Abstract
Objectives
To examine the profiles of K-ras mutations and p16 and preproenkephalin (ppENK) promoter hypermethylation and their associations with cigarette smoking in pancreatic cancer patients.
Methods
In plasma DNA of 83 patients with untreated primary pancreatic ductal adenocarcinoma, DNA hypermethylation was determined by methylation-specific polymerase chain reaction and K-ras codon 12 mutations by enriched-nested polymerase chain reaction followed by direct sequencing. Information on smoking exposure was collected by in-person interview. Pearson χ2 test and Fisher exact test were used in statistical analysis.
Results
K-ras mutations, ppENK, and p16 promoter hypermethylation were detected in 32.5%, 29.3%, and 24.6% of the patients, respectively. Sixty-three percent (52/83) of patients exhibited at least one of the alterations. Smoking was associated with the presence of K-ras mutations (P = 0.003). A codon 12 G-to-A mutation was predominantly observed in regular smokers and in heavy smokers (pack-year of smoking ≥36). Smoking was not associated with p16 or ppENK hypermethylation.
Conclusions
These preliminary observations suggest that plasma DNA might be a useful surrogate in detecting genetic and epigenetic alterations of pancreatic cancer. The findings on the association between K-ras mutation and smoking were in consistency with previous studies. Further studies on environmental modulators of epigenetic changes in pancreatic cancer are warranted.
Keywords: plasma DNA, K-ras mutation, DNA methylation, p16, preproenkephalin, cigarette smoking, pancreatic cancer
Adenocarcinoma of the pancreas is the most common type of pancreatic cancer and is a highly lethal disease among all types of malignancies.1 The profile of genetic and epigenetic alterations of pancreatic cancer has been unveiled by many studies.2,3 Because cigarette smoking is a well established risk factor for pancreatic cancer, elucidation of the relationship between molecular alterations and such environmental factor will not only advance our understanding on the etiology of pancreatic cancer, but will also have implications for the clinical management of this disease.
Mutation activation of the K-ras oncogene is a well established molecular alteration in the pathogenesis of pancreatic ductal adenocarcinoma.4 Aberrant hypermethylation of 5′ CpG islands of tumor suppressor genes has been identified as an alternative mechanism to mutation or deletion in gene inactivation in different types of cancers.5 Alteration of the p16 gene, which is responsible for controlling cell growth, has been well characterized in pancreatic carcinogenesis. The p16 is inactivated in approximately 95% of pancreatic adenocarcinomas and approximately 15% of those were attributable to aberrant promoter methylation.6 The p16 gene is aberrantly methylated in 27% of pancreatic cancer cell lines.7 The preproenkephalin (ppENK) gene encodes the opioid growth factor, which induces apoptosis in lung cancer cell lines, delays cell cycle progression, and exerts a negative growth regulatory effect on various kinds of cancers, including pancreatic cancer.8 The ppENK gene has been shown to be aberrantly methylated in more than 90% of pancreatic carcinomas.3
Both genetic and epigenetic changes can be modulated by environmental factors. Several studies have found that the frequency of K-ras mutations in pancreatic tumor is associated with cigarette smoking9 or alcohol consumption.10 An accumulated body of evidence has shown that smoking affects the methylation of genes involved in cancer development,11,12 but none pertains to pancreatic cancer.
There is strong evidence showing tumor cells release their DNA into circulation and that tumor-derived DNA has potential value in the early detection, monitoring, and prognosis of cancer.13-15 A significantly higher level of plasma cell-free DNA has been found in pancreatic cancer patients.16 Clinically, because of the late appearance of the disease, it is very difficult to accrue pancreatic tumor tissues because curative surgery is only feasible in a small proportion (∼20%) of patients. It would be very valuable if circulating DNA could be used as a surrogate for tumor tissues in detecting molecular markers. Detection of K-ras mutations in circulating DNA has a low sensitivity, but specificity was about 90% for the diagnosis of pancreatic cancer.17 DNA hypermethylation has been detected in the pancreatic juice of patients with pancreatic ductal adenocarcinoma.18 However, to our knowledge, there are no published reports pertaining to the detection of aberrant methylation in the plasma DNA of pancreatic cancer patients.
The major aims of the current study were to (1) determine whether hypermethylation changes can be detected in plasma DNA and to (2) examine whether selected genetic change (K-ras codon 12 mutations) and epigenetic alterations (p16 and ppENK gene hypermethylation) are affected by cigarette smoking. Their relationships to selected clinical characteristics were also investigated.
MATERIALS AND METHODS
Study Subjects and Biologic Specimens
Patients with pancreatic cancer were consecutively recruited from the Gastrointestinal Cancer Clinic at The University of Texas M. D. Anderson Cancer Center from February 2000 to January 2004. Subjects consisted of patients with pathologically confirmed primary pancreatic ductal adenocarcinoma (International Classification of Diseases for Oncology code C25.3; World Health Organization, 2000) who had not received any radiotherapy or chemotherapy before the recruitment. Untreated patients were selected because there is a concern that treatment might affect the biomarkers under investigation. All patients had no prior cancer history. There was no restriction on age, sex, or race. An informed consent was signed for the in-person interview and blood donation. The institutional review board at The University of Texas M. D. Anderson Cancer Center approved the research protocol.
In 9 of 83 patients enrolled in this study, paired pancreatic tumor tissue sections were retrieved from the Department of Pathology at The University of Texas M. D. Anderson Cancer Center. Tumor DNA derived from these sections was analyzed in parallel with plasma DNA as a standard to compare the results generated from these 2 sources.
