Abstract
Phenotypic and genotypic evidence suggests that not all Campylobacter jejuni isolates are pathogenic for humans. We hypothesized that differences in gene content or gene expression alter the degree of pathogenicity of C. jejuni isolates. A C. jejuni isolate (Turkey) recovered from a turkey and a second C. jejuni isolate (CS) recovered from a chicken differed in their degrees of in vitro and in vivo virulence. The C. jejuni Turkey isolate invaded INT 407 human epithelial cells and secreted the Cia (Campylobacter invasion antigen) proteins, while the C. jejuni CS isolate was noninvasive for human epithelial cells and did not secrete the Cia proteins. Newborn piglets inoculated with the C. jejuni Turkey isolate developed more severe clinical signs of campylobacteriosis than piglets inoculated with the C. jejuni CS isolate. Additional work revealed that flagellin was not expressed in the C. jejuni CS isolate. Microarray and real-time reverse transcription-PCR analyses revealed that all flagellar class II genes were significantly downregulated in the C. jejuni CS isolate compared to the C. jejuni Turkey isolate. Finally, nucleotide sequencing of the flgR gene revealed the presence of a single residue that was different in the FlgR proteins of the C. jejuni Turkey and CS isolates. Complementation of the C. jejuni CS isolate with a wild-type copy of the flgR gene restored the isolate's motility. Collectively, these findings support the hypothesis that critical differences in gene content or gene expression can alter the pathogenic potential of C. jejuni isolates.
Campylobacter jejuni strains are genetically diverse. The diversity of this organism has been observed by a number of techniques including pulsed-field gel electrophoresis (PFGE) (82), subtractive hybridization (2), microarray analysis (16, 42, 61, 64, 65), and multilocus sequence typing (MLST) (71). Some differences in genetic content between strains can be explained by the presence of polynucleotide tracts in hypervariable loci, including those encoding the lipo-oligosaccharide (LOS), flagellar biosynthesis, and the polysaccharide capsule (60). Such genetic diversity has been problematic in studies attempting to distinguish pathogenic from nonpathogenic isolates. Aeschbacher and Piffaretti (1) reported that there was no clustering of animal and human isolates based on multilocus enzyme electrophoresis using nine polymorphic loci. Based on this result, the authors concluded that every animal isolate is a potential human pathogen. Manning et al. (47) performed MLST on 266 C. jejuni isolates and found that the populations of C. jejuni veterinary and human isolates overlapped among the 19 clonal complexes identified.
The ability of C. jejuni to cause disease is a complex, multifactorial process. Potential virulence determinants include toxins (7, 11, 26, 34, 40, 46, 48, 63, 69, 79, 85), adherence factors (adhesins) (13, 19, 20, 35, 38, 49, 53, 62), and entry-promoting molecules (invasins) (5, 18, 36, 55, 57, 70, 75). Also of interest are strain-variable genes, which encode factors involved in iron acquisition, DNA restriction/modification, sialylation, flagellar biosynthesis, LOS biosynthesis, and capsular biosynthesis (60, 81). Investigators have proposed that strain-variable genes encode factors that contribute to different disease presentations and enable the organism to survive in unique ecological niches. Regardless, an established set of C. jejuni virulence genes and a determination of their contribution to disease production have yet to emerge.
To better understand the metabolic capacity and virulence properties of C. jejuni, the genome sequence of strain NCTC 11168 was determined (60). Shortly thereafter a comparison of C. jejuni isolates by whole-genome microarray analysis revealed extensive genetic diversity (16). Based on the comparison of 11 C. jejuni isolates, 21% of the genes in the C. jejuni NCTC 11168 strain sequence were proposed to be dispensable as they were either absent or highly divergent among the other isolates included in the study. Dorrell et al. (16) also noted that many of the virulence genes identified to date are conserved among C. jejuni strains.
Published reports suggest that not all C. jejuni strains have the same virulence potential. Everest et al. (18) noted that 86% of Campylobacter isolates from individuals with colitis were able to translocate across Caco-2 polarized cells versus 48% of strains isolated from individuals with noninflammatory disease. Fauchère et al. (20) found that C. jejuni recovered from individuals with fever and diarrhea adhered to cultured cells with greater efficiency than those isolated from asymptomatic individuals. The relative ability of C. jejuni to invade cultured cells also appears to be strain dependent (18, 38, 54). Newell et al. (54) found that environmental isolates were much less invasive for HeLa cells than clinical isolates as determined by immunofluorescence and electron microscopy examination of C. jejuni-infected cells. Moreover, a statistically significant difference in the level of invasion between C. jejuni isolates from individuals with colitis and those from individuals with noninflammatory diarrhea has been observed (18).
