Abstract
Since human immunodeficiency virus (HIV)-specific cell-mediated immune (CMI) responses are critical in the early control and resolution of HIV infection and correlate with postchallenge outcomes in rhesus macaque challenge experiments, we sought to identify a plasmid DNA (pDNA) vaccine design capable of eliciting robust and balanced CMI responses to multiple HIV type 1 (HIV-1)-derived antigens for further development. Previously, a number of two-, three-, and four-vector pDNA vaccine designs were identified as capable of eliciting HIV-1 antigen-specific CMI responses in mice (M. A. Egan et al., Vaccine 24:4510-4523, 2006). We then sought to further characterize the relative immunogenicities of these two-, three-, and four-vector pDNA vaccine designs in nonhuman primates and to determine the extent to which in vivo electroporation (EP) could improve the resulting immune responses. The results indicated that a two-vector pDNA vaccine design elicited the most robust and balanced CMI response. In addition, vaccination in combination with in vivo EP led to a more rapid onset and enhanced vaccine-specific immune responses. In macaques immunized in combination with in vivo EP, we observed a 10- to 40-fold increase in HIV-specific enzyme-linked immunospot assay responses compared to those for macaques receiving a 5-fold higher dose of vaccine without in vivo EP. This increase in CMI responses translates to an apparent 50- to 200-fold increase in pDNA vaccine potency. Importantly, in vivo EP enhanced the immune response against the less immunogenic antigens, resulting in a more balanced immune response. In addition, in vivo EP resulted in an approximate 2.5-log10 increase in antibody responses. The results further indicated that in vivo EP was associated with a significant reduction in pDNA persistence and did not result in an increase in pDNA associated with high-molecular-weight DNA relative to macaques receiving the pDNA without EP. Collectively, these results have important implications for the design and development of an efficacious vaccine for the prevention of HIV-1 infection.
The World Health Organization estimates that in the year 2005, 4.1 million people were newly infected with human immunodeficiency virus type 1 (HIV-1), approximately 38.6 million people were living with HIV-1 or suffering from AIDS, and 2.8 million individuals died as a result of AIDS-related diseases (78). In many developed countries, the morbidity and mortality associated with HIV-1 infection have declined substantially due to the widespread availability and use of highly active antiretroviral therapy (HAART). However, the rate of new HIV infections has not declined substantially, despite prevention programs and HAART availability. In developing countries, where HAART is largely unavailable, social programs intended to reduce the spread of HIV have generally proven insufficient, and the epidemic continues. A safe and effective vaccine for the prevention of HIV infection is urgently needed to slow the epidemic, particularly in developing countries without widespread access to HAART.
In 1990, it was first demonstrated that the direct injection of naked plasmid DNA (pDNA) encoding a foreign antigen into mouse myocytes resulted in uptake of the pDNA by host cells and subsequent expression of the foreign antigen (81). Shortly thereafter, animal studies clearly demonstrated that pDNA vaccines were capable of eliciting humoral (68) and cellular (60, 73) immune responses against the encoded vaccine antigen. Following these pioneering studies, pDNA vaccines have proven to be effective at eliciting HIV antigen-specific immune responses in a wide variety of animal model systems (3, 15, 16) and have been evaluated in a number of phase I clinical trials (8, 9, 11-13, 19, 27, 34-36, 40, 49, 50, 65-67, 69, 72, 74, 75, 77, 84), where they have been found to be well tolerated.
However, despite promising results in preclinical immunogenicity studies, pDNA vaccines have generally been incapable of inducing robust antigen-specific immune responses in clinical trials. One possible explanation for this may be a reduced stimulatory effect of CpG sequences in the plasmid backbone due to the low expression levels of Toll-like receptor 9 in humans (6). Another possible explanation for the poor performance of pDNA vaccines in humans is that the level of protein expression may be insufficient to elicit strong immune responses. Indeed, even for mice, it has been suggested that the amount of protein expressed following pDNA vaccination limits the immune response. A number of methods have therefore been developed to increase the efficiency of pDNA delivery. In vivo electroporation (EP) has proven to be an efficient method for delivering genes into muscle tissue (1, 20). The application of electrical pulses to muscle tissue can enhance cell permeability and facilitate pDNA uptake, resulting in 100- to 1,000-fold increases in protein expression (1, 39, 48, 79) and greatly enhanced humoral and cellular immune responses towards the expressed transgene(s) (4, 5, 10, 30, 44, 47, 55, 61, 64, 71, 79, 83, 85). In addition to increasing expression levels, it has been speculated that in vivo EP may result in mild localized tissue damage resulting in the release of cellular components, which might provide a “danger signal” and thus provide an adjuvant effect (46). Finally, in vivo EP increases transfection of mononuclear cells (21), which could be critical for the induction of an effective immune responses.
Given the importance of the HIV-specific cell-mediated immune (CMI) response in the early control of HIV replication during primary infection (24, 56) and the observed correlation between prechallenge vaccine-elicited CMI responses and postchallenge outcomes in simian/human immunodeficiency virus (SHIV)/rhesus macaque challenge experiments (7, 17), we recently identified several candidate pDNA vaccine designs capable of eliciting robust and balanced CMI responses to multiple HIV-1-derived antigens in mice for further vaccine development (18). To rationally construct candidate vaccines for immunogenicity testing, we first determined the relative immunogenicities of the individual HIV-1-derived vaccine antigens (Env, Gag, Pol, Nef, Tat, and Vif) and the relative strengths of various transcriptional control elements (human cytomegalovirus [HCMV], simian CMV [SCMV], and herpes simplex virus Lap1) in BALB/c mice. Next, a number of one-, two-, three-, and four-vector pDNA vaccine designs were tested for the ability to elicit HIV-1 antigen-specific CMI responses. For these studies, BALB/c mice were immunized with a fixed total pDNA vaccine dose of 100 μg in combination with 25 μg plasmid-based murine interleukin-12 (IL-12) and tested for the induction of HIV-1 antigen-specific CMI responses by gamma interferon (IFN-γ)-specific enzyme-linked immunospot (ELISPOT) analysis. The results of this study indicate that all pDNA vaccine designs were capable of eliciting CMI responses to multiple HIV-1 antigens. As a result of this iterative comparative analysis in mice, we identified a number of top-performing pDNA vaccine candidates capable of eliciting potent and balanced CMI responses to multiple HIV-1-derived antigens. The purpose of the current study was to further characterize the relative immunogenicities of these top-performing two-, three-, and four-vector pDNA vaccine designs in nonhuman primates. In addition, we sought to determine the extent to which in vivo EP could improve the induction of vaccine antigen-specific cell-mediated and humoral immune responses.
MATERIALS AND METHODS
Animals.
For this study, a total of 36 Mamu-A*01-negative captive-bred male rhesus macaques (Macaca mulatta) of Indian origin were used. Macaques were maintained in accordance with the Guide for the Care and Use of Laboratory Animals (43a). In addition, Wyeth Research's Institutional Animal Care and Use Committee approved procedures for the use and care of the macaques.
Quantification of peripheral blood cell subsets by CBC analysis.
The effect of pDNA immunization on various cell subsets in peripheral blood were monitored by complete blood count (CBC) analysis. In addition, absolute peripheral blood CD3+, CD3+CD4+, CD3+CD8+, and CD20+ cell numbers were quantitated by flow cytometry with the following fluorochrome-labeled monoclonal antibodies (BD Pharmingen, San Jose, CA): anti-rhesus macaque CD3-fluorescein isothiocyanate (clone SP34); anti-human CD4-phycoerythrin (PE) (clone M-T477); anti-human CD8-peridinin chlorophyll protein (clone SK1); and anti-human CD20-allophycocyanin (clone L27). Antibodies, diluted per the manufacturer's instructions, were added to 100 μl of EDTA-treated whole blood and incubated for 15 min at room temperature. Subsequently, erythrocytes were lysed by the addition of 450 μl of fluorescence-activated cell sorter (FACS) lysis solution (Becton Dickinson, Franklin Lakes, NJ). Cells were then washed once with 1× phosphate-buffered saline (PBS) containing 0.02% azide and resuspended in 1× PBS containing 1% paraformaldehyde. FACS analysis was performed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) and analyzed using CellQuest software.
pDNAs.
All HIV genes used in this study, including gag p55 (HXB2), pol (HXB2), nef (NL4-3), tat (NL4-3), vif (NL4-3), and env gp160 (from primary isolate 6101 from David Montefiori; gene bank accession no. bankit625244), were RNA optimized. RNA optimization involved the introduction of multiple point mutations within the coding region to inactivate endogenous inhibitory sequences, thus allowing for high-level rev-independent expression, as previously described (57-59). To prepare in-frame gag-pol fusions, the gag terminator codon was removed and mutations were introduced in the natural frameshift region. As an additional safety feature, enzymatic activity associated with the pol gene was inactivated by making the following changes: (i) protease activity was eliminated by introducing a three-amino-acid deletion (Asp25 to Gly27) in the enzyme active site (32, 82); (ii) reverse transcriptase activity was inactivated by deletion of four amino acids (Tyr183 to Asp186) (25, 26); (iii) RNase-activity was abolished by a single amino acid deletion (Glu478) (14, 54); and (iv) integrase function was abolished by deletion of amino acids Asp626, Asp678, and Glu714 (28, 80). The nef, tat, and vif open reading frames (Nef amino acids 4 to 206, Tat amino acids 2 to 80, and Vif amino acids 2 to 192) were fused in frame to generate a single Nef-Tat-Vif fusion protein (NTV). For safety reasons, the ability of Nef to alter T-cell signal transduction pathways and to down regulate cell surface expression of CD4 and major histocompatibility complex class I was inactivated by removal of the myristoylation signal (2). In addition, the ability of Tat to transactivate nuclear gene expression was eliminated by the deletion of two cysteines (Cys30 and Cys34) (51). Other amino acid sequence changes were also made in Nef (A15T, G29R, V33A, and N51T) and Vif (N19R, R23S, R33G, D37G, N48H, K50R, H110Y, K155T, and A167T) to introduce HIV-1 clade B consensus cytotoxic T-lymphocyte (CTL) epitopes (33).
Various pDNA-based expression vectors encoding one or two unique expression cassettes (Table 1) were combined and used as the experimental vaccines. Expression cassettes carrying HIV-1-derived genes were constructed using one of the following three transcriptional unit designs: (i) HCMV immediate-early promoter and bovine growth hormone (BGH) polyadenylation [poly(A)] signal; (ii) HCMV promoter and simian virus 40 poly(A); and (iii) SCMV promoter and BGH poly(A). In addition, expression vectors included a chimeric kanamycin resistance (kmr) gene, an adenylyl 4′-nucleotidyl transferase type 1a gene (52, 62), and a ColE1 bacterial origin of replication required for selection and propagation of bacteria, respectively.
TABLE 1.