Data Collection
A 20-minute in-person interview was conducted to elicit detailed information on the patient’s demographics, medical history (such as diabetes and pancreatitis), family history of cancer (first-degree relatives), and lifetime use of cigarette products. The data collection procedure has been described elsewhere.19 Briefly, patients who have smoked at least 100 cigarettes in their lifetime were classified as ever smokers, otherwise as never smokers. Regular smokers (RSs) were defined as patients who smoked on a regular basis. Never smokers and ever smokers who did not smoke on a regular basis were classified as nonregular smokers (NRSs). Cumulative smoking was evaluated by pack-year, which is a product of the number of packs smoked per day and the number of years of smoking. Light and heavy smokers were classified by the median pack-years of RSs. All patients completed the in-person interview. Information on clinical tumor stage, that is, localized, locally advanced, and metastatic, was obtained from patients’ medical records.
Microdissection of Tumor Cells and DNA Isolation
Two 20-μm serial sections of formalin-fixed, paraffin-embedded tissue from 9 patients were deparaffinized with xylene for 30 minutes (xylene was changed every 10 minutes) and dehydrated in series of ethanol. Slides were then stained with eosin to facilitate microdissection. Pancreatic ductal adenocarcinoma was identified under a microscope by a pathologist (J. Zhu), and the areas of interest were manually microdissected.20 An estimated 10% to 20% of the cells collected were surrounding non-ductal cells. Nontumor tissue was collected in a separate tube. Microdissected tissues were transferred to a 1.5-mL microcentrifuge tube containing 50 μL of 1 × TK buffer (0.05 mmol/L Tris-HCl, pH 8.9; 2 mmol/L EDTA; 1 mmol/L NaCl; 0.5% Tween 20; and 0.2 mg/mL proteinase K) and incubated at 56°C overnight. The tubes were placed in a 100°C block for 10 minutes to inactivate proteinase K (Roche Applied Sciences, Indianapolis, Ind). Genomic DNA was extracted using the Puregene DNA isolation kit following the protocol for DNA isolation from 5 to 10 mg paraffin-embedded tissue (Gentra, Minneapolis, Minn). DNA was quantified by ultraviolet spectrophotometry and stored at 4°C.
Plasma DNA Isolation
Plasma was isolated from freshly drawn blood (within 2 hours of blood collection) by Ficoll-Hypaque (Amersham Biosciences, Piscataway, NJ) density gradients at 2000 rpm for 25 minutes and stored at -80°C until use. Before DNA isolation, plasma was thawed at room temperature and subjected to centrifugation at 1200 rpm for 10 minutes twice. Two milliliters of supernatant was used for DNA isolation using the QIAamp DNA Midi kit (Qiagen, Valencia, Calif) according to the blood and body fluids protocol. The resulting DNA was eluted in 300 μL of sterile double distilled H2O and then concentrated to 100 μL and stored at -20°C.
Bisulfite Treatment and Methylation-Specific Polymerase Chain Reaction
In brief, 500 ng to 1 μg of DNA was first denatured by sodium hydroxide and modified with sodium bisulfite. DNA samples were then purified using Wizard DNA purification resin (Promega, Madison, Wis), treated with sodium hydroxide again, precipitated with ethanol, and resuspended in H2O.21 We used methylation-specific polymerase chain reaction (PCR) to detect the p16 and ppENK gene promoter hypermethylation following the methods described previously.21,22 The primer sequences for the p16 gene methylated reactions were as follows: 5′-TTA TTA GAG GGT GGG GCG GAT CGC-3′ (sense) and 5′-GAC CCC GAA CCG CGA CCG TAA-3′ (antisense), and for the unmethylated reaction, 5′-TTA TTA GAG GGT GGG GTG GAT TGT-3′(sense) and 5′-CAA CCC CAA ACC ACA ACC ATA A-3′(antisense). The primer sequences for the ppENK methylated reaction were as follows: 5′-TGT GGG GAG TTA TCG AGC-3′ (sense) and 5′-GCC TTC GCG AAA AAA ATC G-3′ (antisense), and for the unmethylated reaction, 5′-TTG TGT GGG GAG TTA TTG AGT-3′ (sense) and 5′-CAC CTT CAC AAA AAA AAT CAA TC-3′ (antisense). The methylation-specific PCR (MSP) amplification was carried out in a final volume of 50 μL, containing 3 μL sodium bisulfite-modified DNA, primers (300 ng each), deoxynucleotide triphosphates (dNTP, each at 2.5 mmol/L), MgCl2 (1.5 mmol/L), 2-mercaptoethanol (1 mmol/L), ammonium sulfate (16.6 mmol/L), Tris (6.7 mmol/L, pH 8.8). The reaction was subjected to hot-start at 94°C for 4 minutes before the addition of 1.25 units of Taq polymerase (PGC Scientifics, Frederick, Md). The PCR for the p16 reaction was followed by 55 cycles of 94°C for 45 seconds, the specific annealing temperature for 30 seconds, 72°C for 45 seconds, and a final extension at 72°C for 10 minutes. Annealing temperature was 65°C for the methylated reaction and 60°C for the unmethylated reaction. After hot-start, the PCR for the ppENK reaction was followed by 45 cycles of 95°C for 20 seconds, 62°C for 30 seconds (annealing temperature was the same for the methylated and unmethylated reaction), 72°C for 30 seconds, and a final extension at 72°C for 3 minutes. All PCR reaction was performed using a MJ thermalcycler (MJ Research, Inc, Waltham, Mass). Ten microliters of each PCR product was loaded directly onto nondenaturing 6% polyacrylamide gels, stained with ethidium bromide, and visualized under an ultraviolet illuminator (Fig. 1). The sample was termed methylated if an MSP yielded a visible methylated band. The sample was termed unmethylated if no visible methylated band was present. Negative controls (no DNA loaded), unmethylated allele (human normal lymphocyte DNA) and methylated alleles (CpG genome universal methylated DNA, Chemicon International, Temecular, Calif) were included in each reaction. The unmethylated band was always present, which indicates the presence of normal DNA in plasma. The PCR reaction was conducted at least twice on each modified DNA template to determine the reproducibility of the assay. When inconsistency occurred (ie, methylated band was not consistently visible in duplicates), the assay was repeated starting from the bisulfite treatment step. Two concordant results of 3 measurements were accepted for demonstrating reproducibility. The sensitivity of MSP for p16 methylation was determined using CpG genome universal methylated DNA that was serially diluted in water, mixed with normal lymphocyte DNA, then bisulfite converted and amplified by MSP. The lowest detection limit was 20 ng of universal methylated DNA in 1000 ng lymphocyte DNA, that is, 1 methylated genome in 500 unmethylated genomes (data not shown).
