Abstract
We previously identified a 46-kD protein allergen in latex as having amino acid sequence homology to the patatin gene family. The objective of this study was to characterize this protein by molecular techniques. RNA was isolated from the latex or leaf material from Hevea brasiliensis and from potato tubers. Specific polymerase chain reaction (PCR) primers were designed from the amino acid sequence and reverse transcriptase (RT)-PCR amplified a specific product from latex RNA that was subsequently cloned and sequenced. This product was 1493 bp in length with an 1167 bp open reading frame. The deduced amino acid sequence encodes for a 389 aa protein, pI 4.82 with 43% homology to tobacco patatin. Northern analysis of potato, Hevea leaf, and latex RNA demonstrated the message to be most abundant in latex, weakly present in Hevea leaf, but no hybridization occurred with potato RNA. Patatin has lipid acyl-transferase and PLA2-like activity, suggesting it plays a role as a defence-related protein. Other defence-related proteins in latex such as hevein, glucanase, and hevamine are also allergens. Increased production of defence-related proteins as a result of increased tapping of the rubber trees to meet the demand for latex may explain the increased allergenicity of latex.
Keywords: latex allergy, antigens, IgE antibodies, Hev b7 cDNA
INTRODUCTION
Natural rubber latex allergy is an increasingly common health problem [1]. The major cause of sensitization appears to be exposure to powdered latex gloves produced from natural rubber latex. Proteins found in natural rubber appear to be the responsible agents [2–4]. Once sensitized, individuals can become extremely sensitive, and avoiding natural rubber in modern society becomes a major difficulty [1]. A coexisting problem is a high incidence of food allergy in the latex-sensitive population [5]. Reactivity to certain foods has been reported in > 50% of the latex-allergic individuals [6]. The mechanism for this high degree of reactivity is thought to involve cross-reactive epitopes common to latex and food proteins [7].
Multiple latex proteins have been identified as allergens and several have been cloned and partially characterized. The allergens identified to date are found as soluble C-serum proteins and as proteins that are normally bound to the rubber particles. Several allergens have been given allergen designations by the International Union of Immunological Societies. Hev b1 is a protein that is tightly bound to rubber particles known as rubber elongation factor [8,9]. Hev b2 is a β 1–3 glucanase, a protein highly expressed in latex which appears to have increased allergenic reactivity in spina bifida patients [10,11]. Hev b 3, a 24-kD rubber-bound protein [12], and Hev b 4, a microhelix protein [11], are less well characterized. The rubber-bound proteins, Hev b1 and Hev b3, are more common allergens for spina bifida children, where the major route of exposure to latex appears to be by direct contact with mucous membranes [12]. Hev b 5 is a newly described acidic protein with homology to proteins in kiwi and potato [13,14]. Hev b 6 is the hevein preprotein which appears in multiple forms due to post-translational modification and is a major allergen for health care workers [4,15,16]. The allergenic epitopes in hevein have been mapped to the N-terminal region of the protein [7,16]. Here, we describe the cloning and characterization of a 46-kD protein which binds IgE from latex-allergic patients and has been designated Hev b 7.
MATERIALS AND METHODS
RNA isolation
Total RNA was isolated from potato skins and tubers by acid guanidinium thiocyanate-phenol-chloroform extraction [17]. RNA from Hevea was extracted from leaves or latex obtained from 8-year-old trees (Hevea brasiliensis cv. RRIM C600) growing in our laboratory. RNA was isolated from Hevea leaf tissue by freezing the material in liquid nitrogen, grinding with a mortar and pestle, and extracting the RNA by a modification of the CTAB precipitation method of Murray & Thompson [18]. Rather than using ultracentrifugation after CTAB nucleic acid precipitation, the nucleic acid was recovered by centrifuging at 4000 g for 5 min, followed by LiCl precipitation of the RNA.
RNA was extracted from Hevea latex by a modification of the method of Prescott & Martin [19]. The latex was collected in extraction buffer at pH 9.5 rather than 9.0 [20], phenol/chloroform extracted, and the RNA precipitated with LiCl.
Reverse transcription and polymerase chain reaction
Total RNA from Hevea latex (2.5 μg) was reverse transcribed to cDNA using random hexamer primers and MuLV Reverse Transcriptase (Perkin-Elmer Corp., Foster City, CA). Degenerate polymerase chain reaction (PCR) primers P5 [5′-TTT GGA TCC ACN CA (A/G) GGN AA (A/G) AA (A/G) AT (A/C/T) ACN G-3′] and P7 [5′-TTT GGA TCC (A/G)TA (A/G)AA (A/G)TC (C/T)TT (C/T)TC (A/G)TC (T/C)TT (A/T/G)GC-3′] (where N can be any nucleotide) were synthesized based on the amino acid sequence of the latex protein previously determined by protein sequencing [4], and based on homology to potato patatin. DNA amplification was performed on a Perkin-Elmer thermal cycler, in a 50-μl volume containing 2 mm MgCl2, 5 μm of each primer, 200 μm of each dNTP, 2.5 U of AmpliTaq DNA Polymerase, and 2.5 μg of Hevea latex cDNA. Forty cycles of amplification were performed (94°C for 1 min, 48°C for 3 min, 72°C for 1 min, and a final polymerase extension step at 72°C for 10 min), and the 245-bp PCR product was analysed on a 2% agarose gel (Seakem GTG; FMC Bioproducts, Rockland, ME). The P5xP7 product was inserted into the pCR 2.1 plasmid using TA Cloning (Invitrogen, San Diego, CA). The ligated pCR2.1/P5xP7 was transformed into competent INV ∝ F′ Escherichia coli (Invitrogen), and sequenced using T7 DNA polymerase (Sequenase 2.0; Amersham Corp., Arlington Heights, IL) with universal M13 Reverse Primer and T7 Promoter primer (Invitrogen). The P5xP7 sequence was compared with sequences in Genbank using the BLAST program at the National Center for Biotechnology [21].
