Abstract
Menopause and the associated declines in ovarian function are major health issues for women. Despite the widespread health impact of this process, the molecular mechanisms underlying the aging-specific decline in ovarian function are almost completely unknown. To provide the first gene–protein analysis of the ovarian transition to menopause, we have established and contrasted RNA gene expression profiles and protein localization and content patterns in healthy young and perimenopausal mouse ovaries. We report a clear distinction in specific mRNA and protein levels that are noted prior to molecular evidence of steroidogenic failure. In this model, ovarian reproductive aging displays similarities with chronic inflammation and increased sensitivity to environmental cues. Overall, our results indicate the presence of mouse climacteric genes that are likely to be major players in aging-dependent changes in ovarian function.
INTRODUCTION
The sharp decline in ovarian supply of germ cells and dramatic change in endocrine function is a midlife female aging process leading to menopause. During the transitional or perimenopausal phase, the ovary undergoes accelerated depletion of oocytes coinciding with decreases in fertility and diminished steroidogenic capacity (1). It is not known whether normally occurring ovarian aging is entirely regulated by the loss of germ cells and accompanying steroid-producing follicular cells, or whether the ovary shares generalized or tissue-specific aging pathways observed in other models and tissues (2,3). If this is the case, then identifying the major aging pathways that affect the ovary may have important implications for women during the menopausal transition that go beyond the loss of estrogen. Drawing upon aging research in other tissues and model systems (2,3), ovarian changes at the level of RNA and protein are likely to mark the transition to menopause and thereby provide insight into ovary-dependent health and disease.
To provide the first global assessment of aging pathways in the ovary, we aimed to define general and tissue-specific profiles of ovarian gene expression levels and protein content of ovaries from young (6-week-old) and perimenopausal (8-month-old) C57BL/6 mice. To maximize our ability to identify aging-dependent pathways, we screened for changes in gene expression using a global mouse array and combined the results with an aging-specific mouse DNA array (4) and northern blot assays. This analysis was followed by a cross-examination for changes in protein content and cellular protein location of differentially expressed genes by immunochemistry. We identified genes for which both mRNA and protein accumulate differentially with reproductive age. Our findings argue for the presence of mouse 'climacteric genes' that are likely to be major players in the changes in ovarian function with reproductive aging.
MATERIALS AND METHODS
Animals
The animal protocol used in this study was approved by the National Cancer Institute Animal Safety and Use Committee. Young animals were virgin reproductively mature and cycling 6-week-old female C57BL/6 mice. Climacteric (perimenopaual) animals were cycling and hormonally responsive 8-month-old C57BL/6 female mice that were significantly less active than the young animals and had been retired recently from breeding (5). Young and old mice were each divided into four groups (A, B, C and D) of 6–12 mice each (Figure 1). Identical experiments were done simultaneously in young and old mice. Anestrus was induced by group-housing mice for 1 week and confirmed by conventional vulvovaginal appearance. Estrus was initiated and follicle development was synchronized with a standard treatment of 5 IU equine chorionic gonadotropin followed by 5 IU of human chorionic gonadotropin (hCG) for 48 h. Four hours after hCG and before anticipated ovulation, ovaries were harvested, cleared of adipose and bursa tissue, washed in phosphate-buffered saline (PBS) and pooled. Ovaries from groups A, B and C were divided and processed for RNA isolation or protein analysis and those from group D were processed for immunohistochemistry.
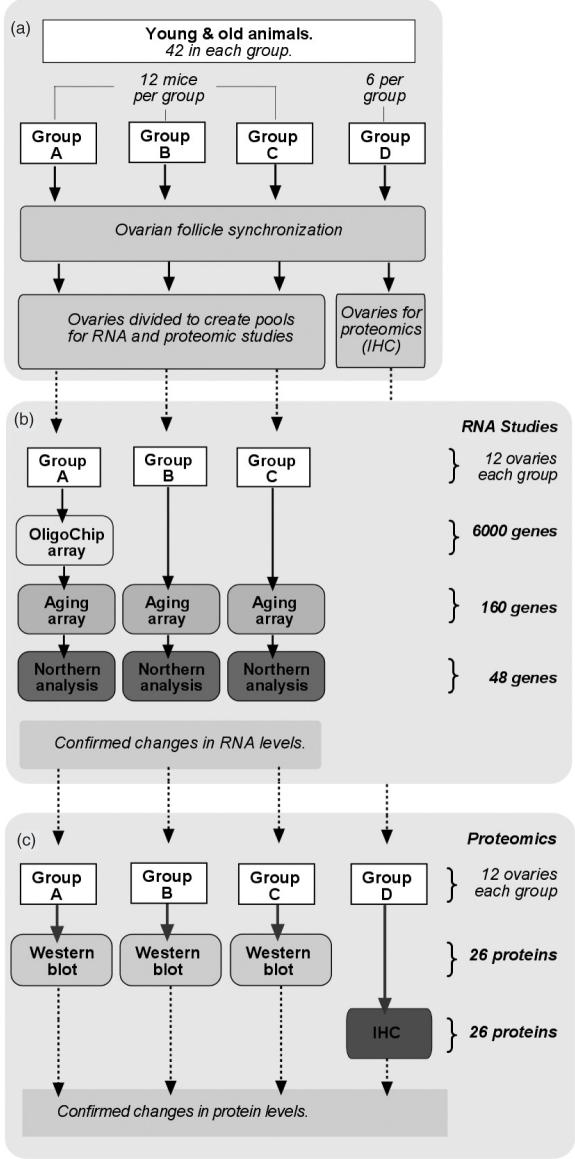
Figure 1.
