Abstract
Hepatitis C virus (HCV) is an RNA virus which is estimated to persistently infect about 170 million people worldwide. After acute infection, there is an initial period during which long-term outcome is decided. There is strong evidence that the cellular immune responses, involving both CD4+ and CD8+ T lymphocytes, are involved at this stage and it is their effectiveness which determines outcome. What is not understood is what determines their effectiveness. The most important component of this is likely to be some aspect of epitope selection, itself dictated by host MHC. Thus, to understand host immunity to HCV, we need to have a detailed understanding of the peptides involved in T lymphocyte responses. In this review, we discuss the peptide epitopes that have been identified so far, and their potential significance. We relate this to a scheme of host defence which may be useful for understanding natural and vaccine-induced immunity.
Keywords: HCV, CD8+ T lymphocyte, CD4+ T lymphocyte, HLA, epitope, immune escape
INTRODUCTION
The virus HCV is a member of the flavivirus family – positive stranded RNA viruses, which include relatives such as viruses causing dengue and yellow fever. Its origin is obscure but certain strains have probably been circulating in human populations for hundreds of years [1]. Unlike many of its relatives, it is able to set up persistent infections. This, coupled with lifestyle changes and a number of iatrogenic disasters, have led to its recent spread worldwide. In the west it is most common amongst IV drug users (both current and those who may have given up decades ago), as well as recipients of blood products in the prescreening era. In some countries, such as Egypt, it appears to have been spread through needles used for medical programs and high rates in other countries may exist for similar reasons [2].
HCV replicates mainly in the liver. Some negative strand RNA, indicative of intracellular replication, has been found in dendritic cells (DCs) [3]. Even in those who clear virus from the blood (spontaneously or after treatment), there is a possibility that some viral RNA persists in the liver – similar to HBV. Direct detection of HCV is currently only possible by PCR, as culture methods from ex vivo samples are not routinely successful, even though in vitro ‘replicon’ (self-replicating RNA constructs) have been established [4].
Clinical features
After inoculation, unlike Hepatitis A and B, the acute illness caused by HCV is not well documented. This is partly because it is genuinely milder, and possibly it is poorly recognized by physicians, and also those in the current western risk groups may not present to hospital. This is unfortunate, as it now appears that early intervention is of benefit [5]. When it has been documented, or in animal models, the following features are apparent reviewed in 2]:
The peak of viraemia may take several weeks to arise [6,7]. The liver inflammation/liver enzyme level in the blood (usually measured as ALT or AST) does not parallel the viral load – consistent with the idea that at this stage much of the liver damage is not caused directly by the virus.
Resolution of the viraemia (accompanied by cellular immune responses) is associated with liver inflammation and in some but not all cases, clinical jaundice. The level of ALT may be 500–2000 IU/l, compared with HBV where it may be 5 or 10 times higher [8–10].
After this period, viraemia either persists or the individual, in about 15% of cases, becomes RNA negative in the blood. There may be a state of ‘instability’ where virus may become undetectable in blood temporarily and then reappears [11].
If viraemia is established, the level of viraemia does not correlate with progression of disease, unlike HIV. Disease progression is measured by the development of liver inflammation, as assessed by blood ALT and histological indices of lymphocytic infiltration and also by creeping fibrosis. The extent of these vary widely between individuals, and apart from a few factors such as alcohol and coinfection (for example with HIV), the basis of this variation is not understood.
In those where disease has progressed, therapy (in the form of interferon-alpha and ribavirin) may lead to long-term clearance of virus from blood, with accompanying improvement in liver histology. The effects of the drugs are not entirely understood, and it is likely that in addition to antiviral activity they influence the immune response, both directly, and indirectly through lowering viral load [12].
Cellular immunology of HCV
HCV, like other viruses, induces multiple immune effector responses, but this review focuses primarily on T lymphocytes. There is good evidence that both CD4+ and CD8+ T lymphocytes play a major role in determining outcome after acute infection and therefore in the long term. This comes from the following observations.
Clearance of acute infection in both man and in chimpanzee models is accompanied by strong CD4+ and CD8+ T cell responses against numerous HCV derived antigens [11,13–16]. The evidence was obtained first for CD4+ T cell responses and initially some specific epitopes were highlighted as potentially ‘protective’ [13]. Although these do appear to be targeted, this is not exclusive and responses to other gene products are also seen [17]. The strength of the CD8+ T cell response against one epitope when measured using a tetramer, may be up to 8% of the total CD8+ T cells, and can include responses to at least 8 separate epitopes [10]. By ELISpot analysis, the CD4+ T cell responses appear to be of a similar magnitude [10].
The timing of these responses appears to correlate with resolution of viraemia in those cases where virus is cleared. The level of activation of HCV-specific T cell responses (assessed by CD38 expression) correlates with the degree of liver inflammation analysed by blood ALT levels [9].
There is an association between possession of specific HLA genes (DRB1*1101 and/or DQ1*0301) and spontaneous clearance of virus [18]. This strongly suggests that selection of particular epitopes is associated with better initial control of viraemia. Those bearing HLA DQ1*0301 (which is in tight linkage disequilibrium with DRB1*1101) were found to be more likely to possess significant HCV-specific CD4+ T cell responses, further evidence that the responses in these individuals are more robust [19].
So much for successful responses – which are in fact the exception. The mechanism for viral persistence, i.e. failure of T cell responses in the majority of patients, is not yet clear. Studies of those who go on to develop persistent infection have highlighted the weak CD4+ T cell responses, although it is not clear yet whether persistence of virus causes attenuation of T cell responses or vice versa [11,19,20]. Re-emergence of CD4+ T cell responses upon clearance of virus with interferon-alpha/ribavirin therapy suggests the latter –, i.e. suppression of T cells by virus may be important [12].
The picture with regards to CD8+ T cell responses is even less clear. CD8+ T cell responses have been observed in the acute phase of infection in those who fail to clear virus at levels of 1–3% of CD8+ lymphocytes against 1–2 separate T cell epitopes [7,9]. Whether these are the main epitopes targeted in these individuals and how, overall, the responses differ in magnitude between clearers and nonclearers is not known. It appears that failure to clear virus is not due to failure to mount any CTL response whatsoever, although, like CD4+ T cell responses, these may be poorly maintained in the face of ongoing viraemia [9]. The overall quality of the response may differ in terms of magnitude or breadth – or, importantly, peptide selection. It is this latter issue that forms the focus of this review.
Peptide epitopes in HCV
The assessment of the exact epitopes used in an individual’s HCV-specific response is likely to be crucial in understanding the overall role of CD8+ and CD4+ T cells in control of disease and in approaching vaccine design. There are two broad reasons for this – one biological (i.e. it affects outcome, see below) and one pragmatic.