Northern blot analysis
Twenty micrograms of total RNA from potato, Hevea leaf, and Hevea latex were separated by electrophoresis in 1.2% agarose/formaldehyde gels and transferred to nylon membranes (Magna, Micron Separations Inc., Westboro, MA). RNA bound to the nylon membranes was UV cross-linked, and prehybridized for 5 h in 30% formamide, 6 × SSC, 5 × Denhardt's, 0.1% SDS, 100 μg/ml salmon sperm DNA. The membranes were probed with a 1460-bp cDNA patatin clone received as a gift from W. Parks (Texas A&M University, College Station, TX) [22], or with the latex P5xP7 cDNA. The cDNA products were purified and 32P-labelled by the random primer labelling technique using Klenow enzyme and ∝-32P-dCTP (Megaprime, Amersham Corp.). The 32P-labelled probes were added to the prehybridization solution and hybridization proceeded overnight at 37°C. The membrane incubated with the patatin cDNA was washed in 6 × SSC, 0.1% SDS at 60°C, while the membrane hybridized with the Hevea P5xP7 cDNA was washed in 2 × SSC, 0.1% at 68°C. The membranes were exposed to x-ray film (XAR 5; Eastman Kodak Co., Rochester, NY) for 24 h with two intensifying screens.
Extension of the cDNA ends of cDNA Hev b 7
cDNA was synthesized from Hevea latex total RNA (1.5 μg) using MMLV reverse transcriptase, a modified oligo (dT) primer (CDS/3′ PCR), and CapSwitch oligonucleotide (5′-TACGGCTGCGAGAAGACGACAGAAGGG-3′ (Clontech Labs, Inc., Palo Alto, CA). The CapSwitch-anchored ss cDNA was amplified by long-distance (LD) PCR using Advantage KlenTaq polymerase, CDS/3′ PCR primer and 5′ PCR primer homologous to CapSwitch (5′-TACGGCTGCGAGAAGACGACA GAA-3′) (Clontech Labs).
The ends of the Hev b7 cDNA were extended by PCR using the amplified CapSwitch-anchored ss cDNA with primers derived from the sequence of P5xP7, the 5′ CapSwitch oligonucleotide, and the CDS/3′ primer. For 5′ extension, primers L4 (5′-CAGGAATGATCCCT CTAATCCCACC-3′) and 5′ CapSwitch oligonucleotide were used. The PCR was performed in a 50-μl volume using 1 μl of the amplified CapSwitch-anchored ss cDNA, 1 μm of each primer, 200 μm of each dNTP, and Advantage KlenTaq polymerase (Clontech Labs). Thirty-five cycles of amplification were performed (95°C 15 s, 68°C 5 min).
For 3′ extension, overlapping primers L1 (5′-GCAGGGACAAGTACCGG-3′) and L2 (5′-GGCTGATAACCACCATGCTTACAG-3′) were designed for use in the PCR with the CDS/3′ PCR primer. PCR was performed as described for the 5′ extension using primers L1 and CDS/3′. This PCR reaction (1 μl) was then used in a nested PCR with primers L2 and CDS/3′ using the same conditions except only 25 cycles of amplification were performed. The PCR products were ligated into the pCR 2.1 plasmid using TA Cloning (Invitrogen), transformed into competent INV ∝ F′ E. coli (Invitrogen), and sequenced with T7 DNA polymerase (Sequenase 2.0; Amersham Corp.) using overlapping forward and reverse primers.
Coding sequence of Hev b 7
Based on the sequence obtained, PCR primers L5 (5′-TTAATGAATTCCGATGGCTACTGGTAGTACTAC-3′), and L6 (5′-TTAATGAATTCTCATTT GAGTTGACGGAGCTTTCG-3′) were synthesized to obtain the entire coding sequence of Hev b 7 in a single clone. Eco RI-sites (underlined) were included for cloning purposes. DNA amplification was carried out on a Perkin-Elmer thermal cycler, in a 100-μl volume containing amplified CapSwitch-anchored ss cDNA from Hevea latex, 2 mm MgCl2, 1 μm of each primer, 200 μm of each dNTP, and Advantage KlenTaq polymerase (Clonetech Labs). Forty cycles of amplification were performed (94°C for 1 min, 48°C for 3 min, 72°C for 1 min, and a final polymerase extension step at 72°C for 10 min). The PCR product was ethanol-precipitated, Eco RI-digested to remove the region of the primers flanking the Eco RI site, and analysed on a 2% agarose gel (Seakem GTG; FMC Bioproducts). The 1182-bp PCR product was excised from the gel and purified with QIAEX II (Qiagen Inc., Chatsworth, CA).
Recombinant DNA cloning procedures
The 1182-bp Hev b 7 PCR product was ligated into the pGEX2T vector, and transformed into JM105 E. coli competent cells according to the manufacturer's directions (Pharmacia Biotech Inc., Piscataway, NJ). Positive clones containing inserts were identified by Eco RI digestion of the plasmids and separation on a 2% agarose gel (Seakem GTG; FMC Bioproducts). Inserts were positively identified by sequencing with T7 DNA polymerase (Sequenase 2.0; Amersham Corp.) using overlapping forward and reverse primers. Plasmid pGEX2T/Hev b 7, which contains full-length Hev b 7 coding sequence fused in-frame with the GST moiety, was used to transform competent BL21 cells.
