite treated as follows. Fifty microliters of the DNA solution was denatured in 0.3 M NaOH for 15 min at 37°C and for 2 min at 95°C and quenched on ice. The bisulfite solution (8.5 g sodium bisulfite [Sigma catalog no. S9000] in 20 ml double-distilled water and 2.25 mM hydroquinone, pH 5) was always prepared freshly in degassed water. Five hundred microliters of the bisulfite solution was added to the denatured DNA, and the reaction was carried out for 17 h at 50°C in the dark. The DNA solution was then purified in a Micrococon YM-100 spin device (1). Alkali desulfonation was carried out directly in the spin device by addition of NaOH to a final concentration of 0.3 N and incubation for 20 min at 37°C. Bisulfite-treated DNA was eluted in 50 μl TE buffer; 1 μl was usually enough for PCR. Primers to amplify bisulfite-treated DNA were designed with the MethPrimer program (21). Nested PCRs in 25 μl were performed with 1 μl of bisulfite-treated DNA, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 1.25 unit of Taq (Sigma, St. Louis, MO), and 0.2 μM primers or by using 2× AmpliTaq Gold PCR master mixture (PE Applied Biosystems, Foster City, CA) in a PerkinElmer GeneAmp PCR System 2400 with the following cycle conditions: 95°C for initial denaturation (5 min); 25 to 45 cycles at 95°C (1 min), 50°C (2 min), and 72°C (2 min); and final extension at 72°C (10 min). Nested PCR was performed under the same conditions with 1/10 of the reaction volume of the first PCR. Amplicons were agarose gel purified and cloned into the pGEM-T vector (Promega, Madison, WI) for the analysis of single DNA molecules. Sequencing was performed on an ABI Prism 377 DNA sequencer with the Taq FS BigDye Terminator cycle sequencing method.
Safeguards in bisulfite sequencing and the interpretation of data.
For all parts of the Ad12 DNA in cell line TR12, at least three DNA clones obtained after bisulfite treatment and PCR amplification of a certain DNA segment were sequenced. In several instances, two or more independent bisulfite experiments were performed to validate the data. Obviously, there was not absolute congruence between independent determinations. PCR amplification and cloning might bias in favor of distinct DNA sequences. Hence, even in heavily methylated segments of the integrated Ad12 DNA, some variability has to be expected with respect to the complete state of DNA methylation. Individual cells, depending on their phase in the cell cycle, could differ in their state of methylation in the integrated Ad12 DNA segments.
When applying the bisulfite sequencing technique, a number of precautions have been taken. During the bisulfite reaction (3, 9), cytosine residues are converted to uracils only in single-stranded DNA. Therefore, complete denaturation of double-stranded DNA samples and prevention of DNA reannealing are essential preconditions for reactions to go to completion. Robust denaturation with 0.3 N NaOH for 15 min at 37°C followed by 2 min at 95°C and subsequent rapid quenching at 0°C facilitate complete conversion. Possibly remaining unreacted cytosines would register as false-positive 5-mC residues, which might occur either in blocks or randomly at single loci. The distribution of nonconverted C residues can be both sequence dependent (12) and independent (30). To control for complete conversion, we initially cleaved the cloned PCR amplicons at the frequent AluI (AGCT) sites. Clones that carried unconverted cytosine residues in that sequence then contained an AluI site and could be cleaved by AluI; these clones were not used further. Differentially methylated DNA molecules will generate molecules with various cytosine contents. Some of these molecules might be preferentially amplified in the PCR (46). To avoid a possible PCR bias, cycle numbers were kept as low as possible. PCR amplicons were subsequently cloned into the pGEM-T vector, and at least three clones were sequenced for each amplicon. In further control experiments, the bisulfite sequencing reactions and PCRs were repeated with the same or with different primer pairs that targeted the same DNA segments. In some experiments, the MethylEasy kit (Human Genetic Signatures, Sydney, Australia) was used.
Micrococcal nuclease treatment of cells after lysolecithin permeabilization.
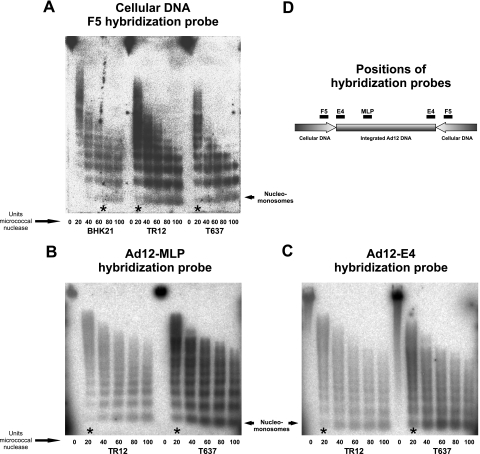
A published procedure (2) was modified. BHK21 cells as the nontransgenic control, T637 cells, or TR12 cells, in each case about 6.6 × 106 cells in a 1-ml suspension, were first permeabilized by treatment with 250 μg of lysolecithin and subsequently treated with increasing amounts of micrococcal nuclease (MBI Fermentas, Vilnius, Lithuania) at 20 to 100 units per 6.6 × 106 cells at 16°C for 15 min (see Fig. 5). Subsequently, DNA was isolated, and fragments were separated by electrophoresis on a 1% agarose gel, transferred by Southern blotting to a positively charged nylon membrane (Hybond XL; Amersham Biosciences, Piscataway, NJ), and hybridized to Ad12 DNA, to different segments of Ad12 DNA, or to the flanking cellular DNA segment F5 (for map positions, see Fig. 5D). The extent of degradation in different parts of the genome has been interpreted as an indicator of chromatin structure.
FIG. 5.
Chromatin organization in Ad12 transgenomes and their vicinity. The degree of chromatin condensation was determined by assessing the susceptibility to micrococcal nuclease (see Materials and Methods) (A) in the F5 cellular DNA segments of BHK21, T637, and TR12 cells and in the Ad12 region MLP (B) and E4 promoter (C) in cell lines T637 and TR12. Micrococcal nuclease units sufficient to generate nucleomonosomes were marked with asterisks. (D) Map positions of hybridization probes. As the amounts of Ad12 DNA integrates present in cell lines T637 and TR12 differ by a factor of about 15, signal intensities are expected to be different when Ad12 hybridization probes are used. The data in Fig. 5A demonstrate that adequate amounts of TR12 DNA had been employed in the experiments shown in this figure. In the panel A experiments, an F5 hybridization probe from hamster cellular DNA was used. TR12 and T637 DNAs contain the same equivalents of cellular DNA segments, like F5, but differ by a factor of about 15 in the content of Ad12 DNA sequences. Therefore, the blots of TR12 DNA are expected to exhibit weaker Ad12 DNA signals (B and C).
ChIP.