FIGURE 1.

Examples of aberrant methylation of p16 and ppENK genes in plasma DNA of pancreatic cancer patients. m indicates methylated; MC, methylated allele control; NC, negative control, distilled water; um, unmethylated; UMC, unmethylated allele control.
Analysis of K-ras Codon 12 Mutations
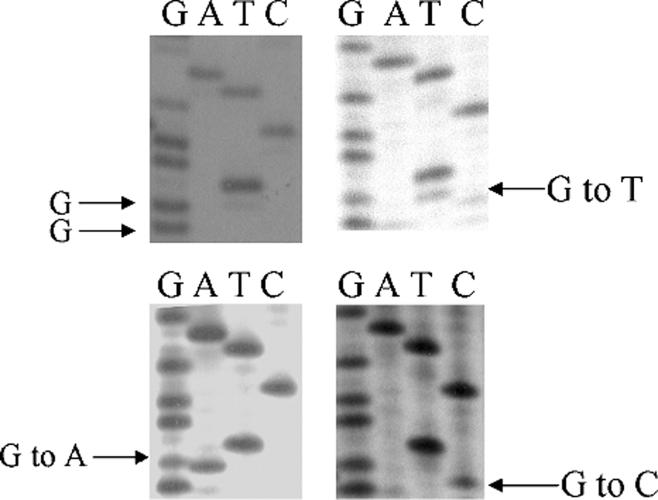
K-ras codon 12 mutations were detected by enrichednested PCR-restriction fragment length polymorphism and direct DNA sequencing analysis as previously described with little modification.23 Two sets of primers were used for the first PCR and the second PCR amplification. The primer sequence for the first PCR were: 5′-ACT GAA TAT AAA CTT GTG GTA GTT GGA CCT-3′ (sense) and 5′-TCA AAG AAT GGT CCT GGA CC-3′ (antisense). For the second PCR, the sequence of sense primer was the same as the first PCR and the sequence for the antisense primer was 5′-CTC TAT TGT TGG ATC ATA TT-3′. The first PCR was performed in a total volume of 50 μL containing: 39.75 μL of plasma DNA, 0.1 μmol/L of each primer, 200 μmol/L of each dNTP, 1.25 units of DNA polymerase (PGC Scientifics, Frederick, Md), 1 × PCR buffer with 1.5 mmol/L of MgCl2. The PCR conditions consisted of 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. The product was digested with 20 units of BstNI (New England Biolab, Beverly, Mass) under 65°C for 7 hours. The product was resolved on 3% agarose gel. The second PCR reaction was performed in a total volume of 50 μL containing 1 μL of first PCR reaction product, 0.1 μmol/L of each primer, 200 Kmol/L of each dNTP, 1.25 units of DNA polymerase, and 1 × PCR ×buffer with 1.5 mmol/L of MgCl2. The PCR condition consists of 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. The PCR product was digested with 20 units of BstNI under 65°C for 7 hours. The second PCR product (enriched PCR product) was subjected to direct sequencing after being excised from 2% 3:1 Nusieve agarose gel (Cambrex, Rockland, Me) using the Qiaquick Gel Extraction Kit following the manufacture’s instructions (Qiagen, Valencia, Calif). The excised PCR product was sequenced using Thermo Sequenase Radiolabeled Terminator Cycle Sequencing Kit (USB, Cleveland, Ohio).33 P-labeled terminators were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). Before sequencing, the reaction product was subjected to denaturing at 72°C for 7 minutes. After electrophoresis, the gel was dried and exposed to the film for 24 hours. The sensitivity of detection was 1 mutant allele in 100 normal alleles.23 The analysis was randomly duplicated in 5% of samples, beginning from the PCR step, depending on the availability of DNA. The samples that harbored mutations were verified by duplicate direct sequencing (Fig. 2).
FIGURE 2.
Autoradiograph shows K-ras codon 12 mutations as detected by direct sequencing. Because plasma DNA contains normal DNA, the wild-type allele is present.