On the pragmatic level, too many studies have focused on too few epitopes. Even those using a larger number of epitopes have tended to limit themselves to those restricted by HLA-A2. There is clear evidence from detailed studies of individual patients in acute and chronic disease that this approach misses many responses [21]. This means that it becomes difficult to draw conclusions as to the overall role of CTL responses from clinico-pathological studies – as we have not obtained in each individual an accurate assessment of their magnitude. As an example, a single ‘clearer’ who made 8 separate CTL responses made only one to a previously mapped epitope (restricted by HLA-A2), and even this was not the dominant HLA-A2 restricted response [10], and similar findings are obtained in studies of those persistently infected [21]. This is similar to the situation in HIV where an epitope in p17 gag (SLYNTVATL) has been found to be frequently targeted in HLA-A2 positive individuals, but during acute disease, it is rarely seen – other epitopes are likely to be much more important at this critical stage [22].
Thus, it is likely that there are, as for HIV, a reasonably large number of epitopes available for targeting by any individual, dependent on their HLA type, and it is not possible to assess the complete response using only a restricted number of peptides. It may not be possible in each case to map the entire array of responses, as this is labour intensive and expensive in terms of peptides, as well as being hampered by the relatively weak responses seen in many patients. Therefore, what is minimally required, is a ‘customised’ mapping for each patient using peptides based on the full HLA type and viral genotype. This is not perfect, but should improve the yield of responses obtained and provide a better sense of the breadth and relative importance of the T lymphocyte response. The process should improve over time as individual peptides are mapped and confirmed between groups, provided a clear and up-to-date database is available.
HCV-specific epitopes – the evidence base
We have compiled a table of HLA Class I-restricted HCV epitopes to enable researchers in the field to establish panels of peptides suitable for their patients – and thus address the issue of the role of HCV-specific T cells comprehensively (Table 1). Table 1a shows the currently published peptides restricted by HLA-A2. The figure above the table illustrates the position of these epitopes, which are spread throughout the genome without any obvious clustering. Many of these were generated by computer predictions and have been subsequently validated in patients. However, many have also not been observed frequently or using ex vivo assays. The use of ex vivo assays is relevant since responses which only appear after multiple restimulations may also be seen in HCV negative individuals and their in vivo relevance is less concrete than those seen using direct ex vivo tetramer or ELISpot assays [25,45]. Thus, from this large panel of peptides only a handful are likely to be significant in terms of providing an ‘anchor’ for a successful T cell response. In our hands, three peptides, NS31073–81, NS3 1406–14, and NS5b 2594–2604, appear to be the most commonly recognized and have been identified using ex vivo assessments. The first of these may be cross-reactive with an epitope from influenza, although initiation of strong activated responses in several patients with acute disease suggest that the cells are HCV-specific in vivo[60].
Table 1.
| Protein | Amino acid position | HLA-restriction | Amino acid sequence | Technique | Reference |
|---|---|---|---|---|---|
| (a) HCV CTL epitopes restricted by HLA-A2 | |||||
| Core | 35–44 | YLLPRRGPRL | A,B | [20,23–34] | |
| YLLPSRGPKL | A | [32] | |||
| Core | 90–98 | GLGWVGWLL | A | [31] | |
| Core | 132–140 | DLMGYIPLV | A,B,C | [23,24,29–31,34–39] | |
| ALMGYIPLV | A | [36] | |||
| DLMGYIPAV | A | [36] | |||
| 131–140 | ADLMGYIPLV | A,B | [20,25–28,40] | ||
| Core | 156–165 | RVLEDGVNYA | A | [31] | |
| Core | 177–185 | FLLALLSCL | A | [31] | |
| Core | 178–187 | LLALLSCLTV | A,B | [24–26,41] | |
| LLALLSCLTI | A,B | [20] | |||
| E1 | 220–227 | ILHTPGCV | A,B | [38,40] | |
| E1 | 257–266 | QLRRHIDLLV | A,B | [27,28,40] | |
| AIRRHVDLLV | B | [20] | |||
| E1 | 285–293 | FLVSQLFTF | B | [39] | |
| E1 | 363–371 | SMVGNWAKV | A,B | [38,40] | |
| E1 | 398–407 | SLASLFTQGA | A | [42] | |
| E2 | 401–411 | SLLAPGAKQNV | A,B | [38,40,41] | |
| E2 | 614–622 | RLWHYPCTV | A,B | [39] | |
| E2 | 686–694 | ALSTGLIHL | A | [23] | |
| E2 | 723–732 | LLFLLLADA | A,B | [29,34] | |
| E2 | 725–734 | FLLLADARV | A,B | [23,29,30,34] | |
| NS3 | 1073–1081 | CINGVCWTV | A,B,C | [9,10,21,25–28,32,33,40,41,43–47] | |
| CVNGVCWTV | A,B | [20] | |||
| CVVGVCWTA | A | [32] | |||
| NS3 | 1131–1139 | YLVTRHADV | A,B | [29,34] | |
| NS3 | 1169–1177 | LLCPAGHAV | A | [25,27,28,40] | |
| LLCPSGHVV | A | [20] | |||
| NS3 | 1287–1296 | TGAPVTYSTY | A | [48] | |
| NS3 | 1406–1415 | KLVALGINAV | A,B,C | [9,25–28,32,33,40,41,43,47,49] | |
| KLSGLGINAV | B | [7] | |||