Chromatin immunoprecipitation (ChIP) analyses were performed following established protocols (Upstate, Lake Placid, NY) with some modifications. Cells were grown to 90 to 95% confluence, and proteins were cross-linked by adding formaldehyde directly to the cell culture medium at a final concentration of 1% and by incubation for 10 min at 37°C. After fixation, cells were rinsed thoroughly with ice-cold phosphate-buffered saline without Ca2+ and Mg2+ and were scraped off the plastic support in micrococcal nuclease buffer (150 mM sucrose, 80 mM KCl, 30 mM HEPES, pH 7.4, 5 mM MgCl2, 2.5 mM CaCl2) at a concentration of 1 × 106 cells per ml. Cells were permeabilized with lysolecithin at a final concentration of 250 μg/ml. Chromatin was digested to nucleomonosomes by addition of 100 U of micrococcal nuclease (MBI Fermentas, Vilnius, Lithuania) and incubation for 10 min at 37°C. The reaction was stopped by adding 20 μl of 0.5 M EDTA. Cells were pelleted (700 × g, 5 min, 4°C) and resuspended in 200 μl lysis buffer (1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 10 mM Tris-HCl [pH 8.1], 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin A), followed by incubation on ice for 10 min. Cell debris was pelleted for 15 min at 5,300 × g and 4°C. The supernatant was diluted 1:100 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 167 mM NaCl, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1) containing protease inhibitors as above. Two 20-μl samples were taken as input and reverse cross-linked by the addition of 1 μl 5 M NaCl and incubation for 4 h at 65°C. Immunoprecipitation, washing steps, and reverse cross-linking were done by following the manufacturer's instructions (Upstate, Lake Placid, NY). A nonantibody control was included to assess nonspecific antibody binding. DNA was recovered by phenol-chloroform extraction and ethanol precipitation. The DNA was resuspended in 50 μl TE buffer; 1 to 2 μl of this solution was used for one qPCR.
RESULTS
Organization and sequence of the Ad12 transgenome in revertant cell line TR12.
We have analyzed in detail the structure of the Ad12 transgenome in the TR12 revertant, with a limited amount of foreign DNA, instead of in the parent cell line, T637, with a much more complex array of about 15 copies of Ad12 DNA. The maps of the adenoviral genome in Fig. 1B and 2 localize restriction sites and hybridization probes used in the analyses of the integrated Ad12 DNA in cell lines T637 and TR12.
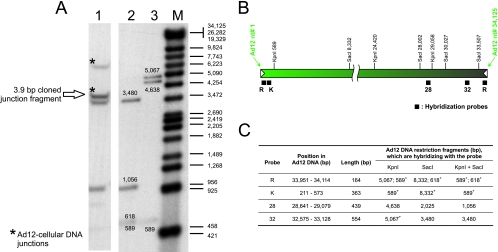
FIG. 1.
Identification of the off-size fragment covering the junction between Ad12 DNA in TR12 cells (A) and localization of the DNA probes within the Ad12 genome (B). (A) Lane 1, TR12 genomic DNA (30 μg), KpnI and SacI cleavage; lanes 2 and 3, Ad12 DNA (100 pg), KpnI and SacI (lane 2) and KpnI (lane 3) cleavage; M, lambda DNA/Eco130I (StyI) and MluI, marker 17 (MBI Fermentas, Vilnius, Lithuania), 3′ end labeled with [α-32P]dCTP. DNA was transferred to a positively charged membrane, hybridized to a mixture of probes R, 32, and 28, which were labeled with [α-32P]dCTP by random priming. (B) Nucleotide numbering of the Ad12 genome is from reference 38. KpnI and SacI sites that are relevant for mapping the integrated DNA are indicated. Filled bars beneath the map of Ad12 DNA show the positions of hybridization probes. Inverted terminal repeats of Ad12 DNA are depicted by white arrowheads. (C) Description of the probes. Probe R was copied from the inverted repeat sequence at the right end of Ad12 DNA; 153 bp (bp 12 to 164 of Ad12 DNA) are homologous to the left end, and 11 bp (bp 165 to 175 of Ad12 DNA) are nonhomologous. The fragments marked with an asterisk were expected to generate off-size bands due to Ad12 termini involved in junctions with cellular DNA or with adjacent Ad12 DNA copies. The inverted terminal repeat of Ad12 DNA is 161 bp (38, 44).
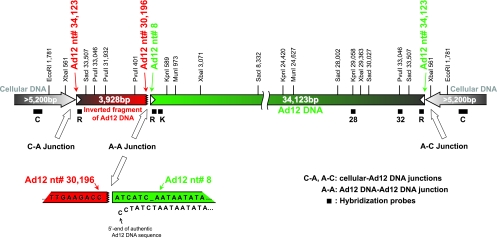
FIG. 2.
Scheme of the Ad12 transgenome and its integration site in cell line TR12. This map is based on a number of restriction endonuclease and Southern blot hybridization experiments as well as partial nucleotide sequence determinations of recloned fragments of the integrated Ad12 DNA. Filled bars beneath the map of the transgenome show the positions of hybridization probes (see also Fig. 1B). Probe C, for the cellular DNA flanking the integrated Ad12 genomes, was excised as a 1,165-bp XbaI fragment from the plasmid subclone F7 (18). Inverted terminal repeats of Ad12 DNA are depicted by white arrowheads. The green bar represents a full-length copy of Ad12 DNA, and the red bar is a truncated second, flip-flopped copy derived from the right terminus of Ad12 DNA. The cellular DNA segments of at least 5.2 kbp adjacent to the transgenome seem to be identical on both flanks and palindromic. Numbers refer to nucleotides in the authentic Ad12 DNA sequence (38) or in the adjacent cellular DNA (18). At the bottom, the magnified structure and sequence of the A-A junction (jagged line) are presented in comparison to the authentic sequence at the 5′ end of Ad12 DNA.
KpnI and SacI cleavage of TR12 genomic DNA identified two Ad12-specific off-size DNA fragments (Fig. 1A) that hybridized to a mixture of the R, 32, and 28 probes or of the 32 and R probes (not shown). Both in TR12 and T637 cells, the ∼7-kbp off-size band (Fig. 1A, lane 1) most likely spans the previously cloned and sequenced 5,250-bp cellular DNA fragment from the right terminal junction of the integrated Ad12 genome (18). This DNA segment does not contain KpnI or SacI sites. The origin of the ∼4-kbp off-size band (Fig. 1A) was not apparent from earlier data. None of the other probes shown in Fig. 1B recognized the same off-size fragments (data not shown).