Statistical Analysis
STATA intercooled 9.0 software (Stata Corporation, College Station, Tex) was used for data analysis. Two-tailed P ≤ 0.05 were considered statistically significant. Pearson χ2 test was used to evaluate the association between each marker and other variables. Fisher exact test was used when the number in any cell was less than 5. K-ras codon 12 status was categorized as 0 (wild type), 1 (G-to-A mutation), 2 (G-to-T mutation), and 3 (G-to-C mutation). Mutants were defined by combing 3 types of mutations together. Some biomarker assays were unsuccessful because of inadequate quality or quantity of plasma DNA. The missing data rate was 7.2% (n = 6), 9.6% (n = 8), and 31.3% (n = 26) for K-ras mutation, ppENK, and p16 methylation, respectively. The missing data rate was high for the p16 gene because it was the last marker to be evaluated, at which point the DNA had been exhausted for some samples. Forty-three patients (51.8%) had a complete set of all 3 biomarkers informative.
RESULTS
Table 1 shows the major characteristics of the 83 patients. The median age was 61 years (range, 35-79 years). Non-Hispanic whites comprised 85.5% of the patients. A total of 61.4% of the patients reported having a family history of cancer among their first-degree relatives; 25.3% and 10.8% of the patients, respectively, reported having a history of type II diabetes or pancreatitis; 19.3%, 44.6%, and 36.1% of the patients had localized, locally advanced, and metastatic disease, respectively. The median value of DNA yield was 250 ng/mL (range, 28.5-857.75 ng/mL) plasma. The DNA yield did not differ significantly by tumor stage. Hypermethylation of the p16 and ppENK genes was detected in 60% (3/5) and 80% (4/5) of the plasma samples taken from patients whose tumors harbored the same methylation, respectively. No methylated band was present in the plasma sample of the patients whose corresponding tumor DNA had no methylation in the p16 and ppENK promoter, which suggested that the specificity of methylation detection using plasma DNA was 100%.
TABLE 1.
Characteristics of Patients and Frequency of Alterations
| Frequency |
|||||||
|---|---|---|---|---|---|---|---|
| Patients Characteristics (N = 83) | K-ras* Mutations 25/77 (32.5) | P† | ppENK Methylation 22/75 (29.3) | P† | p16 Methylation 14/57 (24.6) | P† | |
| Age at diagnosis (yrs) | |||||||
| <54 | 20 (24.1) | 8/19 (42.1) | 0.79 | 6/20 (30.0) | 0.70 | 4/12 (33.3) | 0.20 |
| 54-61 | 20 (24.1) | 7/20 (35.0) | 6/16 (37.5) | 5/12 (41.7) | |||
| 61-69 | 23 (27.7) | 6/19 (31.6) | 4/20 (20.0) | 3/19 (15.8) | |||
| <69 | 20 (24.1) | 4/19 (21.0) | 6/19 (31.6) | 2/14 (14.3) | |||
| Sex | |||||||
| Women | 33 (39.8) | 12/31 (38.7) | 0.46 | 9/30 (30.0) | 1.00 | 5/21 (23.8) | 0.92 |
| Men | 50 (60.2) | 13/46 (28.3) | 13/45 (28.9) | 9/36 (25.0) | |||
| Race | |||||||
| Non-Hispanic white | 71 (85.5) | 22/65 (33.9) | 0.03 | 19/64 (29.7) | 0.42 | 11/49 (22.4) | 0.17 |
| Hispanic | 8 (9.6) | 0/8 (0.0) | 2/8 (25.0) | 3/5 (60.0) | |||
| African Americans | 4 (4.9) | 3/4 (75.0) | 1/3 (33.3) | 0/3 (0.0) | |||
| Family history of cancer in first-degree relatives | |||||||
| No | 32 (38.6) | 8/30 (26.7) | 0.39 | 7/28 (25.0) | 0.61 | 7/21 (33.3) | 0.24 |
| Yes | 51 (61.4) | 17/47 (36.2) | 15/47 (31.9) | 7/36 (19.4) | |||
| Type II diabetes | |||||||
| No | 62 (74.7) | 19/58 (32.7) | 1.00 | 16/56 (28.6) | 0.78 | 12/45 (26.7) | 0.48 |
| Yes | 21 (25.3) | 6/19 (31.6) | 6/19 (31.6) | 92/12 (16.7) | |||
| History of pancreatitis | |||||||
| No | 74 (89.2) | 23/68 (33.8) | 0.46 | 20/67 (29.8) | 0.70 | 12/50 (24.0) | 1.00 |
| Yes | 9 (10.8) | 4/9 (44.4) | 2/8 (25.0) | 2/7 (28.6) | |||
| Tumor stage | |||||||
| Localized | 16 (19.3) | 3/15 (20.0) | 0.33 | 3/14 (21.4) | 0.48 | 5/11 (45.4) | 0.12 |
| Locally advanced | 37 (44.6) | 11/36 (30.6) | 12/33 (36.4) | 6/23 (26.1) | |||
| Metastatic | 30 (36.1) | 11/26 (42.3) | 7/28 (25.0) | 3/23 (13.0) | |||
Values are expressed as n (%).
Includes all types of mutations.
Pvalue for χ2test or Fisher exact test.