Design of study to elucidate the effects of pDNA vaccine design and in vivo EP on the resulting vaccine-specific immune response in rhesus macaques
| Group (n) | Macaque no. | Vector dose (mg) | Vector [encoded antigen(s)]a |
|---|---|---|---|
| 2d (6) | A1N093 | ||
| A1N098 | |||
| A1N111 | 4.25 | WLV-151 (HCMV-Gag/Pol) | |
| A2N040 | 4.25 | WLV-159 (SCMV-Env and HCMV-NTV) | |
| A2N043 | 1.5 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| A2N065 | |||
| 3a (6) | A1N020 | ||
| A1N104 | |||
| A2N009 | 2.8 | WLV-151 (HCMV-Gag/Pol) | |
| A2N024 | 2.8 | WLV-106 (HCMV-Env) | |
| A2N027 | 2.8 | WLV-147 (HCMV-NTV) | |
| A2N028 | 1.5 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| 3c (6) | A1N089 | ||
| A1N097 | |||
| A2N029 | 2.8 | WLV-159 (SCMV-Env and HCMV-NTV) | |
| A2N042 | 2.8 | WLV-008 (HCMV-Gag) | |
| A2N060 | 2.8 | WLV-146 (HCMV-Pol) | |
| A2N070 | 1.5 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| 3cE (6) | A1N078 | ||
| A1N086 | 0.56 | WLV-159 (SCMV-Env and HCMV-NTV) | |
| A2N010 | 0.56 | WLV-008 (HCMV-Gag) | |
| A2N014 | 0.56 | WLV-146 (HCMV-Pol) | |
| A2N036 | 0.30 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| A2N054 | |||
| 4a (6) | A2N092 | ||
| A2N053 | 2.13 | WLV-008 (HCMV-Gag) | |
| A2N077 | 2.13 | WLV-106 (HCMV-Env) | |
| A2N093 | 2.13 | WLV-146 (HCMV-Pol) | |
| A2N088 | 2.13 | WLV-147 (HCMV-NTV) | |
| A2N094 | 1.5 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| Control (6) | A1N040 | ||
| A1N080 | |||
| A1N112 | 8.5 | WLV-001a (empty vector control) | |
| A1N118 | 1.5 | WLV-104 (HCMV-IL-12 p35 and SCMV-IL-12 p40) | |
| A2N037 | |||
| A2N049 |
HCMV, human cytomegalovirus immediate-early promoter; Gag/Pol, made by fusion of the HIV-1 HXB2 gag-pol genes. SCMV, simian cytomegalovirus immediate-early promoter; Env, HIV-16101 Env gp160; NTV, made by fusion of the HIV-1 NL4-3 nef, tat, and vif genes.
For each pDNA expression vector, in vitro expression of the encoded HIV-1 antigen(s) was confirmed by Western blotting after transient transfection of human rhabdomyosarcoma (RD) cells (ATCC, Manassas, VA) as previously reported (18).
An expression plasmid encoding rhesus IL-12 was codelivered as a molecular adjuvant. The dual-promoter IL-12 expression vector carries the rhesus IL-12 p35 and p40 genes. The IL-12 p35 subunit is expressed under the control of the HCMV immediate-early promoter and the simian virus 40 poly(A) signal, while the IL-12 p40 subunit is expressed under control of the SCMV promoter and the BGH poly(A) signal. Production of rhesus IL-12 was confirmed after transient transfection of RD cells and screening of cell supernatants for cytokine expression, using an anti-human IL-12 p70 capture enzyme-linked immunosorbent assay (ELISA; Endogen, Woburn, MA) (data not shown). Bioactivity of the plasmid-expressed rhesus IL-12 was confirmed by assaying supernatants from transiently transfected RD cells for the capacity to induce IFN-γ secretion in resting rhesus peripheral blood lymphocytes (PBLs) (data not shown).
Expression plasmids for inoculation of macaques were produced by Puresyn, Inc. (Malvern, PA). Plasmids were propagated in Escherichia coli, isolated from cells by alkaline lysis, purified by column chromatography, and formulated individually at a concentration of 2.5 mg/ml in isotonic citrate buffer (29.3 mM sodium citrate, 0.67 mM citric acid, 150 mM NaCl, 0.34 mM EDTA, pH 6.4 to 6.7) containing 0.25% bupivacaine to allow for the formation of DNA-bupivacaine complexes (45). Final pDNA preparations were shown to consist of >90% supercoiled pDNA, and residual endotoxin was shown to be <30 endotoxin units/mg DNA (data not shown).
Vaccinations.
Immediately prior to inoculation, the appropriate pDNA expression vectors were mixed and administered by intramuscular (i.m.) injection (groups 2d, 3a, 3c, 4a, and controls) into both deltoid and quadriceps muscles (1.0 ml/injection site, 2.5 mg DNA per site), using an 18-gauge needle and a 3-ml syringe. For macaques receiving the vaccine in combination with EP (group 3cE), i.m. injections and in vivo EP were performed using a combined injection/EP device where the injection needles also served as electrodes. The prototype device used in this study was a semiautomatic version of the injector previously described by Tjelle et al. (70). pDNA was injected through two needles (21-gauge) during insertion into the muscle and was thus distributed along the entire needle path, ensuring a good match between the pDNA and the electric field. Immediately following injection, six 20-ms pulses at 250 mA of constant current (corresponding to a nominal field strength of approximately 70 V/cm) were applied.
IFN-γ ELISPOT assay.
Ninety-six-well flat-bottomed ELISPOT plates (Millipore, Bedford, MA) were coated overnight with a mouse anti-human IFN-γ monoclonal antibody (clone 27; BD-Pharmingen, San Diego, CA) at a concentration of 1 μg/ml, after which the plates were washed three times with 1× PBS and then blocked for 2 h with PBS containing 5% heat-inactivated fetal bovine serum (FBS). Heparinized rhesus macaque whole blood was collected at various time points after immunization, and PBLs were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation, resuspended in complete R05 culture medium (RPMI 1640 medium supplemented with 5% FBS, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, 1 mM sodium pyruvate, 1 mM HEPES, 100 mM nonessential amino acids), and shipped via overnight courier. Within 24 h of blood draw, the isolated macaque PBLs were washed once with complete R05 culture medium and resuspended in complete R05 culture medium containing either 50 μg/ml phytohemagglutinin mucoprotein (Sigma), peptide pools (15-mers overlapping by 11 amino acids; final peptide concentration, 1 μM [each]) spanning HIV-1HXB2 Gag p55, Pol, Nef, Tat, and Vif and HIV-16101 Env gp160, or medium alone. The input cell number was 2 × 105 PBLs per well (2 × 106 PBLs/ml), and cells were assayed in duplicate wells. Cells were incubated for 18 to 24 h at 37°C and then removed from the ELISPOT plate by first being washed with deionized water and then being washed six times with 1× PBS containing 0.25% Tween 20 and three additional times with 1× PBS. Thereafter, plates were treated with a rabbit polyclonal anti-human IFN-γ biotinylated detection antibody (0.2 μg/well) (Biosource, Camarillo, CA) diluted with 1× PBS containing 1% bovine serum albumin (BSA) and were incubated at room temperature for 2 h. ELISPOT plates were then washed 10 times with 1× PBS containing 0.25% Tween 20, treated with 100 μl per well of streptavidin-alkaline phosphatase conjugate (0.4 mg/ml stock solution; Southern Biotech, Birmingham, AL) diluted 1:500 with 1× PBS containing 5% FBS and 0.005% Tween 20, and incubated for an additional 1 h at room temperature. Unbound conjugate was removed by rinsing the plate 10 times with 1× PBS containing 0.25% Tween 20. A chromogenic substrate (100 μl/well) (one-step nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate [NBT/BCIP]; Pierce, Rockford, IL) was then added for 3 to 5 min before being rinsed away with water, after which the plates were air dried and the resulting spots counted using an immunospot reader (CTL Inc., Cleveland, OH). Peptide-specific IFN-γ ELISPOT responses were considered positive if the responses (minus the medium background) were >3-fold above the medium response and ≥50 spot-forming cells (SFC)/106 PBLs.
Preparation of bead-depleted PBLs.
CD4+ or CD8+ cells were depleted from unfractionated PBLs by using magnetic polystyrene beads coated with anti-human CD4- or CD8-specific mouse monoclonal antibodies per the manufacturer's instructions (Dynal Biotech, Oslo, Norway). Briefly, freshly isolated rhesus PBLs were washed and resuspended to a final concentration of 2 × 106 cells/ml in ice-cold 1× PBS containing 2% FBS. Dynal microbeads were washed three times with 1× PBS containing 2% FBS, added to unfractionated PBLs at a 5:1 bead-to-cell ratio, and incubated for 1 hour at 4°C on a rotating/tilting apparatus. After incubation, the bead-cell suspension was placed in a magnetic column, and the flowthrough containing either CD4+ or CD8+ cell-depleted PBLs was removed, washed once with complete culture medium supplemented with 5% FBS, and resuspended to the original volume with complete culture medium supplemented with 5% FBS. An equal volume of unfractionated, bead-depleted PBLs was used directly in the ELISPOT assay.
The efficiency of CD4+ and CD8+ cell subset depletion and the precise number of CD4+ and CD8+ cells added to the ELISPOT plate were subsequently quantified by flow cytometry. Briefly, bead-depleted PBLs were washed once with 1× PBS containing 2% FBS and stained for 15 min at room temperature with the following monoclonal antibodies (BD Pharmingen, San Jose, CA): anti-rhesus macaque CD3-fluorescein isothiocyanate (clone SP34), anti-human CD4-PE (clone M-T477), anti-human CD8- peridinin chlorophyll protein (clone SK1), and anti-human CD20-allophycocyanin (clone L27). Cells were then washed once with 1× PBS containing 2% FBS and 0.02% azide and resuspended in 1× PBS containing 1% paraformaldehyde. FACS analysis was performed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) and analyzed using CellQuest software. The percentage of CD4+ or CD8+ cell depletion was routinely >95% (data not shown).
Antigen-specific antibody titers by ELISA.
For the determination of HIV-1 viral lysate- and HIV-1 Env-specific antibody titers, ELISA plates were coated for 18 h at 4°C with purified HIV-1IIIB viral lysate (20 ng/well; Immunodiagnostics, Woburn, MA) or 20 ng/well HIV-16101 Env gp120 diluted in carbonate-bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6). The plates were then washed five times with 1× PBS containing 0.1% Tween 20 and blocked for 2 h at room temperature with 1× PBS containing 0.1% Tween 20 and 3% BSA. Serum samples, diluted with 1× PBS containing 1% BSA and 0.1% Tween 20, were added to the ELISA plates at a starting dilution of 1:64 and further diluted twofold across the plates. Plates were kept at 4°C overnight, after which they were washed five times with 1× PBS containing 0.1% Tween 20; a biotin-conjugated primary antibody specific for monkey immunoglobulin G (IgG; diluted 1:30,000 with 1× PBS supplemented with 0.1% Tween 20 and 1% BSA) (Accurate Scientific, Westbury, NY) was then added (100 μl/well) for 2 h at room temperature, followed by a 1-h incubation at room temperature with 100 μl/well of a streptavidin-horseradish peroxidase-conjugated anti-biotin antibody (500-units/ml stock, diluted 1:10,000 with 1× PBS supplemented with 0.1% Tween 20 and 1% BSA; Roche Immunochemical, Indianapolis, IN) used to detect antigen-specific immunoglobulin. Finally, ELISA plates were developed by the addition of 100 μl/well of TMB (3,3′,5,5′-tetramethylbenzidine; Sigma). The antigen-specific antibody titer was defined as the reciprocal of the last serum dilution giving an optical density at 450 nm (OD450) greater than that for the same macaque's naïve serum (i.e., week 0) plus 3 standard deviations.