Flow diagram of experimental design. (a) Forty-two young (6 weeks) and 42 old (8 months) mice were divided into four groups (A, B, C and D) of 6 or12 mice. Animals in all four groups were identically treated for synchronized ovarian follicle development. For each animal the ovaries were divided such that one was included in pools for RNA analysis and the other in pools for proteomic analysis. Both ovaries from group D mice were used for immunohistochemistry (IHC). (b) RNA was extracted from group A ovaries (n = 12) and specific differences in RNA levels in young and old pools were first tested by the OligoChip oligonucleotide array (6000 genes) in duplicate reactions. RNA pools A, B and C were also tested in three independent experiments using six identical aging-specific oligonucleotide arrays (160 genes) in duplicate reactions. RNA's with significant differences in oligonucleotide hybidization reactions between young and old samples were then tested by northern analysis (48 genes) in groups A, B and C. (c) The genes with confirmed differences at the level of RNA were evaluated for differences in protein levels. Western blots for the proteins of these genes were performed on protein pools A, B and C in three independent experiments using duplicate blot reactions. These proteins were also probed in situ by immunofluorescence using triplicate frozen tissue sections from the ovaries in group D.
Oligonucleotide LabChip array
We analyzed total RNA isolated from two pools (young and old) of 12 ovaries from 12 young and old animals, respectively (Group A, Figure 1). Two identical microarrays (Agilent 2100) were used to compare both pools. RNA was quantified and the quality of these RNA samples was examined using RNA 6000 Nano LabChip® Kit on Agilent 2100 Bioanalyzer Nanochip to ensure the integrity of RNA samples before use. The RNA samples from each experimental group (young or old) were pooled, thus creating RNA mixes (ovary-young and ovary-old) that were further used for gene expression analyses. Cyanine 3- or 5-labeled CTP (10.0 mM) were purchased from Perkin–Elmer/NEN Life Science. Fluorescently labeled cRNA targets were generated from 500 ng of total RNA for each reaction using the Agilent Fluorescent Linear Amplification Kit following the protocol described in the user's manual. Each RNA mix was labeled with both cyanine 3 and cyanine 5 to allow the performing of dye reversal experiments to minimize the impact of dye bias.
Hybridization was performed using the Agilent's in situ Hybridization Plus kit with 1 μg labeled cRNA as recommended by the supplier. The arrays were scanned using the Agilent dual-laser DNA microarray scanner and the data were then extracted from images by Agilent's Feature Extraction software 6.1. Genes were defined as significant when significantly under- or over-expressed in young versus old ovary, as determined by the Significance Analysis of Microarrays (SAM) software (http://www-stat.stanford.edu/~tibs/SAM/) which contains a sliding scale for false discovery rate (FDR) of significantly upregulated genes (6). The output criteria from two microarrays selected for SAM included 2-fold or greater levels (P < 0.05) in the old ovary tissues as compared with young ovary tissues, and a significance threshold expected to produce a median FDR of <10 genes. A 3-fold difference in hybridization levels (determined by Agilent) in young compared with old was selected as a threshold for significance and the corresponding genes were further studied.
Mouse aging-specific oligonucleotide array
We analyzed three sets of 12 pooled young and 12 pooled old ovaries independently (Groups A, B and C, Figure 1). The RNA pool used for the Agilent microarray experiment (RNA pool A) was included to verify consistencies between array experiments. RNA was used to prepare cDNA as described earlier (4). 32P-labeled cDNA (25 ng) was hybridized to an aging-specific mouse oligo DNA array containing probes for 160 genes as described previously (4). Each hybridization reaction was performed in duplicate. Twelve identical DNA membranes were used for the comparative hybridizations. The array contains 65 genes that are not presented in the LabChip array from Agilent. Spot reading was performed using a Phosphorimager (Molecular Dynamics) and data were analyzed using an overlay grid to allow calculation of signal density using ImageQuant (Amersham Biosciences) software. Data were imported into Excel (Microsoft) and analyzed as described previously (4).
Virtual northern analysis
Virtual northern analysis was performed to confirm level changes in RNA for genes with consistent 3-fold or greater differences in expression between young and old ovaries as described previously (4). For a single hybridization 2 μg of total RNA was used. As above, 12 pooled young and 12 pooled old ovaries were used for total RNA isolation (Qiagen) and repeated in three replicate sets of experiments. This included testing one RNA pool (pool A) that had been used in the arrays which were also analyzed for consistency. Hybridization reading and data analyses were performed as described in Materials and Methods.
Protein analysis by western blot
Whole ovaries pooled from three pools of 12 young and old ovaries were homogenized at 4°C in buffer containing 20 mM HEPES, 50 mM NaCl, 1 mM EDTA, 5 mM DTT and protease inhibitors (Protease inhibitor cocktail; Sigma). SDS (1%) was added to the homogenates and after centrifugation (50 000 g for 20 min at 4°C) the resulting supernatants were collected for protein analysis. Protein lysates (100 μg) were analyzed for presence of specific proteins by western blotting. Exposed films were scanned using a UMAX 1100 Powerlook scanner and Adobe Photoshop. Protein concentration was determined using BSA as a standard (Pierce kit). Duplicate confirmatory western blots were performed.