The KpnI and SacI TR12 DNA fragments between 3.5 and 4.5 kbp were isolated and cloned into the pBS(+) plasmid vector. Two colonies, among 150,000, that hybridized to Ad12 probes R and 32 and that proved to be identical upon sequencing were identified. The cloned fragment was 3.9 kbp and, as shown by sequencing, comprised Ad12 DNA from bp 33507 (SacI site at the right end) to bp 30196, the bridging sequence ATCATC, and bp 8 to 589 (KpnI site) from the left end of Ad12 DNA (Fig. 2). In comparison to the published Ad12 DNA sequence (38), only three single-nucleotide exchanges were observed, G to T in Ad12 DNA positions 32173 and 32229 and G to A in position 32254. These mutations were localized in the 5′-terminal part of the coding sequence for the Ad12 34-kDa E4 protein. Nucleotide sequencing revealed that in TR12 cells both termini of the integrated Ad12 genome represented right terminal sequences of the viral DNA that were organized in flip-flopped repeats. Assuming that two right-terminal Ad12 nucleotides were missing also on the left end of the Ad12 transgenome, as shown for its right end (18), the length of the right-terminal fragment of the viral sequence in the transgenome was calculated to be 3,928 bp (Fig. 2).
A portion of 12.4 kbp from the integrated Ad12 genome was resequenced by standard methods in order to investigate to what extent the transgenomic DNA sequence might differ from that of virion DNA. In the genome segment analyzed, only 10 nucleotide alterations (0.08%) were found compared to the published nucleotide sequence of the virion DNA (38). Furthermore, successful bisulfite sequencing (see Fig. 4) also implies that the Ad12 sequence in the integrate is identical to that of virion DNA. Of course, in bisulfite sequencing the conversion of all unmethylated cytosines to thymidines limits interpretations of the original Ad12 sequence.
FIG. 4.
Methylation profile of the Ad12 transgenome in cell line TR12. The methylation pattern was investigated at single-CpG resolution using the bisulfite conversion of genomic DNA. Both ends of the adenoviral integrate are unmethylated, and the remainder of the integrated viral DNA is heavily methylated with a small number of unmethylated CpG sites. Some CpG sites show variable methylation status. Unmethylated CpGs are indicated by open squares and methylated ones by filled squares. CpG sites with variable methylation status are assigned diagonally half-filled symbols. The results of all PCR clones that were sequenced are shown. Each horizontal array of symbols corresponds to one PCR clone. Individual CpGs are numbered; on top of these numbers Ad12 genome nucleotide positions of the PCR fragments are shown. Ad12 gene groups are indicated with gray bars. The methylation status of the rightward-transcribed strand of the transgenome has been determined throughout. Since the right- and left-terminal transgenome segments containing 134 CpGs are in inverted orientations, the primers employed for PCR encompass both DNA complements in these regions.
We conclude that in the revertant cell line TR12 the one complete Ad12 genome, from nucleotides 8 to 34123, represents genuine Ad12 DNA. The few nucleotide exchanges might be considered natural fluctuations in adenoviral genomes. The first three nucleotides of the integrated complete adenoviral DNA copy differ from the original virion sequence. In addition, the seventh nucleotide of the virion sequence is missing in the integrated Ad12 DNA (Fig. 2, scheme at bottom). Nucleotide 1 of the full-length copy of Ad12 DNA is linked to nucleotide 30196 of a flip-flopped right terminal fragment from Ad12 DNA with a length of 3,928 nucleotide pairs, an authentic palindrome of the Ad12 sequence. Both termini of the integrated adenoviral DNA then represent bona fide right terminal Ad12 sequences that are linked to cellular DNA (Fig. 2).
Structure of the junction between hamster genomic DNA and Ad12 DNA in TR12 cells.
An Ad12 DNA-cellular DNA junction from cell line T637 had been cloned and sequenced earlier (18). Nucleotide sequencing data on the junction sequence in cell line TR12 demonstrated that this sequence was identical to the previously published one from cell line T637. Hence, the site of Ad12 DNA integration had remained unchanged in cell line TR12. We now tried to characterize the opposite cellular DNA-Ad12 DNA junction in TR12 cells (Fig. 3) and to validate data on the unchanged integration site.
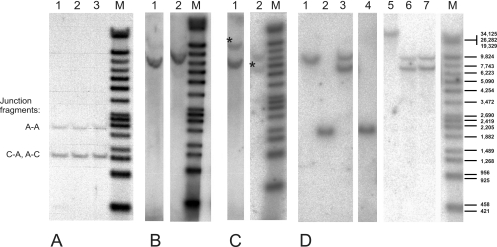
FIG. 3.
Analyses of the junctions between hamster genomic DNA and Ad12 DNA in TR12 and T637 cells. (A) DNA probe R. Lanes 1 to 3 show results for three independent isolates of TR12 genomic DNA (30 μg) cleaved with PvuII. The sequence of the cloned A-A and A-C junctions (Fig. 2) (18) predicted that probe R would hybridize with a 2,137-bp PvuII fragment from the A-A junction and a 1,260-bp fragment from the A-C junction. These two fragments were indeed observed. However, there was no additional fragment originating from the C-A junction. The 1,260-bp band produced a hybridization signal twice as strong as the 2,137-bp band, since it likely comprised both the identical A-C and C-A fragments. (B) DNA probe K. Lanes 1 and 2 show results for TR12 genomic DNA (30 μg), with EcoRI and MunI (lane 1) and XbaI (lane 2) cleavage. The predicted lengths of the EcoRI/MunI and XbaI fragments that span both the C-A and A-A junctions and hybridize to the K probe, are 6,685 bp and 7,563 bp, respectively. This calculation was based on the assumption that the cellular preinsertion site sequence is duplicated and inverted on the left side of the Ad12 transgenome. The observed fragment sizes are in agreement with these calculations. (C) DNA probe R. Lanes 1 and 2 show results for TR12 genomic DNA (30 μg), with EcoRI and MunI (lane 1) and XbaI (lane 2) cleavage. In addition to the C-A and A-A fragments (Fig. 2), probe R hybridized to an 11,227-bp EcoRI/MunI fragment and a 5,321-bp Xba fragment from the A-C junction (marked with asterisks). The lengths of these fragments were derived from the previously published sequence (18). (D) DNA probe C. Lanes 1 and 2, BHK21 genomic DNA (30 μg); lanes 3 to 6, TR12 genomic DNA (30 μg); lane 7, T637 genomic DNA (30 μg). Lanes 1, 3, 6, and 7 show results for SacI cleavage; lanes 2 and 4 show results for EcoRI cleavage; and lane 5 shows results for KpnI cleavage. Lanes M, lambda DNA/Eco130I (StyI) and MluI, marker 17 (MBI Fermentas, Vilnius, Lithuania), 3′ end labeled with [α-32P]dCTP. The lengths of the marker fragments are indicated on the right.