| KLVSLGVNAV | A | [32] | |||
| KLSGLGLNAV | A | [32] | |||
| KLVALGVNAV | A | [32] | |||
| NS3 | 1585–1593 | YLVAYQATV | A,B | [23,30,34] | |
| NS4 | 1661–1669 | VLVGGVLAA | A,B | [29,34] | |
| NS4 | 1666–1675 | VLAALAAYCL | A | [30,41] | |
| NS4 | 1769–1777 | HMWNFISGI | A,B | [23,29,30,34] | |
| NS4 | 1789–1797 | SLMAFTAAV | A,B | [25,27,28,40] | |
| SLMAFTASV | A | [20] | |||
| NS4 | 1807–1816 | LLFNILGGWV | A,B,C | [9,20,23–30,32,34,35,40] | |
| VFFNILGGWV | A | [32] | |||
| NS4 | 1851–1859 | ILAGYGAGV | A,B,C | [20,23,24,27–30,34,40] | |
| NS4 | 1915–23 | WMNRLIAFA | B | [29] | |
| NS4 | 1987–95 | VLDSFKTWL | A | [41] | |
| NS5 | 2141–2149 | LLREEVSFRV | A | [41] | |
| NS5 | 2221–2231 | SPDAELIEANL | A | [41,46] | |
| NS5 | 2252–2260 | ILDSFDPLV | A,B | [25,26,40] | |
| NS5 | 2578–2587 | RLIVFPDLGV | B | [34,40] | |
| NS5 | 2594–2602 | ALYDVVTKL | A,B,C | [10,20,41] | |
| NS5 | 2727–2735 | GLQDCTMLV | A,B | [24,27,28,32,33,40] | |
| GLQDCTMFV | A | [20] | |||
| (b) HCV CTL epitopes restricted by other alleles | |||||
| Core | 2–9 | A11 | STNPKPQK | A | [26,50] |
| 1–9 | MSTNPKPQK | A | [21] | ||
| Core | 28–36 | B60 | GQIVGGVYL | A | [51] |
| Core | 41–49 | B7 | GPRLGVRAT | A,B | [21,40,44,45,50] |
| Core | 43–51 | A3 | RLGVRATRK | A,B | [34,52] |
| Core | 51–59 | A3 | KTSERSQPR | A,B | [34,52] |
| Core | 88–96 | B44 | NEGCGWMGW | A | [53–56] |
| Core | 111–119 | B7 | DPRRRSRNL | B | [40] |
| Core | 169–177 | B7 | LPGCSFSIF | A | [52] |
| E1 | 234–242 | B35 | NASRCWVAM | A | [21,45,57] |
| E1 | 289–297 | A3 | QLFTFSPRR | A,B | [29,34,52] |
| E2 | 460–469 | B53 | RPLTDFDQGW | A | [21,44] |
| E2 | 489–496 | B51 | YPPKPCGI | A,B | [21,50] |
| E2 | 497–507 | B35 | VPASQVCGPVY | A | [58] |
| E2 | 530–539 | B60 | GENDTDVFVL | A | [46] |
| E2 | 569–578 | B50 | CVIGGAGNNT | A | [21,50] |
| E2 | 621–628 | A11 | TINYTIFK | A | [21,44–46] |
| E2 | 632–641 | A3 | RMYVGGVEHR | A,B | [29,34,52] |
| E2 | 654–662 | B60 | LEDRDRSEL | A | [46] |
| NS2 | 827–834 | A29 | MALTLSPY | A | [21] |
| NS2 | 831–840 | A25 | LSPYYKRYIS | A,B | [10] |
| NS2 | 838–846 | A23 | YISWCLWWL | A | [21] |
| 838–845 | YISWCLWW | A | [44] | ||
| NS2 | 957–964 | B37 | RDWAHNGL | A | [49] |
| NS3 | 1031–1039 | A24 | AYSQQTRGL | A | [59] |
| NS3 | 1069–1077 | B35 | LPGCSFSIF | A | [58] |
| NS3 | 1100–1108 | A24 | MYTNVDQDL | A | [35] |
| NS3 | 1261–1270 | A3 | TLGFGAYMSK | A | [21,46] |
| 1262–1270 | LGFGAYMSK | A,B | [29,34,52] | ||
| NS3 | 1359–1367 | B35 | HPNIEEVAL | A | [58] |
| NS3 | 1391–1399 | A3 | LIFCHSKKK | A,B | [29,34,52] |
| NS3 | 1395–1403 | B8 | HSKKKCDEL | A,B,C | [9,21,40,44–46] |
| NS3 | 1402–1410 | B8 | ELAAKLVGL | A | [49] |
| NS3 | 1531–1539 | B35 | TPAETTVRL | A | [58] |
| NS3 | 1611–1618 | B8 | LIRLKPTL | A | [46] |
| NS3 | 1636–1643 | A11 | TLTHPVTK | A | [21,46] |
| NS4 | 1744–1754 | A25 | EVIAPAVQTNW | A,B | [10] |
| NS4 | 1758–1766 | A25 | ETFWAKHMW | A,B | [10] |
| NS4 | 1858–1867 | A3 | GVAGALVAFK | A,B | [34,52] |
| 1859–1867 | VAGALVAFK | A,B | [29,34,52] | ||
| NS4 | 1941–48 | B38 | AARVTAIL | A | [46] |
| NS4 | 1966–76 | B37 | SECTTPCSGSW | A,B | [10] |
| NS4 | 2000–2008 | B35 | LPKLPGVPF | A | [58] |
| NS5 | 2152–2160 | B60 | HEYPVGSQL | A | [46] |
| NS5 | 2161–2171 | B35 | PCEPEPDVAVL | A | [46] |
| 2163–2171 | EPEPDVAVL | A | [58] | ||
| NS5 | 2218–2226 | B38 | NHDSPDAEL | A | [46] |
| NS5 | 2225–2233 | A25 | ELIEANLLW | A,B | [10] |
| NS5 | 2266–2275 | B60 | REISVPAEIL | A | [21] |
| NS5 | 2510–2518 | A3 | SLTPPHSAK | A | [21,46] |
| NS5 | 2588–2596 | A3 | RVCEKMALY | A | [21,44] |
| NS5 | 2629–2637 | B57 | KSKKTPMGF | A | [21] |
| NS5 | 2794–2802 | A3 | HDGAGKRVY | A | [26] |
| NS5 | 2794–2804 | B38 | HDGAGKRVYYL | A | [21,46] |
| NS5 | 2819–2828 | A25 | TARHTPVNSW | A,B | [10] |
| NS5 | 3003–3011 | A31 | VGIYLLPNR | A | [21] |
Table 1b shows peptides derived from studies of other MHC restrictions. This list is proportionately much smaller considering the relative number of alleles involved and this reflects the effort spent in defining new epitopes in these patients, rather than the importance of such responses. It is not clear whether, unlike HLA-A2, one peptide may be immunodominant if a particular restriction element is present as for HLA-B27 in HIV [61]. Much more work needs to be done in this area before any strong conclusions can be drawn. Work using tetramers derived from HLA-B8 and -B7 restricted peptides has given similar results to A2-based tetramers, i.e. most persistently infected patients show weak or absent responses [9,62].
Table 2 shows the variability of some of these epitopes, dependent on viral genotype. Clearly, viral genotype and even subtype needs to be taken into account when identifying the appropriate peptide to use for study. This may be difficult in cases where virus has been cleared from the blood and also in situations, which may not be uncommon, where several viral strains may have been previously encountered. It should be stressed that these variations only apply to bulk sequences from different genotypes and within one individual, viral sequences may vary – potentially under CTL selection pressure.