Immunofluorescence
Surgically dissected ovaries from six young and six old mice were snap frozen and stored at −80°C in Tissue Freezing Medium (Triangle Biomedical Sciences, Inc.). Sections (5 μm) were fixed in acetone and 10% buffered formalin and stained with hemotoxylin and eosin. For immunofluorescence, sections were fixed in 4% paraformaldehyde, washed with PBS, 0.05% Tween, 0.05% saponin and briefly treated with 0.5% Triton X-100 prior incubation with protein-specific antibodies. Sections were incubated with secondary antibodies conjugated to FITC or Alexa488, Alexa596 (Molecular Probes), Cy3 or Cy5 (Sigma) and visualized using a Zeiss Axioskop 2 microscope, Zeiss Axiocam and Photoshop. As negative control sections were incubated with the secondary antibodies alone (data not shown). For autofluorescence studies, fresh cryosections were directly visualized using a Zeiss Axioskop 2 microscope and Zeiss Axiocam. All antibody experiments were performed in duplicate with young and old ovary sections from different animals.
RESULTS
To provide a global screen of changes in gene expression as ovarian function declines, we contrasted morphological structure, specific RNA and protein levels between isolated ovaries of estrus-synchronized young and old mice. The flow diagram of the experimental design is presented in Figure 1. Young 6-week-old mice were cycling and reproductive competent compared with perimenopausal 8-month-old mice that were still cycling and hormonally responsive but were reproductively compromised by age-dependent infertility (5).
Morphological changes with ovarian reproductive aging
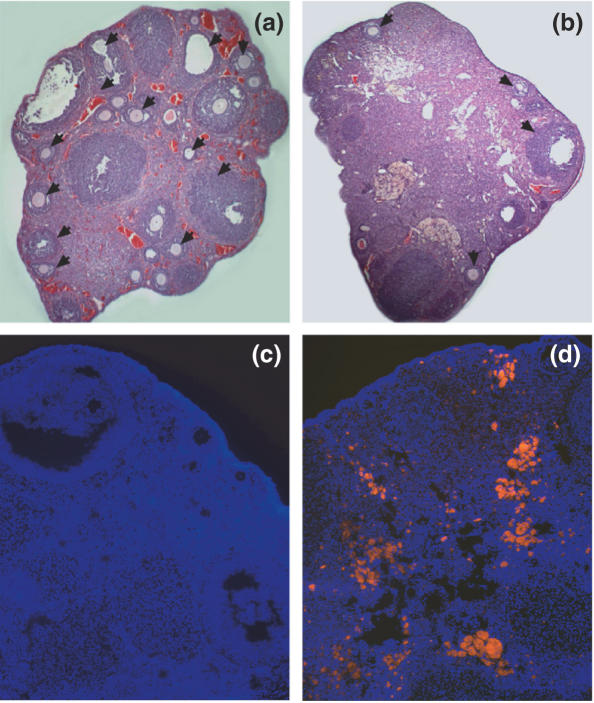
All ovaries consistently contained mature antral follicles (Figure 2a and b). Morphological analysis of hematoxylin and eosin stained cryosections revealed a gross reduction in the total number mature follicles and an increase in intervening stromal tissue in retired breeder ovaries (Figure 2b). Fluorescent microscopy of ovarian cryosections revealed the presence of a significant amount of autofluorescent inclusions (4) in the aged ovaries (Figure 2d). A significantly smaller amount of autofluorescent inclusions were observed in young ovaries (Figure 2c).
Figure 2.
Microscopic analysis of young and aged ovaries. (a) Hematoxylin and eosin stained sections of young ovaries and (b) old ovaries. Black arrows mark the follicles. Autofluorescence of young (c) and old ovarian cryosections (d) visualized with excitation filter in the the green (560–580 nm) spectral regions. Cellular DNA was fluorescently stained with Hoechst 33342 (blue) and visualized with the excitation filter in the blue (350–461 nm) spectral regions. Pictures were taken at magnification ×600.
Comparative genomic analysis of healthy young and perimenopausal ovaries
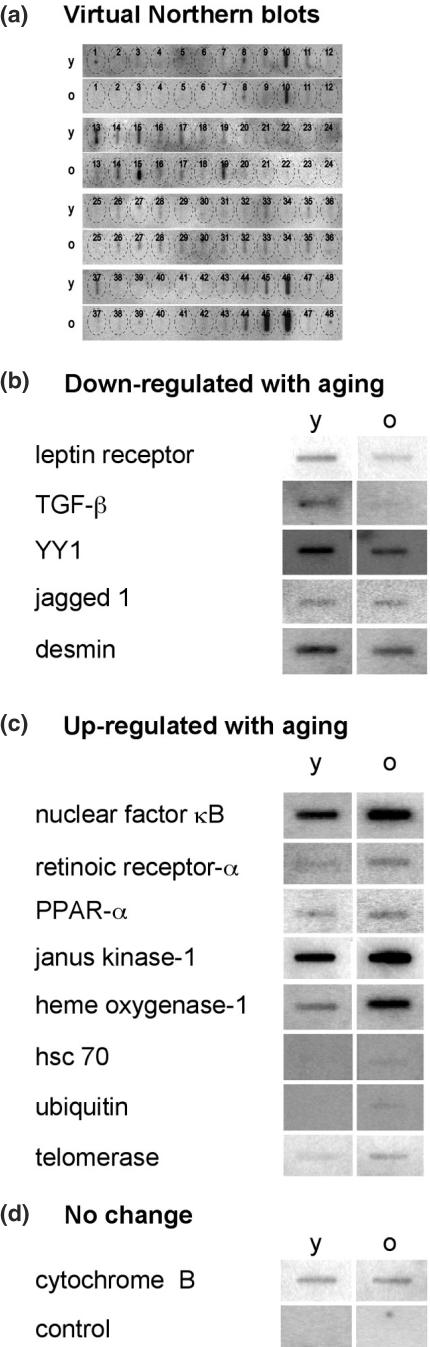
Initially, we compared RNA levels from both ages in two sets of hybridization analyses with a gene array containing 6000 mouse genes. Following duplicate sets of independent hybridization experiments, we confirmed decreased levels of 42 transcripts with aging and increased levels of 68 transcripts. We performed an additional gene expression analysis using a mouse oligonucleotide array developed for aging-related differentially expressed genes (Figure 3a and b). The array contains probes for 160 genes identified previously to change level in the progression of cardiac aging (4). Several genes on this array are not presented in the large LabChip array. With the aging-specific array, 26 transcripts were confirmed to decrease with aging and 31 increased. Of these 57 transcripts, 32 were confirmed by both arrays. To further validate the arrays data we analyzed mRNA levels in young and older ovaries by virtual northern blotting for 52 selected available gene probes corresponding to those transcripts confirmed previously to be expressed differentially with aging. Figure 4a displays an example of the northern slot blot assays performed. Five clones were confirmed to be present at a significantly lower level in the older relative to young ovaries including leptin receptor, TGFβ, transcription factor Yin Yang 1 (YY1), jagged 1 and desmin (Figure 4b and Table 1). Increased levels in older ovaries were found for eight genes including NFκB, retinoic receptor, Jak1, heme-oxygenase-1, ubiquitin and telomerase (Figure 4c and Table 1). No change was observed for cytochrome B and its mRNA level was used as a control for equal RNA loading in the experiments (Figure 4d).
