Abstract
The leucine-rich acidic nuclear protein (LANP) belongs to the INHAT family of corepressors that inhibits histone acetyltransferases. The mechanism by which LANP restricts its repression to specific genes is unknown. Here, we report that LANP forms a complex with transcriptional repressor E4F and modulates its activity. As LANP interacts with ataxin 1—a protein mutated in the neurodegenerative disease spinocerebellar ataxia type 1 (SCA1)—we tested whether ataxin 1 can alter the E4F–LANP interaction. We show that ataxin 1 relieves the transcriptional repression induced by the LANP–E4F complex by competing with E4F for LANP. These results provide the first functional link, to our knowledge, between LANP and ataxin 1, and indicate a potential mechanism for the transcriptional aberrations observed in SCA1.
Keywords: SCA1, ataxin 1, LANP, E4F, INHAT
Introduction
The leucine-rich acidic nuclear protein (LANP; also known as ANP32a and pp32) is a nucleoplasmic shuttling protein involved in several processes such as regulation of gene expression, RNA transport, apoptosis, intracellular signalling and cytoskeletal dynamics (reviewed by Matilla & Radrizzani, 2005). In the nucleus, the best-characterized feature of LANP is its ability to bind to amino-terminal histone tails (Seo et al, 2001; Schneider et al, 2004). Therefore, LANP masks histones from histone acetyltransferases (HATs), inhibits histone acetylation and represses transcription (Kutney et al, 2004). As transcription itself is highly orchestrated, we reasoned that transcriptional modulators such as LANP must also be regulated—particularly, with regard to targeting appropriate genes. Hence, we sought to identify factors that recruit LANP to specific promoters. Here, we report one such factor, the transcriptional repressor p120E4F (henceforth called E4F), a ubiquitously expressed member of the Gli–Kruppel family (Raychaudhuri et al, 1987). Specifically, we show that LANP interacts with E4F, potentiates E4F-induced repression and is required for its full inhibitory capacity.
Finally, we tested the implications of our findings in the context of the neurodegenerative disorder spinocerebellar ataxia type 1 (SCA1). SCA1 is caused by a glutamine repeat expansion in the protein ataxin 1, an expansion that increases its tendency to misfold and accumulate, eventually causing toxicity by a gain-of-function mechanism (Opal & Zoghbi, 2002). Ataxin 1 binds to LANP (Matilla et al, 1997); therefore we postulated that the accumulated ataxin 1 modulates the repression of LANP target genes. This idea is particularly attractive, given that transcriptional misregulation is the earliest pathological change identified in SCA1 mice (Lin et al, 2000; Serra et al, 2004). We show that expanded ataxin 1 relieves E4F-mediated transcriptional repression, providing the first functional link, to our knowledge, between LANP and ataxin 1.
Results And Discussion
Identification of E4F as an LANP interactor
To understand the targeting of LANP to specific genes, we carried out a yeast two-hybrid screen to identify LANP interactors. As described previously (Opal et al, 2003), we identified the microtubule-associated protein MAP1B as an LANP-binding protein—this is consistent with the role of LANP as a modulator of microtubule function (Ulitzur et al, 1997). In addition, we identified the transcriptional repressor E4F (NM007892; Raychaudhuri et al, 1987; Fernandes & Rooney, 1997) and a few other proteins that are important in transcriptional repression: histone deacetylase 2 (HDAC2), Sin3-associated polypeptide 30 kDa (SAP30) and SET-binding protein 1 (SEB1). We focused on the LANP–E4F interaction because it provides a mechanism by which LANP can be recruited to the promoters of specific genes.
We confirmed the E4F–LANP interaction by showing that E4F does not interact with nonspecific baits (data not shown). Next, by using truncation mutants, we delimited the interacting regions of LANP and E4F (Fig 1). A minor deletion in the carboxyl terminus of E4F (E4F1–614) does not abrogate the interaction. However, further deletion of the C-terminal domain (E4F1–358), which truncates the last four zinc-finger motifs, abolishes LANP binding. We also found that removing the major portions of the two N-terminal zinc-finger domains (E4F 188-C) did not affect LANP–E4F binding. It is not clear why deleting some domains of E4F makes the interaction more robust than that of full-length E4F; however one possibility is that these domains provide steric hindrance to binding.