Recombinant Hev b 7 production and expression
The GST Gene Fusion System (Pharmacia Biotech Inc.) was used to express rHev b 7 following the procedure modifications of Frangioni & Neal [23]. An overnight culture of BL21/pGEX2T/Hev b 7 was diluted 1:10, grown at 37°C to an optical density (OD)600 of 1.0, and induced with 0.1 mm IPTG for 3 h at 37°C. The culture was centrifuged and the pellet resuspended in STE with 100 μg/ml lysozyme. Following a 15-min incubation on ice, dithiothreitol and Sarkosyl were added to 5 mm and 1.5% final volumes, respectively. The sample was sonicated for 2 min on ice in a water bath sonicator, centrifuged, and Triton-X (2% final volume) was added to the supernatant. Glutathione agarose beads were added to the supernatant and incubated for 20 min at 4°C. The beads were washed five times with cold PBS. The fusion protein (Hev b 7/GST) was eluted from an aliquot of the beads by adding elution buffer (100 mm Tris pH 9.0, 150 mm NaCl, 20 mm reduced glutathione, 2% n-octyl-glucoside, 5 mm dithiothreitol) and incubating at room temperature for 15 min with shaking. To cleave GST from Hev b 7, an aliquot of the eluted Hev b 7/GST fusion protein was thrombin-digested overnight (thrombin digest). To obtain purified rHev b 7, the beads were thrombin-digested overnight at room temperature, centrifuged, and the supernatant containing rHev b 7 was dialysed against water, lyophilized, and resuspended in 100 mm Tris pH 9. Total protein concentrations were determined by the Coomassie Protein Assay (Pierce, Rockford, IL)
Western blot analysis
The fusion protein (2 μg/lane), thrombin digest (4 μg/lane), and rHev b 7 (2 μg/lane) were separated by SDS–PAGE on 15% gels, after denaturation in electrophoresis sample buffer containing 0.0625 m Tris–HCl pH 6.8, 10% glycerol, 2% SDS, and 5% β-mercaptoethanol. The proteins were transferred to nitrocellulose membrane (Protran, 0.1 μm; Schleicher and Schuell, Keene, NH) according to the method of Towbin et al. [24], and stained with 0.5% Ponceau S to visualize the protein standards (Pharmacia Biotech Inc.). The membrane was blocked for 1 h in 5% bovine serum albumin (BSA) in PBS, and incubated overnight in E. coli- adsorbed patient sera [7] pooled from five skin test-positive latex-allergic healthcare workers (1:10, v/v in PBS). The membrane was washed, incubated for 1 h in monoclonal anti-human IgE (clone GE-1, 1:1000; Sigma), washed, and incubated for 1 h in anti-mouse IgG conjugated to alkaline phosphatase (1:5000; Sigma). After washing, the membrane was developed in the alkaline phosphatase substrate NBT/BCIP (Promega Corp., Madison, WI). Each antiserum was preadsorbed for 30 min with 1/10th volume of 0.2 mg/ml E. coli extract (Promega Corp.). Western blot analysis was also performed using anti-GST antibody according to the manufacturer's instructions (Pharmacia Biotech Inc.). Inhibition experiments using non-ammoniated latex proteins (NAL) separated by SDS–PAGE were performed as previously described [6]. The sera were preincubated with various concentrations of rHev b 7 or GST (control) for 30 min before addition to the membrane.
RESULTS
Isolation of clone cDNA Hev b 7
Using PCR, the cDNA for the latex protein allergen was obtained and designated as Hev b 7. Oligonucleotide primers were constructed for use with Hevea latex cDNA based on the published DNA sequence of the potato patatin gene, and the previously determined N-terminal amino acid sequence for the intact Hev b 7 protein and a 30-kD cyanogen bromide cleavage product [6]. Several degenerate primer pairs were initially utilized and one pair, P5/P7, produced a product of the expected size (245 bp). The product was sequenced and a nucleotide sequence homology search [21] performed which showed it to be homologous to the patatin gene family. Based on a MacVector nucleotide alignment with potato patatin mRNA (Potpatb accession no. M21879), the Hevea sequence P5xP7 was 67% homologous.
The Hev b 7 P5xP7 PCR product was purified, labelled with ∝32P-CTP, and used as a probe in Northern analysis of total RNA from potato, Hevea leaf and latex (Fig. 1). The Hev b 7 probe did not hybridize to potato RNA, hybridized weakly to Hevea leaf tissue RNA, and quite strongly to Hevea latex RNA, indicating that Hev b 7 is much more prevalent in Hevea latex than in Hevea leaf.
Fig. 1.
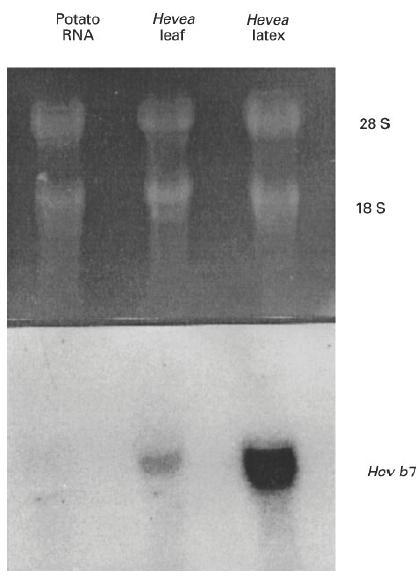
Hev b 7 Northern blot. Total RNA (20 μg) from potato, Hevea leaf or latex was separated in 1.2% agarose/formaldehyde gel and transferred to nylon membrane. The RNA was UV cross-linked to the membrane and probed for the Hev b 7 gene product. Top panel represents the ethidium bromide-stained gel and the bottom panel represents the blot probed with Hev b 7. The Hevea P5xP7 cDNA clone was used as a probe for Hev b 7.