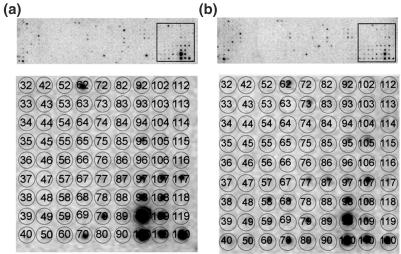
Figure 3.
DNA array to confirm differential gene expression in young and old ovaries. Identical DNA array membranes containing probes for 440 aging-related mouse genes were probed with individual 33P-labeled cDNA libraries prepared from (a) young and (b) aged ovaries. Lower panels of (a) and (b) show an enlargement of the framed membrane portions in the upper panels. Matrix overlay maps the position of the nucleotides for each gene. Positions 62, Leptin; 79, CD36 antigen; 117, unknown EST are examples of decreased levels with aging. Positions 58, peroxisome proliferators activated receptor-a; 105, transcription factor NFκB are examples of increased levels with aging.
Figure 4.
Autoradiograms of representative virtual northern blots that confirm changes in mRNA levels in young and old ovaries. (a) One of the three confirmatory virtual northern blots with a complete set of 48 genes probed in young and old samples. The matrix overlay maps the position of each reaction. Three independent northern blot experiments produced consistent results (SD = 3%). (b) Examples of genes that are downregulated in perimenopausal ovaries compared with young ovaries: leptin receptor at position 37 in (a); desmin at position 10 in (a); TGF-β at position 24. (c) Examples of genes that are upregulated in perimenopausal ovaries: heme-oxygenase-1, position 45 in (a); nuclear factor kB, position 46; retinoic receptor-α, position 19; janus kinase 1, position 32; telomerase, position 44. (d) An example of a gene with no change in level with aging, cytochrome B, position 3 in (a) and the control. Hybridization paper was blotted with oligonucleotides (1 μg) that are specific for different genes and then hybridized with 32P-labeled ssDNA from young (y) and old (o) mouse ovaries. Genes to which the probes correspond, are shown to the left of the autoradiograms. The level of cytochrome B mRNA did not change significantly with aging and was used as an internal control. Hybridization in the absence of oligonucleotides was used as a negative control (control).
Table 1.
Genes with age-dependent changes in mRNA and protein levels in mouse ovaries
| Accession | Gene | RNA | Protein | ||
|---|---|---|---|---|---|
| Level | Method | Level | Method | ||
| Steroids and growth factors | |||||
| TC188770 | OB Leptin | D | 2,3 | NC | 4 |
| M13177 | Transforming growth factor B | D | 2,3 | NC | 4 |
| NM_023135 | Estrogen sulfotransferase | U | 1 | D | 4,5 |
| D000659 | Aromatase | U | 1 | NC | 4,5 |
| NM_008829 | Progesterone receptor | U | 1 | NC | 4 |
| Transcription factors | |||||
| NM_009537 | Yin Yang 1 | D | 2,3 | D | 4,5* |
| AF022363 | SP1 | D | 3 | D | 4,5* |
| M57999 | Nuclear factor κb | U | 2,3 | U | 5** |
| X57528 | Retinoic acid receptor α | U | 2,3 | U | 5** |
| BC062915 | GATA-3 | U | 1,3 | D | 5 |
| NM_011144 | Peroxisome proliferatory-activated receptor α | U | 1,2,3 | NC | 4,5 |
| Cell signaling | |||||
| BC037598 | GDP dissociation inhibitor 1 | D | 1,3 | D | 4,5* |
| AA152708 | Jagged1 | D | 1,2,3 | NC | 4,5 |
| NM_008716 | Notch 3 | D | 1,3 | NC | 4,5 |
| UO7951 | GDP dissociation inhibitor 2 | U | 1 | U | 4** |
| NM_153400 | Purinergic receptor P2X2 | U | 1 | U | 4,5** |
| BC031297 | Janus kinase 1 | U | 2,3 | D | 4,5 |
| Stress-induced | |||||
| NM_010442 | Heme oxygenase 1 | U | 2,3 | U | 4,5** |
| U73744 | Heat shock cognate 70 | U | 2,3 | U | 4** |
| M35021 | Heat shock protein 70 | U | 3 | NC | 4 |
| Other | |||||
| L22550 | Desmin | D | 1,2,3 | D | 5* |
| X82786 | Ki-67 cell proliferation antigen | D | 1 | D | 5* |
| NM_016960 | Small inducible cytokine subfamily A20 | U | 1 | U | 5** |
| AF051911 | Telomerase reverse transcriptase | U | 2,3 | U | 5** |
| BC100341 | Ubiquitin | U | 2,3 | D | 4,5 |
Genes with age-dependent differential expression at the level of mRNA were identified by (1) mouse LabChip array; (2) virtual northern analysis; and (3) DNA aging-specific gene array. Genes included above were confirmed to change with aging >3-fold in duplicate assays. These genes were then screened for differential protein content by western blotting (4) and (or) localization by immunofluorescence (5). From left to right, the table columns are the GenBank accession number, encoded gene, upregulation (U) or downregulation (D) with aging, methodology used to quantify and compare mRNA levels and protein content. Single asterisk marks genes downregulated both at the level of RNA and protein and double asterisk marks genes upregulated both at the level of RNA and protein.