When TR12 genomic DNA was cut with PvuII and the fragments were hybridized with the R probe (Fig. 3A), only two bands, instead of three as to be expected for three different junctions (C-A, A-A, and A-C), were apparent. The lengths of the hybridizing fragments agreed with the calculated lengths of the fragments due to the sequenced interviral (A-A) and viral-cellular DNA (A-C) junctions. In all experiments, the signal of the latter PvuII fragment was about twice as strong as the former. Hence the right and left terminal junctions (A-C and C-A) might be identical and therefore yielded twice the amount of the junction PvuII fragments (1,270 bp; Fig. 3A) and double-strength signals with the R probe. Restriction analyses of TR12 DNA with BglII, BstEII, BstXI, EcoRI, KpnI, SacI, and SwaI (data not shown) followed by hybridization with probes C and R supported the notion that the cellular DNA flanking the Ad12 transgenome on either side represented identical inverted repeats. Probe C is part of the previously cloned cellular DNA (18). This model was further tested by comparative hybridization experiments with probes R and K (Fig. 2 and 3). For more-detailed analyses, restriction enzymes that cut within the previously sequenced part of the flanking cellular DNA and in the Ad12 genome but not in the inverted repeat of the transgenome were selected. Probe K (Fig. 1C) hybridized to the fragments that spanned junctions A-A and C-A but not to the A-C junction. Probe R annealed to fragments from all three junctions (Fig. 2). In TR12 DNA, probe K hybridized to an EcoRI/MunI fragment with a calculated length of 6,685 bp (Fig. 3B) and probe R to the same fragment and to a longer one of 11,227 bp (Fig. 3C). Similarly, probe K recognized an XbaI fragment with a calculated length of 7,563 bp and probe R the same fragment plus a shorter one of 5,321 bp (Fig. 3B and C).
These data indicate that in cell line TR12 at least 1,781 bp (EcoRI site in the flanking cellular DNA) (18) are derived from the cellular preinsertion site sequence duplicated and inverted on both sides of the Ad12 transgenome.
SacI restriction of TR12 genomic DNA and hybridization of the DNA fragments with probe C indicated that the inverted part of the cellular genome on both sides of the Ad12 integrate probably exceeded 5,200 bp (Fig. 3D). SacI did not cut in this sequence. An ∼10-kbp SacI DNA fragment and a 2,283-bp EcoRI fragment hybridized to the C probe also in DNA from the nontransgenic BHK21 cell line (Fig. 3D). The same 10-kbp SacI DNA fragment was found in the T637 and TR12 genomes (Fig. 3D). Apparently, Ad12 DNA remained integrated at the same site of the preintegration sequence after excision of the majority of the viral genomes. Only one additional 7-kbp SacI band was detected with probe C in the genomic DNA from T637 and TR12 cells (Fig. 3D, lanes 3, 6, and 7), and this band yielded a stronger hybridization signal than the 10-kbp band in BHK21 DNA. The double intensity of this hybridization signal and the absence of additional off-size fragments supported the interpretation that the integration of the viral genome was accompanied by the duplication and inversion of a cellular DNA stretch of at least 5.2 kbp in the preinsertion region.
In earlier work on the cloning and sequencing of the junctions between Ad12 DNA and cellular DNA in cell line T637, all attempts to clone a left terminal Ad12 DNA fragment with an abutting cellular DNA segment had failed (18). It is now apparent that a left terminal Ad12 DNA fragment linked to cellular DNA does not exist in cell lines T637 and TR12. Thus, the cellular DNA sequence of at least 5,251 nucleotides with only 13 CpG dinucleotides appears to be linked to a right terminal Ad12 DNA segment on either side of the integrate in both cell lines (Fig. 2).
The data then support a model of multiple copies of Ad12 DNA being arranged continuously in the T637 genome with linkage of an identical, palindromic cellular DNA sequence to the right termini of Ad12 DNA sequences at either end of the integrate. It is unknown exactly how the multiple copies of Ad12 DNA are internally arranged in cell line T637. The data from previous studies suggest that the 15 copies of Ad12 DNA might be separated from one another by short segments of cellular DNA or of rearranged viral DNA (35, 39). Possibly, the bulk of the integrate was excised exactly at the site of junction between the left terminus of one “intact” Ad12 genome linked to cellular DNA and the right terminus of the flip-flopped Ad12 genome at the opposite end of the entire integrate. The cellular DNA sequences flanking the Ad12 DNA integrate in cell line TR12 represent inverted identical repeats with a length of at least 5.2 kbp.
Methylation profile in the transgenomic Ad12 DNA in the revertant cell line TR12.
The patterns of methylation in the integrated Ad12 DNA in cell line TR12 were determined by using the bisulfite sequencing method (3, 9), which recognizes all 5-mC residues in a DNA sequence irrespective of the presence of methylation-sensitive restriction sites. We also compared the methylation profiles in selected segments of the integrated Ad12 DNA in cell line TR12 with those in the parent cell line T637.
The Ad12 genome contains 1,500 CpGs, and the inverted 3.9-kbp fragment derived from the right terminus of Ad12 DNA carries 134 CpGs (38). All 1,634 CpGs in cell line TR12 have been analyzed for their methylation status. The scheme in Fig. 4 presents a summary of the data. Each numbered square stands for a CpG and its methylation status. The numbers have been assigned to CpGs and are not related to the nucleotide numbers of Ad12 DNA (38). Except for 118 unmethylated sites, all CpGs in the Ad12 DNA in cell line TR12 are methylated; 81 CpGs have remained partly unmethylated. The latter CpGs appear to be mosaic, i.e., methylated in some cells, unmethylated in others. The unmethylated CpGs are located in blocks mainly at the termini of the integrated DNA. However, there are individual, consistently unmethylated CpGs, sometimes occurring in clusters inside the main part of and spread over the entire integrated Ad12 genome (Fig. 4).
Micrococcal nuclease sensitivity of Ad12 transgenomes and their vicinity in cell lines T637 and TR12.
It was of interest to investigate the micrococcal nuclease sensitivity of the Ad12 integrates and of their flanking cellular sites in intact T637 and TR12 cells. BHK21 cells as the nontransgenic control, T637 cells, or TR12 cells in suspension were first permeabilized by treatment with 250 μg of lysolecithin and subsequently treated with micrococcal nuclease at between 20 and 100 units per 6.6 × 106 cells at 16°C for 15 min (Fig. 5). DNA fragments were isolated and analyzed by electrophoresis on a 1% agarose gel, Southern blotting, and hybridization to different segments of Ad12 DNA or to flanking cellular DNA segment F5 (map positions are in Fig. 5D). The extents of degradation in different parts of the genomes were interpreted as a reflection of certain aspects of chromatin structure and stability. Signal intensities of nucleomonosomes in F5 cellular DNA generated with 20 units of micrococcal nuclease in cell lines T637 and TR12 were comparable to those elicited with 60 to 80 units in BHK21 cells (Fig. 5A). The F5 cellular DNA sequence that adjoins the Ad12 integrate thus proved to be more sensitive to digestion with micrococcal nuclease in transgenic cell lines T637 and TR12 than in the parent cell line, BHK21. When Ad12 DNA probes were used to assess the extent of micrococcal nuclease digestion in cell lines T637 and TR12, a distinct nucleosome arrangement was equally discernible also in the Ad12 integrate (Fig. 5B and C).