Using the sequence of P5xP7, the cDNA ends were extended by PCR using cDNA from Hevea latex as a template. Primers were constructed from the 245-bp sequence and used with primers from CloneTech's CapFinder cloning kit to construct a series of overlapping clones which encompass the entire cDNA sequence of Hev b 7 (Fig. 2). The sequences of the PCR products obtained using primer pairs 5′ CapSwitch oligonucleotide/L4, P5/P7, and L2/CDS/3′ PCR Primer (CloneTech Labs) were aligned to obtain the complete Hev b 7 sequence from the 5′ Cap to the oligo dT (Fig. 3). The overlapping clones were sequenced, and a consensus sequence determined. Minor differences were observed as variations in one of the three L2xdT clones sequenced. From the determined sequence, oligonucleotide primers (L5/L6) were designed for PCR to obtain a single product spanning the complete Hev b 7 coding sequence.
Fig. 2.
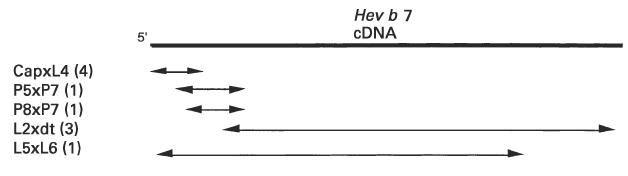
Diagram of overlapping clones obtained for the Hev b 7 cDNA. The numbers behind the clone designations indicate the number of clones that were sequenced.
Fig. 3.

Nucleotide and deduced amino acid sequence of Hev b 7 cDNA. The consensus nucleotide sequence with deduced amino acid sequence (below) was determined by alignment of the various clones using MacVector. The underlined nucleotide sequences represent oligos used in the polymerase chain reaction (PCR). The peptide sequences (dotted underline) were observed by aa sequence analysis [6]. The ATG start codon, TGA stop codon and AATAAA polyadenylation signals are indicated in bold. The sequence data are available from GenBank under accession no. U80598.
Characterization of Hev b 7 cDNA
BLAST analysis of the nucleotide sequence revealed homology to patatins in tobacco and potato. Based on MacVector alignment, the nucleotide sequence of Hev b7 was 51% homologous to potato and 54% homologous to tobacco patatins. The deduced amino acid sequence revealed an open reading frame of 1167 nucleotides which translated into a peptide of 389 amino acids. The first seven amino acids appeared to be cleaved by post-translational modification, as they were not identified by N-terminal aa sequence analysis of the mature protein [4,6], but they did not appear as a typical signal peptide. The Hev b 7 amino acid sequence aligned with tobacco patatin (NTU68484) using the MacVector PAM 250 matrix, showed 43% identities with 66% homology using conservative changes (Fig. 4) and 40% identities and 60% homology using conservative changes compared with potato (Potpatb no. M21879). The calculated molecular mass of the molecule is 42 995 D with a pI of 4.82. There are two potential N glycosylation sites at amino acid positions 238 and 289.
Fig. 4.

Comparison of the Hev b 7 amino acid sequence with the patatin homologue from Nicotiana tobacum (NTU68484). Identical matches are represented by a vertical bar. +, a conservative change in amino acids. Gaps (−) introduced to maximize alignment. Alignment analysis was performed using the MacVector PAM 250 matrix. The leucine at the start of the mature protein is indicated in bold.
Expression of the recombinant Hev b 7 protein
The coding sequence of cDNA Hev b 7 was expressed as part of the pGEX2T fusion protein, Hev b 7/GST. The procedure of Frangioni & Neal [23] was used to improve solubility of the fusion protein. By SDS–PAGE, the molecular mass of Hev b 7/GST fusion product was ≈ 72 000. Cleavage of Hev b 7/GST with thrombin yielded two bands with molecular mass of ≈ 46 000 (rHev b 7) and 29 000 (GST) (Fig. 5). The yield of affinity-purified rHev b 7 was 1.5 mg/l of broth.
Fig. 5.
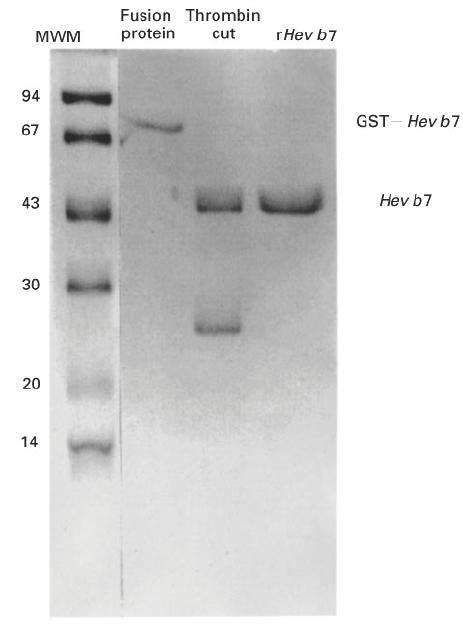
SDS–PAGE analysis of recombinant Hev b 7 products. Recombinant fusion protein GST–Hev b 7, the recombinant protein cut with thrombin, and the purified rHev b 7 were analysed by SDS–PAGE on 15% gels. The gel was stained with coomassie blue. Molecular mass was estimated using marker proteins (left lane) obtained from Pharmacia.