Proteomics analyses of differentially expressed genes in young and perimenopausal ovaries to confirm changes at protein level and tissue distribution
We verified the protein level for the differentially expressed genes by applying western blot with total ovarian lysates and immunofluorescence on frozen tissue sections with protein-specific antibodies. The results of the western blot analyses are displayed in Figure 5. With aging, total protein content of estrogen sulfotransferase, transcription factors YY1, SP1 and GATA3, the cell signaling factors GDP dissociation inhibitor-1 as well as Janus kinase 1, and the post-translational protein modifying ubiquitin decreased (Figure 5a). Stress-induced factor heme-oxygenase-1, the cell signaling factors GDP dissociation inhibitor-2 and purinergic receptor P2X2 protein levels increased with aging (Figure 5c). The remaining proteins corresponding to genes with changes in mRNA expression level demonstrated no significant change with aging or were undetectable (examples shown in Figure 5c).
Figure 5.
Western blots of total protein lysates from young and old mouse ovaries. (a) Proteins with reduced content in old (o) ovaries compared with young ovaries (y); (b) proteins with higher level in young ovaries; and (c) no change in protein content. Proteins were visualized with protein-specific antibodies as indicated at the left of the panels. The size (kDa) of the protein marker is indicated at the right of the panels. All lanes received 100 μg of total ovarian lysate. The immunological reactions are visualized by HSP-coupled secondary antibodies.
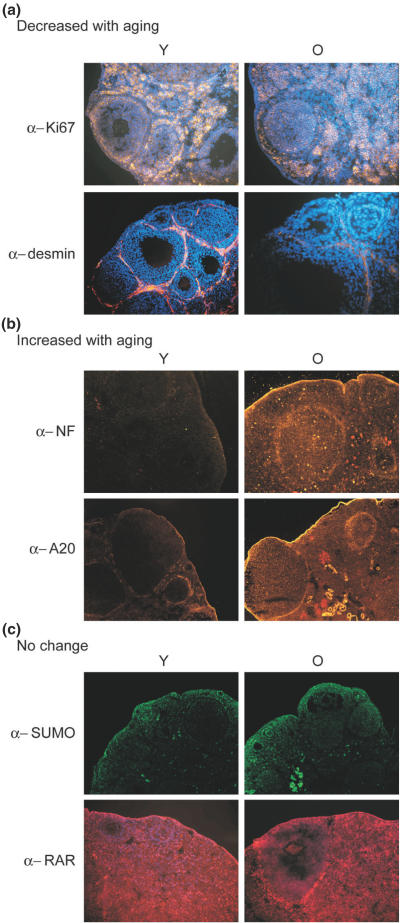
Immunofluorescent histochemical analyses confirmed aging-related decreases in protein levels of transcription factors YY1, SP1 and GATA3, the signal transduction kinase Jak1, ubiquitin, proliferation marker Ki67 and desmin all with similar patterns of cellular and tissue localization. Staining with antibodies against the stress response enzyme heme-oxygenase-1, transcription factors NFκB, retinoic receptor α, A20, the nucleotide cell signaling molecules GDP dissociation inhibitor-2, the purinergic receptor P2X2 and telomerase reverse transcriptase (TERT) revealed increases in protein content. Examples of the 26 proteins screened by immunohistochemistry demonstrating confirmatory increased or decreased levels are shown in Figure 6. Also observed were decreased histochemical levels of estrogen sulfotransferase, which previously demonstrated increased RNA levels. Differential staining of young oocytes was notable for estrogen sulfotransferase EST and ubiquitin-specific antibodies. EST in old ovaries was prominently localized to the surface epithelium. We noted a prominent increased localization of NFκB (Figure 6b) and HO-1 in the follicular layer and the ovarian surface epithelium of old ovaries. This pattern was shared by zinc finger protein A20 (Figure 6b).
Figure 6.
Immunofluorescence microscopy. Ovarian cryosections were incubated with protein-specific antibodies and fluorescently tagged secondary antibodies (green or red). Nuclear DNA was stained with Hoechst 33342 (blue). (a) Proteins that decrease level with aging; (b) proteins with increased level in young ovaries; and (c) no change in protein level. The identity of the protein-specific antibodies is indicated at the left. The immunofluorescent staining of young (Y) and old (O) is indicated at the top of the panels. Images at (a) and (c) are taken at ×600 magnification. They are presented as an overlay of the protein staining layer on Hoechst-stained DNA image. The images in (b) are taken at ×200 magnification without Hoechst staining.