Figure 1.
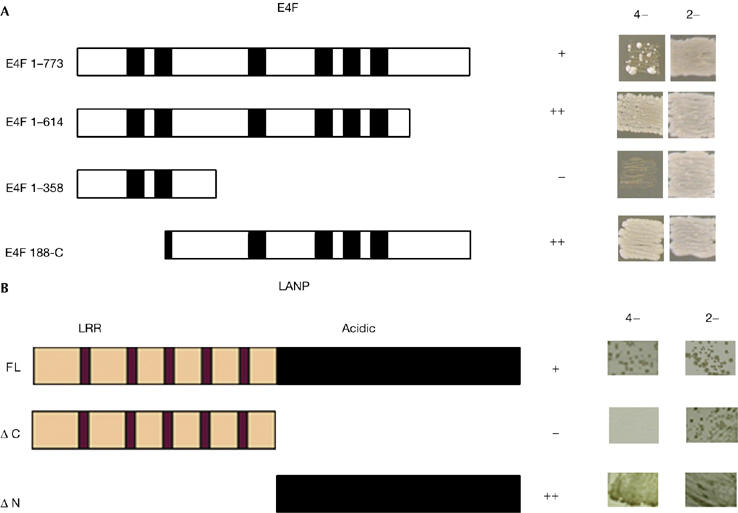
Delimiting the interaction between E4F and LANP. (A) Interaction of full-length bait LANP with prey E4F constructs (full-length E4F (E4F 1–773) or deletion constructs as indicated, with zinc-finger motifs shown as black boxes). (B) Full-length (FL), amino-terminal (ΔN) and carboxy-terminal (ΔC) terminal deletion constructs of LANP were tested as interacting baits against full-length E4F as prey. Interaction was scored on the basis of relative growth on auxotrophic (4−) selection media (−Trp/−Leu/−His/−Ade); the presence of both bait and prey plasmids is recorded by growth on nonselective (2−) media (−Trp/−Leu). LANP, leucine-rich acidic nuclear protein; LRR, leucine-rich-repeat domain.
With respect to LANP, it is the C-terminal acidic domain of LANP that is necessary and sufficient for E4F binding (Fig 1B). Incidentally, it is this highly conserved acidic tail that interacts with basic histones to inhibit HAT activity (Seo et al, 2002).
LANP associates with E4F in mammalian cells
To determine whether LANP and E4F associate in mammalian cells, we tested the colocalization of these proteins by using confocal microscopy. In HeLa cells, endogenous LANP and E4F colocalize with overlapping staining patterns (Fig 2A–E). To confirm the interaction of E4F and LANP, we carried out co-immunoprecipitation in HeLa cells transfected with Myc-tagged LANP and S-tagged E4F constructs (Fig 2F,G). It is important to note that we also precipitated endogenous LANP in N2A cells, a neuronal cell line with a high concentration of LANP (Fig 2H). This was accomplished by expressing S-tagged E4F and using S-beads to precipitate the E4F–LANP complex. Not surprisingly, we found it difficult to precipitate endogenous E4F, as E4F is expressed at low levels (Fernandes & Rooney, 1997).
Figure 2.