Detection of rhesus IL-12-binding and neutralizing antibody.
A commercially available human IL-12 ELISA kit (Pierce Endogen) was used to detect anti-IL-12 antibody in serum. Briefly, microplate strips precoated with anti-human IL-12 antibody were incubated with 20 ng/well of rhesus IL-12 for 2 h at 37°C. The IL-12-loaded strips were then washed and incubated with test serum serially diluted with 1× PBS containing 1% BSA and 0.1% Tween 20 overnight at 4°C. A biotin-conjugated primary antibody specific for monkey IgG diluted in 1% BSA-1× PBS at 1:30,000 (Accurate Scientific, Westbury NY), followed by a streptavidin-horseradish peroxidase-conjugated anti-biotin antibody (500-units/ml stock, diluted 1:10,000 with 1× PBS supplemented with 0.1% Tween 20 and 1% BSA; Roche Immunochemical, Indianapolis, IN), was added to the plates. The strips were then developed with TMB (Sigma), and the OD450 was read. The anti-rhesus IL-12 antibody titer was defined as the reciprocal of the last serum dilution giving an OD450 greater than that for the same macaques's naïve serum (i.e., week 0) plus 3 standard deviations.
Serum samples containing anti-rhesus IL-12 antibodies were further screened for the ability to block IFN-γ secretion by an IL-12-responsive cell line. Briefly, test serum was diluted 1:10 in assay medium (RPMI 1640 with glutamine; Mediatech, Herndon, VA) supplemented with 10% FBS (Invitrogen, Grand Island, NY) and then serially diluted 1:3 in a 96-well plate. To the diluted serum samples, an equal volume (50 μl) of assay medium containing 0.12 ng/ml rhesus IL-12 was added and incubated for 1 h at room temperature. NK-92MI cells (ATCC, Manassas, VA) were then added in a 100-μl volume to each well of the 96-well plate containing the serum-IL-12 mixture and incubated overnight at 37°C in a 5% CO2 incubator. IL-12-dependent secretion of IFN-γ into the culture supernatant was quantified using a commercially available IFN-γ ELISA kit (BD Biosciences, San Diego, CA) per the manufacturer's instructions.
HIV neutralizing antibody assays.
Neutralization was measured as a function of the reduction in luciferase reporter gene expression after a single round of infection in TZM-bl cells as described previously (38, 42). TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. Briefly, 200 50% tissue culture infective dose of virus was incubated with a serial threefold dilution of each serum sample in triplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottomed culture plates. Freshly trypsinized cells (10,000 cells in 100 μl of growth medium containing 75 mg/ml DEAE-dextran) were added to each well. One set of control wells received cells plus virus (virus control), and another set received cells only (background control). After a 48-h incubation, 100 μl of cells was transferred to a 96-well black solid plate (Costar) for measurements of luminescence, using Bright Glo substrate solution as described by the supplier (Promega). Assay stocks of Env-pseudotyped viruses were prepared by transfection in 293T cells and were titrated in TZM-bl cells as described previously (38). An assay stock of HIV-1MN was prepared in H9 cells (43) and titrated in TZM-bl cells.
Collection of injection site muscle biopsies.
Seventy days after the final pDNA immunization, macaques were anesthetized, and single injection site muscle biopsies were collected. Briefly, an incision (approximately 6 to 8 cm) was made over the quadriceps femoris. A muscle biopsy was then taken from the middle of the muscle belly by incising the quadriceps femoris, sparing the ligamentous attachment and not completely transecting the muscle belly. The size of the muscle biopsy approximated a width of 1 to 2 cm, a length of 2 to 4 cm, and the full depth of the muscle belly. All injection site tissue samples were snap frozen in a dry ice-ethanol bath and stored at −70°C or on dry ice prior to genomic DNA extraction.
During the collection of injection site muscle biopsies, stringent measures were put into place to prevent contamination. To avoid cross-contamination of injection site muscle tissue between macaques, biopsies were collected with the macaques placed on single-use absorbent bench paper. After collection of the biopsy tissue, the procedure table was cleaned thoroughly with a 10% bleach solution and draped with fresh absorbent bench paper. Surgical instruments were used for a single macaque, and the instruments were cleaned thoroughly with a 10% bleach solution and rinsed twice with sterile water prior to being reused. Surgical gloves were always changed between macaques.
Genomic DNA isolation.
Genomic DNA was extracted from injection site muscle tissue by using a PUREGENE tissue DNA isolation kit per the manufacturer's instructions. Briefly, a small piece of injection site tissue (∼16 to 20 mg) was first cut from the biopsy sample and sliced into smaller pieces. The sliced tissue sample was then incubated with the cell lysis solution and proteinase K overnight at 55°C. The tissue lysate was further treated with RNase, and protein was removed by precipitation with the protein precipitation solution. Genomic DNA was then precipitated with isopropanol and resuspended in DNA hydration solution (10 mM Tris, 1 mM EDTA, pH 7.5). The concentration of purified DNA was quantitated by measuring the absorbance at 260 nm. The quality of the extracted DNA was evaluated by 1% agarose gel electrophoresis using Tris-borate-EDTA buffer (TBE; 0.089 M Tris, 0.089 M borate, 2 mM EDTA, pH 8.3).
Quantitation of plasmid copies associated with genomic DNA by Q-PCR.
Two independent quantitative PCR (Q-PCR) assays using HCMV promoter- or BGH poly(A)-specific primer-probe pairs were performed on each tissue DNA sample in triplicate, using 0.5 to 1.0 μg of extracted DNA as the template. The sequences of the forward primer, reverse primer, and probe were selected using Primer Express primer design software (Applied Biosystems). The internal fluorogenic probe was labeled with the 6-carboxyfluorescein (FAM) reporter dye (Applied Biosystems). The primer and probe sequences specific for the HCMV promoter were as follows: forward primer BREL 697, 5′-CATCTACGTATTAGTCATCGCTATTACCAT-3′; reverse primer BREL 698, 5′-TGGAAATCCCCGTGAGTCA-3′; and probe BREL 086, 5′-FAM-ACATCAATGGGCGTGGATAGCGGT-3′. The primer and probe sequences specific for the BGH poly(A) region were as follows: forward primer BREL 699, 5′-TCTAGTTGCCAGCCATCTGTTGT-3′; reverse primer BREL 700, 5′-TGGGAGTGGCACCTTCCA-3′; and probe BREL 087, 5′-FAM-TCCCCCGTGCCTTCCTTGACC-3′.
The linearized plasmid WLV008M was used for both the positive control and the DNA standard. Each tissue DNA sample was spiked with 100 copies of the positive control plasmid to verify the absence of PCR inhibitors in the tissue extract. In addition, each set of PCRs also included a no-template control to verify the absence of contamination in the assay reagents. Standard curves for both Q-PCRs covered the range from 10 to 1 × 106 copies.
PCR amplification and fluorescence detection were performed using the ABI Prism 7700 sequence detection system. After amplification, the average cycle threshold (CT) for each sample was calculated, and the quantity of pDNA present in the biopsy tissue was extrapolated from the standard curve. The sensitivity of detecting pDNA was routinely 10 copies/μg DNA.
Quantitation of plasmid copies associated with HMW DNA by Q-PCR.
High-molecular-weight (HMW) DNA was extracted from 200 to 700 mg of injection site muscle biopsy tissue to obtain enough genomic DNA to quantify the level of pDNA associated with HMW DNA. Free pDNA was separated from HMW DNA by using conventional electrophoresis on 20- by 25-cm agarose gels. A series of four purification runs using two different electrophoresis procedures was used, alternating 0.5% agarose gel and 1% agarose gel. The 0.5% agarose gels were run in Tris-acetate-EDTA buffer (0.04 M Tris-acetate, 1 mM EDTA, pH 8.2) at 40 V for 22.5 h. The 1% agarose gels were run in TBE (0.089 M Tris, 0.089 M borate, 2 mM EDTA, pH 8.3) at 50 V for 45 h. Both gels were run in electrophoresis buffer supplemented with 0.2 μg/ml ethidium bromide and circulated using a medium-flow peristaltic pump.
For the first electrophoresis run, approximately 40 μg of unpurified injection site tissue DNA was loaded into two wells of a 0.5% agarose gel. To avoid potential cross-contamination among individual macaque samples, only one sample was run in each gel box. After electrophoresis, HMW genomic DNA was excised as one band across the gel. To minimize DNA loss and shearing that occurs during electroelution and subsequent reconcentration of DNA, the excised band was directly subjected to the next round of electrophoresis without elution. The gel fragment containing the partially purified HMW DNA was placed upside down on a clean gel plate, and the next gel was poured around it.
The HMW genomic DNA bands in the last 1% agarose-TBE gel were excised as individual blocks, and HMW DNA was extracted using a QIAquick gel extraction kit (QIAGEN Inc.) per the manufacturer's instructions. Control experiments demonstrated that this agarose gel purification scheme can effectively separate 100 copies of “spiked” pDNA from genomic DNA (data not shown). The yield and concentration of the agarose gel-purified HMW DNA were determined by measuring the absorbance at 260 nm. Both HCMV Q-PCR and BGH Q-PCR were performed in duplicate, using 0.2 μg purified HMW DNA as the template, under the PCR conditions noted above.
Statistical analysis.
Analysis of variance of IFN-γ ELISPOT data was performed with SAS, version 8.2, software, using the type 1 GENMOD procedure with negative binomial distribution. Log10-transformed ELISA and neutralizing antibody data were analyzed using SAS, version 8.2, software, and the least-square means of the groups were compared using the mixed procedure. In all cases, P values of <0.05 indicated that the tests were statistically significant.
RESULTS
In this study, we sought to further compare the relative abilities of several candidate pDNA vaccine designs to elicit humoral immune and CMI responses to multiple HIV-1-derived antigens in rhesus macaques. Previously, we had shown that the various pDNA vaccine designs under study were similarly immunogenic in a mouse model (18). As a follow-on to these earlier studies, we sought to identify a single pDNA vaccine design capable of eliciting robust and balanced humoral and cellular immune responses for further clinical development. In addition, we sought to determine the relative ability of in vivo EP to augment the induction of antigen-specific humoral and cellular immune responses.
Study design.
For this study, 36 outbred male rhesus macaques were used as outlined in Table 1. All macaques were negative for the Mamu-A*01 class I allele by PCR using sequence-specific primers (23); however, the macaques were not screened for the presence of other Mamu-A or Mamu-B alleles. Vaccinated macaques were immunized with a fixed total dose of vaccine pDNA (8.5 mg; groups 2d, 3a, 3c, and 4a) in combination with 1.5 mg of the dual-promoter pDNA encoding rhesus IL-12 p40 and p35. Control macaques were primed with 8.5 mg of empty expression vector DNA in combination with 1.5 mg plasmid IL-12. For macaques receiving the pDNA vaccine in combination with in vivo EP (group 3cE), one-fifth of the total vaccine dose (1.7 mg vaccine pDNA in combination with 0.3 mg IL-12 pDNA) was used. The various plasmid IL-12-supplemented DNA vaccines were administered by i.m. injection using a needle and syringe, followed by in vivo EP (group 3cE only), on a schedule of 0, 4, and 8 weeks.