The remaining proteins examined with aging-related changes at RNA level demonstrated no perceptible level change by western blot and immunohistochemistry. As an example in Figure 6c, the retinoic acid receptor (RAR) alpha staining with marked localization in the antral follicle thecal layer in both young and old ovaries is shown.
The results from our genomic and protein comparative studies are summarized in Table 1. Additional mRNAs were found to change level with aging, but their proteins remained essentially at the same level for both ages.
DISCUSSION
Ovarian aging resembles the cellular aging of other mammalian and invertebrate systems to some extent. The significant accumulation of autofluorescent inclusions observed in aged ovaries is consistent with the accumulation of lipofuscin in aged cardiac muscle (4). Increased lipofuscin likely typifies a cumulative response to lipid peroxidation and oxidative stress (7) and was observed prominently in the stroma and perivascular tissue of the ovarian medulla and hilum of older ovaries. Our results demonstrate significant follicular depletion and decreased generalized proliferation with aging. Further, intervening stroma was increased in all regions of older ovaries. Our results indicate significantly reduced level of desmin mainly in the perifollicular stroma in old ovaries. Lower desmin content would ultimately result in reduced cellular contractility (8,9) leading to reduced follicle contraction and changes in the atretic process (1). The ovarian architectural aging response may be characterized by stromal remodeling concurrent with loss of cortical follicular units and the accumulation of autofluorescent inclusions.
Based on substantial clinical data from women with age-related ovarian failure, it is believed that changes in steroid and growth factor-related mechanisms may be causative or consequential factors in ovarian aging (10). The present hypotheses of human ovarian aging propose significant changes in steroid content and activity. Surprisingly, our data did not reveal changes in steroid-producing proteins and growth factors. We observed upregulation of mRNA levels of EST, aromatase, progesterone receptor and TGF-β without coincident changes at the level of protein. We observed downregulation of leptin at the level of mRNA but not at the level of protein. Our findings may indicate that appreciable aging changes in the ovary are operative prior to evidence of steroidogenic failure. Alternatively, aging could change the stability of these proteins, involving the heat-shock protein–ubiquitin–proteasome pathway through the enhanced level of Hsp70 (11). Furthermore, a growing body of evidence supports a role for variation of the rates of mRNA decay as an important process in aging and that the response to stress is in part mediated by stabilizing or destabilizing cofactors such as heat-shock factors. For example, differential stability of aromatase mRNA transcripts under stress conditions has been demonstrated in ovarian cell culture lines (12).
We noted that ovaries in older animals had considerable upregulation of stress response factors Hsp70, Hsc70 and Hsp32 (HO-1), all of which are members of inflammatory and oxidative stress response aging pathways. A change in Hsp70, Hsc70 and Hsp32 is a common feature of aging in skeletal muscle, cardiac muscle and brain (4,13,14). Hsp70 functions as a critical chaperone for steroid receptors and forms receptor–steroid complexes with Hsp90 and p23 that are capable of binding to hormone response elements on DNA to regulate transcription (15). In human ovarian granulosa cells, heat shock induces a robust Hsp70 response (16). Further, Hsp70 is capable of halting ovarian follicular apoptosis by blocking the activity of caspase proteases or inhibiting chaperone-mediated protein import to the mitochondria (17). Our observation of an age-dependent increase of heme-oxygenase-1 (HO-1, Hsp32) suggests that upregulation of protective oxidative stress mechanisms is a feature of early ovarian aging. HO-1 is inducible by heme compounds, heavy metals, sulfhydryl reagents and hydrogen peroxide. It functions to remove lipophilic pro-oxidant heme molecules from the system, generating antioxidant biliverdin molecules and mediating an important cellular defense against oxygen free-radical damage. Our finding of increased expression of the pro-inflammatory transcription factor NFκB in conjunction with increased HO-1 indicates that mechanistic changes are occurring in inflammatory pathways (18) during mammalian ovarian aging. We also observed aging-related increases in A20, a zinc finger protein that is induced by NFκB and itself downregulates NFκB-dependent chronic inflammation via ubiquitin signaling pathways (19). In addition, we observed changes in expression of RARα and PPARα, both of which are potential modulators of inflammation involving the action of NFκB and ubiquitin-proteasome degradation (20,21). The upregulated expression of heat-shock proteins and associated inflammatory stress response factors could significantly contribute to changes in ovarian sensitivity to environment.
Aging might either induce repression or fail to trigger pathways that lead to activation of specific transcription promoters. The levels of transcription factors could also be modulated by the synergistic action of different signaling pathways. Transcription factors YY1 and Sp1 decline and NFκB mRNAs increase with ovarian aging. YY1 is a zinc finger transcription factor that influences expression of a wide variety of genes including HSP70 and HO-1. YY1 interacts with transcription regulators such as Sp1 and p300 (22). YY1 and NFκB compete for binding sites on a cytokine response unit (23). Together transcription factors YYI, NFκB and Sp1 have been implicated in aging mechanisms involving responses to reactive oxygen species and inflammatory cytokines (24). Our findings of downregulation of YY1 and Sp1 and upregulation of NFκB suggest that transcriptional regulatory responses to oxidative stress may be important features of the aging ovarian system and that these processes may be targets for modulation of aging-dependent decline of ovarian function.