Immunoreactivity of Hev b 7/GST
The Hev b 7/GST fusion product, thrombin digest, and purified rHev b 7 were separated by SDS–PAGE and blotted to nitrocellulose. The blot was probed for IgE binding with sera pooled from five latex-allergic patients known to react with several latex proteins including the 46-kD allergen (Fig. 6). The 46 000 rHev b 7, and 72 000 Hev b 7/GST fusion product were reactive with patient IgE, while the 29 000 GST was negative. The IgE binding indicates that rHev b 7 is the 46 000 allergen previously identified in NAL. A similar blot was probed with anti-GST (Pharmacia Biotech Inc.) as a control and the 46 000 Hev b 7 was negative while the 72 000 Hev b 7/GST and the 29 000 GST reacted with the anti-GST (Fig. 6). Pooled sera from three atopic non-latex-allergic patients with high IgE levels [7] served as a negative control and failed to recognize any protein bands (data not shown).
Fig. 6.
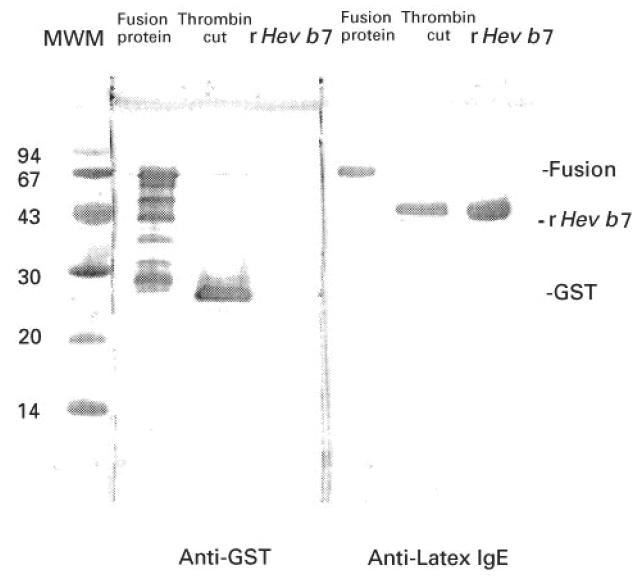
Immunoblot demonstrating anti-latex IgE binding by rHev b 7. Recombinant fusion protein (2 μg; GST/Hev b 7), fusion protein cut with thrombin (4 μg), and purified Hev b 7 (2 μg) were separated on 15% SDS–PAGE gels and transferred to nitrocellulose. The blot was blocked with 5% bovine serum albumin (BSA) and incubated with pooled sera from five latex-allergic patients. IgE binding was detected by monoclonal anti-IgE and visualized with an alkaline phosphatase-labelled anti-mouse IgG. Anti-GST (left panel) recognized the fusion protein and several additional proteins (lane 1) and the GST in the thrombin cut (lane 2) but not the rHev b 7 (lane 3). Patient IgE (right panel) recognized the fusion protein (lane 4) and the rHev b 7 (lane 6) but not GST (lane 5).
To demonstrate further that the IgE was recognizing the latex patatin homologue in NAL, an inhibition Western blot was performed. Pooled latex-allergic patient sera were first incubated with increasing amounts of the recombinant Hev b 7 protein. Figure 7 shows that allergic sera could be completely inhibited from reacting with the 46-kD allergen in NAL using rHev b 7. Other IgE binding proteins in the NAL preparation were not inhibited by rHev b 7, demonstrating specificity for Hev b 7. GST, used as a control, failed to inhibit patient IgE from binding to the 46-kD allergen.
Fig. 7.
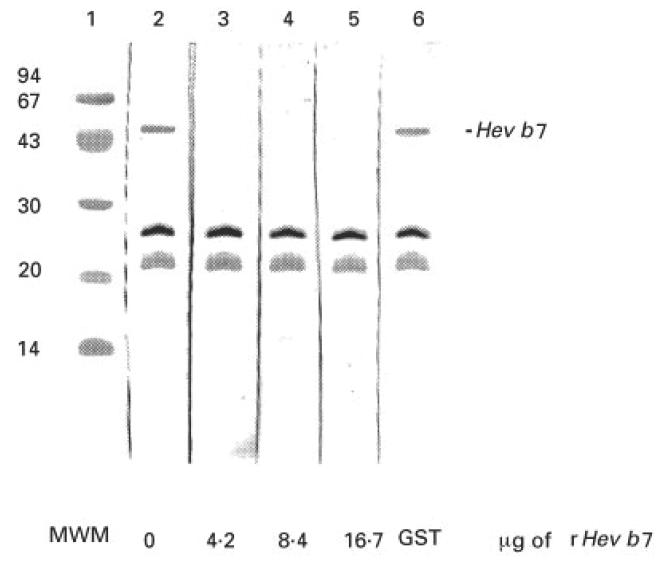
Inhibition immunoblot to demonstrate IgE recognition of 46-kD allergen in non-ammoniated latex (NAL). NAL (25 μg/well) was separated on 15% SDS–PAGE and transferred to nitrocellulose. Patient sera (1:10 dilution) were inhibited with the indicated amounts of rHev b 7 for 30 min before addition to the membrane. Several IgE binding proteins in the NAL are not inhibited by rHev b 7, while the 46-kD allergen is completely inhibited. No inhibition is observed with GST (lane 6) used as a control protein.