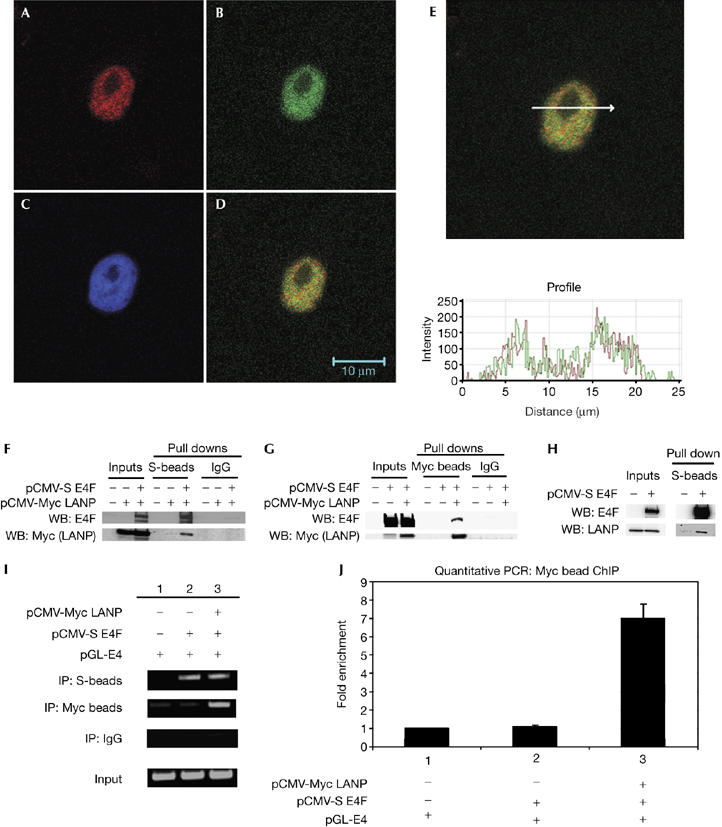
LANP and E4F interact in mammalian cells. (A–E) Confocal immunofluorescence of HeLa cells showing that endogenous LANP and E4F colocalize. Stained for (A) E4F, (B) LANP, (C) DAPI (nuclear), and (D) A and B merged. (E) Fluorescence intensity profile over a random cross-section of the merged image in (D) shows a correlation of intensities of fluorophores used to visualize E4F and LANP. (F,G) Co-precipitation of S-tagged E4F and Myc-tagged LANP in HeLa cells. Western blots (WB) showing the proteins in the immunoprecipitates. (H) In N2A cells, endogenous LANP is precipitated with S-beads when S-tagged E4F is expressed. (I) Chromatin immunoprecipitation (IP) showing that LANP–Myc and S-E4F occupy E4 promoter. (J) Real-time PCR to quantify the amount of pGL-E4 immunoprecipitated with LANP. DAPI, 4,6-diamidino-2-phenylindole; LANP, leucine-rich acidic nuclear protein.
To test whether LANP affects E4F-induced repression, we used the previously characterized reporter, pGL-E4. This reporter—generated by placing a firefly luciferase cassette downstream of the adenovirus E4 promoter—has been important in understanding E4F function as a repressor (Fernandes & Rooney, 1997; Sandy et al, 2000; Fajas et al, 2001). Furthermore, because plasmid-based reporter constructs have been excellent tools for studying promoter occupancy, we also used this reporter to describe the mechanistic details of transcriptional repression (Ishizuka & Lazar, 2003; Lavrrar & Farnham, 2004).
We transfected HeLa cells with pGL-E4 in the presence or absence of S-tagged E4F and Myc-tagged LANP, and carried out chromatin immunoprecipitation (ChIP) of E4F or LANP. The E4F target promoter region was enriched when either LANP or E4F was precipitated, indicating that both E4F and LANP occupy the E4 promoter (Fig 2I). The ChIP with LANP was quantified by real-time PCR (Fig 2J). Without exogenous E4F, the amount of LANP occupying the E4F target promoter was not significantly different from the background (supplementary Fig 1 online), indicating that LANP recruitment to the E4 promoter is E4F dependent.
E4F and LANP synergistically repress transcription
As expected, the pGL-E4 reporter produces a baseline luminescence that decreases in a dose-related manner on transfection with E4F in CV-1 fibroblasts (Fig 3A). We also observed repression when LANP was expressed at higher levels, providing evidence that LANP can repress the E4 reporter construct (Fig 3A). To detect a functional synergism between E4F and LANP, we expressed E4F and LANP at levels (25 ng) at which they individually cause minimal repression. However, when combined, the two proteins increased the level of repression beyond a simple additive effect (Fig 3A, right, the relevant histogram is marked with an asterisk). This synergism provides functional evidence of the E4F–LANP interaction. The extent of this synergism becomes less obvious as the amount of LANP is increased, indicating that the system can be saturated. We have also found that depleting endogenous E4F, by using small interfering RNA (siRNA), significantly relieves LANP-mediated repression of pGL-E4 luciferase activity (supplementary Fig S2 online), providing further evidence that LANP requires E4F to be recruited.