Table 1 shows the frequency of 3 alterations in patients by different characteristics. The frequency of K-ras mutations and ppENK and p16 hypermethylation was 32.5%, 29.3%, and 24.6%, respectively. A total of 62.6% (52/83) of patients expressed at least one of these alterations. None of the patients harbored all 3 alterations. There was no significant difference in the frequency of these alterations by age, sex, family history of cancer, history of pancreatitis or type II diabetes, and tumor stage. The prevalence of p16 gene hypermethylation was not significantly different between patients who were younger than 61 years and those older than 61 years (P = 0.21, Fisher exact test). However, the frequency of K-ras mutation differed by race. None of the Hispanic patients and 3 of 4 African American patients carried the codon 12 mutations (P = 0.03). K-ras mutations were more frequently detected among patients with metastatic tumor than among patients with localized tumor (42.3% vs 20.0%). The occurrence of p16 hypermethylation was more frequently found among patients with localized tumors than those with metastatic tumors (45.4% vs 13.0%). However, these differences were not statistically significant (Table 1).
The distribution of age, sex, race, and tumor stage did not differ significantly between RSs and NRSs (data not shown). Table 2 presents the frequency of alterations by smoking status. Three types of K-ras mutations were detected in 25 of 77 patients, including 18 G(G)T to G(A)T mutation (Gly to Asp), 5 G(G)T to G(T)T mutation (Gly to Val), and 2 (G)GT to (C)GT mutation (Gly to Arg), which accounted for 72.0%, 20.0%, and 8.0% of all mutations, respectively. The K-ras mutation spectrum was significantly different between the RSs and the NRSs (P = 0.003, Fisher exact test). The frequency of the G(G)T to G(A)T transition mutation was 31.9% in the RSs and 10.0% in the NRSs, whereas the G(G)T to G(T)T transversion mutation was exclusively detected in the NRSs. The K-ras mutation spectrum was also associated with pack-years of smoking. The G(G)T to G(A)T mutation was detected in 10.0%, 27.3%, and 36.0% of the NRSs, the RSs who had less than 36 or equal to or greater than 36 pack-years of smoking, respectively (P = 0.03, Fisher exact test). The frequency of ppENK and p16 hypermethylation was comparable between the RSs and the NRSs and also did not differ by pack-years of smoking (all P ≥ 0.5). There was no significant association between these alterations and current smoking status (never, former, and current; data not shown).
TABLE 2.
Frequency of Alteration in Plasma DNA of Pancreatic Cancer Patients by Smoking Status
| Pack-Yrs‡ |
||||||||
|---|---|---|---|---|---|---|---|---|
| All* (N = 83) | NRSs (n = 33) | RSs (n = 50) | 0 |
1-36 |
≥36 |
|||
| Biomarkers | P† | (n = 33) | (n = 24) | (n = 26) | P† | |||
| K-ras codon 12 | ||||||||
| Wild type | 52 (67.5) | 21 (70.0) | 31 (66.0) | 0.003 | 21 (70.0) | 15 (68.1) | 16 (64.0) | 0.03 |
| G-to-A mutation | 18 (23.4) | 3 (10.0) | 15 (31.9) | 3 (10.0) | 6 (27.3) | 9 (36.0) | ||
| G-to-T mutation | 5 (6.5) | 5 (16.7) | 0 (0.0) | 5 (16.7) | 0 (0.0) | 0 (0.0) | ||
| G-to-C mutation | 2 (2.6) | 1 (3.3) | 1 (2.1) | 1 (3.3) | 1 (4.6) | 0 (0.0) | ||
| p16 | ||||||||
| Unmethylated | 43 (75.4) | 15 (75.0) | 28 (75.7) | 0.60 | 15 (75.0) | 11 (68.8) | 17 (80.9) | 0.80 |
| Methylated | 14 (24.6) | 5 (25.0) | 9 (24.3) | 5 (25.0) | 5 (31.2) | 4 (19.1) | ||
| ppENK | ||||||||
| Unmethylated | 53 (70.7) | 20 (69.0) | 33 (71.7) | 0.50 | 20 (69.0) | 16 (72.7) | 17 (70.8) | 0.96 |
| Methylated | 22 (29.3) | 9 (31.0) | 13 (28.3) | 9 (31.0) | 6 (27.3) | 7 (29.2) | ||
Values are expressed as n (%).
The numbers do not add up because of missing data.
P value for Pearson χ2test or Fisher exact test.
Pack-year was categorized by the median of RSs.
DISCUSSION
In the current study, we have shown that gene promoter hypermethylation can be detected in the plasma DNA of patients with pancreatic cancer. We confirmed that K-ras codon 12 mutations were associated with cigarette smoking in patients. However, we failed to show any association between smoking and p16 or ppENK hypermethylation. These observations support a role of cigarette smoking-induced gene mutation in pancreatic carcinogenesis and that plasma DNA might have a value as a surrogate for tumor tissues in detecting genetic and epigenetic alterations in pancreatic cancer.
The frequency of K-ras mutations tended to be higher in the patients who smoked regularly than the patients who did not. A series of studies have shown that K-ras mutation detected in paraffin-embedded pancreatic tumor tissue can be modulated by environmental factors,24 including coffee drinking,25 organochlorine compounds,26 occupational exposure to hydrocarbon solvents,27 and alcohol consumption.10 These studies indicated that environmental factors could play a role in pancreatic carcinogenesis through modulation of K-ras oncogene activation. In particular, cigarette smoking, the only established environmental risk factor of pancreatic cancer, has been linked to the presence of K-ras mutation in 89 tumor tissues9 and in nonneoplastic exocrine pancreatic lesions of 39 cancer-free male heavy smokers.28 Some studies, however, did not show that K-ras mutation rate differed in ever smokers and never smokers.29 In the current study, we also found the association between smoking status and K-ras mutation spectrum. The G-to-A transition mutation at codon 12 was predominantly detected in the RSs. This observation supports the etiologic role of the N-nitroso compounds in smoking-related pancreatic cancer as suggested by animal studies.30,31 Aromatic amine and alkylating agents can activate K-ras through G-to-A transition.32,33 Interestingly, patients who had high cumulative exposure to smoking tended to have a higher frequency of G-to-A mutations than those who had lower exposure. However, this observation was based on a small sample size and needs to be confirmed.