DISCUSSION
Sensitivity to latex products is of growing medical importance as 5–17% of healthcare workers are affected in some fashion [1]. Characterization of the latex-allergic response has shown that it is not directed against a single allergen but to many different proteins. Hev b 7 is the newest member of a growing list of cloned latex-derived proteins shown to elicit an allergic response. We initially characterized the molecule by N-terminal peptide sequencing, which revealed a strong homology to the patatin gene family [6]. In an attempt to obtain the cDNA for this protein, we used Solanum tuberosum patatin cDNA as a probe for Northern blot analysis of RNA from potato, Hevea leaf and Hevea latex. The patatin cDNA hybridized to potato RNA, but did not hybridize to Hevea leaf or latex, even under low stringency conditions (data not shown). Successful cloning and sequencing of Hev b 7 required PCR using degenerate primers and revealed a previously unknown latex patatin homologue as another allergen.
Sequence analysis of the Hev b 7 clones reveals a cDNA structure with characteristics typical of plant cDNAs. Following the translational ATG start (69–71), there is an uninterrupted open reading frame of 1167 bp which is terminated by a single TGA codon. Two putative polyadenylation signals are present at 45 and 60 bp downstream of the stop codon. The position and sequence of these signals are consistent with published results showing that plant genes characteristically contain two polyadenylation sites [25]. The first is located between 25 and 45 bp following the termination codon and the second 15–35 bp after that, with the preferred signal being AATAAA. Comparison of nucleotide sequences of Hev b7 cDNA and potato patatin reveals that the coding sequences are 51% homologous. We sequenced three L2xdT clones each spanning > 80% of the cDNA and seven nucleotide differences were found between the sequenced clones, suggesting that Hev b 7 may be a member of a multigene family. This would not be unexpected, as patatins are a gene family that consists of 16–18 members per haploid genome [26]. Alternatively, the variation seen may be the result of Taq polymerase errors. Definitive proof will require the sequencing of Hev b 7 genomic clones and analysis of non-coding regions of DNA flanking the gene.
The finding that Hev b 7 shows homology to a potato protein is not surprising. Two other latex allergens Hev b 6 [7] and Hev b 5 [13] have also been shown to share homology with potato proteins. Potato is a common cross-reacting food for latex-allergic patients [6]. The cDNAs for both patatin and Hev b 7 code for a protein of ≈ 43 kD. Both are soluble proteins. The calculated pI of Hev b 7 (4.82) is very close to that determined for patatin (4.75) [27]. This acidic isoelectric point is characteristic of the great majority of proteins from latex when analysed by 2D SDS–PAGE [20,28]. The translated product of the Hev b 7 open reading frame and the published potato patatin amino acid sequence are 40% identical. Allowing for conserved amino acid changes this homology rises to 60%. The greatest degree of homology between the two proteins lies at the 5′ end of the gene and may reflect a conservation of function between the two proteins. The first 23 amino acids of the patatin molecule constitute a signal peptide and are removed to yield the mature protein with a molecular mass of 40 kD [29]. While we found no evidence for the presence of a signal peptide in Hev b 7, it is interesting to note that N-terminal amino acid sequencing of the Hev b 7 starts at the leucine of position 8 of the cDNA sequence. When patatin and Hev b 7 are aligned to maximize homology, this leucine is located one position away from the cleavage site of the signal peptide in patatin. It is possible that the loss of these amino acids is the result of partial protease digestion. Since Hev b 7 does not appear to possess a classic signal peptide, processing of the 5′ end of the molecule may be required for enzyme function or compartmentation. Alternatively, the lack of a signal peptide may be the result of cloning a truncated member of the Hev b 7 family.
Patatin is the major tuber protein isolated from potato (S. tuberosum). It is a 40-kD glycoprotein that has acyl transferase as well as PLA2 activity [27,30]. It should be noted that NAL proteins were found to possess PLA2 activity as well (P. Vadus, personal communication). We have not yet correlated this activity with the Hev b 7 product, but experiments are underway to test this hypothesis. PLA2 is thought to function in plants by activating lipoxygenase to generate lipid peroxides that results in the accumulation of the antibacterial phytoalexins [31]. Another role of this acyl hydrolase activity may be to produce waxy substances to block infections by pathogens [31]. The similarity between these Hevea and potato proteins may reflect the fact that these plant species share a common mechanism of defence against pathogens [32].
The presence of a 46-kD latex allergen has been noted in several studies [28,33–35]. We also reported that it is one of several allergens recognized by the sera of healthcare workers [4]. Interestingly, in one study immunoblots of 10 of 12 allergic patient sera reacted with proteins with both a pI of 4.8 and a molecular mass of 46 kD [28]. While Hev b 7 possesses both of these properties it is possible that there are additional allergens with each of these properties. Sera from latex-allergic patients used in Western blot analysis recognized the non-glycosylated rHev b 7, which would suggest that the allergen is a protein and does not require the presence of carbohydrate. It is possible that glycosylation of Hev b 7 may produce additional antigenic or allergenic epitopes, but our results with rHev b 7 suggest that the amino acid sequence alone is sufficient for IgE binding. This further suggests that the allergenic epitope(s) are composed of linear amino acid determinants. Additionally, our results demonstrate that the IgE antibodies recognize the reduced form of the allergen. This is consistent with results showing that allergic sera reacted to a 46-kD allergen, whether reduced or non-reduced [28].