Table 2.
HCV CTL epitope sequences. Dashes indicate identity with the model peptide sequence (HCV-1 isolate–PubMed P26664)
| HLA Restriction | B7 | A11 | A2 | B8 | A2 | A2 | A2 |
|---|---|---|---|---|---|---|---|
| (Location) | (core 41–49) | (E2621–628) | (NS31073–1081) | (NS31395–1403) | (NS31406–1415) | (NS41807–1816) | (NS5B 2594–2602) |
| Sequence | GPRLGVRAT | TINYTIFK | CINGVCWTV | HSKKKCDEL | KLVALGINAV | LLFNILGGWV | ALYDVVTKL |
| P26664 (1a) | ––––––––– | –––––––– | ––––––––– | ––––––––– | –––––––––– | –––––––––– | ––––––––– |
| AAB67038 (1a) | ––––––––– | –––––––– | ––––––––– | ––––––––– | –––––––––– | –––––––––– | ––––––S–– |
| P27958 (1a) | ––––––––– | –––––––– | ––––––––– | ––––––––– | –––––––––– | –––––––––– | ––––––S–– |
| P26663 (1b) | ––––––––P | –V–F–––– | –V––––––– | ––––––––– | –––SG––––– | –––––––––– | ––––––ST– |
| P26662 (1b) | ––––––––– | –V–F–V–– | –V––––––– | ––––––––– | –––TG–L––– | –––––––––– | ––––––ST– |
| Q00269 (1b) | ––T–––––– | –V–F–––– | –V––––––– | ––––––––– | –––SG––––– | –––––––––– | ––––––ST– |
| P29846 (1b) | ––––––––– | –V–F–––– | ––––––––– | ––––––––– | –––S–––H–– | –––––––––– | ––––––ST– |
| BAA14035 (1b) | ––––––––– | –––F–––– | –V––––––– | ––––––––– | ––TG––L––– | –––––––––– | ––––––ST– |
| BAA09075 (1b) | ––––––––– | –V–F–––T | –V––––––– | ––––––––– | ––S–––V––– | –––––––––– | ––––––ST– |
| CAA03854 (1b) | ––––––––L | –V–F–––– | –V––––––– | ––––––––– | –––SG–L––– | –––––––––– | ––––––ST– |
| CAB41951 (1b) | ––––––––– | –V–F–––– | –V––––––– | ––––––––– | Q–SS––V–– | –––––––––– | ––––––ST– |
| AAD50312 (1b) | ––––––––– | AV–F–––– | –V––––––– | ––––––––– | –––SG–L–– | –––––––––– | ––––––ST– |
| BAB18814 (1b) | ––––––––– | –V–F–––– | –V––––––– | ––––––––– | ––SS––L–– | –––––––––– | ––––––ST– |
| AAL00900 (1b) | ––––––––– | –V–F–––– | –V––––––– | ––––––––– | ––SG––L–– | –––––––––– | ––––––ST– |
| CAB53095 (1b) | ––––––––– | –V–F–––– | –V––––––– | ––––––––– | ––SG––L–– | –––––––––– | ––––––ST– |
| P26660 (2) | ––––––––– | –V–––––– | T–S––L––– | ––––––––– | A–RGM–L–– | I–L––––––L | ––––ITQ–– |
| BAA88057 (2) | ––––––––– | –––F––H– | T–S–IL––– | ––––––––– | A–RGM–L–– | I–L––M–––L | ––––ITQ–– |
| JQ1303 (2a) | ––––––––– | –V–––––– | T–S––L––– | ––––––––– | A–RGM–L–– | I–L––––––L | ––––ITQ–– |
| BAB32876 (2a) | ––––––––– | –V–F–––– | S–S––L––– | ––––––––– | A–RGM–L–– | I–L––––––L | –––––TQ–– |
| AAF25613 (2a) | ––––––––– | –––––––– | S-S––L––– | ––––––––– | A–RGM–L–– | I–L––––––L | –––––TQ–– |
| P26661 (2b) | ––––––––– | –V–F–––– | S-S––L––– | ––––––––– | A–RGM–L–– | I–L––M–––L | ––––IAQ–– |
| AAF59945 (2b) | ––––––––– | NV–F–––– | S-S––L––– | ––R–––––– | A–RGM–L–– | I–L––M–––L | ––––IAQ–– |
| BAB08107 (2b) | ––––––––– | –V–F–––– | S-S––L––– | ––––––––– | A–RGM–L–– | I–L––M–––L | ––––IAQ–– |
| BAA08911 (2c) | ––––––––A | –V––––F– | S-S––L––– | ––––––––– | A–RGM–L–– | I–L––M–––L | ––––ITQ–– |
| BAA04609 (3a) | ––––––––– | –VDFRL–– | TVG––I–––– | ––––––––I | ––RGM–L–– | TM–––––––– | –––––IQ–– |
| BAA06044 (3a) | ––––––––– | –V–F–L–– | TVG––M––– | –––––––KM | ––RGM–L–– | TM–––––––– | –––––IQR– |
| AAC03058 (3a) | ––––––C–– | –V–F–L–– | TVG––T–––– | ––––––––I | ––RGM–L–– | TM–––––––– | –––––IQ–– |
| BAA08372 (3b) | ––Q––––EV | –V–FS––– | TVG––M––– | –––E––––– | ––RGM–V–– | TM–––––––– | –––––IQ–– |
| CAA72338 (4a) | ––––––––– | –A–FSV–N | AV–––M––– | ––––––––– | Q–TS––L––– | –––––––––– | ––H––IK–T |
| CAA73640 (5a) | ––K–––––– | –V–––––– | –––––M––L | ––––––––– | Q–TS––V––– | –––––––––– | –––––AQ–– |
| AAC61696 (5a) | ––––––––– | –L–––––– | –––––M––– | ––––––––– | Q–TS––V––– | –––––––––– | ––––IAQ–– |
| CAA72801 (6a) | ––––––––– | –V–F–LH– | S––––M-––– | ––––––––– | ––KS––L––– | I–L––––––I | –––––TQ–– |
| BAA09890 (10a) | ––K–––––V | –V–––––– | SVG––M––– | ––––––––– | Q–TS––V––– | –––––M–––– | –––––IQ–– |
All the epitopes illustrated do show significant capacity for variation, although the extent varies between epitopes. Thus, targeting of a completely conserved region which cannot mutate at all for reasons for structure or function, may not be an option in vaccine design. However, whether an epitope mutates or not depends on the intensity of CTL selection, its breadth, and crucially, the viral load. Therefore, the issue of epitope selection as a determinant of outcome may be very complex (see below).