Figure 3.
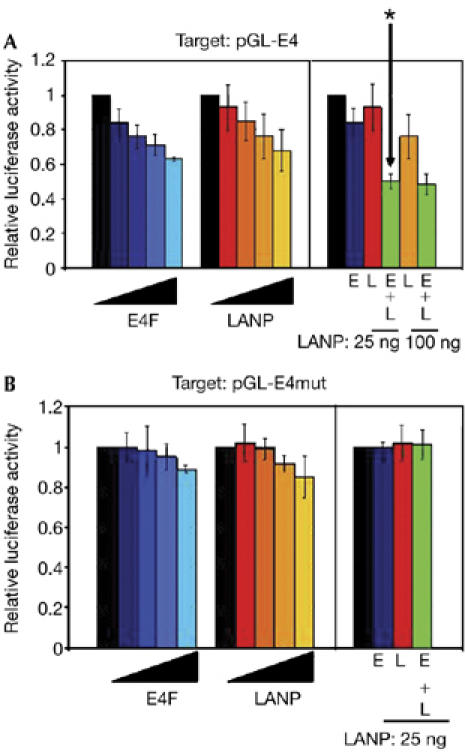
LANP and E4F synergistically repress the E4 promoter. The effects of E4F and LANP on (A) pGL-E4 and (B) pGL-E4 mut promoter activity in CV-1 cells. The left side of each panel shows the activity expressed from pGL-E4 alone (black bars) or with increasing amounts (25, 50, 100, 150 ng) of E4F or LANP expression plasmids (colour shaded). The right side of each panel shows the effect of coexpression of E4F and LANP on pGL-E4 activity. The amount of E4F (E) and/or LANP (L) expression plasmid in each transfection is indicated by colour. The asterisk indicates synergistic repression. LANP, leucine-rich acidic nuclear protein.
To control repression on a nonspecific promoter, we mutated the two E4F-binding sites on pGL-E4mut to generate the mutant reporter pGL-E4 (Fernandes & Rooney, 1997). When assayed against pGL-E4mut, E4F and LANP caused significantly lower repression than observed with the wild-type E4 promoter (Fig 3B, left side), without a synergistic effect (Fig 3B, right side).
LANP depletion relieves E4F inhibition
To investigate the extent to which LANP is necessary for E4F activity, we evaluated the contribution of endogenous LANP to E4F activity. We used two siRNA constructs (#SP1 and #3) that targeted distinct regions of the LANP message to reduce the levels of LANP in N2A cells (Fig 4A). The level of LANP depletion and E4F overexpression is shown by western blots (Fig 4B). It is important to note that depleting LANP does not affect the levels of E4F. Repression of pGL-E4 by E4F was significantly relieved when LANP was depleted (Fig 4A), indicating that endogenous LANP is required for the inhibitory effect of E4F.
Figure 4.