Many proteins in latex have been characterized as defence proteins. This is not surprising, as the function of latex is to wall off the site of a wound and to prevent pathogen invasion. Hevein is a defence-related protein from Hevea possessing a chitin binding domain. It is an allergen in latex that shares a great deal of aa homology with the wound-induced proteins (WIN) from potato [36,37]. Expressed in tomatoes it increased the resistance of the plants to fungal pathogens [38]. Another allergen, hevamine, is a 29-kD protein with chitinase and lysozyme-like activities [4,39]. We observed the level of expression of Hev b 7 to be much greater in latex than leaf tissue, and this is also true for hevein [40]. In general, defence-related genes show a 10–50-fold greater expression in lacticifers than leaves [10,20,39].
The defence-related proteins may have precipitated the current epidemic of latex allergy. In the mid 1980s, universal precautions dramatically increased the use of latex gloves, resulting in an increased demand for the liquid latex rather than solid rubber. To meet the demand for liquid latex, latex suppliers (i) tapped dormant trees, (ii) increased the use of yield stimulants, and (iii) increased the frequency of tapping (D. Shaw, personal communication). As a result of this added stress, the Hevea trees may have increased their production of defence-related proteins [41]. Increased levels of these protein allergens may have increased the allergenicity of the latex at a time when latex glove use increased in both the number of gloves worn and the duration of time for which the gloves were worn. Increased exposure to these allergens is a likely explanation for the outbreak of latex allergy.
Acknowledgments
The authors thank Lee Meyer for technical assistance and Mee-Len Chye for helpful discussions. This work was supported by research grants from Regent Hospital Products (Norcross, GA) and The Office of Naval Research Grant no. N00014-95-1-1278.
References
- 1.Sussman GL, Beezhold DH. Safe use of latex rubber. Ann Intern Med. 1995;122:43–46. doi: 10.7326/0003-4819-122-1-199501010-00007. [DOI] [PubMed] [Google Scholar]
- 2.Makinen-Kiljunen S, Turjanmaa K, Palosuo T, Reunala T. Characterization of latex antigens and allergens in surgical gloves and natural rubber by immunoelectrophoretic methods. J Allergy Clin Immunol. 1992;90:230–5. doi: 10.1016/0091-6749(92)90076-e. [DOI] [PubMed] [Google Scholar]
- 3.Slater JE, Trybul DE. Latex antigens. J Allergy Clin Immunol. 1994;93:825–30. doi: 10.1016/0091-6749(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 4.Beezhold DH, Chang N-S, Kostyal DA, Sussman G. Identification of a 46 kilodalton latex protein allergen in healthcare workers. Clin Exp Immunol. 1994;98:408–13. doi: 10.1111/j.1365-2249.1994.tb05505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco C, Carrillo T, Castillo R, Quiralte J, Cuevas M. Latex allergy: clinical features and cross-reactivity with fruits. Ann Allergy. 1994;73:309–14. [PubMed] [Google Scholar]
- 6.Beezhold DH, Sussman GL, Liss GM, Chang N-S. Latex allergy can induce clinical reactions to specific foods. Clin Exp Allergy. 1996;26:416–22. [PubMed] [Google Scholar]
- 7.Beezhold DH, Kostyal DA, Sussman GL. IgE epitope analysis of the hevein preprotein; a major latex allergen. Clin Exp Immunol. 1997;108:114–21. doi: 10.1046/j.1365-2249.1997.d01-983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baur X, Chen Z, Rozynek P, Duser M, Raulf-Heimsoth M. Cross-reacting IgE antibodies recognizing latex allergens, including Hev b 1 as well as papain. Allergy. 1995;50:604–9. doi: 10.1111/j.1398-9995.1995.tb01208.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, van Kampen V, Raulf-Heimsoth M, Baur X. Allergenic and antigenic determinants of latex allergen Hev b 1: peptide mapping of epitopes recognized by human, murine and rabbit antibodies. J Clin Exp Allergy. 1996;26:406–15. [PubMed] [Google Scholar]
- 10.Chye M-L, Cheung K-Y. β-1,3-Glucanase is highly expressed in laticifers of Hevea brasiliensis. Plant Mol Bio. 1995;29:397–402. doi: 10.1007/BF00043663. [DOI] [PubMed] [Google Scholar]
- 11.Sunderasan E, Hamzah S, Hamid S, Ward WA, Yeang HY, Cardosa MJ. Allergenic proteins of Hevea brasiliensis latex fractions. J Nat Rubb Res. 1995;10:82–99. [Google Scholar]
- 12.Yeang HY, Cheong KF, Sunderasan E, Hamzah S, Chew NP, Hamid S, Hamilton RG, Cardosa MJ. The 14.6 kd rubber elongation factor (Hev B 1) and 24 kd (Hev B 3) rubber particle proteins are recognized by IgE from patients with spina bifida and latex allergy. J Allergy Clin Immunol. 1996;98:628–39. doi: 10.1016/s0091-6749(96)70097-0. [DOI] [PubMed] [Google Scholar]
- 13.Slater J, Vedvick T, Arthur-Smith A, Trybul D, Kekwick R. Identification, cloning and sequence of a major allergen (Hev b 5) from natural rubber latex (Hevea brasiliensis) J Biol Chem. 1996;271:25394–9. doi: 10.1074/jbc.271.41.25394. [DOI] [PubMed] [Google Scholar]
- 14.Akasawa A, Hsieh L-S, Martin BM, Liu T, Lin Y. A novel acidic allergen, Hev b 5, in latex. J Biol Chem. 1996;271:25389–93. doi: 10.1074/jbc.271.41.25389. [DOI] [PubMed] [Google Scholar]