These tables should be regarded as ‘work in progress’. Such a table could never be used to fully predict the response of an individual patient, but it could potentially cover the most important responses, once comprehensive mapping has been performed in many more individuals. Further tables for CD4+ T cell responses are also required, although beyond the scope of this review.
Success versus failure of T cell responses: the potential role of epitope selection
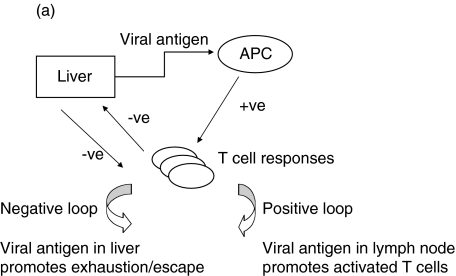
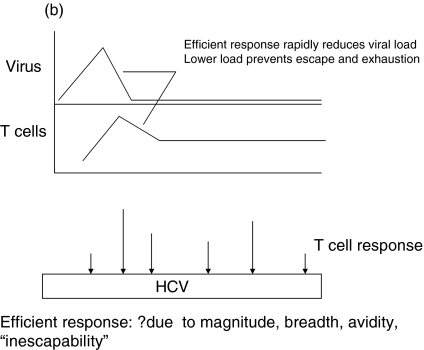
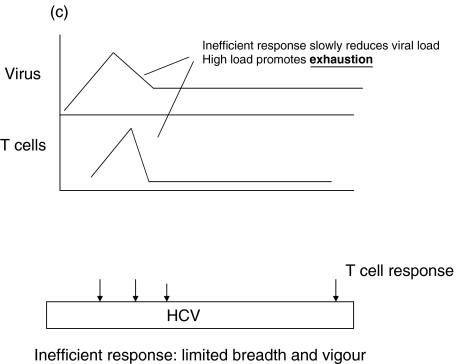
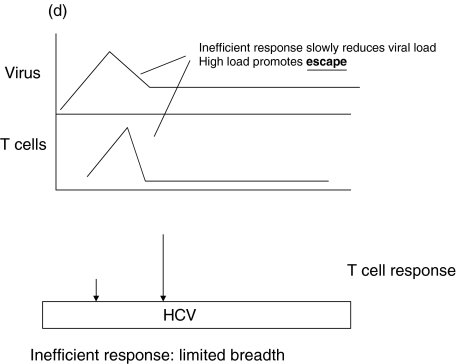
A simple model for the role of epitope selection in determining outcome in HCV is as follows (Fig. 1). After infection, viral replication takes place within the liver and at this stage is influenced by host innate immune responses, including interferon alpha and intrahepatic lymphocyte populations, such as NK and NKT cells [63,64]. Viral antigen is presented within the liver and in lymphoid organs. Induced responses include CD4+ and CD8+ T cells, the exact responses dictated by host MHC and incoming viral genotype. The magnitude of the responses depends not only on the exact epitopes targeted, but also on initial viral kinetics [65]. Induced T cell populations migrate to the liver, where – in concert with innate responses – they serve to reduce viral load (Positive loop in Fig. 1]. At the same time, viral replication – either through T cell ‘exhaustion’ [66], a tolerogenic effect of the liver environment [67] or through action of a specific viral gene product such as core, will tend to attenuate T cell responses. The phenomenon of temporarily reduced interferon gamma secretion in HCV-specific populations during the period of most intense activation (‘stunning’), has been observed by two groups [7,9]. This may be related to the phenomenon of exhaustion as it is commonly seen in murine models in the ‘pre-exhausted’ phase, before responses appear altogether [68,69]. Although the exact mechanism is not clear, this can be observed even if CD4+ T cell responses are intact, and it is likely that this is indicative of the ‘negative’ loop of Fig. 1. The outcome of this race – i.e which loop becomes dominant – depends on peptide epitopes in three ways (illustrated in Fig. 1b-d).
Fig. 1.
A simple model for the role of T cell epitope selection in HCV outcome. (a) T cell responses are induced optimally in lymphoid tissue and migrate to the liver to suppress viral replication (positive loop). The liver environment, acccompanied by a high viral load serves to attenuate T cell responses (negative loop). High viral loads will also tend to promote escape. (b) An optimal ‘efficient’ response rapidly reduces viral load and thus accentuates the positive over the negative loop. The mechanism behind ‘efficiency’ is not well defined, but includes breadth, vigour, avidity (i.e. the ability to recognize infected cells sensitively), good effector function and targeting of ‘constrained’ epitopes. (c) An inefficient response fails to lower viral load rapidly and enters the negative loop. In this example the responses lack vigour or avidity, and are then attenuated further in the face of maintained viral loads. (d) An inefficient response fails to lower viral load early and allows for the emergence of escape variants. This will be promoted if the response is targeted at epitopes which can escape rapidly, or if the response is very highly focused.
Epitope selection dictates the ‘efficiency’ of the responses within the liver, which in turn depends on the ability of T cells to recognize low levels of antigen in cells – ideally before they generate new virions [70]. Certain epitope(s) listed in Table 1, or yet to be discovered, may favour this process, whilst others, although providing adequate MHC binding and presentation to T cells, may be less efficient. Unlike HIV, there are no early regulatory genes, such as tat, which provide obvious targets in this respect, but there may be differences between gene products.
Epitope selection may determine how easily escape occurs at the level of an individual peptide. If escape occurs in variable viruses, it is highly epitope dependent, as is clear from detailed studies of HLA-B27-restricted epitopes in HIV [71–73]. Some of these epitopes may fall in regions that are ‘constrained’ or stable within the virus, due to conserved structure or function and multiple ‘compensatory’ mutations may need to accumulate, which does not occur readily. Thus, a clear understanding of the epitopes targeted and their escape ‘potential’ is needed – certain responses may be inherently more or less ‘escapable’.
Epitope selection will dictate the breadth of the response, which crucially affects the ability of replicating virus to escape T cell responses. A response which is highly vigorous and efficient may rapidly lose its efficacy in vivo if viral load is not quickly contained, as this will generate escape mutation, which has been very clearly illustrated in the LCMV and SIV challenge systems [74,75]. How many effective epitopes need to be targeted is an important question, and one that is critical to vaccine design. In the LCMV model the answer appears to be three, although kinetics of this system are much faster than HCV [76].
Once escape has occurred, it is difficult for cellular responses to regain control due to the phenomena of T cell exhaustion (clonal deletion in the face of continuing viral loads) [66,77] and original sin (inability to generate new responses against emerging variants) [78]. Thus, once this situation is established, chronicity becomes inevitable [79].