Safety and tolerability of various pDNA vaccines administered by standard needle and syringe.
To monitor the safety and tolerability of the cytokine-enhanced pDNA vaccines, macaques were assessed by daily observations. In addition, macaques were monitored weekly by physical examinations and body weight measurements and for changes in peripheral blood cell subsets by CBC analysis. Over the course of the vaccination regimen, all macaques appeared normal and had body weights that were stable or increased during the study interval. In addition, macaques immunized with the candidate pDNA vaccines by i.m. injection using a standard needle and syringe maintained stable peripheral blood bulk cell subsets (i.e., total white blood cell, total lymphocyte, and CD20+ cell counts), as determined by CBC analysis (data not shown).
Comparative analysis of antigen-specific CMI responses elicited by various pDNA vaccine designs.
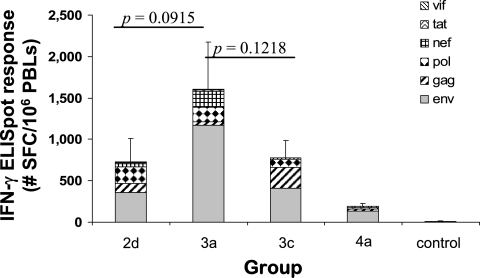
Two weeks after the final pDNA immunization, PBLs were collected from the immunized macaques and screened for the presence of HIV-1 antigen-specific CMI responses by IFN-γ ELISPOT analysis (Fig. 1). The three-vector vaccine design represented by group 3a elicited the greatest total HIV-specific IFN-γ ELISPOT response (mean, 1,607 SFC/106 PBLs), with the vaccine designs represented by groups 3c and 2d eliciting somewhat lower total HIV-specific IFN-γ ELISPOT responses (mean, 779 and 729 SFC/106 PBLs, respectively). However, the reduced responses elicited by the pDNA vaccine designs represented by groups 2d and 3c were not significantly different from that for group 3a. The lowest total HIV-specific IFN-γ ELISPOT response (mean, 191 SFC/106 PBLs) was seen within the group receiving the four-vector vaccine design represented by group 4a. The relative ranking of the various pDNA vaccine designs in terms of the ability to elicit total antigen-specific IFN-γ ELISPOT responses (i.e., 3a > 2d = 3c > 4a) remained consistent regardless of whether the IFN-γ ELISPOT responses were measured after the first, second, or third immunization (data not shown).
FIG. 1.
HIV-1 antigen-specific CMI responses after pDNA immunization. Macaques were immunized i.m. at 0, 4, and 8 weeks with the pDNA expression vectors indicated in Table 1 in combination with plasmid rhesus IL-12. Two weeks after the final immunization, PBLs were collected and tested for HIV-1 Env-, Gag-, Pol-, Nef-, Tat-, and Vif-specific IFN-γ secretion by ELISPOT assay. Bars represent the average total HIV-specific IFN-γ ELISPOT responses (n = 6), with the standard errors of the means indicated.
Over the course of the study, all of the group 2d, 3a, 3c, and 4a immunized macaques (24/24 [100%]) developed Env-specific IFN-γ ELISPOT responses, with the highest Env-specific response seen in the group of macaques receiving the three-vector vaccine design represented by group 3a (Fig. 1). A majority (14/24 [∼58%]) of the macaques developed Gag-specific IFN-γ ELISPOT responses, with the highest Gag-specific response and the highest Gag response frequency seen in the group of macaques receiving the three-vector vaccine design represented by group 3c. The vaccine design represented by group 3a, which consistently elicited the highest total HIV-specific ELISPOT response, was the least effective at eliciting a Gag-specific ELISPOT response. HIV Pol-specific IFN-γ ELISPOT responses were detected in 10 of 24 (∼42%) immunized macaques, with the lowest Pol-specific response frequency seen in group 4a. IFN-γ ELISPOT responses to NTV were consistently the lowest. Only 7 of 24 (∼29%) immunized macaques developed Nef-specific IFN-γ ELISPOT responses, with the highest response frequency (4/6) seen in group 3a. IFN-γ ELISPOT responses specific to the Tat and Vif peptide pools remained <100 SFC/106 PBLs throughout the course of the experiment.
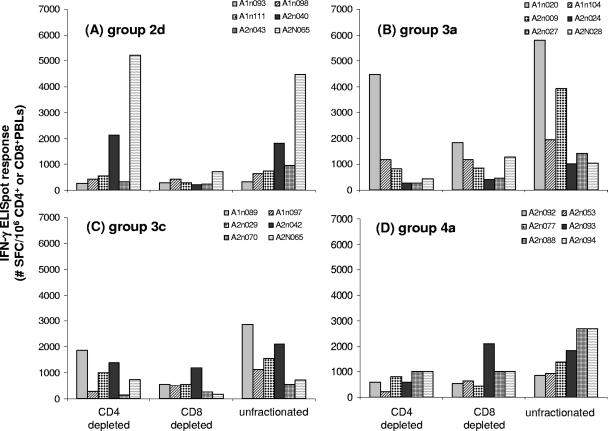
A detailed analysis of the HIV-1 antigen-specific IFN-γ ELISPOT responses at week 10 in PBLs depleted of either CD4+ or CD8+ cells revealed that the various pDNA vaccine designs tended to elicit both CD4+ and CD8+ cells capable of secreting IFN-γ (Fig. 2). In most cases, when the ELISPOT response was adjusted for the precise number of CD4+ or CD8+ cells in the assay, total HIV-1 antigen-specific responses were found to be distributed similarly among CD8+ and CD4+ lymphocytes, as the SFC activity in PBLs depleted of CD4+ cells was often equivalent to the response seen in PBLs depleted of CD8+ cells.
FIG. 2.
Total HIV-specific CMI responses in CD4+ and CD8+ cell-depleted PBLs. The graphs show total HIV peptide pool-specific IFN-γ ELISPOT responses at week 10 in unfractionated, CD4+ or CD8+ cell-depleted PBLs from macaques immunized with pDNA vaccines plus plasmid IL-12. Responses were normalized to the precise number of CD3+ CD4+ or CD3+ CD8+ cells used in the assay, as determined by flow cytometry after bead depletion.
Comparative analysis of serum antibody responses elicited by various pDNA vaccine designs.
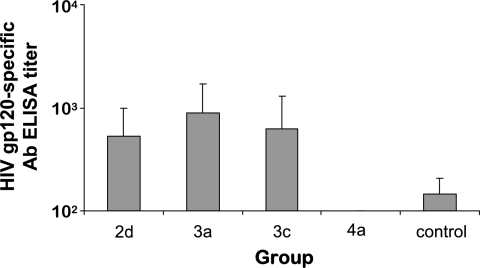
Immunized rhesus macaques were also monitored for the induction of serum antibody responses to HIV-16101 Env gp120 protein by ELISA (Fig. 3). At week 10, 2 weeks after the final pDNA immunization, serum anti-HIV Env antibody responses were similar in macaques immunized with the various two- and three-vector pDNA designs, but gp120-specific serum antibody responses in group 4a remained undetectable (Fig. 3).
FIG. 3.
Serum antibody responses to monomeric HIV-16101 Env gp120 in rhesus macaques after pDNA immunization. Two weeks after the final pDNA immunization, geometric mean HIV-16101 Env gp120-specific IgG antibody titers (± standard errors) were determined by ELISA. Antibody end-point titers are reported as the highest reciprocal dilutions of sera giving OD450 readings threefold higher than those for week 0 (preimmune) samples. HIV-16101 gp120-specific antibody titers for group 4a were below the limit of detection (i.e., <1:100).
The induction of anti-rhesus IL-12 serum antibodies was also monitored by ELISA. Significantly elevated serum anti-IL-12 IgG antibody responses relative to those in preimmune serum samples were routinely detected in only a single group 3a macaque (A1N020; anti-IL-12 titers of 450, 50, and 450 at weeks 6, 10, and 16, respectively), while sporadic serum anti-IL-12 IgG responses were detected in two other macaques, namely, group 3a macaque A2N024 (anti-IL-12 titer of 450 at week 16) and control immunized macaque A1N112 (anti-IL-12 titer of 450 at week 6) (data not shown). Importantly, these low-level anti-IL-12 serum antibodies were found to be nonneutralizing in that they were unable to block the ability of exogenous IL-12 protein to stimulate an IL-12-responsive cell line (data not shown).
Safety and tolerability of pDNA vaccination in combination with in vivo EP.
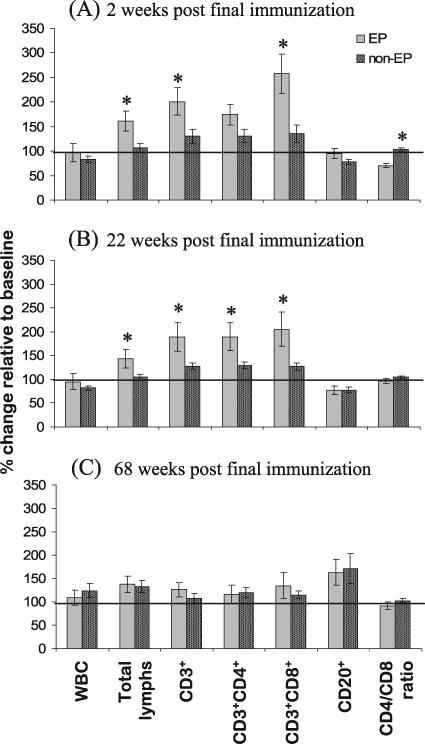
To monitor the relative safety of pDNA vaccination with in vivo EP, macaques were assessed by daily observations and by weekly physical examinations and body weight measurements. Over the course of the vaccination regimen, all macaques appeared normal, and they had stable or increased body weights during the study interval (data not shown). In addition, immunized macaques were closely monitored by CBC analysis to determine if pDNA immunization in combination with in vivo EP was associated with changes in peripheral blood cell subsets (Fig. 4). As shown in Fig. 4A, 2 weeks after the final pDNA immunization, macaques receiving the pDNA immunizations in combination with in vivo EP demonstrated substantial increases in peripheral blood total CD3+ and CD3+ CD8+ lymphocytes as well as a significantly decreased CD4/CD8 ratio (P < 0.05) relative to those macaques receiving pDNA immunization in the absence of in vivo EP. This increase in peripheral blood CD3+ and CD3+CD8+ lymphocytes seen in macaques receiving pDNA immunization in combination with in vivo EP persisted for at least 22 weeks after the final immunization (Fig. 4B) but resolved by 68 weeks after the final immunization (Fig. 4C).
FIG. 4.
Changes in CBC parameters associated with in vivo EP. At baseline (week −2) and 2, 22, and 68 weeks after the final immunization (weeks 10, 30, and 76), macaques were screened for changes in peripheral blood cell subsets by CBC analysis. Data are reported as percent changes at (A) week 10, (B) week 30, and (C) week 76 relative to baseline (± standard errors) for macaques receiving pDNA vaccination with in vivo EP (EP; n = 6) versus macaques receiving pDNA vaccination without in vivo EP (non-EP; n = 30). *, statistically significant difference (P < 0.05) between EP and non-EP groups. WBC, white blood cells.