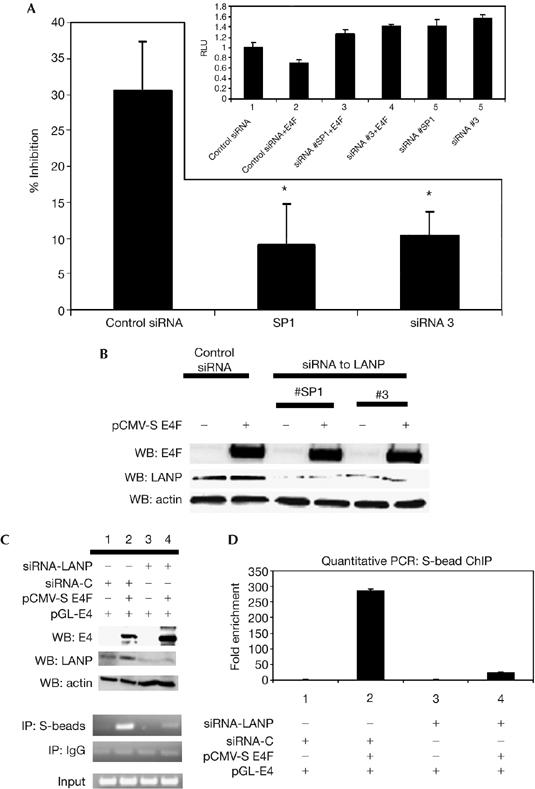
LANP is required for E4F-mediated repression. (A) N2A cells were transiently transfected with E4F, pGL-E4 and siRNA targeting LANP (or control siRNA). Expressed in terms of percentage inhibition, it shows that E4F causes significantly less repression when LANP is depleted (P<0.05 for siRNA#1 and siRNA#3). The inset shows the data presented in the form of relative luciferase activity. Depletion of LANP by two distinct siRNA targeting LANP (#SP1 and #3) relieved exogenous E4F repression (lanes 3 and 4 compared with lane 2; P<0.05), and also relieved baseline repression of the reporter by targeting LANP complexed to endogenous E4F (lanes 5 and 6 compared with lane 1; P<0.05). The asterisks indicate statistical significance P<0.05. (B) Western bloting (WB) shows the level of overexpression of E4F (approximately 30-fold) and the depletion of LANP (greater than 60%). (C) Reducing LANP decreases E4F binding to the E4 promoter. Chromatin immunoprecipitation (IP) with S-beads was carried out on HeLa cells transfected with siRNA targeting LANP (or control siRNA) and S-E4F. Western blots show the level of E4F overexpression and LANP depletion. (D) Quantitative PCR of (C). LANP, leucine-rich acidic nuclear protein; RLU, relative luciferase units; siRNA, short interfering RNA.
One mechanism by which the depletion of LANP relieves E4F inhibition could be that LANP promotes the binding of E4F to its target promoter. To test this hypothesis, we carried out ChIP of E4F in the presence or absence of LANP. As shown in Fig 4C,D, the amount of E4F target promoter bound to E4F is reduced when LANP is depleted (compare histograms 2 and 4 in Fig 4D). These results indicate that LANP promotes E4F binding to its target promoter.
Mutant ataxin 1 relieves E4F-induced inhibition
SCA1 is characterized by early transcriptional derangements that presage ataxia. We hypothesized that some of the transcriptional defects caused by mutant ataxin 1 might result from the misregulation of LANP. To address this possibility, we tested whether mutant ataxin 1 modulates the transcriptional repression of the E4F–LANP complex. Co-transfection of mutant ataxin 1 (ataxin 1-84Q) significantly relieves the E4F-induced repression of pGL-E4 (Fig 5A; lane 3). This repression of E4F could be restored by the addition of exogenous LANP (Fig 5A; lane 4), indicating that ataxin 1 titrates LANP away from E4F. We also found a substantial relief of E4F repression with wild-type ataxin 1 (data not shown), which is not surprising given that high levels of wild-type ataxin 1 can induce pathology in animal models (Fernandez-Funez et al, 2000).
Figure 5.
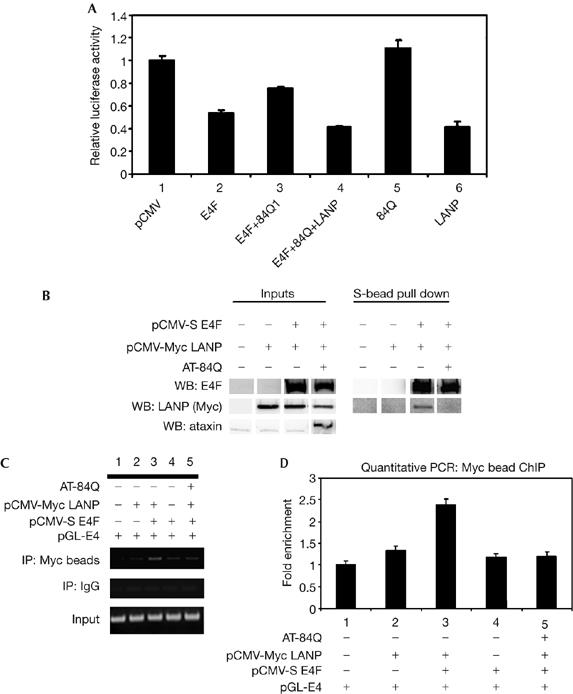
Ataxin 1 relieves the transcriptional repression induced by the LANP–E4F complex. (A) Ataxin 1 relieves E4F-induced inhibition. N2A cells were transfected with the indicated constructs. E4F-induced repression relieved by ataxin 1 (compare histograms 3 and 2; P<0.05) could be restored by the addition of exogenous LANP (compare histograms 4 and 3; P<0.05). Ataxin 1 and LANP alone are shown as controls in histograms 5 and 6. (B) The amount of LANP bound to E4F is reduced in the presence of mutant ataxin 1. Co-precipitation of LANP with S-beads was carried out in HeLa cells transfected with the indicated constructs. (C) Chromatin immunoprecipitation (IP) with a Myc antibody that precipitates Myc-tagged LANP. (D) Quantitative PCR of (C). LANP, leucine-rich acidic nuclear protein; WB, western blot.