CONCLUSIONS
We present here a discussion of the effectiveness of T cell responses in HCV. It is now clear that cellular immune responses play an important role in determining outcome, and what forms the important topic of interest now is why some responses are more effective than others. The main conclusion is that this issue cannot be satisfactorily addressed until more data is available on the exact peptides used by individuals making cellular immune responses against their own ‘endogenous’ virus. However, a database of peptides is now emerging that should allow better insights into this complex process. Viral persistence does not depend entirely on T cell epitope selection, as numerous other host and viral factors are involved which are of major importance in controlling viral load. It may not depend entirely on mutational escape, as other issues affecting the efficiency of T cell responses are also likely to be important. However, the simple model presented here suggests this is a major component, and one clearly relevant to infections with other variable pathogens, such as HIV.
Acknowledgments
This work was sponsored by the Wellcome trust and also by the EU (QLK2-CT-1999–00356), the Doris Duke foundation and the Deutsche Forschungsgemein. We are grateful to Annie Lorton and Jane Collier for continuing help with clinical studies and Rodney Phillips for ongoing support in the Peter Medawar Building.
REFERENCES
- 1.Pybus OG, Charleston MA, Gupta S, Rambaut A, Holmes EC, Harvey PH. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323–5. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- 2.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 3.Bain C, Fatmi A, Zoulim F, Zarski JP, Trepo C, Inchauspe G. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology. 2001;120:512–24. doi: 10.1053/gast.2001.21212. [DOI] [PubMed] [Google Scholar]
- 4.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–3. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 5.Jessner W, Gschwantler M, Steindl-Munda P, et al. Primary interferon resistance and treatment response in chronic hepatitis C infection: a pilot study. Lancet. 2001;358:1241–2. doi: 10.1016/S0140-6736(01)06356-5. [DOI] [PubMed] [Google Scholar]
- 6.Farci P. Hepatitis C virus. The importance of viral heterogeneity. Clin Liver Dis. 2001;5:895–916. doi: 10.1016/s1089-3261(05)70200-2. [DOI] [PubMed] [Google Scholar]
- 7.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 9.Lechner F, Gruener NH, Urbani S, et al. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000;30:2470–87. doi: 10.1002/1521-4141(200009)30:9<2479::AID-IMMU2479>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 10.Lechner F, Wong DK, Dunbar PR, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4 (+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117:933–41. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 12.Cramp ME, Rossol S, Chokshi S, Carucci P, Williams R, Naoumov NV. Hepatitis C virus-specific T-cell reactivity during interferon and ribavirin treatment in chronic hepatitis C. Gastroenterology. 2000;118:346–55. doi: 10.1016/s0016-5085(00)70217-4. [DOI] [PubMed] [Google Scholar]
- 13.Diepolder HM, Zachoval R, Hoffman RM, Wierenga EA, Santantonio T, Jung MC, Eichenlaub D, Pape GR. Possible mechanism involving T-lymphocyte response to nonstructural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–7. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 14.Cooper S, Erickson AL, Adams EJ, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–49. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 15.Naoumov NV. Hepatitis C virus-specific CD4 (+) T cells. Gastroenterology. 1999;117:1012–4. doi: 10.1016/s0016-5085(99)70361-6. [DOI] [PubMed] [Google Scholar]
- 16.Eckels DD, Wang H, Bian TH, Tabatabai N, Gill JC. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev. 2000;174:90–7. doi: 10.1034/j.1600-0528.2002.017403.x. 10.1034/j.1600-065x.2000.174001090.x. [DOI] [PubMed] [Google Scholar]
- 17.Missale G, Bertoni R, Lamonaca V, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98:706–14. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thursz M, Yallop R, Goldin R, Trepo C, Thomas HC. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Res Lancet. 1999;354:2119–24. doi: 10.1016/s0140-6736(99)91443-5. [DOI] [PubMed] [Google Scholar]
- 19.Harcourt G, Hellier S, Bunce M, Satsangi J, Collier J, Chapman R, Phillips R, Klenerman P. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat. 2001;8:174–9. doi: 10.1046/j.1365-2893.2001.00289.x. 10.1046/j.1365-2893.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- 20.Takaki A, Wiese M, Maertens G, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578–82. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 21.Wong DK, Dudley DD, Afdhal NH, et al. Liver-derived CTL in hepatitis C virus infection. breadth and specificity of responses in a cohort of persons with chronic infection. J Immunol. 1998;160:1479–88. [PubMed] [Google Scholar]
- 22.Goulder PJ, Altfeld MA, Rosenberg ES, et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–94. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander J, Del Guercio MF, Fikes JD, Chesnut RW, Chisari FV, Chang KM, Appella E, Sette A. Recognition of a novel naturally processed, A2 restricted, HCV-NS4 epitope triggers IFN-gamma release in absence of detectable cytopathicity. Hum Immunol. 1998;59:776–82. doi: 10.1016/s0198-8859(98)00080-9. [DOI] [PubMed] [Google Scholar]
- 24.Battegay M, Fikes J, Di Bisceglie AM, et al. Patients with chronic hepatitis C have circulating cytotoxic T cells which recognize hepatitis C virus-encoded peptides binding to HLA-A2.1 molecules. J Virol. 1995;69:2452–70. doi: 10.1128/jvi.69.4.2462-2470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerny A, McHutchison JG, Pasquinelli C, et al. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J Clin Invest. 1995;95:521–30. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cucchiarini M, Kammer AR, Grabscheid B, et al. Vigorous peripheral blood cytotoxic T cell response during the acute phase of hepatitis C virus infection. Cell Immunol. 2000;203:111–23. doi: 10.1006/cimm.2000.1683. 10.1006/cimm.2000.1683. [DOI] [PubMed] [Google Scholar]