- 15.Alenius H, Kalkkinen N, Lukka M, Reunala T, Turjanmaa K, Makinen-Kiljunen S, Yip E, Palosuo T. Prohevein from the rubber tree (Hevea brasiliensis) is a major latex allergen. Clin Exp Allergy. 1995;24:659–65. doi: 10.1111/j.1365-2222.1995.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee B, Want X, Kelley K, Fink J, Sussman G, Kurup U. IgE from latex-allergic patients binds to clonal and expressed B cell epitopes of Prohevein. J Immunol. 1997;159:5724–32. [PubMed] [Google Scholar]
- 17.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 18.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–5. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott A, Martin C. A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Reporter. 1987;4:219–24. [Google Scholar]
- 20.Kush A, Goyvaerts E, Chye M-L, Chua NH. Lacticifer-specific gene expression in hevea brasiliensis (rubber tree) Proc Natl Acad Sci USA. 1990;87:1787–90. doi: 10.1073/pnas.87.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Mignery GA, Pikaard GS, Hannapei DJ, Park WD. Isolation and sequence analysis of cDNAs for the major potato protein patatin. Nucleic Acids Res. 1984;12:7987–8000. doi: 10.1093/nar/12.21.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frangioni J, Neal B. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–87. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 24.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–6. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messing J, Geraghty D, Heidecker G, Hu N-T, Kridl J, Rubenstein I. Plant gene structure. In: Kosuge T, Meredith CP, Hollender A, editors. Genetic engineering of plants: an agricultural perspective. New York: Plenum Press; 1983. [Google Scholar]
- 26.Mignery GA, Pikaard CS, Park WD. Molecular characterization of the patatin multigene family of potato. Gene. 1988;62:27–44. doi: 10.1016/0378-1119(88)90577-x. [DOI] [PubMed] [Google Scholar]
- 27.Senda K, Yoshioka H, Doke N, Kawakita K. A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiol. 1996;37:347–53. doi: 10.1093/oxfordjournals.pcp.a028952. [DOI] [PubMed] [Google Scholar]
- 28.Akasawa A, Hsieh L-S, Lin Y. Serum reactivities to latex proteins (Hevea brasiliensis) J Allergy Clin Immunol. 1995;95:1196–205. doi: 10.1016/s0091-6749(95)70076-5. [DOI] [PubMed] [Google Scholar]
- 29.Roshal S, Schmidt R, Schell J, Willmitzer L. Isolation and characterization of a gene from Solanum tuberosum encoding patatin, the major storage protein of potato tubers. Mol Gen Genet. 1986;203:2144–220. [Google Scholar]
- 30.Racusen D. Lipid acyl hydrolase of patatin. Can J Bot. 1984;62:154–64. [Google Scholar]
- 31.Bostock RM, Kuc JA, Laine RA. Eicosapentaenoic and arachidonic acids from Phytohthora infestans elicit fungitoxic sesquiterpenes in the potato. Science. 1981;212:67–69. doi: 10.1126/science.212.4490.67. [DOI] [PubMed] [Google Scholar]
- 32.Bostock RM, Sterner BA. Perspective on wound healing in resistance to pathogens. Annu Rev Phytopathol. 1989;27:343–71. [Google Scholar]
- 33.Akasawa A, Hsieh L-S, Lin Y. Comparison of latex specific IgE binding among nonammoniated latex, ammoniated latex, and latex glove allergenic extracts by ELISA and immunoblot inhibition. J Allergy Clin Immunol. 1996;97:1116–20. doi: 10.1016/s0091-6749(96)70266-x. [DOI] [PubMed] [Google Scholar]
- 34.Aamir R, Safadi GS, Melton AL, Wagner WO, Pien LC, Cornish K, Battisto JR. Shared IgE-binding sites among separated components of natural rubber latex. Allergy Immunol. 1996;111:48–54. doi: 10.1159/000237345. [DOI] [PubMed] [Google Scholar]
- 35.Tomazic VJ, Withrow TJ, Hamilton RG. Characterization of the allergen(s) in latex protein extracts. J Allergy Clin Immnunol. 1995;96:635–42. doi: 10.1016/s0091-6749(95)70262-8. [DOI] [PubMed] [Google Scholar]
- 36.Lee H-I, Broekaert WF, Raikhel NV. Co- and post translational processing of the hevein preprotein of latex of the rubber tree (Hevea brasiliensis) J Biol Chem. 1991;266:15944–8. [PubMed] [Google Scholar]
- 37.Kieliszewski MJ, Showalter AM, Leykam JF. Potato lectin: a modular protein sharing sequence similarities with the extensin family, the hevein lectin family, and snake venom disintegrins (platelet aggregation inhibitors) Plant J. 1994;5:849–61. doi: 10.1046/j.1365-313x.1994.5060849.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee H-I, Raikhel NV. Prohevein is poorly processed but shows enhanced resistance to a chitin-binding fungus in transgeneic tomato plants. Braz J Med Biol Res. 1995;28:743–50. [PubMed] [Google Scholar]
- 39.Yagami T, Sato M, Nakamura A, Shono M. One of the rubber latex allergens is a lysozyme. J Allergy Clin Immunol. 1995;96:677–86. doi: 10.1016/s0091-6749(95)70267-9. [DOI] [PubMed] [Google Scholar]
- 40.Broekaert W, Lee H-I, Chua N-H. Wound-induced accumulation of mRNA containing a hevein sequence in laticifers of rubber tree (Hevea brasiliensis) Proc Natl Acad Sci USA. 1990;87:7633–7. doi: 10.1073/pnas.87.19.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Churngchow N, Suntaro A, Wititsuwannakul R. β-1,3-glucanase isozymes from the latex of Hevea brasiliensis. Phytochem. 1995;39:505–9. [Google Scholar]