We observed alterations in protein members of the Notch signaling pathway, which is generally associated with changes in cellular activation and proliferation. Downregulation of Jagged1 and Notch3 mRNA levels is associated with ovarian aging. The Notch pathway has been implicated in mediating folliculogenesis in the mammalian ovary (25). In Drosophila, activation of Notch and Jak/STAT signaling pathways are critical in regulating polarity in early oogenesis via communication between somatic follicular cells and germ-line cells such that disruption of this pathway leads to defective oocyte development (26). An anti-aging regenerative role for Notch signaling has been suggested in muscle and other tissues (27). If tissue regeneration is an important response to the cumulative aging in the ovary due to repeated cycles of folliculogenesis and regression, Notch pathways may be important mediators of aging-related stromal remodeling.
Ovarian-specific responses to aging involving nucleotide sensitivity and signaling pathways could explain the functional decline in endocrine function and fertility. Our results indicate heightened expression of purinergic receptor P2X2 at both mRNA and protein levels in older ovaries. Elevated local ATP released during biological processes of cell death, noxious stimuli and inflammation stimulates purinergic P2X receptors. P2X receptors appear to be regulated in response to cellular stresses, which may be a direct effect of stress-induced changes in ATP levels (28). Our finding of increased P2X2 with aging suggests an altered ovarian responsiveness to ATP and this increase may be ovarian specific as decreases in P2X receptors have been observed with aging in other tissues. The P2X2 receptor is a glycoprotein ligand-gated channel with functional capacity and current conduction correlated with its level of glycosylation. Interestingly, we have observed alterations in protein glycosylation with ovarian aging (data not shown). However, whether increased P2X2 receptor protein level via glycosylation-related proteolytical stability is a feature of the aging response in the ovary remains to be explored. Nevertheless, increased receptor level may significantly contribute to changes in ovarian mechanisms of adaptation and sensitivity to environment.
Alterations in nucleotide cell signaling mediators, guanine (GDP) dissociation inhibitors 1 and 2 (GDI-1 and GDI-2), can be demonstrated as well. GDI-1 levels decrease with ovarian aging while GDI-2 increases. GDIs bind to and inhibit GDP release from signaling proteins in Rho and Rac families thereby mediating pathways involved in proliferation, differentiation, apoptosis, cytoskeletal organization and cellular trafficking (29). Modification of GDI through nitration is prominent finding in aging skeletal muscle (30) suggesting a role for GDI metabolism in aging. Rho-GDI functions as a transactivator for nuclear steroid receptors and upregulates transcriptional activity of estrogen, androgen and glucocorticoid receptors transcriptional activity (31). This mechanism could provide a link between changes in GDI signaling, nucleotide sensitivity and steroid responsiveness in aging ovarian tissue.
In summary, our results demonstrate that early mouse aging and waning reproductive capacity alters ovarian mRNA and protein levels. Although the primary regulation of reproductive aging in rodents and humans may differ, in our climacteric model we demonstrate clear architectural evidence of ovarian aging including follicular depletion and stromal remodeling. Such changes are typical also for the human ovarian response to aging supporting the possible validity of this model for human perimenopausal ovarian aging. How exactly the mouse model differs from human ovarian aging, is yet to be established. While the depletion of germ cells is accepted as a major determinant of ovarian aging, our gene expression profiling suggests that reproducible changes in gene expression and protein processing coincide with aging-dependent follicular loss and functional decline. Interestingly, the gene expression patterns observed in this climacteric model are evident before coexistent molecular evidence for steroidogenic failure. The specific pattern of gene expression in ovarian aging differs from muscle, brain and cardiac aging indicating a tissue-specific aging response. However, the general functional pathways implicated during early loss of ovarian capacity resemble aging pathways conserved in model organisms. In particular, our data suggest that activation of pro-inflammatory and anti-proliferative pathways is a prominent feature of ovarian aging. Increased sensitivity to the environment via nucleotide signaling pathways may be another important adaptation to aging in the ovary. Such pathways could profoundly influence the early pathologic changes in the ovaries observed in premature and aging-related ovarian failure. How exactly the subset of mRNAs is selected for expression changes with ovarian aging and whether these changes modulate aging-dependent germ cell loss remains to be determined.