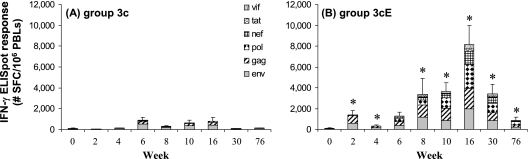
HIV-specific CMI responses elicited by pDNA immunization with and without in vivo EP.
The group 3c macaques, immunized with 8.5 mg total vaccine pDNA by standard needle and syringe, and the group 3cE macaques, immunized with one-fifth the total vaccine pDNA dose (1.7 mg) by standard needle and syringe followed by in vivo EP, were monitored for the induction of HIV-1 Env, Gag, Pol, Nef, Tat, and Vif peptide pool-specific CMI responses by IFN-γ ELISPOT assay (Fig. 5). The results from the IFN-γ ELISPOT assay show that compared to macaques administered the three-vector multiantigen pDNA delivered by standard needle and syringe (Fig. 5A), macaques immunized with one-fifth the amount of pDNA in combination with in vivo EP demonstrated a significant increase in total HIV-specific IFN-γ SFC activity (Fig. 5B). Peak total HIV-specific ELISPOT responses (mean, 893 IFN-γ SFC/106 PBLs; range, 308 to 2,073 IFN-γ SFC/106 PBLs) were seen in the group 3c macaques at week 6, 2 weeks after the first booster immunization, and these responses were not increased appreciably by the second booster immunization at week 8. In contrast, the second booster immunization at week 8 significantly elevated the HIV-specific ELISPOT responses in the group 3cE macaques, and this response (mean, 8,141 IFN-γ SFC/106 PBLs; range, 6,613 to 11,840 IFN-γ SFC/106 PBLs) peaked at week 16.
FIG. 5.
HIV-1 antigen-specific CMI responses following pDNA immunization, with and without in vivo EP. Macaques were immunized at 0, 4, and 8 weeks with the three-vector pDNA vaccine design 3c in combination with plasmid rhesus IL-12 by standard i.m. injection (A) or by i.m. injection followed by in vivo EP (B). At various time points, PBLs were collected and tested for HIV-1 Env-, Gag-, Pol-, Nef-, Tat-, and Vif-specific IFN-γ secretion by ELISPOT assay. Bars represent the average total HIV-specific IFN-γ ELISPOT responses (n = 6), with the standard errors of the means indicated. *, statistically significant difference (P < 0.05) between group 3c and group 3cE at the indicated time point.
Over the course of the study, none of the group 3c immunized macaques developed measurable Nef-, Tat- or Vif-specific IFN-γ ELISPOT responses. In contrast, a majority of the group 3cE macaques developed significant (i.e., >100 SFC/106 PBLs) IFN-γ ELISPOT responses to Nef (five of six), Tat (four of six), and Vif (four of six).
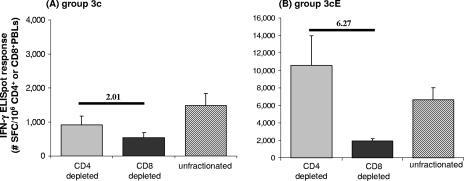
An analysis of the HIV antigen-specific IFN-γ ELISPOT responses at week 10 in PBLs depleted of either CD4+ or CD8+ cells revealed that the three-vector pDNA vaccine design delivered by standard needle and syringe tended to elicit both CD4+ and CD8+ HIV peptide pool-specific responses (Fig. 6A), and these antigen-specific responses were found to be similarly distributed among CD8+ and CD4+ lymphocytes, as SFC activity in PBLs depleted of CD4+ cells was indistinguishable from the response seen in PBLs depleted of CD8+ cells. Delivering the three-vector pDNA vaccine design in combination with in vivo EP increased both CD4+ and CD8+ HIV peptide pool-specific responses (Fig. 6B). However, these antigen-specific responses were found to be heavily biased towards CD8+ lymphocytes, as SFC activity in PBLs depleted of CD4+ cells was significantly elevated relative to the response seen in PBLs depleted of CD8+ cells.
FIG. 6.
Total HIV-specific CMI responses in CD4+ and CD8+ cell-depleted PBLs after pDNA immunization alone or in combination with in vivo EP. The graphs show average total HIV peptide pool-specific IFN-γ ELISPOT responses (± standard errors) at week 10 in unfractionated, CD4+ or CD8+ cell-depleted PBLs from macaques immunized with pDNA vaccines plus plasmid IL-12 alone (A) or followed by in vivo EP (B). Responses were normalized to the precise number of CD3+ CD4+ or CD3+ CD8+ cells used in the assay, as determined by flow cytometry after bead depletion. The ratio of the response seen in CD4+ cell-depleted PBLs to the response seen in CD8+ cell-depleted PBLs is indicated.
HIV-specific humoral immune responses elicited by pDNA immunization with and without in vivo EP.
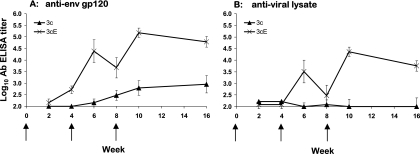
The group 3c macaques, immunized by standard needle and syringe, and the group 3cE macaques, immunized by standard needle and syringe followed by in vivo EP, were monitored for the induction of serum HIV-16101 Env gp120- and HIV-1IIIB viral lysate-specific humoral immune responses by standard ELISAs (Fig. 7). Macaques receiving the pDNA vaccine in combination with in vivo EP demonstrated a significant increase in total HIV-16101 Env gp120-specific serum IgG antibody titers relative to those macaques receiving a fivefold higher dose of pDNA in the absence of in vivo EP (Fig. 7A). Peak serum anti-gp120 IgG ELISA titers (geometric mean, 151,638) were seen in the group 3cE macaques at week 10, 2 weeks after the second booster immunization. In contrast, a >2-log10 reduction in peak serum anti-gp120 IgG ELISA titers was seen in the group 3c macaques, and these responses peaked at 1:900 (week 16) (Fig. 7A).
FIG. 7.
Serum HIV-16101 Env gp120- and HIV-1IIIB viral lysate-specific humoral immune responses in rhesus macaques after pDNA immunization, with and without in vivo EP. Geometric mean HIV-16101 Env gp120-specific IgG antibody titers (± standard errors) (A) and HIV-1IIIB viral lysate-specific IgG antibody titers (± standard errors) (B) were determined by ELISA. Antibody end-point titers are reported as the highest reciprocal dilution of sera giving OD450 readings threefold higher than those for week 0 (preimmune) samples. Arrows indicate the timing of pDNA immunization.
A similar trend was observed with regards to the induction of HIV-1IIIB viral lysate-specific serum IgG antibodies (Fig. 7B). All (six of six) group 3cE macaques had seroconverted by week 10, 2 weeks after the final pDNA immunization, with a geometric mean anti-viral lysate-specific antibody IgG titer of 1:22,712. In contrast, at week 10, none of the group 3c macaques demonstrated a measurable (>1:100) antiviral lysate-specific antibody titer (Fig. 7B).
All immunized macaques were screened for serum antibody responses capable of neutralizing the homologous primary isolate HIV-16101 in vitro. Two weeks after the final pDNA immunization, all macaques were negative for antibodies capable of neutralizing the primary isolate HIV-16101 (data not shown).
pDNA persistence and association with HMW DNA following immunization with and without in vivo EP.
The persistence of total vaccine pDNA at the injection site and the association of this pDNA with agarose gel-purified HMW genomic DNA were examined in all group 3c and 3cE macaques as well as a select number of control immunized macaques 70 days after the final pDNA immunization, using BGH- and HCMV-specific primer-probe pairs (Table 2). Using the BGH-specific primer/probe set, vaccine pDNA was found to persist within the injection site muscle tissues of all immunized macaques 70 days after the final injection. Group 3c and control macaques had, on average, 3,577 (range, 918 to 7,206) and 9,610 (range, 1,135 to 16,113) copies of vaccine pDNA per μg of injection site tissue. In contrast, group 3cE macaques, which received one-fifth the total pDNA vaccine dose in combination with in vivo EP, demonstrated a significantly lower level of pDNA persistence (947 [range, 217 to 1,601] copies/μg injection site tissue). A similar profile of pDNA persistence was found following the analysis with the HCMV-specific primer/probe set.
TABLE 2.
Persistence of pDNA in injection site DNA and association with HMW DNA
| Group | Macaque no. | BGH-specific primer/probe set
|
HCMV-specific primer/probe set
|
||
|---|---|---|---|---|---|
| pDNA persistencea (plasmid copies/μg injection site DNA) | pDNA association with genomic DNAb (plasmid copies/0.2 μg HMW DNA) | pDNA persistencea (plasmid copies/μg injection site DNA) | pDNA association with genomic DNAb (plasmid copies/0.2 μg HMW DNA) | ||
| 3c | A1N089 | 3,166 | <10 | 129 | <10 |
| A1N097 | 3,682 | <10 | 1,490 | <10 | |
| A2N029 | 918 | 35 | 125 | 34 | |
| A2N042 | 1,388 | <10 | 318 | 36 | |
| A2N060 | 5,101 | <10 | 892 | 23 | |
| A2N070 | 7,206 | <10 | 2,710 | <10 | |
| Mean ± SE | 3,577 ± 958 | 14 ± 4 | 944 ± 414 | 21 ± 5 | |
| 3cE | A1N078 | 507 | <10 | 258 | <10 |
| A1N086 | 217 | <10 | 118 | <10 | |
| A2N010 | 1,601 | <10 | 285 | 15 | |
| A2N014 | 1,043 | <10 | 869 | 13 | |
| A2N036 | 735 | <10 | 165 | <10 | |
| A2N054 | 1,576 | <10 | 89 | 48 | |
| Mean ± SE | 947** ± 231 | 10 | 297* ± 119 | 18 ± 6 | |
| Control | A1N118 | 11,581 | <10 | 5,455 | <10 |
| A2N037 | 1,135 | 13 | 580 | <10 | |
| A2N049 | 16,113 | <10 | 4,103 | 216 | |
| Mean ± SE | 9,610 ± 4,435 | 11 ± 1 | 3,379 ± 1,453 | 79 ± 69 | |
The level of vaccine pDNA associated with unpurified injection site tissue DNA was examined 70 days after the final immunization, using a BGH- or HCMV-specific primer-probe pair as described in Materials and Methods. *, significantly reduced (P < 0.05) relative to controls; **, significantly reduced (P < 0.05) relative to group 3c and controls.
The level of vaccine pDNA associated with agarose gel-purified HMW genomic DNA was examined 70 days after the final immunization, using a BGH- or HCMV-specific primer-probe pair as described in Materials and Methods.
Following agarose gel purification of HMW genomic DNA, a low level of vaccine pDNA was found in association with HMW genomic DNA in only 2 of 15 immunized macaques. Interestingly, the macaques found to harbor vaccine pDNA in association with HMW genomic DNA were not within the experimental group vaccinated in combination with in vivo EP.