We conclude that the Ad12 transgenome shows nucleosome arrangement in both cell lines. The adjacent nonrepetitive F5 cellular DNA sequence in the Ad12 transgenic cells exhibits increased micrococcal nuclease sensitivity due to altered nucleosome structure compared to the progenitor BHK21 cell line. Foreign DNA insertion thus seems to have affected the chromatin structure at the insertion site.
Methylation profiles in selected segments in the transgenomic Ad12 DNA in cell line T637.
Earlier analyses with the methylation-detecting restriction endonuclease pair HpaII and MspI had shown the integrated Ad12 DNA in cell line T637 to be less heavily methylated than in the revertant cell line TR12 (27). For selected segments of the integrated Ad12 DNA in cell line T637, methylation profiles were now also assessed by bisulfite sequencing. Juxtaposition of the data from cell lines T637 and TR12 revealed that Ad12 DNA in the TR12 revertant was indeed more intensely methylated in Ad12 genome segments E1A, E1B, and E4 than in cell line T637. Methylation levels in the MLP region were about the same in both cell lines (Fig. 6A). We cannot distinguish between the possibility of all 15 copies in cell line T637 having been methylated to similar extents or of only one or a few of these copies having been hypermethylated and thus having been preserved in revertant cell line TR12.
FIG. 6.
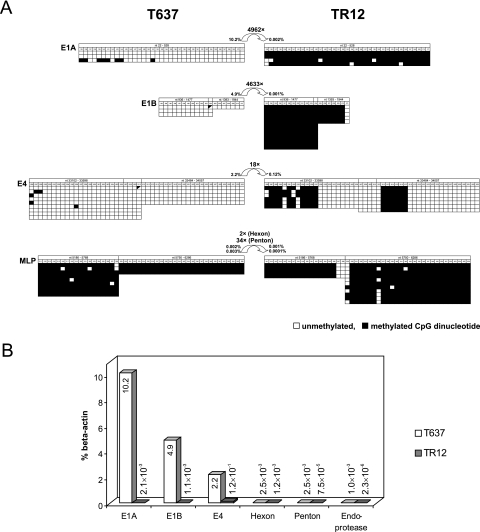
Juxtaposition of methylation patterns in selected Ad12 promoter segments (A) and adenoviral transcription levels in cell lines T637 and TR12 (B). (A) The methylation patterns of Ad12 promoter regions E1A, E1B, major late, and E4 in cell lines T637 and TR12 are depicted. Unmethylated CpGs are indicated by open squares and methylated ones by filled squares. CpG sites with variable methylation status are assigned diagonally half-filled symbols. Curved arrows indicate the difference (n-fold) in transcription levels between the two cell lines. Percentages are transcription levels in comparison to β-actin levels. (B) Transcription levels of selected Ad12 genes have been calculated as percentages of β-actin levels as determined by qPCR.
Transcription of integrated Ad12 genes and relation to DNA methylation profiles in cell lines T637 and TR12.
The E1A and E1B promoter regions are almost completely methylated in all CpG dinucleotides in revertant cell line TR12 but not methylated in cell line T637. The MLPs in both cell lines are practically completely methylated (Fig. 6A). The CpGs in the E4 promoter region in cell line T637 are not methylated and in cell line TR12 are only partly methylated. In integrated adenovirus genomes, the levels of gene transcription and promoter methylation are frequently inversely correlated (4, 25, 43, 45). Hence, it was of interest to examine Ad12 gene transcription in cell lines T637 and TR12.
Total RNA was extracted and reverse transcribed; cDNA was then analyzed by qPCR. Transcriptional activities of the Ad12 genes were related to the amounts of β-actin mRNA present in cell lines T637 and TR12. Except for the E4 region, which continued to be transcribed at moderate levels in cell line TR12, all other Ad12 genes investigated were almost completely silenced in cell line TR12, although tiny amounts of RNA from the regions E1A, E1B, hexon, penton, and endoprotease could reliably be detected by the very sensitive qPCR method (Fig. 6B). In agreement with the absence or low levels of DNA methylation in the Ad12 segments E1A, E1B, and E4 in cell line T637, substantial amounts of RNA were transcribed from these regions, although considerably less than from the cellular β-actin gene (Fig. 6B). The ratio of 18:1 in the amounts of E4 RNA transcribed from the unmethylated and hypomethylated promoters in both cell lines T637 and TR12, respectively, might be explained by differences in gene dosage. Cell line T637 carried about 15 copies versus 2 copies of the E4 region in the revertant cell line TR12.
In cell line T637, the unmethylated E1A, E1B, and E4 promoter regions allowed the efficient transcription of these segments from the integrated Ad12 DNA (Fig. 6B). The sensitive qPCR technique revealed very small amounts of late transcripts from the Ad12 hexon, penton, and viral endoprotease genes (Fig. 6B) in cell line T637. These late genes were previously thought to be silenced in cell line T637 and in several other Ad12-transformed or Ad12-induced tumor cells (28, 34).
Histone modifications in the integrated Ad12 genomes.
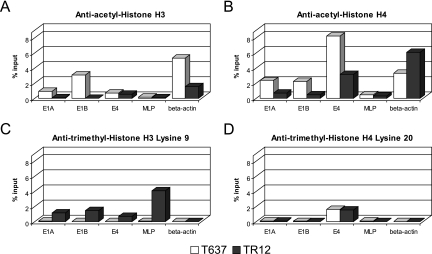
By applying the ChIP technique as described in Materials and Methods, we determined histone modifications in the E1A, E1B, E4, and MLP regions in the Ad12 integrate in T637 and TR12 cells. The cellular β-actin gene served as an internal control (Fig. 7). Two histone markers were chosen for active genes, acetylated histones H3 and H4 (33), and two for inactive genes, histone H3 trimethylated at lysine 9 and histone H4 trimethylated at lysine 20 (20). Histone H3 and H4 acetylation levels of promoter regions corresponded to gene activities in cell lines T637 and TR12 (Fig. 7A and B), although exact quantitative comparisons cannot be drawn between levels of transcription and extent of histone acetylation. Histone H3 acetylation could not be detected for the viral E1A, E1B, and major late promoters in cell line TR12, and histone H4 acetylation was more prominent in cell line T637 than in line TR12. Histone H3 was acetylated in the E4 promoter region of both cell lines to about the same extent (Fig. 7A).