The current study failed to show that cigarette smoking was associated with either p16 or ppENK hypermethylation. In contrast, both experimental studies34-36 and clinical studies using human tissue sample37-40 have shown significant associations between carcinogen exposure, including tobacco compounds and their derivatives, and gene promoter methylation in cancer patients. In 1995, Costa41 proposed a model for the epigenetic mechanism of nickel compounds. Based on the model, nickel compounds induce an increase in chromatin condensation, causing neighboring genes (such as tumor suppressor genes) that are actively expressed in euchromatin to be condensed into heterochromatin. The involvement of DNA cytosine methyltransferase causes subsequent gene promoter hypermethylation, which eventually silences the genetic activity that might be essential for maintenance of genomic stability.41 Given existing evidence, it is still important to investigate the effect of environmental exposure on gene promoter hypermethylation in pancreatic cancer. The lack of association in our study may be explained by non-differential misclassification of methylated or unmethylated alleles in the RSs and the NRSs. Because plasma DNA is a mixture of neoplastic and nonneoplastic DNA, MSP might not be sensitive enough to pick up the methylated allele when copy number is low. In addition, the bisulfite modification can result in DNA breakage and thus lowering the sensitivity of MSP relative to ordinary PCR.18 However, there is no reason to believe that the false-negative results were related to the smoking status of the patients because the exposure status was unknown to the laboratory personnel doing the assay. Such non-differential misclassification would bias the association toward null. Alternatively, our results suggest that mechanisms other than smoking, such as intrinsic tumor microenvironment and dietary factors, may play a role in determining the profile of gene promoter hypermethylation.
Determining whether there is an association between the tested markers and clinical outcome was not the major goal of the current study because we had a limited number of patients who had localized tumors. Nevertheless, our study showed that K-ras mutations were more frequently detected in patients with late-stage disease than in patients with localized tumors. Several previous studies also found that the occurrence of K-ras mutation in circulating DNA was correlated with late tumor stage42 and large tumor size.43 Patients with metastatic disease may have tumors with a larger volume, and therefore, more abundant tumor DNA in circulation. Promoter hypermethylation of the p16 and ppENK genes seems to be more frequently detected in patients with localized tumors than those with more advanced tumors. This is consistent with previous findings that p16 alteration is an early event in pancreatic carcinogenesis.44,45 Indeed, both p16 and ppENK hypermethylation have been detected in precancerous lesions in the pancreas.22 The prognostic value of promoter hypermethylation of tumor suppressor gene such as p16 in pancreatic cancer is not clear and needs further investigation.
Early diagnosis is critical to improve the survival of patients. Because both K-ras mutations and gene methylation occur in the early stage of carcinogenesis, detection of these alterations in the plasma/serum of cancer patients may serve as an attractive noninvasive tool for early detection.46 However, K-ras mutation in serum alone is not a satisfactory marker in early detection of pancreatic cancer.47 Recently, Marchase et al48 concluded that K-ras mutation is not useful in the early detection of pancreatic cancer because K-ras abnormality could not be identified in serum-derived DNA of patients whose tumors harbored the abnormalities. This observation was in contrast with many other previous studies42,43,49 that detected K-ras mutation in circulating DNA with varied detection rates. The discrepancy may be caused by differences in study sample (plasma or serum), sample storage and process, sensitivity of the mutation detection assay, and patient group. Generally, plasma DNA is considered to be more representative of tumor status and is enriched for tumor DNA than serum DNA.49-51 In the current study, the sensitivity for any one of the markers is low, with frequencies ranging between 24% and 32%. When combining the 3 markers, the detection rate increased to 63%. In a subset of 43 patients who had all 3 biomarkers measured, 67% (29/43) of the patients expressed at least 1 alteration. Even in patients with localized tumors, 56% expressed at least 1 alteration. Therefore, combination of biomarkers representing different biologic pathways may be valuable in early detection of pancreatic cancer, thus warrants further investigation. Apparently, it is important to evaluate these markers in age-matched cancer-free controls or patients with chronic pancreatitis as well. In particular, p16 promoter hypermethylation has been detected in normal human mammary epithelia.52
There are some limitations in this study. First, the use of plasma DNA has its own limitations. Although much attention has been focused on circulating DNA, its origin and mechanisms of release and clearance are unclear, and we have no solid evidence to show that plasma DNA comes from pancreatic tumors. Second, although comparison of p16 methylation profiles in paired tumor and plasma DNA from 9 patients showed 70% sensitivity and 100% specificity in detecting methylated alleles using plasma DNA, we were unable to either retrieve more tumor samples or to include patients without cancer to validate these findings. Thus, the true sensitivity and specificity of the assay need to be further determined in future studies. Third, low sensitivity could be one of the reasons that we did not observe any association between methylation and smoking. Furthermore, missing data and small numbers in certain analyses may all contribute to the lack of association between smoking and methylation patterns. On the other hand, some significant observations may be caused by random bias. Nevertheless, the frequency of K-ras mutations in the plasma DNA of the current study (32.5%) is within the range reported in other studies using plasma DNA of patients with pancreatic cancer (25%-35%).49,53 More advanced techniques and the enrichment of tumor-derived DNA may reduce the likelihood of both false-negative and false-positive results.54 Recently, a method has been developed to detect mutant templates in a ratio of 1:10,000 wild-type alleles.55 Such highly sensitive techniques, together with the application of multiple markers, may hold the hope of early detection of pancreatic cancer using circulating DNA.