- 27.Rehermann B, Chang KM, McHutchinson J, Kokka R, Houghton M, Rice CM, Chisari FV. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. J Virol. 1996;70:7092–102. doi: 10.1128/jvi.70.10.7092-7102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehermann B, Chang KM, McHutchison JG, Kokka R, Houghton M, Chisari FV. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Invest. 1996;98:1432–40. doi: 10.1172/JCI118931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scognamiglio P, Accapezzato D, Casciaro MA, et al. Presence of effector CD8+ T cells in hepatitis C virus-exposed healthy seronegative donors. J Immunol. 1999;162:6681–9. [PubMed] [Google Scholar]
- 30.Wentworth PA, Sette A, Celis E, et al. Identification of A2-restricted hepatitis C virus-specific cytotoxic T lymphocyte epitopes from conserved regions of the viral genome. Int Immunol. 1996;8:651–9. doi: 10.1093/intimm/8.5.651. [DOI] [PubMed] [Google Scholar]
- 31.Jackson M, Smith B, Bevitt DJ, Steward M, Toms GL, Bassendine MF, Diamond AG. Comparison of cytotoxic T-lymphocyte responses to hepatitis C virus core protein in uninfected and infected individuals. J Med Virol. 1999;58:239–46. doi: 10.1002/(sici)1096-9071(199907)58:3<239::aid-jmv9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 32.Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, Chisari FV. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest. 1997;100:2376–85. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang KM, Thimme R, Melpolder JJ, et al. Differential CD4 (+) and CD8 (+) T-cell responsiveness in hepatitis C virus infection. Hepatology. 2001;33:267–76. doi: 10.1053/jhep.2001.21162. [DOI] [PubMed] [Google Scholar]
- 34.Prezzi C, Casciaro MA, Francavilla V, et al. Virus-specific CD8 (+) T cells with type 1 or type 2 cytokine profile are related to different disease activity in chronic hepatitis C virus infection. Eur J Immunol. 2001;31:894–906. doi: 10.1002/1521-4141(200103)31:3<894::aid-immu894>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 35.Ito A, Kanto T, Kuzushita N, et al. Generation of hepatitis C virus-specific cytotoxic T lymphocytes from healthy individuals with pep-tide-pulsed dendritic cells. J Gastroenterol Hepatol. 2001;16:309–16. doi: 10.1046/j.1440-1746.2001.02383.x. 10.1046/j.1440-1746.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- 36.Sarobe P, Pendleton CD, Akatsuka T, Lau D, Engelhard VH, Feinstone SM, Berzofsky JA. Enhanced in vitro potency and in vivo immunogenicity of a CTL epitope from hepatitis C virus core protein following amino acid replacement at secondary HLA-A2.1 binding positions. J Clin Invest. 1998;102:1239–48. doi: 10.1172/JCI3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirai M, Okada H, Nishioka M, et al. An epitope in hepatitis C virus core region recognized by cytotoxic T cells in mice and humans. J Virol. 1994;68:3334–42. doi: 10.1128/jvi.68.5.3334-3342.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirai M, Arichi T, Nishioka M, et al. CTL responses of HLA-A2.1-transgenic mice specific for hepatitis C viral peptides predict epitopes for CTL of humans carrying HLA-A2.1. J Immunol. 1995;154:2733–42. [PubMed] [Google Scholar]
- 39.Sarobe P, Huarte E, Lasarte JJ, Lopez-Diaz de Cerio A, Garcia N, Borras-Cuesta F, Prieto J. Characterization of an immunologically conserved epitope from hepatitis C virus E2 glycoprotein recognized by HLA-A2 restricted cytotoxic T lymphocytes. J Hepatol. 2001;34:321–9. doi: 10.1016/s0168-8278(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 40.Gruener NH, Gerlach TJ, Jung MC, et al. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. J Infect Dis. 2000;181:1528–36. doi: 10.1086/315450. [DOI] [PubMed] [Google Scholar]
- 41.Urbani S, Uggeri J, Matsuura Y, Miyamura T, Penna A, Boni C, Ferrari C. Identification of immunodominant hepatitis C virus (HCV) -specific cytotoxic T-cell epitopes by stimulation with endogenously synthesized HCV antigens. Hepatology. 2001;33:1533–43. doi: 10.1053/jhep.2001.25091. 10.1053/jhep.2001.25091. [DOI] [PubMed] [Google Scholar]
- 42.Tsai SL, Chen YM, Chen MH, et al. Hepatitis C virus variants circumventing cytotoxic T lymphocyte activity as a mechanism of chronicity. Gastroenterology. 1998;115:954–65. doi: 10.1016/s0016-5085(98)70268-9. [DOI] [PubMed] [Google Scholar]
- 43.He XS, Rehermann B, Lopez-Labrador FX, et al. Quantitative analysis of hepatitis C virus-specific CD8 (+) T cells in peripheral blood and liver using peptide-MHC tetramers. Proc Natl Acad Sci U S A. 1999;96:5692–7. doi: 10.1073/pnas.96.10.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koziel MJ, Dudley D, Afdhal N, Grakoui A, Rice CM, Choo QL, Houghton M, Walker BD. HLA class-I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Invest. 1995;96:2311–21. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koziel MJ, Wong DK, Dudley D, Houghton M, Walker BD. Hepatitis C virus-specific cytolytic T lymphocyte and T helper cell responses in seronegative persons. J Infect Dis. 1997;176:859–66. doi: 10.1086/516546. [DOI] [PubMed] [Google Scholar]
- 46.Wong DK, Dudley DD, Dohrenwend PB, Lauer GM, Chung RT, Thomas DL, Walker BD. Detection of diverse hepatitis C virus (HCV)-specific cytotoxic T lymphocytes in peripheral blood of infected persons by screening for responses to all translated proteins of HCV. J Virol. 2001;75:1229–35. doi: 10.1128/JVI.75.3.1229-1235.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruener NH, Lechner F, Jung MC, et al. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J Virol. 2001;75:5550–8. doi: 10.1128/JVI.75.12.5550-5558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurokohchi K, Akatsuka T, Pendleton CD, Takamizawa A, Nishioka M, Battegay M, Feinstone SM, Berzofsky JA. Use of recombinant protein to identify a motif-negative human cytotoxic T-cell epitope presented by HLA-A2 in the hepatitis C virus NS3 region. J Virol. 1996;70:232–40. doi: 10.1128/jvi.70.1.232-240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giuggio VM, Bonkovsky HL, Smith J, Rothman AL. Inefficient recognition of autologous viral sequences by intrahepatic hepatitis C virus-specific cytotoxic T lymphocytes in chronically infected subjects. Virology. 1998;251:132–40. doi: 10.1006/viro.1998.9401. [DOI] [PubMed] [Google Scholar]
- 50.Koziel MJ, Dudley D, Afdhal N, Choo QL, Houghton M, Ralston R, Walker BD. Hepatitis C virus (HCV) -specific cytotoxic T lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522–32. doi: 10.1128/jvi.67.12.7522-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaneko T, Nakamura I, Kita H, Hiroishi K, Moriyama T, Imawari M. Three new cytotoxic T cell epitopes identified within the hepatitis C virus nucleoprotein. J General Virol. 1996;77(6):1305–9. doi: 10.1099/0022-1317-77-6-1305. [DOI] [PubMed] [Google Scholar]