DISCUSSION
In this study, a number of two-, three-, and four-vector pDNA vaccine designs carrying HIV env, gag, pol, nef, tat, and vif were tested for the ability to elicit HIV-1 antigen-specific CMI and humoral immune responses in rhesus macaques. The immunogenicities of the candidate vaccine designs were compared by using a fixed vaccine dose in an effort to mimic the clinical situation, where the total vaccine one can deliver is often limited by tolerability, formulation, or injection volume. The results of this study indicated that all of the two- and three-vector vaccine designs were capable of eliciting measurable total HIV-specific CMI responses. The greatest total HIV-specific CMI response was elicited by the three-vector vaccine design evaluated within group 3a, but the difference in total HIV-1 antigen-specific IFN-γ ELISPOT activities between the various two- and three-vector vaccine designs did not achieve statistical significance. Despite similar levels of total HIV-specific immune responses, individual antigen-specific CMI responses were found to vary widely and were most affected by the promoter used and whether the antigen was expressed alone or as part of a larger fusion protein. Overall, the results indicated that a two-vector pDNA vaccine design consisting of pDNAs WLV151 (HCMV gag-pol) and WLV159 (HCMV nef-tat-vif and SCMV env) appeared to elicit the most robust and balanced CMI responses to multiple HIV-1-derived antigens in rhesus macaques.
In an effort to identify a pDNA vaccine delivery technology that could effectively increase the efficiency of pDNA delivery in vivo, we sought to determine the extent to which in vivo EP could improve the induction of vaccine antigen-specific CMI and humoral immune responses. The results of this study also demonstrated that vaccination in combination with in vivo EP led to faster-onset and stronger cellular responses measured by the IFN-γ ELISPOT assay. For example, 8 and 22 weeks after the final pDNA immunization, there were 10- and 40-fold increases in HIV-specific ELISPOT responses in the group 3cE macaques compared to the group 3c macaques receiving the same pDNA vaccine without in vivo EP. Given the fact that the macaques immunized in combination with in vivo EP received one-fifth the total pDNA dose, this translates into an apparent 50- or 200-fold increase in pDNA vaccine potency, respectively. Importantly, in vivo EP was especially efficient at enhancing the CMI response against the less immunogenic antigens (Nef-Tat-Vif), resulting in a more balanced immune response overall.
Recently, Lichterfeld et al. conducted a longitudinal analysis of HIV-1-specific IFN-γ ELISPOT responses directed against the entire expressed HIV-1 clade B consensus sequence in patients with primary and chronic HIV-1 infections (31). The systematic analysis of CMI responses to Env, Gag, Pol, and Nef in these HIV-infected individuals provides a benchmark with which to compare the responses seen in the current study. Lichterfeld et al. reported that acutely infected individuals generally displayed weak IFN-γ ELISPOT responses (∼250 SFC/106 peripheral blood mononuclear cells [PBMC]) and that these responses were predominantly targeted to Nef. Individuals with early HIV infection had cumulative Env-, Gag-, Pol-, and Nef-specific IFN-γ ELISPOT responses of ∼400 SFC/106 PBMC. In contrast, patients with chronic HIV-1 infection displayed cumulative Env-, Gag-, Pol-, and Nef-specific IFN-γ ELISPOT responses of ∼7,500 SFC/106 PBMC. The data from the current study indicate that the level and breadth of the IFN-γ ELISPOT responses in macaques immunized with the candidate three-vector pDNA vaccine in combination with plasmid IL-12 and in vivo EP are virtually identical to those of the IFN-γ ELISPOT responses seen in chronically infected individuals.
In the current study, in vivo EP also resulted in a dramatic 2- to 3-log increase in serum antibodies capable of binding HIV-16101 gp120. Unfortunately, we failed to detect serum antibodies with functional activity capable of neutralizing the homologous primary isolate. The inability of the current pDNA vaccine strategy to elicit serum antibodies with functional neutralizing activity was most likely due to issues related to Env immunogen design. The pDNA expression vectors tested in the current study all encoded the same wild-type clade B primary isolate (HIV-16101) gp160, which was not modified in any way to improve the induction of neutralizing antibodies.
Previously, we demonstrated with a rhesus macaque model that the codelivery of plasmid-based IL-12 with a simian immunodeficiency virus (SIV) Gag-expressing plasmid resulted in a significant improvement in Gag-specific CMI and humoral immune responses relative to macaques receiving the SIV gag pDNA alone (53). In the current study, macaques immunized with the three-vector multiantigen pDNA, with and without in vivo EP, all received plasmid IL-12 as an adjuvant. Based on previously published work, it is reasonable to assume that in vivo EP increased the transfection efficiency of the IL-12-encoding plasmid, resulting in a 10- to 100-fold increase in IL-12 production within the injection site tissue. However, given the current study design, it is unclear to what extent the increased expression of the biologically active IL-12 was responsible for the increased antigen-specific immunity seen following in vivo EP. Previously, we showed for rhesus macaques that SIV Gag-specific CMI and humoral immune responses were increased by the codelivery of a 1.5-mg dose of plasmid IL-12, but these responses were not further increased by a threefold higher dose (5.0 mg) of plasmid IL-12 (53). These data suggest that in the current experiment, the increased IL-12 expression as a result of in vivo EP may not be the principal factor driving the increased antigen-specific immune response. As a result, it is possible that the increase in pDNA vaccine delivery and subsequent antigen expression, the increased transfection of mononuclear cells, and an increased inflammatory response realized by in vivo EP will obviate the need for the codelivery of plasmid-based cytokines.
A number of studies performed in larger animal models (4, 55, 70, 71) and nonhuman primates (30, 41, 44, 83) have demonstrated that in vivo EP can markedly augment vaccine antigen-specific humoral immune and CMI responses. However, many of these studies used in vivo EP conditions where the applied voltage was 150 to 250 V/cm. Furthermore, electrical pulses were delivered to the tissue by making use of multiple electrode arrays consisting of as many as four to six needles. For vaccine delivery into humans, especially in a prophylactic setting, the tolerability of in vivo EP will undoubtedly be an important issue. Here we demonstrate for nonhuman primates that a dramatic increase in pDNA vaccine immunogenicity can be achieved with in vivo EP performed using a minimalistic electrode configuration of a two-needle array and mild electrical conditions (voltages of <70 V/cm). Similar conditions were found to be tolerable in unanesthetized human volunteers (22) and are currently being deployed in the first human clinical trial using EP for repeated delivery of a pDNA vaccine in cancer patients.
Consistent with previously published work (29, 37, 76), this study shows that a significant amount of extrachromosomal vaccine pDNA remains within muscle tissue 70 days after the final immunization. Interestingly, macaques immunized in combination with in vivo EP, despite demonstrating a profound increase in vaccine-specific immune responses, suggesting improved pDNA delivery, demonstrated a significant reduction in pDNA persistence relative to macaques immunized with a fivefold higher dose of pDNA vaccine in the absence of in vivo EP.
Despite these high levels of extrachromosomal vaccine pDNA, only a subset of immunized macaques were found to harbor pDNA (13 to 216 copies/μg HMW DNA) associated with genomic DNA after four rounds of agarose gel purification. Using the Q-PCR methods reported here, the association of the pDNA with the HMW genomic DNA fraction is consistent with, but does not prove, integration of pDNA within genomic DNA. These results are consistent with previously published work (29, 37, 63) suggesting that the frequency of integration following i.m. injection is negligible. Nevertheless, the low level of pDNA association with HMW genomic DNA seen in the current nonhuman primate study is estimated to be at least 3 orders of magnitude below the frequency of spontaneous gene-inactivating mutations (29).
It was previously shown for mice that immunization with a pDNA encoding a self antigen in combination with in vivo EP led to higher levels of pDNA persistence at the injection site and increased pDNA integration into chromosomal DNA compared to mice immunized with an equal dose of pDNA in the absence of in vivo EP (76). Despite this increase in pDNA association with chromosomal DNA, the frequency of integration was at least 3 orders of magnitude below the spontaneous rate of gene-inactivating mutations (76). Nevertheless, these results have led to concerns regarding the safety of in vivo EP for pDNA vaccine delivery.
In the current study, we found that in contrast to the results generated in the mouse model, in vivo EP was associated with a significant reduction in pDNA persistence at the injection site, and perhaps more importantly, in vivo EP did not result in an increase in pDNA associated with HMW genomic DNA relative to macaques receiving the pDNA without in vivo EP. The reason for these differing results is unclear, but the difference is likely due to several factors. In the current study, the significant reduction in pDNA vaccine persistence at the injection site could potentially be explained by the fact that a fivefold lower pDNA vaccine dose was administered to the group receiving the vaccine in combination with in vivo EP. We find this explanation unlikely, as in vivo EP most likely increased cellular uptake of the pDNA into myocytes at least 10-fold, as demonstrated in numerous small animal studies (1, 39, 48, 79). A more plausible explanation for the observed reduction in pDNA vaccine persistence is that macaques immunized in combination with in vivo EP generated increased vaccine-specific cellular immune responses, resulting in a more efficient clearance of myocytes expressing vaccine-encoded antigens. This interpretation is consistent with the observation that macaques immunized with control plasmid did not mount a cellular immune response and showed an increased level of pDNA persistence.
These results indicate that in vivo EP appears to be safe and can dramatically improve the delivery and immunogenicity of a multivector, multiantigen pDNA vaccine in nonhuman primates. Collectively, these data have important implications for the design and development of an efficacious therapeutic vaccine for the treatment of HIV-1 infection.
Acknowledgments
We thank Jane Fontenot and the veterinary staff from the New Iberia Research Center (New Iberia, LA) for their expert assistance with animal care and handling and Wang-Ting Hsieh from BioReliance for expert assistance with the Q-PCR assays.