FIG. 7.
Chromatin immunoprecipitation of modified histones in Ad12 promoter regions in cell lines T637 and TR12. Binding of modified histones to selected Ad12 promoter regions (E1A, E1B, E4, and major late) and the housekeeping β-actin gene was assessed by ChIP. Cross-linked chromatin was precipitated with antibodies against acetylated histone H3 (A), acetylated histone H4 (B), histone H3 trimethylated at lysine 9 (C), and histone H4 trimethylated at lysine 20 (D). The extent of histone binding to promoter regions was determined by qPCR and displayed as percent precipitation of input chromatin.
Frequently, the trimethylation modifications lysine 9 on histone H3 and lysine 20 on H4 correlate with inactive genes (20). In the housekeeping β-actin gene, histone H3 lysine 9 (Fig. 7C) and H4 lysine 20 trimethylation was undetectable (Fig. 7C and D). In cell line T637, the E1A, E1B, E4, and major late promoters were devoid of histone H3 lysine 9 trimethylation, and these promoters were active, with minimal activity of the MLP. In the revertant cell line TR12, E1A, E1B, and MLP were inactive, while the E4 promoter showed some activity (Fig. 6B). Significant histone H3 lysine 9 methylation levels in these promoters corresponded to their state of inactivity, except for the E4 region, which retained some activity (Fig. 7C). Interestingly trimethylated histone H4 lysine 20 status could be observed only for the E4 promoter region in both cell lines (Fig. 7D), although this promoter was active in both cell lines.
DISCUSSION
Foreign DNA insertion into mammalian genomes has important consequences for genome stability as well as for the methylation and transcription profiles of the transgenomes. Foreign DNA integration plays a major role in many fields of molecular biology and medicine, in gene therapeutic experimental regimens of cells and organisms, in knock-in and knockout experiments, and in the generation of transgenic organisms. Aside from the obvious effect of DNA sequence perturbations at the site of foreign DNA integration, other less frequently considered effects on the recipient genome have been observed. The insertion of retroviral (16) or adenoviral DNA (23) leads to alterations of DNA methylation in the cellular DNA sequences immediately abutting the foreign DNA integrate. However, alterations in methylation patterns are not restricted to the targeted recipient site. Changes in DNA methylation patterns in cellular genes, and particularly in endogenous retrotransposons, have been described also at loci remote from the immediate region of foreign DNA integration (13, 24, 32).
Structure of integrate: palindromic viral plus adjacent cellular sequences.
In some of the Ad12-induced hamster tumor cell lines or in Ad12-transformed cell lines, Ad12 DNA was localized in the vicinity of repetitive cellular DNA sequences, e.g., the IAP (internal A particle) retrotransposons (22). Integration of Ad12 DNA into repetitive DNA might not be detrimental to cell survival, since essential cellular functions are less likely to be affected by this insertion, while targeted unique sequences could lead to gene inactivation upon integration. Cells with essential transcription profiles intact might then be selected for survival in culture. The following results argue for a palindromic array of both integrated Ad12 and recipient cellular DNA sequences in cell lines T637 and TR12: (i) restriction/Southern blot data for adjacent cellular DNA (Fig. 3) and (ii) the flip-flop structure of the second right terminal fragment of Ad12 DNA (Fig. 2).
Excision of transgenic Ad12 DNA sequences.
The excision mechanism acting on foreign transgenomes is unknown. There is evidence that palindromic sequences might be involved in the excision of amplified viral DNA from cell line T637. The excision of viral DNA from cellular DNA, as it occurs during reversion, has been successfully mimicked by autoincubation of isolated nuclei from T637 cells (8). Could this excision apparatus have discerned between more heavily and less completely methylated transgenomic DNA? The cellular DNA at either flank of the Ad12 DNA integrate presents a palindromic DNA sequence of at least 5.2 kbp (Fig. 2 and 3). When cell line TR12 was generated from line T637, by the excision of a large part of the viral integrate (Fig. 2), the abutting cellular DNA sequences appeared to remain unaltered.
De novo methylation and stability of foreign genomes.
De novo methylation of integrated Ad12 DNA sequences was detected early in the course of studies on viral DNA integration patterns (42, 43, 45). The Ad12 transgenomic sequence is more extensively methylated in the revertant TR12 than in its parent cell line, T637 (27) (this study). Thus, the most stably integrated parts of the transgenome are hypermethylated. The Ad12 DNA sequences still persisting in the revertant TR12 could have been hypermethylated already in cell line T637 or could have assumed this hypermethylated status following the excision of the bulk of the Ad12 genomes. Obviously, in cell line T637, we cannot separately identify the methylation profile of individual integrated Ad12 DNA molecules. The possibility exists that hypermethylated DNA, perhaps due to its altered degree of compaction, could resist excision from and reside more stably in the recipient genome.
De novo methylation is of interest beyond integrated viral genomes. The state of foreign transgenomes in long-term-cultivated mammalian cells might be comparable to the state of repetitive DNA sequences. During mammalian development, methylation patterns are erased and subsequently reestablished (31) by a mechanism akin to de novo methylation.
Selectivity of de novo methylation.
Upon insertion into a mammalian genome, foreign DNA frequently becomes de novo methylated by an unknown mechanism. The nucleotide sequence of the integrate, its transcriptional activity related to promoter strength (14), and the cellular DNA sequences in the environment of the foreign integrate could be decisive factors that affect de novo methylation. In cell line T637 and in its revertant TR12, the E4 regions, both in the intact viral genome and in the inverted right terminal segment, remain hypomethylated or even unmethylated. Since both sequences are palindromic and also abut the same cellular DNA sequence due to the latter′s inverted repeat structure, either the inserted viral DNA sequences or the CpG-poor (18) cellular DNA sequences adjacent to the site of integration might be responsible for the de novo methylation of the integrated Ad12 DNA. Of course, the absence of methylation and continued genetic activity of parts of a transgenome can be explained biologically by the requirement for the products from these viral gene segments and the selective advantage they bestow upon the transformed cell line in culture.
Even in the most completely methylated Ad12 DNA segments in cell line TR12, islands of consistently unmethylated CpGs have remained (Fig. 4). There is no obvious explanation for this topical absence of 5-mC residues in a sea of completely methylated CpG dinucleotides. Local structural parameters and/or altered interactions with specific proteins could be plausible speculations.
Sensitivity to micrococcal nuclease: chromatin structure.