- 52.Chang KM, Gruener NH, Southwood S, Sidney J, Pape GR, Chisari FV, Sette A. Identification of HLA-A3 and – B7-restricted CTL response to hepatitis C virus in patients with acute and chronic hepatitis C. J Immunol. 1999;162:1156–64. [PubMed] [Google Scholar]
- 53.Ando K, Hiroishi K, Kaneko T, et al. Perforin, Fas/Fas ligand, and TNF-alpha pathways as specific and bystander killing mechanisms of hepatitis C virus-specific human CTL. J Immunol. 1997;158:5283–91. [PubMed] [Google Scholar]
- 54.Kita H, Moriyama T, Kaneko T, Okamoto H, Hiroishi K, Ohnishi S, Imawari M. HLA B44-restricted cytotoxic T lymphocyte responses to the peptides of HCV nucleoprotein residues 81–100 in patients with chronic hepatitis C. J Gastroenterol. 1995;30:809–12. doi: 10.1007/BF02349654. [DOI] [PubMed] [Google Scholar]
- 55.Kita H, Hiroishi K, Moriyama T, Okamoto H, Kaneko T, Ohnishi S, Yazaki Y, Imawari M. A minimal and optimal cytotoxic T cell epitope within hepatitis C virus nucleoprotein. J General Virol. 1995;76(12):3189–93. doi: 10.1099/0022-1317-76-12-3189. [DOI] [PubMed] [Google Scholar]
- 56.Hiroishi K, Kita H, Kojima M, et al. Cytotoxic T lymphocyte response and viral load in hepatitis C virus infection. Hepatology. 1997;25:705–12. doi: 10.1002/hep.510250336. [DOI] [PubMed] [Google Scholar]
- 57.Koziel MJ, Dudley D, Wong JT, Dienstag J, Houghton M, Ralston R, Walker BD. Intrahepatic cytotoxic T lymphocytes specific for hepatitis C virus in persons with chronic hepatitis. J Immunol. 1992;149:3339–44. [PubMed] [Google Scholar]
- 58.Ibe M, Sakaguchi T, Tanaka K, et al. Identification and characterization of a cytotoxic T cell epitope of hepatitis C virus presented by HLA-B*3501 in acute hepatitis. J General Virol. 1998;79(7):1735–44. doi: 10.1099/0022-1317-79-7-1735. [DOI] [PubMed] [Google Scholar]
- 59.Kurokohchi K, Arima K, Nishioka M. A novel cytotoxic T-cell epitope presented by HLA-A24 molecule in hepatitis C virus infection. J Hepatol. 2001;34:930–5. doi: 10.1016/s0168-8278(01)00041-1. [DOI] [PubMed] [Google Scholar]
- 60.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J Virol. 2001;75:11392–400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nixon DF, Townsend AR, Elvin JG, Rizza CR, Gallwey J, McMichael AJ. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–7. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 62.Lechner F, Sullivan J, Spiegel H, et al. Why do cytotoxic T lymphocytes fail to eliminate hepatitis C virus? Lessons from studies using major histocompatibility complex class I peptide tetramers. Philos Trans R Soc Lond B Biol Sci. 2000;355:1085–92. doi: 10.1098/rstb.2000.0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karadimitris A, Gadola S, Altamirano M, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci U S A. 2001;98:3294–8. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valiante NM, D’Andrea A, Crotta S, Lechner F, Klenerman P, Nuti S, Wack A, Abrignani S. Life, activation and death of intrahepatic lymphocytes in chronic hepatitis C. Immunol Rev. 2000;174:77–89. doi: 10.1034/j.1600-0528.2002.017417.x. 10.1034/j.1600-065x.2000.174001077.x. [DOI] [PubMed] [Google Scholar]
- 65.Bocharov G, Klenerman P, Ehl S. Predicting the dynamics of antiviral cytotoxic T-cell memory in response to different stimuli: cell population structure and protective function. Immunol Cell Biol. 2001;79:74–86. doi: 10.1046/j.1440-1711.2001.00985.x. [DOI] [PubMed] [Google Scholar]
- 66.Moskophidis D, Laine E, Zinkernagel RM. Peripheral clonal deletion of antiviral memory CD8+ T cells. Eur J Immunol. 1993;23:3306–11. doi: 10.1002/eji.1830231237. [DOI] [PubMed] [Google Scholar]
- 67.Crispe IN. Death and destruction of activated T lymphocytes. Immunol Res. 1999;19:143–57. doi: 10.1007/BF02786483. [DOI] [PubMed] [Google Scholar]
- 68.Zajac AJ, Vance RE, Held W, Sourdive DJ, Altman JD, Raulet DH, Ahmed R. Impaired anti-viral T cell responses due to expression of the Ly49A inhibitory receptor. J Immunol. 1999;163:5526–34. [PubMed] [Google Scholar]
- 69.Ou R, Zhou S, Huang L, Moskophidis D. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal exhaustion of cytotoxic T cells. J Virol. 2001;75:8407–23. doi: 10.1128/JVI.75.18.8407-8423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klenerman P, Meier UC, Phillips RE, McMichael AJ. The effects of natural altered peptide ligands on the whole blood cytotoxic T lymphocyte response to human immunodeficiency virus. Eur J Immunol. 1995;25:1927–31. doi: 10.1002/eji.1830250720. [DOI] [PubMed] [Google Scholar]
- 71.Goulder PJ, Phillips RE, Colbert RA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–7. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 72.Kelleher AD, Long C, Holmes EC, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–86. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goulder PJ, Brander C, Tang Y, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–8. doi: 10.1038/35085576. 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 74.Pircher H, Moskophidis D, Rohrer U, Burki K, Hengartner H, Zinkernagel RM. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629–33. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 75.Allen TM, O’Connor DH, Jing P, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–90. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 76.Moskophidis D, Zinkernagel RM. Immunobiology of cytotoxic T-cell escape mutants of lymphocytic choriomeningitis virus. J Virol. 1995;69:2187–93. doi: 10.1128/jvi.69.4.2187-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ciurea A, Klenerman P, Hunziker L, Horvath E, Senn BM, Ochsenbein AF, Hengartner H, Zinkernagel RM. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc Natl Acad Sci U S A. 2000;97:2749–54. doi: 10.1073/pnas.040558797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klenerman P, Zinkernagel RM. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–5. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 79.Klenerman P, Lechner F, Kantzanou M, Ciurea A, Hengartner H, Zinkernagel R. Viral escape and the failure of cellular immune responses. Science. 2000;289:2003. doi: 10.1126/science.289.5487.2003a. 10.1126/science.289.5487.2003a. [DOI] [PubMed] [Google Scholar]