This work was supported by HIV-1 Vaccine Design and Development Team contracts NIH/NIAID HVDDT NO1-AI-25458 and NO1-AI-50010 from the National Institutes of Health.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Aihara, H., and J. Miyazaki. 1998. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 16:867-870. [DOI] [PubMed] [Google Scholar]
- 2.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853-864. [DOI] [PubMed] [Google Scholar]
- 3.Babiuk, L. A., S. van Drunen Littel-van den Hurk, and S. L. Babiuk. 1999. Immunization of animals: from DNA to the dinner plate. Vet. Immunol. Immunopathol. 72:189-202. [DOI] [PubMed] [Google Scholar]
- 4.Babiuk, S., M. E. Baca-Estrada, M. Foldvari, M. Storms, D. Rabussay, G. Widera, and L. A. Babiuk. 2002. Electroporation improves the efficacy of DNA vaccines in large animals. Vaccine 20:3399-3408. [DOI] [PubMed] [Google Scholar]
- 5.Bachy, M., F. Boudet, M. Bureau, Y. Girerd-Chambaz, P. Wils, D. Scherman, and C. Meric. 2001. Electric pulses increase the immunogenicity of an influenza DNA vaccine injected intramuscularly in the mouse. Vaccine 19:1688-1693. [DOI] [PubMed] [Google Scholar]
- 6.Barouch, D. H. 2006. Rational design of gene-based vaccines. J. Pathol. 208:283-289. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, J. D., M. A. Chattergoon, K. E. Ugen, A. Shah, M. Bennett, A. Cohen, S. Nyland, K. E. Lacy, M. L. Bagarazzi, T. J. Higgins, Y. Baine, R. B. Ciccarelli, R. S. Ginsberg, R. R. MacGregor, and D. B. Weiner. 1999. Enhancement of cellular immune response in HIV-1 seropositive individuals: a DNA-based trial. Clin. Immunol. 90:100-107. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, J. D., A. D. Cohen, S. Vogt, K. Schumann, B. Nath, L. Ahn, K. Lacy, M. L. Bagarazzi, T. J. Higgins, Y. Baine, R. B. Ciccarelli, R. S. Ginsberg, R. R. MacGregor, and D. B. Weiner. 2000. Vaccination of seronegative volunteers with a human immunodeficiency virus type 1 env/rev DNA vaccine induces antigen-specific proliferation and lymphocyte production of beta-chemokines. J. Infect. Dis. 181:476-483. [DOI] [PubMed] [Google Scholar]
- 10.Buchan, S., E. Gronevik, I. Mathiesen, C. A. King, F. K. Stevenson, and J. Rice. 2005. Electroporation as a “prime/boost” strategy for naked DNA vaccination against a tumor antigen. J. Immunol. 174:6292-6298. [DOI] [PubMed] [Google Scholar]
- 11.Calarota, S., G. Bratt, S. Nordlund, J. Hinkula, A. C. Leandersson, E. Sandstrom, and B. Wahren. 1998. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet 351:1320-1325. [DOI] [PubMed] [Google Scholar]
- 12.Calarota, S. A., A. C. Leandersson, G. Bratt, J. Hinkula, D. M. Klinman, K. J. Weinhold, E. Sandstrom, and B. Wahren. 1999. Immune responses in asymptomatic HIV-1-infected patients after HIV-DNA immunization followed by highly active antiretroviral treatment. J. Immunol. 163:2330-2338. [PubMed] [Google Scholar]
- 13.Conry, R. M., D. T. Curiel, T. V. Strong, S. E. Moore, K. O. Allen, D. L. Barlow, D. R. Shaw, and A. F. LoBuglio. 2002. Safety and immunogenicity of a DNA vaccine encoding carcinoembryonic antigen and hepatitis B surface antigen in colorectal carcinoma patients. Clin. Cancer Res. 8:2782-2787. [PubMed] [Google Scholar]
- 14.Davies, J. F. II, Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 15.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 16.Egan, M. A., W. A. Charini, M. J. Kuroda, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan, M. A., S.-Y. Chong, S. Megati, D. C. Montefiori, N. F. Rose, M. Sidhu, J. Quiroz, E. B. Schadeck, G. Pavlakis, D. B. Weiner, J. K. Rose, Z. R. Israel, S. A. Udem, and J. H. Eldridge. 2005. Priming with plasmid DNAs expressing IL-12 and SIV Gag protein enhances the immunogenicity and efficacy of an experimental AIDS vaccine based on recombinant vesicular stomatitis virus. AIDS Res. Hum. Retrovir. 21:629-643. [DOI] [PubMed] [Google Scholar]
- 18.Egan, M. A., S. Megati, V. Roopchand, D. Garcia-Hand, A. Luckay, S.-Y. Chong, M. Rosati, S. Sackitey, D. B. Weiner, and B. K. Felber. 2006. Rational design of a plasmid DNA vaccine capable of eliciting cell-mediated immune responses to multiple HIV antigens in mice. Vaccine 24:4510-4523. [DOI] [PubMed] [Google Scholar]
- 19.Epstein, J. E., Y. Charoenvit, K. E. Kester, R. Wang, R. Newcomer, S. Fitzpatrick, T. L. Richie, N. Tornieporth, D. G. Heppner, C. Ockenhouse, V. Majam, C. Holland, E. Abot, H. Ganeshan, M. Berzins, T. Jones, C. N. Freydberg, J. Ng, J. Norman, D. J. Carucci, J. Cohen, and S. L. Hoffman. 2004. Safety, tolerability, and antibody responses in humans after sequential immunization with a PfCSP DNA vaccine followed by the recombinant protein vaccine RTS,S/AS02A. Vaccine 22:1592-1603. [DOI] [PubMed] [Google Scholar]
- 20.Gehl, J., and L. M. Mir. 1999. Determination of optimal parameters for in vivo gene transfer by electroporation, using a rapid in vivo test for cell permeabilization. Biochem. Biophys. Res. Commun. 261:377-380. [DOI] [PubMed] [Google Scholar]
- 21.Gronevik, E., S. Tollefsen, L. I. Sikkeland, T. Haug, T. E. Tjelle, and I. Mathiesen. 2003. DNA transfection of mononuclear cells in muscle tissue. J. Gene Med. 5:909-917. [DOI] [PubMed] [Google Scholar]
- 22.Kjeken, R., T. Tjelle, D. Kvale, and I. Mathiesen. 2004. Clinical evaluation of pain and muscle damage induced by electroporation of skeletal muscle in humans. Mol. Ther. 9:S60 [Google Scholar]
- 23.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 24.Koup, R., J. Safrit, Y. Cao, C. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larder, B. A., S. D. Kemp, and D. J. M. Purifoy. 1989. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc. Natl. Acad. Sci. USA 86:4803-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larder, B. A., D. J. Purifoy, K. L. Powell, and G. Darby. 1987. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature 327:716-717. [DOI] [PubMed] [Google Scholar]
- 27.Le, T. P., K. M. Coonan, R. C. Hedstrom, Y. Charoenvit, M. Sedegah, J. E. Epstein, S. Kumar, R. Wang, D. L. Doolan, J. D. Maguire, S. E. Parker, P. Hobart, J. Norman, and S. L. Hoffman. 2000. Safety, tolerability and humoral immune responses after intramuscular administration of a malaria DNA vaccine to healthy adult volunteers. Vaccine 18:1893-1901. [DOI] [PubMed] [Google Scholar]
- 28.Leavitt, A., L. Shiue, and H. Varmus. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 268:2113-2119. [PubMed] [Google Scholar]
- 29.Ledwith, B. J., S. Manam, P. J. Troilo, A. B. Barnum, C. J. Pauley, T. G. Griffiths II, L. B. Harper, C. M. Beare, W. J. Bagdon, and W. W. Nichols. 2000. Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 43:258-272. [DOI] [PubMed] [Google Scholar]
- 30.Li, Z., H. Zhang, X. Fan, Y. Zhang, J. Huang, Q. Liu, T. E. Tjelle, I. Mathiesen, R. Kjeken, and S. Xiong. 2006. DNA electroporation prime and protein boost strategy enhances humoral immunity of tuberculosis DNA vaccines in mice and non-human primates. Vaccine 24:4565-4568. [DOI] [PubMed] [Google Scholar]
- 31.Lichterfeld, M., X. G. Yu, D. Cohen, M. M. Addo, J. Malenfant, B. Perkins, E. Pae, M. N. Johnston, D. Strick, T. M. Allen, E. S. Rosenberg, B. Korber, B. D. Walker, and M. Altfeld. 2004. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS 18:1383-1392. [DOI] [PubMed] [Google Scholar]
- 32.Loeb, D. D., R. Swanstrom, L. Everitt, M. Manchester, S. E. Stamper, and C. A. Hutchison III. 1989. Complete mutagenesis of the HIV-1 protease. Nature 340:397-400. [DOI] [PubMed] [Google Scholar]
- 33.Los Alamos National Laboratory. 2003. HIV immunology and HIV/SIV vaccine databases. LA-UR 04-8162. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM.
- 34.MacGregor, R. R., J. D. Boyer, R. B. Ciccarelli, R. S. Ginsberg, and D. B. Weiner. 2000. Safety and immune responses to a DNA-based human immunodeficiency virus (HIV) type I env/rev vaccine in HIV-infected recipients: follow-up data. J. Infect. Dis. 181:406. [DOI] [PubMed] [Google Scholar]
- 35.MacGregor, R. R., J. D. Boyer, K. E. Ugen, K. E. Lacy, S. J. Gluckman, M. L. Bagarazzi, M. A. Chattergoon, Y. Baine, T. J. Higgins, R. B. Ciccarelli, L. R. Coney, R. S. Ginsberg, and D. B. Weiner. 1998. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J. Infect. Dis. 178:92-100. [DOI] [PubMed] [Google Scholar]
- 36.Mancini-Bourgine, M., H. Fontaine, D. Scott-Algara, S. Pol, C. Brechot, and M. L. Michel. 2004. Induction or expansion of T-cell responses by a hepatitis B DNA vaccine administered to chronic HBV carriers. Hepatology 40:874-882. [DOI] [PubMed] [Google Scholar]
- 37.Martin, T., S. E. Parker, R. Hedstrom, T. Le, S. L. Hoffman, J. Norman, P. Hobart, and D. Lew. 1999. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum. Gene Ther. 10:759-768. [DOI] [PubMed] [Google Scholar]
- 38.Mascola, J. R., P. D'Souza, P. Gilbert, B. H. Hahn, N. L. Haigwood, L. Morris, C. J. Petropoulos, V. R. Polonis, M. Sarzotti, and D. C. Montefiori. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathiesen, I. 1999. Electropermeabilization of skeletal muscle enhances gene transfer in vivo. Gene Ther. 6:508-514. [DOI] [PubMed] [Google Scholar]
- 40.Miller, A. M., V. Ozenci, R. Kiessling, and P. Pisa. 2005. Immune monitoring in a phase 1 trial of a PSA DNA vaccine in patients with hormone-refractory prostate cancer. J. Immunother. 28:389-395. [DOI] [PubMed] [Google Scholar]
- 41.Mir, L. M., M. F. Bureau, J. Gehl, R. Rangara, D. Rouy, J.-M. Caillaud, P. Delaere, D. Branellec, B. Schwartz, and D. Scherman. 1999. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc. Natl. Acad. Sci. USA 96:4262-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montefiori, D. C. 2004. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays, p. 12.11.1-12.11.15. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, New York, NY. [DOI] [PubMed]
- 43.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academic Press, Washington, DC.