In summary, we detected K-ras codon 12 mutations and aberrant hypermethylation using plasma DNA in a series of pancreatic cancer patients. We confirmed the previous observation showing that cigarette smoking was significantly associated with K-ras mutation. In addition, we found that G(G)T to G(A)T transition mutation is more frequently seen in the RSs and those with a higher level of cigarette exposure than in the NRSs. This finding agrees with animal studies that suggest that the N-nitroso compounds in cigarette smoke can induce specific activating mutation in the K-ras gene and play a role in human pancreatic carcinogenesis. On the other hand, we failed to show any association between cigarette smoking and ppENK and p16 gene methylation. Our observation supports the feasibility of using plasma DNA as a surrogate to detect molecular alterations in pancreatic cancer patients. Future research is needed to demonstrate the interaction between environmental modulators and epigenetic events.
ACKNOWLEDGMENTS
The authors thank Rabia Khan, Kaustubh Mestry, Ajay Nooka, and Hui Liu for their contributions to patient recruitment and data collection. The authors also thank all physicians and clinical staff at the Gastrointestinal Cancer Clinic at The University of Texas M. D. Anderson Cancer Center who helped with patient recruitment. We appreciated the laboratory support provided by Ping Chang, Yanan Li, and Yingqiu Du. The authors thank Dr JeanPierre Issa (Department of Leukemia of this institute) for his comments.
Footnotes
This study was supported by the National Institutes of Health grant CA98380, National Institute of Environmental Health Sciences grant P30 ES07784, National Institutes of Health Cancer Center Support (Core) grant CA16672, and a grant from The Lockton Pancreatic Cancer Research Funds.
REFERENCES
- 1.ACS . Cancer Facts and Figures: 2006. American Cancer Society; Atlanta, GA: 2006. [Google Scholar]
- 2.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 3.Ueki T, Toyota M, Skinner H, et al. Identification and characterization of differentially methylated CpG islands in pancreatic carcinoma. Cancer Res. 2001;61:8540–8546. [PubMed] [Google Scholar]
- 4.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 5.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 6.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 7.Moore PS, Sipos B, Orlandini S, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 8.Zagon IS, Roesener CD, Verderame MF, et al. Opioid growth factor regulates the cell cycle of human neoplasias. Int J Oncol. 2000;17:1053–1061. doi: 10.3892/ijo.17.5.1053. [DOI] [PubMed] [Google Scholar]
- 9.Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 10.Malats N, Porta M, Corominas JM, et al. Ki-ras mutations in exocrine pancreatic cancer: association with clinico-pathological characteristics and with tobacco and alcohol consumption. PANK-ras I Project Investigators. Int J Cancer. 1997;70:661–667. doi: 10.1002/(sici)1097-0215(19970317)70:6<661::aid-ijc6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 11.Marsit CJ, Karagas MR, Danaee H, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 12.Toyooka S, Suzuki M, Tsuda T, et al. Dose effect of smoking on aberrant methylation in non-small cell lung cancers. Int J Cancer. 2004;110:462–464. doi: 10.1002/ijc.20125. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 14.Anker P, Mulcahy H, Chen XQ, et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh CJ, Klump B, Holzmann K, et al. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998;58:3942–3945. [PubMed] [Google Scholar]
- 16.Giacona MB, Ruben GC, Iczkowski KA, et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 17.Maire F, Micard S, Hammel P, et al. Differential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNA. Br J Cancer. 2002;87:551–554. doi: 10.1038/sj.bjc.6600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukushima N, Walter KM, Uek T, et al. Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice. Cancer Biol Ther. 2003;2:78–83. doi: 10.4161/cbt.183. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Jiao L, Li Y, et al. Polymorphisms of cytochrome P4501A2 and N-acetyltransferase genes, smoking, and risk of pancreatic cancer. Carcinogenesis. 2006;27:103–111. doi: 10.1093/carcin/bgi171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coombs NJ, Gough AC, Primrose JN. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res. 1999;27:e12. doi: 10.1093/nar/27.16.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukushima N, Sato N, Ueki T, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee SK, Makdisi WF, Weston AP, et al. A two-step enriched-nested PCR technique enhances sensitivity for detection of codon 12 K-ras mutations in pancreatic adenocarcinoma. Pancreas. 1997;15:16–24. doi: 10.1097/00006676-199707000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Porta M, Ayude D, Alguacil J, et al. Exploring environmental causes of altered ras effects: fragmentation plus integration. Mol Carcinog. 2003;36:45–52. doi: 10.1002/mc.10093. [DOI] [PubMed] [Google Scholar]