In this study, total Ad12 DNA, selected Ad12 DNA fragments, and the F5 segment of cellular DNA adjacent to the insertion site (18) have been used as probes in micrococcal nuclease-treated cells upon their permeabilization by lysolecithin treatment. In the hamster cell lines T637 and TR12, the F5 cellular DNA is more susceptible to micrococcal nuclease digestion than in BHK21 cells (Fig. 5A). Apparently, upon the insertion of viral DNA, the cellular chromatin structure becomes destabilized, as can be determined by this method to a certain extent. In both Ad12 transgenic cell lines, Ad12 DNA (data not shown) and in particular its E4 and MLP fragments (Fig. 5B and C) appear equally digestible with micrococcal nuclease. The micrococcal nuclease data demonstrate that the integrated Ad12 DNA has assumed nucleosome structure in cell lines T637 and TR12.
Low levels of transcription in hypermethylated Ad12 DNA segments.
Hypermethylation of the E1A and E1B regions in the integrated Ad12 DNA in the revertant TR12 cells (Fig. 6A) is associated with and may cause their transcriptional down-regulation. For the E1A and E1B regions of the integrated Ad12 DNA, the amount ratios of transcripts in cell lines T637 and TR12 are 4,962:1 and 4,633:1, respectively (Fig. 6B). On the basis of gene dosage effects alone, a ratio of 15:1 would have been expected. Thus, gene silencing in cell line TR12 must be largely due to the epigenetic modifications of complete DNA methylation (Fig. 4 and 6) and increased histone deacetylation (Fig. 7A and B) in these regions. Gene dosage, however, could be the explanation for the larger amounts of E4 transcripts in cell line T637 compared to TR12. Results for histone modifications (Fig. 7) are in qualitative agreement with the transcriptional profiles of the Ad12 genes in both cell lines.
Even under the control of the completely methylated MLP in cell lines T637 and TR12, spurious amounts of Ad12 hexon, penton, and endoprotease transcripts were detected in cell lines T637 and TR12 (Fig. 6B). Late Ad12 gene transcription from the hypermethylated MLP might be attributable to transient hemimethylation of the MLP in the Ad12 integrates during DNA replication and cell division or to residual activities of fully methylated promoters that can be detected only by the very sensitive methods employed here.
Persistent activity of genes from the Ad12 E4 region.
In the adenovirion genome, E4 is located between map units 91.3 and 99.1. It has been predicted that the E4 sequence encodes seven different polypeptides, and six of these have been found in infected cells. E1A activates the E4 promoter, which remains active also in the late phase of infection. Information available on the E4 functions comes mainly from work on Ad2 and Ad5. Many E4 proteins are multifunctional viral regulators. Although the activity of the E1 genes in the Ad12 transgenome integrated in the revertant cell line TR12 has been reduced to a minimum, the E4 genes continue to be transcribed at about the same level as in the parent cell line, T637. Adenovirus E4 genes have been shown to modulate transcription, the cell cycle, cell signaling, and DNA repair. The E4 functions are also thought to affect transgene persistence, RNA splicing and processing, late protein synthesis, and the transition from the early to late stages of infection (48, 49). The influence of the E4 genes on transgene persistence is not understood; nevertheless, E4 activities may have played a role in stabilizing the residual Ad12 transgenome in cell line TR12 and/or in excising the majority of the Ad12 DNA from the genome of cell line T637. There is evidence that the E4 gene products from Ad12 have transformation potential; the E1 gene products alone are not sufficient to induce tumors in newborn hamsters (36). Thus, the availability of E4 functions would help in maintaining the transformed phenotype of cell line TR12 and contribute to the selection for viability of this revertant cell line.
Acknowledgments
I.M. was the recipient of fellowships from the Alexander von Humboldt-Stiftung and Deutscher Akademischer Austauschdienst, Bonn, Germany. N.H. was supported by amaxa GmbH, Cologne, Germany, throughout his predoctoral training. This research was financed by the Center for Molecular Medicine Cologne (TP13), by amaxa GmbH, Cologne, and by the Institute for Clinical and Molecular Virology, Erlangen University Medical School.
In the bisulfite sequencing studies, Lars Corzelius and Anja Naumann rendered invaluable help. We thank Jeannine Gerhardt, Vanderbilt University, Nashville, TN, for advice on the ChIP technique.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Boyd, V. L., and G. Zon. 2004. Bisulfite conversion of genomic DNA for methylation analysis: protocol simplification with higher recovery applicable to limited samples and increased throughput. Anal. Biochem. 326:278-280. [DOI] [PubMed] [Google Scholar]
- 2.Boyes, J., and G. Felsenfeld. 1996. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 15:2496-2507. [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerfler, W. 1983. DNA methylation and gene activity. Annu. Rev. Biochem. 52:93-124. [DOI] [PubMed] [Google Scholar]
- 5.Doerfler, W. 1991. Patterns of DNA methylation—evolutionary vestiges of foreign DNA inactivation as a host defense mechanism—a proposal. Biol. Chem. Hoppe-Seyler 372:557-564. [PubMed] [Google Scholar]
- 6.Doerfler, W. 2006. De novo methylation, long-term promoter silencing, methylation patterns in the human genome, and consequences of foreign DNA insertion. Curr. Top. Microbiol. Immunol. 301:125-175. [DOI] [PubMed] [Google Scholar]
- 7.Eick, D., and W. Doerfler. 1982. Integrated adenovirus type 12 DNA in the transformed hamster cell line T637: sequence arrangements at the termini of viral DNA and mode of amplification. J. Virol. 42:317-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eick, D., B. Kemper, and W. Doerfler. 1983. Excision of amplified viral DNA at palindromic sequences from the adenovirus type 12-transformed hamster cell line T637. EMBO J. 2:1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groneberg, J., D. Sutter, H. Soboll, and W. Doerfler. 1978. Morphological revertants of adenovirus type 12-transformed hamster cells. J. Gen. Virol. 40:635-645. [DOI] [PubMed] [Google Scholar]
- 11.Günthert, U., M. Schweiger, M. Stupp, and W. Doerfler. 1976. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc. Natl. Acad. Sci. USA 73:3923-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, J., C. Stirzaker, and S. J. Clark. 1998. Cytosines adjacent to methylated CpG sites can be partially resistant to conversion in genomic bisulfite sequencing leading to methylation artifacts. Anal. Biochem. 264:129-132. [DOI] [PubMed] [Google Scholar]
- 13.Heller, H., C. Kämmer, P. Wilgenbus, and W. Doerfler. 1995. Chromosomal insertion of foreign (adenovirus type 12, plasmid, or bacteriophage lambda) DNA is associated with enhanced methylation of cellular DNA segments. Proc. Natl. Acad. Sci. USA 92:5515-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertz, J., G. Schell, and W. Doerfler. 1999. Factors affecting de novo methylation of foreign DNA in mouse embryonic stem cells. J. Biol. Chem. 274:24232-24240. [DOI] [PubMed] [Google Scholar]