- 44.Otten, G., M. Schaefer, B. Doe, H. Liu, I. Srivastava, J. Z. Megede, D. O'Hagan, J. Donnelly, G. Widera, and D. Rabussay. 2004. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine 22:2489-2493. [DOI] [PubMed] [Google Scholar]
- 45.Pachuk, C. J., R. B. Ciccarelli, M. Samuel, M. E. Bayer, R. D. Troutman, D. V. Zurawski, J. I. Schauer, T. J. Higgins, D. B. Weiner, D. M. Sosnoski, V. R. Zurawski, and C. Satishchandran. 2000. Characterization of a new class of DNA delivery complexes formed by the local anesthetic bupivacaine. Biochim. Biophys. Acta 1468:20-30. [DOI] [PubMed] [Google Scholar]
- 46.Prud'homme, G. J., Y. Glinka, A. S. Khan, and R. Draghia-Akli. 2006. Electroporation-enhanced nonviral gene transfer for the prevention or treatment of immunological, endocrine and neoplastic diseases. Curr. Gene Ther. 6:243-273. [DOI] [PubMed] [Google Scholar]
- 47.Quaglino, E., M. Iezzi, C. Mastini, A. Amici, F. Pericle, E. Di Carlo, S. M. Pupa, C. De Giovanni, M. Spadaro, C. Curcio, P. L. Lollini, P. Musiani, G. Forni, and F. Cavallo. 2004. Electroporated DNA vaccine clears away multifocal mammary carcinomas in Her-2/neu transgenic mice. Cancer Res. 64:2858-2864. [DOI] [PubMed] [Google Scholar]
- 48.Rizzuto, G., M. Cappelletti, D. Maione, R. Savino, D. Lazzaro, P. Costa, I. Mathiesen, R. Cortese, G. Ciliberto, R. Laufer, N. La Monica, and E. Fattori. 1999. Efficient and regulated erythropoietin production by naked DNA injection and muscle electroporation. Proc. Natl. Acad. Sci. USA 96:6417-6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rottinghaus, S. T., G. A. Poland, R. M. Jacobson, L. J. Barr, and M. J. Roy. 2003. Hepatitis B DNA vaccine induces protective antibody responses in human non-responders to conventional vaccination. Vaccine 21:4604-4608. [DOI] [PubMed] [Google Scholar]
- 50.Roy, M. J., M. S. Wu, L. J. Barr, J. T. Fuller, L. G. Tussey, S. Speller, J. Culp, J. K. Burkholder, W. F. Swain, R. M. Dixon, G. Widera, R. Vessey, A. King, G. Ogg, A. Gallimore, J. R. Haynes, and D. Heydenburg Fuller. 2000. Induction of antigen-specific CD8+ T cells, T helper cells, and protective levels of antibody in humans by particle-mediated administration of a hepatitis B virus DNA vaccine. Vaccine 19:764-778. [DOI] [PubMed] [Google Scholar]
- 51.Sadaie, M. R., J. Rappaport, T. Benter, S. F. Josephs, R. Willis, and F. Wong-Staal. 1988. Missense mutations in an infectious human immunodeficiency viral genome: functional mapping of tat and identification of the rev splice acceptor. Proc. Natl. Acad. Sci. USA 85:9224-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadaie, Y., K. C. Burtis, and R. H. Doi. 1980. Purification and characterization of a kanamycin nucleotidyltransferase from plasmid pUB110-carrying cells of Bacillus subtilis. J. Bacteriol. 141:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schadeck, E. B., M. Sidhu, M. A. Egan, S.-Y. Chong, P. Piacente, A. Masood, D. Garcia-Hand, S. Cappello, V. Roopchand, and S. Megati. 2006. A dose sparing effect by plasmid encoded IL-12 adjuvant on a SIVgag-plasmid DNA vaccine in rhesus macaques. Vaccine 24:4677-4687. [DOI] [PubMed] [Google Scholar]
- 54.Schatz, O., F. V. Cromme, F. Gruninger-Leitch, and S. F. J. Le Grice. 1989. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 257:311-314. [DOI] [PubMed] [Google Scholar]
- 55.Scheerlinck, J.-P. Y., J. Karlis, T. E. Tjelle, P. J. A. Presidente, I. Mathiesen, and S. E. Newton. 2004. In vivo electroporation improves immune responses to DNA vaccination in sheep. Vaccine 22:1820-1825. [DOI] [PubMed] [Google Scholar]
- 56.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 57.Schneider, R., M. Campbell, G. Nasioulas, B. Felber, and G. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, S., M. Campbell, G. Nasioulas, J. Harrison, B. K. Felber, and G. N. Pavlakis. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 66:7176-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 66:150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sedegah, M., R. Hedstrom, P. Hobart, and S. L. Hoffman. 1994. Protection against malaria by immunization with plasmid DNA encoding circumsporozoite protein. Proc. Natl. Acad. Sci. USA 91:9866-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selby, M., C. Goldbeck, T. Pertile, R. Walsh, and J. Ulmer. 2000. Enhancement of DNA vaccine potency by electroporation in vivo. J. Biotechnol. 83:147-152. [DOI] [PubMed] [Google Scholar]
- 62.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheets, R. L., J. Stein, T. S. Manetz, C. Duffy, M. Nason, C. Andrews, W. P. Kong, G. J. Nabel, and P. L. Gomez. 2006. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, severe acute respiratory syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol. Sci. 91:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smorlesi, A., F. Papalini, A. Amici, F. Orlando, S. Pierpaoli, C. Mancini, and M. Provinciali. 2006. Evaluation of different plasmid DNA delivery systems for immunization against HER2/neu in a transgenic murine model of mammary carcinoma. Vaccine 24:1766-1775. [DOI] [PubMed] [Google Scholar]
- 65.Swain, W. E., D. Heydenburg Fuller, M. S. Wu, L. J. Barr, J. T. Fuller, J. Culp, J. Burkholder, R. M. Dixon, G. Widera, R. Vessey, and M. J. Roy. 2000. Tolerability and immune responses in humans to a PowderJect DNA vaccine for hepatitis B. Dev. Biol. 104:115-119. [PubMed] [Google Scholar]
- 66.Tacket, C. O., M. J. Roy, G. Widera, W. F. Swain, S. Broome, and R. Edelman. 1999. Phase 1 safety and immune response studies of a DNA vaccine encoding hepatitis B surface antigen delivered by a gene delivery device. Vaccine 17:2826-2829. [DOI] [PubMed] [Google Scholar]
- 67.Tagawa, S. T., P. Lee, J. Snively, W. Boswell, S. Ounpraseuth, S. Lee, B. Hickingbottom, J. Smith, D. Johnson, and J. S. Weber. 2003. Phase I study of intranodal delivery of a plasmid DNA vaccine for patients with stage IV melanoma. Cancer 98:144-154. [DOI] [PubMed] [Google Scholar]
- 68.Tang, D. C., M. De Vit, and S. A. Johnston. 1992. Genetic immunization is a simple method for eliciting an immune response. Nature 356:152-154. [DOI] [PubMed] [Google Scholar]
- 69.Timmerman, J. M., G. Singh, G. Hermanson, P. Hobart, D. K. Czerwinski, B. Taidi, R. Rajapaksa, C. B. Caspar, A. Van Beckhoven, and R. Levy. 2002. Immunogenicity of a plasmid DNA vaccine encoding chimeric idiotype in patients with B-cell lymphoma. Cancer Res. 62:5845-5852. [PubMed] [Google Scholar]
- 70.Tjelle, T. E., R. Salte, I. Mathiesen, and R. Kjeken. 2006. A novel electroporation device for gene delivery in large animals and humans. Vaccine 24:4667-4670. [DOI] [PubMed] [Google Scholar]
- 71.Tollefsen, S., M. Vordermeier, I. Olsen, A. K. Storset, L. J. Reitan, D. Clifford, D. B. Lowrie, H. G. Wiker, K. Huygen, G. Hewinson, I. Mathiesen, and T. E. Tjelle. 2003. DNA injection in combination with electroporation: a novel method for vaccination of farmed ruminants. Scand. J. Immunol. 57:229-238. [DOI] [PubMed] [Google Scholar]
- 72.Triozzi, P. L., W. Aldrich, K. O. Allen, R. R. Carlisle, A. F. LoBuglio, and R. M. Conry. 2005. Phase I study of a plasmid DNA vaccine encoding MART-1 in patients with resected melanoma at risk for relapse. J. Immunother. 28:382-388. [DOI] [PubMed] [Google Scholar]
- 73.Ulmer, J. B., J. J. Donnelly, S. E. Parker, G. H. Rhodes, P. L. Felgner, V. J. Dwarki, S. H. Gromkowski, R. R. Deck, C. M. De Witt, A. Friedman, et al. 1993. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259:1745-1749. [DOI] [PubMed] [Google Scholar]
- 74.Wang, R., D. L. Doolan, T. P. Le, R. C. Hedstrom, K. M. Coonan, Y. Charoenvit, T. R. Jones, P. Hobart, M. Margalith, J. Ng, W. R. Weiss, M. Sedegah, C. de Taisne, J. A. Norman, and S. L. Hoffman. 1998. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science 282:476-480. [DOI] [PubMed] [Google Scholar]
- 75.Wang, R., J. Epstein, F. M. Baraceros, E. J. Gorak, Y. Charoenvit, D. J. Carucci, R. C. Hedstrom, N. Rahardjo, T. Gay, P. Hobart, R. Stout, T. R. Jones, T. L. Richie, S. E. Parker, D. L. Doolan, J. Norman, and S. L. Hoffman. 2001. Induction of CD4(+) T cell-dependent CD8(+) type 1 responses in humans by a malaria DNA vaccine. Proc. Natl. Acad. Sci. USA 98:10817-10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, Z., P. J. Troilo, X. Wang, T. G. Griffiths, S. J. Pacchione, A. B. Barnum, L. B. Harper, C. J. Pauley, Z. Niu, L. Denisova, T. T. Follmer, G. Rizzuto, G. Ciliberto, E. Fattori, N. L. Monica, S. Manam, and B. J. Ledwith. 2004. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 11:711-721. [DOI] [PubMed] [Google Scholar]
- 77.Weber, R., W. Bossart, R. Cone, R. Luethy, and K. Moelling. 2001. Phase I clinical trial with HIV-1 gp160 plasmid vaccine in HIV-1-infected asymptomatic subjects. Eur. J. Clin. Microbiol. Infect. Dis. 20:800-803. [DOI] [PubMed] [Google Scholar]
- 78.WHO. 2006. Overview of the global AIDS epidemic, 2006 report on the global AIDS epidemic. WHO, Geneva, Switzerland.
- 79.Widera, G., M. Austin, D. Rabussay, C. Goldbeck, S. W. Barnett, M. Chen, L. Leung, G. R. Otten, K. Thudium, et al. 2000. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J. Immunol. 164:4635-4640. [DOI] [PubMed] [Google Scholar]
- 80.Wiskerchen, M., and M. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolff, J. A., R. W. Malone, P. Williams, W. Chong, G. Acsadi, A. Jani, and P. L. Felgner. 1990. Direct gene transfer into mouse muscle in vivo. Science 247:1465-1468. [DOI] [PubMed] [Google Scholar]
- 82.Wu, X., H. Liu, H. Xiao, J. Conway, and J. Kappes. 1996. Inhibition of human and simian immunodeficiency virus protease function by targeting Vpx-protease-mutant fusion protein into viral particles. J. Virol. 70:3378-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao, Y.-G., B. Peng, H. Deng, G. Chen, F. Yang, M. Shao, H. Lu, Y. Li, J. Peng, L. Xu, and Y. Xu. 2006. Anti-HBV immune responses in rhesus macaques elicited by electroporation mediated DNA vaccination. Vaccine 24:897-903. [DOI] [PubMed] [Google Scholar]
- 84.Zlotocha, S., M. J. Staab, D. Horvath, J. Straus, J. Dobratz, K. Oliver, S. Wasielewski, D. Alberti, G. Liu, G. Wilding, J. Eickhoff, and D. G. McNeel. 2005. A phase I study of a DNA vaccine targeting prostatic acid phosphatase in patients with stage D0 prostate cancer. Clin. Genitourin. Cancer 4:215-218. [DOI] [PubMed] [Google Scholar]
- 85.Zucchelli, S., S. Capone, E. Fattori, A. Folgori, A. Di Marco, D. Casimiro, A. J. Simon, R. Laufer, N. La Monica, R. Cortese, and A. Nicosia. 2000. Enhancing B- and T-cell immune response to a hepatitis C virus E2 DNA vaccine by intramuscular electrical gene transfer. J. Virol. 74:11598-11607. [DOI] [PMC free article] [PubMed] [Google Scholar]