To test whether mutant ataxin 1 and E4F compete for the available LANP, we carried out co-precipitations of E4F and LANP in the presence or absence of mutant ataxin 1 (Fig 5B). The amount of LANP bound to E4F was significantly reduced when ataxin 1 was present, supporting our finding that ataxin 1 successfully competes with E4F for LANP. This indicates that the amount of LANP present at the E4F-regulated promoter is decreased in the presence of mutant ataxin 1. To address this possibility, we carried out ChIP of LANP in the presence or absence of mutant ataxin 1 (Fig 5C). We found that the amount of LANP bound to the E4 promoter was significantly reduced in the presence of expanded ataxin 1 (Fig 5D, compare lanes 3 and 5).
Together, our findings have important implications. First, our results indicate a model in which inhibitor of histone acetyltransferase (INHAT) activity is recruited in a promoter-specific manner by transcription factors such as E4F. These results are reminiscent of findings where NIR—a recently described novel INHAT repressor—is recruited in a sequence-specific manner (by the tumour suppressor p53; Hublitz et al, 2005). Second, our data show that LANP is required for the stable binding of E4F to its target regions, indicating that INHAT corepressors are necessary for the optimal binding and repression by transcriptional repressors.
Our efforts to explore these issues further in the context of SCA1 are hampered by the limited knowledge of E4F targets. Indeed, the only known target of E4F, cyclin A—a regulator of the cell cycle (Fajas et al, 2001)—is unlikely to have a role in SCA1, as this disease affects nondividing neurons. Nonetheless, there is significant evidence that LANP might modulate the tumorigenic potential of cells by affecting the cyclin A promoter (Bai et al, 2001). In the context of tumorigenesis, it is also possible that LANP might interfere with non-transcriptional functions of E4F that revolve around cell proliferation such as its interactions with p14ARF, the retinoblastoma tumour suppressor and p53—a protein that it also ubiquitinates (Fajas et al, 2000, 2001; Sandy et al, 2000; Rizos et al, 2003; Le Cam et al, 2006).
Analysis of E4F knockout mice (embryonic lethal at embryonic day 7.5) indicates that E4F is also involved in mitotic progression in early embryonic cell cycles by a non-transcriptional role that influences the mitotic spindle (Le Cam et al, 2004). As LANP-null mice are viable and fertile (Opal et al, 2004), it seems unlikely that LANP is important in these early non-transcriptional functions of E4F. However, one cannot exclude the possibility that a relative lack of phenotype in LANP-null mice originates from a redundancy at an organismal level provided by close homologues of LANP (Matilla & Radrizzani, 2005).