- 15.Hösel, M., D. Webb, J. Schröer, B. Schmitz, and W. Doerfler. 2001. The overexpression of the adenovirus type 12 pTP or E1A gene facilitates Ad12 DNA replication in nonpermissive BHK21 hamster cells. J. Virol. 75:16041-16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jähner, D., and R. Jaenisch. 1985. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivation. Nature 315:594-597. [DOI] [PubMed] [Google Scholar]
- 17.Kämmer, C., and W. Doerfler. 1995. Genomic sequencing reveals absence of DNA methylation in the major late promoter of adenovirus type 2 DNA in the virion and in productively infected cells. FEBS Lett. 362:301-305. [DOI] [PubMed] [Google Scholar]
- 18.Knoblauch, M., J. Schröer, B. Schmitz, and W. Doerfler. 1996. The structure of adenovirus type 12 DNA integration sites in the hamster cell genome. J. Virol. 70:3788-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koetsier, P. A., J. Schorr, and W. Doerfler. 1993. A rapid optimized protocol for downward alkaline Southern blotting of DNA. BioTechniques 15:260-262. [PubMed] [Google Scholar]
- 20.Lachner, M., R. J. O′Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 21.Li, L. C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427-1431. [DOI] [PubMed] [Google Scholar]
- 22.Lichtenberg, U., C. Zock, and W. Doerfler. 1987. Insertion of adenovirus type 12 DNA in the vicinity of an intracisternal A particle genome in Syrian hamster tumor cells. J. Virol. 61:2719-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenberg, U., C. Zock, and W. Doerfler. 1988. Integration of foreign DNA into mammalian genome can be associated with hypomethylation at site of insertion. Virus Res. 11:335-342. [DOI] [PubMed] [Google Scholar]
- 24.Müller, K., H. Heller, and W. Doerfler. 2001. Foreign DNA integration. Genome-wide perturbations of methylation and transcription in the recipient genomes. J. Biol. Chem. 276:14271-14278. [DOI] [PubMed] [Google Scholar]
- 25.Munnes, M., and W. Doerfler. 1997. DNA methylation in mammalian genomes: promoter activity and genetic imprinting, p. 435-446. In R. Dulbecco (ed.), Encyclopedia of human biology, vol. 3. Academic Press, San Diego, CA. [Google Scholar]
- 26.Orend, G., M. Knoblauch, C. Kämmer, S. T. Tjia, B. Schmitz, A. Linkwitz, G. Meyer zu Altenschildesche, J. Maas, and W. Doerfler. 1995. The initiation of de novo methylation of foreign DNA integrated into a mammalian genome is not exclusively targeted by nucleotide sequence. J. Virol. 69:1226-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orend, G., M. Knoblauch, and W. Doerfler. 1995. Selective loss of unmethylated segments of integrated Ad12 genomes in revertants of the adenovirus type 12-transformed cell line T637. Virus Res. 38:261-267. [DOI] [PubMed] [Google Scholar]
- 28.Ortin, J., K. H. Scheidtmann, R. Greenberg, M. Westphal, and W. Doerfler. 1976. Transcription of the genome of adenovirus type 12. III. Maps of stable RNA from productively infected human cells and abortively infected and transformed hamster cells. J. Virol. 20:355-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pena, R. N., J. Webster, S. Kwan, J. Korbel, and B. A. Whitelaw. 2004. Transgene methylation in mice reflects copy number but not expression level. Mol. Biotechnol. 26:215-220. [DOI] [PubMed] [Google Scholar]
- 30.Ramsahoye, B. H., D. Biniszkiewicz, F. Lyko, A. P. Bird, and R. Jaenisch. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 97:5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razin, A., C. Webb, M. Szyf, J. Yisraeli, A. Rosenthal, T. Naveh-Many, N. Sciaky-Gallili, and H. Cedar. 1984. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc. Natl. Acad. Sci. USA 81:2275-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remus, R., C. Kämmer, H. Heller, B. Schmitz, G. Schell, and W. Doerfler. 1999. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J. Virol. 73:1010-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 34.Schirm, S., and W. Doerfler. 1981. Expression of viral DNA in adenovirus type 12-transformed cells, in tumor cells, and in revertants. J. Virol. 39:694-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schröer, J., I. Hölker, and W. Doerfler. 1997. Adenovirus type 12 DNA firmly associates with mammalian chromosomes early after virus infection or after DNA transfer by the addition of DNA to the cell culture medium. J. Virol. 71:7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiroki, K., S. Hashimoto, I. Saito, Y. Fukui, H. Kato, and H. Shimojo. 1984. Expression of the E4 gene is required for establishment of soft-agar colony-forming rat cell lines transformed by the adenovirus 12 E1 gene. J. Virol. 50:854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 38.Sprengel, J., B. Schmitz, D. Heuss-Neitzel, C. Zock, and W. Doerfler. 1994. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J. Virol. 68:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabel, S., W. Doerfler, and R. R. Friis. 1980. Integration sites of adenovirus type 12 DNA in transformed hamster cells and hamster tumor cells. J. Virol. 36:22-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoker, M., and I. Macpherson. 1964. Syrian hamster fibroblast cell line BHK21 and its derivatives. Nature 203:1355-1357. [DOI] [PubMed] [Google Scholar]
- 41.Strohl, W. A., A. S. Rabson, and H. Rouse. 1967. Adenovirus tumorigenesis: role of the viral genome in determining tumor morphology. Science 156:1631-1633. [DOI] [PubMed] [Google Scholar]
- 42.Sutter, D., M. Westphal, and W. Doerfler. 1978. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell 14:569-585. [DOI] [PubMed] [Google Scholar]
- 43.Sutter, D., and W. Doerfler. 1980. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc. Natl. Acad. Sci. USA 77:253-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolun, A., P. Alestrom, and U. Pettersson. 1979. Sequence of inverted terminal repetitions from different adenoviruses: demonstration of conserved sequences and homology between SA7 termini and SV40 DNA. Cell 17:705-713. [DOI] [PubMed] [Google Scholar]
- 45.Vardimon, L., R. Neumann, I. Kuhlmann, D. Sutter, and W. Doerfler. 1980. DNA methylation and viral gene expression in adenovirus-transformed and -infected cells. Nucleic Acids Res. 8:2461-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warnecke, P. M., C. Stirzaker, J. R. Melki, D. S. Millar, C. L. Paul, and S. J. Clark. 1997. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 25:4422-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisenberger, D. J., M. Campan, T. I. Long, M. Kim, C. Woods, E. Fiala, M. Ehrlich, and P. W. Laird. 2005. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 33:6823-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitzman, M. D. 2005. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front. Biosci. 10:1106-1117. [DOI] [PubMed] [Google Scholar]
- 49.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686-7696. [DOI] [PubMed] [Google Scholar]
- 50.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]