- 25.Porta M, Malats N, Guarner L, et al. Association between coffee drinking and K-ras mutations in exocrine pancreatic cancer. PANKRAS II Study Group. J Epidemiol Community Health. 1999;53:702–709. doi: 10.1136/jech.53.11.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porta M, Malats N, Jariod M, et al. Serum concentrations of organochlorine compounds and K-ras mutations in exocrine pancreatic cancer. PANKRAS II Study Group. Lancet. 1999;354:2125–2129. doi: 10.1016/s0140-6736(99)04232-4. [DOI] [PubMed] [Google Scholar]
- 27.Alguacil J, Porta M, Malats N, et al. Occupational exposure to organic solvents and K-ras mutations in exocrine pancreatic cancer. Carcinogenesis. 2002;23:101–106. doi: 10.1093/carcin/23.1.101. [DOI] [PubMed] [Google Scholar]
- 28.Berger DH, Chang H, Wood M, et al. Mutational activation of K-ras in nonneoplastic exocrine pancreatic lesions in relation to cigarette smoking status. Cancer. 1999;85:326–332. doi: 10.1002/(sici)1097-0142(19990115)85:2<326::aid-cncr9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 29.Nagata Y, Abe M, Motoshima K, et al. Frequent glycine-to-aspartic acid mutations at codon 12 of c-Ki-ras gene in human pancreatic cancer in Japanese. Jpn J Cancer Res. 1990;81:135–140. doi: 10.1111/j.1349-7006.1990.tb02539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujii H, Egami H, Chaney W, et al. Pancreatic ductal adenocarcinomas induced in Syrian hamsters by N-nitrosobis(2-oxopropyl)amine contain a c-Ki-ras oncogene with a point-mutated codon 12. Mol Carcinog. 1990;3:296–301. doi: 10.1002/mc.2940030510. [DOI] [PubMed] [Google Scholar]
- 31.Engelbergs J, Thomale J, Rajewsky MF. Role of DNA repair in carcinogen-induced ras mutation. Mutat Res. 2000;450:139–153. doi: 10.1016/s0027-5107(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 32.Anderson K, Potter J, Mack T. Pancreatic cancer. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. Oxford University Press; New York, NY: 1996. pp. 725–771. [Google Scholar]
- 33.Li D, Jiao L. Molecular epidemiology of pancreatic cancer. Int J Gastrointest Cancer. 2003;33:3–14. doi: 10.1385/IJGC:33:1:3. [DOI] [PubMed] [Google Scholar]
- 34.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56:3655–3658. [PubMed] [Google Scholar]
- 35.Bai H, Gu L, Zhou J, et al. p16 hypermethylation during gastric carcinogenesis of Wistar rats by N-methyl-N′-nitro-N-nitrosoguanidine. Mutat Res. 2003;535:73–78. doi: 10.1016/s1383-5718(02)00288-7. [DOI] [PubMed] [Google Scholar]
- 36.Pulling LC, Vuillemenot BR, Hutt JA, et al. Aberrant promoter hypermethylation of the death-associated protein kinase gene is early and frequent in murine lung tumors induced by cigarette smoke and tobacco carcinogens. Cancer Res. 2004;64:3844–3848. doi: 10.1158/0008-5472.CAN-03-2119. [DOI] [PubMed] [Google Scholar]
- 37.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 38.Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 39.Eguchi K, Kanai Y, Kobayashi K, et al. DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res. 1997;57:4913–4915. [PubMed] [Google Scholar]
- 40.Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–3424. [PubMed] [Google Scholar]
- 41.Costa M. Model for the epigenetic mechanism of action of nongenotoxic carcinogens. Am J Clin Nutr. 1995;61:666S–669S. doi: 10.1093/ajcn/61.3.666S. [DOI] [PubMed] [Google Scholar]
- 42.Castells A, Puig P, Mora J, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17:578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 43.Mulcahy HE, Lyautey J, Lederrey C, et al. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res. 1998;4:271–275. [PubMed] [Google Scholar]
- 44.Hustinx SR, Leoni LM, Yeo CJ, et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol. 2005;18:959–963. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 45.Rosty C, Geradts J, Sato N, et al. p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am J Surg Pathol. 2003;27:1495–1501. doi: 10.1097/00000478-200312000-00001. [DOI] [PubMed] [Google Scholar]
- 46.Baylin SB, Belinsky SA, Herman JG. Aberrant methylation of gene promoters in cancer-concepts, misconcepts, and promise. J Natl Cancer Inst. 2000;92:1460–1461. doi: 10.1093/jnci/92.18.1460. [DOI] [PubMed] [Google Scholar]
- 47.Goggins M. Molecular markers of early pancreatic cancer. J Clin Oncol. 2005;23:4524–4531. doi: 10.1200/JCO.2005.19.711. [DOI] [PubMed] [Google Scholar]
- 48.Marchese R, Muleti A, Pasqualetti P, et al. Low correspondence between K-ras mutations in pancreatic cancer tissue and detection of K-ras mutations in circulating DNA. Pancreas. 2006;32:171–177. doi: 10.1097/01.mpa.0000202938.63084.e3. [DOI] [PubMed] [Google Scholar]
- 49.Sorenson GD. Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin Cancer Res. 2000;6:2129–2137. [PubMed] [Google Scholar]
- 50.Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004;22:4157–4164. doi: 10.1200/JCO.2004.11.123. [DOI] [PubMed] [Google Scholar]
- 51.Lee TH, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41:276–282. doi: 10.1046/j.1537-2995.2001.41020276.x. [DOI] [PubMed] [Google Scholar]
- 52.Holst CR, Nuovo GJ, Esteller M, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 53.Uemura T, Hibi K, Kaneko T, et al. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J Gastroenterol. 2004;39:56–60. doi: 10.1007/s00535-003-1245-1. [DOI] [PubMed] [Google Scholar]
- 54.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 55.Luo JD, Chan EC, Shih CL, et al. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res. 2006;34:e12. doi: 10.1093/nar/gnj008. [DOI] [PMC free article] [PubMed] [Google Scholar]