The observation that ataxin 1 modulates repression induced by LANP has important ramifications for SCA1. In addition to the effects of ataxin 1 on LANP at the E4 promoter, we have found that ataxin 1 fused to the Gal4 DNA-binding domain can harness the repressive functions of LANP to a Gal-4-responsive promoter (M.C. & P.O., unpublished data). Thus, by relieving repression on some targets and potentiating repression on others, the mutant ataxin 1–LANP interaction might cause a mixed picture of transcriptional aberrations, consistent with the reported up- and downregulation of specific transcripts in SCA1 mice (Lin et al, 2000; Serra et al, 2004). Furthermore, such a scenario might explain why LANP knockout mice do not mirror the SCA1 phenotype (Opal et al, 2004). Our observations also add to growing evidence that ataxin 1 has an important role in transcriptional regulation. For example, ataxin 1 has been shown to interact with the transcriptional regulators: silencing mediator of retinoid and thyroid receptor (SMRT)/nuclear receptor corepressor (NcoR), Histone deacetylase 3 (HDAC3), Growth factor independence 1 (Gfi1), Tat interactive protein, 60 kDa (Tip60)/Retinoid-related orphan receptor α (ROR-α) and capicua (Tsai et al, 2004; Tsuda et al, 2005; Lam et al, 2006; Serra et al, 2006). The consequences of these interactions are likely to be complex with different transcriptional aberrations integrating to yield the disease phenotype. This might be another reason why targeting individual corepressors in mouse models is not sufficient to mimic SCA1. We are now initiating studies to link misregulation in gene expression to specific corepressors and transcription factors using whole-genome approaches. Such approaches are likely to be crucial in understanding the pathogenesis of SCA1.
Methods
Yeast two-hybrid assay. We carried out a yeast two-hybrid screen using full-length mouse LANP as bait to interrogate a mouse brain complementary DNA library as described previously (Opal et al, 2003; also see the supplementary information online).
Cells. The human cervical carcinoma HeLa cell line, the African green monkey CV1 cell line and the mouse neuroblastoma N2A cell line were cultured at 37°C in a humidified, 5% (v/v) CO2 atmosphere in DMEM supplemented with heat-inactivated fetal bovine serum (10% v/v), 0.1 mM MEM nonessential amino acids, 1 mM sodium pyruvate, 100 U/ml penicillin G and 100 mg/ml streptomycin.
Protein immunoprecipitations. HeLa cells (2 × 106 cells/dish) were transfected using Effectene (Qiagen, Valencia, CA, USA) with 2 μg of pCMV-Myc LANP and pCMVs-E4F 2.5K (with empty vector (pCDNA-3) to keep the total DNA constant at 4 μg). At 36 h after transfection, cells were lysed and co-precipitation was carried out with S-beads or Myc antibody linked to protein G beads (also see the supplementary information online).
Chromatin immunoprecipitations. HeLa cells (4 × 106) were transfected with the indicated constructs. At 24 h after transfection, cells were crosslinked in 1% formaldehyde and lysed. Chromatin samples were precleared with protein A agarose/salmon sperm DNA and then subjected to precipitation with the S-beads or Myc antibody with G-sepharose beads.
After washing and decrosslinking, DNA was extracted with phenol–chloroform, precipitated with ethanol and the pellet was resuspended in 30 μl Tris–EDTA. A 1 μl portion of resuspended DNA was used for PCR (also see the supplementary information online).
Luciferase assays. Luciferase assays were carried out using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA); pRL-CMV (CMV IE promoter-driven Renilla luciferase reporter construct; Promega) was included in all transfections as an internal control (also see the supplementary information online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
supplementary Fig 1
supplementary Fig 2
Acknowledgments
P.O. thanks A.R. Kini, R. Kular, K. Rundell, D. Chakravarti, S. Natesan and P. Cheresh for advice. P.O. was funded by the National Organization for Rare Disorders (NORD), the National Ataxia Foundation, the American Cancer Society and the National Institute for Neurological Disorders and Stroke (NINDS) (K08/K02); R.J.R. was funded by an American Cancer Society (ACS) grant. M.C. was funded by a National Ataxia Foundation Grant.