Acknowledgments
We thank Benjamin Sachs, Department of Obstetrics and Gynecology, Beth Israel Deaconess Medical Center for the support and suggestions, Wenchao Song for providing anti-estrogen sulfotransferase antibody and Marlon Stoeckius for the autofluorescence analysis. Funding to pay the Open Access publication charges for this article was provided by Department of Obstetrics and Gynecology, Beth Israel Deaconess Medical Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ottolenghi C., Uda M., Hamatani T., Crisponi L., Garcia J., Ko M., Pilia G., Sforza C., Schelssinger D., Forabosco A. Aging of oocyte, ovary, and human reproduction. Ann. NY Acad. Sci. 2004;1034:117–131. doi: 10.1196/annals.1335.015. [DOI] [PubMed] [Google Scholar]
- 2.Tissenbaum H.A., Guarente L. Model organisms as a guide to mammalian aging. Dev. Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 3.McCarroll S.A., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nature Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 4.Bodyak N., Kang P.M., Hiromura M., Sulijoadikusumo I., Horikoshi N., Khrapko K., Usheva A. Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Res. 2002;30:3788–3794. doi: 10.1093/nar/gkf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gosden R.G., Laing S.C., Felicio L.S., Nelson J.F., Finch C.E. Imminent oocyte exhaustion and reduced follicular recruitment mark the transition to acyclicity in aging C57BL/6J mice. Biol. Reprod. 1983;28:255–60. doi: 10.1095/biolreprod28.2.255. [DOI] [PubMed] [Google Scholar]
- 6.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szweda P.A., Camouse M., Lundberg K.C., Oberley T.D., Szweda L.I. Aging, lipofuscin formation, and free radical-mediated inhibition of cellular proteolytic systems. Ageing Res. Rev. 2003;2:383–405. doi: 10.1016/s1568-1637(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 8.Milner D.J., Weitzer G., Tran D., Bradley A., Capetanaki Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J. Cell Biol. 1996;134:1255–1270. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motta P.M., Familiari G. Occurrence of a contractile tissue in the theca externa of atretic follicles in the mouse ovary. Acta Anat. (Basel) 1981;109:103–114. doi: 10.1159/000145372. [DOI] [PubMed] [Google Scholar]
- 10.Klein N.A., Battaglia D.E., Woodruff T.K., Padmanabhan V., Giudice L.C., Bremner W.J., Soules M.R. Ovarian follicular concentrations of activin, follistatin, inhibin, insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-2 (IGFBP-2), IGFBP-3, and vascular endothelial growth factor in spontaneous menstrual cycles of normal women of advanced reproductive age. J. Clin. Endocrinol. Metab. 2000;85:4520–4525. doi: 10.1210/jcem.85.12.7056. [DOI] [PubMed] [Google Scholar]
- 11.Laroia G., Cuesta R., Brewer G., Schneider R.J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 12.Hanoux V., Bouraima H., Mittre H., Feral C., Benhaim A. Differential regulation of two 3′ end variants of P450 aromatase transcripts and of a new truncated aromatase protein in rabbit preovulatory granulosa cells. Endocrinology. 2003;144:4790–4798. doi: 10.1210/en.2003-0260. [DOI] [PubMed] [Google Scholar]
- 13.Lee C.K., Weindruch R., Prolla T.A. Gene-expression profile of the ageing brain in mice. Nature Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 14.McArdle A., Dillmann W.H., Mestril R., Faulkner J.A., Jackson M.J. Overexpression of HSP70 in mouse skeletal muscle protects against muscle damage and age-related muscle dysfunction. FASEB J. 2004;18:355–357. doi: 10.1096/fj.03-0395fje. [DOI] [PubMed] [Google Scholar]
- 15.Freeman B.C., Yamamoto K.R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 16.Kim A.H., Khanna A., Aten R.F., Olive D.L., Behrman H.R. Cytokine induction of heat shock protein in human granulosa-luteal cells. Mol. Hum. Reprod. 1996;2:549–554. doi: 10.1093/molehr/2.8.549. [DOI] [PubMed] [Google Scholar]
- 17.Yoon S.J., Choi K.H., Lee K.A. Nitric oxide-mediated inhibition of follicular apoptosis is associated with HSP70 induction and Bax suppression. Mol. Reprod. Dev. 2002;61:504–510. doi: 10.1002/mrd.10033. [DOI] [PubMed] [Google Scholar]
- 18.Lavrovsky Y., Song C.S., Chatterjee B., Roy A.K. Age-dependent increase of heme oxygenase-1 gene expression in the liver mediated by NFκB. Mech. Ageing Dev. 2000;114:49–60. doi: 10.1016/s0047-6374(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 19.Wertz I.E., O'Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–649. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 20.Vanden Berghe W., Vermeulen L., Delerive P., De Bosscher K., Staels B., Haegeman G. A paradigm for gene regulation: inflammation, NF-κB and PPAR. Adv. Exp. Med. Biol. 2003;544:181–196. doi: 10.1007/978-1-4419-9072-3_22. [DOI] [PubMed] [Google Scholar]
- 21.Bastien J., Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Thomas M.J., Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 23.Lu S.Y., Rodriguez M., Liao W.S. YY1 represses rat serum amyloid A1 gene transcription and is antagonized by NF-κB during acute-phase response. Mol. Cell. Biol. 1994;14:6253–6263. doi: 10.1128/mcb.14.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adrian G.S., Seto E., Fischbach K.S., Rivera E.V., Adrian E.K., Herbert D.C., Walter C.A., Weaker F.J., Bowman B.H. YY1 and Sp1 transcription factors bind the human transferrin gene in an age-related manner. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51:B66–B75. doi: 10.1093/gerona/51a.1.b66. [DOI] [PubMed] [Google Scholar]
- 25.Johnson J., Espinoza T., McGaughey R.W., Rawls A., Wilson-Rawls J. Notch pathway genes are expressed in mammalian ovarian follicles. Mech. Dev. 2001;109:355–361. doi: 10.1016/s0925-4773(01)00523-8. [DOI] [PubMed] [Google Scholar]
- 26.Grammont M., Irvine K.D. Organizer activity of the polar cells during Drosophila oogenesis. Development. 2002;129:5131–5141. doi: 10.1242/dev.129.22.5131. [DOI] [PubMed] [Google Scholar]
- 27.Conboy I.M., Conboy M.J., Smythe G.M., Rando T.A. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 28.North R.A. Molecular physiology of P2X receptors. Physiol. Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 29.Olofsson B. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 1999;11:545–554. doi: 10.1016/s0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 30.Kanski J., Alterman M.A., Schoneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic. Biol. Med. 2003;35:1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 31.Su L.F., Wang Z., Garabedian M.J. Regulation of GRIP1 and CBP coactivator activity by Rho GDI modulates estrogen receptor transcriptional enhancement. J. Biol. Chem. 2002;277:37037–37044. doi: 10.1074/jbc.M111607200. [DOI] [PubMed] [Google Scholar]