References
- Bai J, Brody JR, Kadkol SS, Pasternack GR (2001) Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 20: 2153–2160 [DOI] [PubMed] [Google Scholar]
- Fajas L, Paul C, Zugasti O, Le Cam L, Polanowska J, Fabbrizio E, Medema R, Vignais ML, Sardet C (2000) pRB binds to and modulates the transrepressing activity of the E1A-regulated transcription factor p120E4F. Proc Natl Acad Sci USA 97: 7738–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Paul C, Vie A, Estrach S, Medema R, Blanchard JM, Sardet C, Vignais ML (2001) Cyclin A is a mediator of p120E4F-dependent cell cycle arrest in G1. Mol Cell Biol 21: 2956–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes ER, Rooney RJ (1997) The adenovirus E1A-regulated transcription factor E4F is generated from the human homolog of nuclear factor phiAP3. Mol Cell Biol 17: 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P et al. (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408: 101–106 [DOI] [PubMed] [Google Scholar]
- Hublitz P et al. (2005) NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev 19: 2912–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA (2003) The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol 23: 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutney SN, Hong R, Macfarlan T, Chakravarti D (2004) A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression. J Biol Chem 279: 30850–30855 [DOI] [PubMed] [Google Scholar]
- Lam YC et al. (2006) ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127: 1335–1347 [DOI] [PubMed] [Google Scholar]
- Lavrrar JL, Farnham PJ (2004) The use of transient chromatin immunoprecipitation assays to test models for E2F1-specific transcriptional activation. J Biol Chem 279: 46343–46349 [DOI] [PubMed] [Google Scholar]
- Le Cam L, Lacroix M, Ciemerych MA, Sardet C, Sicinski P (2004) The E4F protein is required for mitotic progression during embryonic cell cycles. Mol Cell Biol 24: 6467–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L et al. (2006) E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Lin X, Antalffy B, Kang D, Orr HT, Zoghbi HY (2000) Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat Neurosci 3: 157–163 [DOI] [PubMed] [Google Scholar]
- Matilla A, Radrizzani M (2005) The Anp32 family of proteins containing leucine-rich repeats. Cerebellum 4: 7–18 [DOI] [PubMed] [Google Scholar]
- Matilla A, Koshy B, Cummings CJ, Isobe T, Orr HT, Zoghbi HY (1997) The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1. Nature 389: 974–978 [DOI] [PubMed] [Google Scholar]
- Opal P, Zoghbi HY (2002) The role of chaperones in polyglutamine disease. Trends Mol Med 8: 232–236 [DOI] [PubMed] [Google Scholar]
- Opal P, Garcia JJ, Propst F, Matilla A, Orr HT, Zoghbi HY (2003) Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem 278: 34691–34699 [DOI] [PubMed] [Google Scholar]
- Opal P, Garcia JJ, McCall AE, Xu B, Weeber EJ, Sweatt JD, Orr HT, Zoghbi HY (2004) Generation and characterization of LANP/pp32 null mice. Mol Cell Biol 24: 3140–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P, Rooney R, Nevins JR (1987) Identification of an E1A-inducible cellular factor that interacts with regulatory sequences within the adenovirus E4 promoter. EMBO J 6: 4073–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Diefenbach E, Badhwar P, Woodruff S, Becker TM, Rooney RJ, Kefford RF (2003) Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition. J Biol Chem 278: 4981–4989 [DOI] [PubMed] [Google Scholar]
- Sandy P, Gostissa M, Fogal V, Cecco LD, Szalay K, Rooney RJ, Schneider C, Del Sal G (2000) p53 is involved in the p120E4F-mediated growth arrest. Oncogene 19: 188–199 [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Weise C, Kouzarides T (2004) Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem 279: 23859–23862 [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104: 119–130 [DOI] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D (2002) Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem 277: 14005–14010 [DOI] [PubMed] [Google Scholar]
- Serra HG, Byam CE, Lande JD, Tousey SK, Zoghbi HY, Orr HT (2004) Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum Mol Genet 13: 2535–2543 [DOI] [PubMed] [Google Scholar]
- Serra HG et al. (2006) RORα-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell 127: 697–708 [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM (2004) Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc Natl Acad Sci USA 101: 4047–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H et al. (2005) The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122: 633–644 [DOI] [PubMed] [Google Scholar]
- Ulitzur N, Humbert M, Pfeffer SR (1997) Mapmodulin: a possible modulator of the interaction of microtubule- associated proteins with microtubules. Proc Natl Acad Sci USA 94: 5084–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
supplementary Fig 1
supplementary Fig 2