Poly et al. (65) compared two unrelated C. jejuni strains (ATCC 43431 and NCTC 11168) by a shotgun genomic DNA microarray technique. Not surprisingly, a large number of genes unique to ATCC 43431 encoded proteins involved in LOS and capsular synthesis. Interestingly, 36 of 130 DNA fragments unique to ATCC 43431 showed similarity to DNA from the Helicobacter hepaticus Imp locus, located on a putative pathogenicity island. In other organisms, the Imp locus has been proposed to modulate host responses (9). More recently, Poly et al. (64) compared the genomic content of C. jejuni strain 81-176 with that of C. jejuni strain NCTC 11168 and found divergence at LOS, capsular biosynthesis, and restriction/modification system loci.
In this study, we assessed the putative virulence properties of C. jejuni isolates recovered from poultry. More specifically, we analyzed the virulence attributes of two C. jejuni isolates, designated Turkey and CS, because they were indistinguishable by PFGE, MLST, and genomotyping. The C. jejuni Turkey isolate showed greater pathogenic potential than the C. jejuni CS isolate based on both in vitro and in vivo assays. The lack in pathogenic potential of the C. jejuni CS isolate, relative to the C. jejuni Turkey isolate, was attributed to a loss in the synthesis of a functional flagellum. The flagellum and associated motility are a known C. jejuni virulence determinant (8, 51, 53, 77, 78, 84). In addition, host cell invasion and the secretion of the Cia (Campylobacter invasion antigen) proteins require a functional flagellar export apparatus (39). We identified a point mutation in the flgR gene of the C. jejuni CS isolate. The flgR gene in C. jejuni encodes the response regulator of the FlgS/FlgR two-component signal transduction system that participates in flagellar biosynthesis. Phosphorylation of the C-terminal output domain of the FlgR response regulator protein presumably results in its binding to DNA, whereby it interacts with σ54 to initiate the expression of flagellar class II genes. The class II flagellar genes encode proteins that are required for the assembly of a functional flagellum (32, 33).
While other investigators have examined differences in C. jejuni isogenic strains or passage variants, we are unaware of another study undertaken to examine the virulence attributes of naturally occurring clonally matched isolates. We identified a difference in the nucleotide sequences of the flgR genes in C. jejuni Turkey and CS isolates, which resulted in the nonmotile and avirulent phenotype of the C. jejuni CS isolate. This mutation appears to provide the isolate a fitness advantage in the environment. More specifically, the C. jejuni CS isolate exhibited greater resistance to the antimicrobial activity of the bile salt deoxycholate and is thus able to reach a greater cell density in deoxycholate-supplemented medium than a genotypically matched isolate. Further, the C. jejuni Turkey isolate harboring a mutation in flgR mirrored the phenotype of the C. jejuni CS isolate.
(A portion of this work was presented at 105th General Meeting of the American Society for Microbiology, Atlanta, GA, 5 to 9 June 2005, and at the Conference of Research Workers in Animal Diseases, Chicago, IL, 13 to 15 November 2004.)
MATERIALS AND METHODS
Culture of bacterial isolates.
Two C. jejuni human clinical isolates (C. jejuni F38011 and 81-176) and six C. jejuni poultry isolates were used throughout this study. Five of the six C. jejuni poultry isolates were recovered from chicken rinses (CS, S1, S2B, S3, and SC), and one isolate was recovered from the liver of a turkey (Turkey) exhibiting clinical signs of poultry enteritis. All C. jejuni isolates were cultured at 37°C under microaerobic conditions (5% O2, 10% CO2, 85% N2) on Mueller-Hinton (MH) agar plates supplemented with 5% citrated bovine blood (MH-blood agar). Cultures were subcultured to a fresh plate every 24 to 48 h. In addition to the six C. jejuni isolates used throughout this study, we also screened an additional 99 C. jejuni poultry isolates for motility. These isolates used were recovered from either poultry processing plants located in Kansas (A1a, A2a, A3a, A4a, A5a, A7a, A8a, A9a, A11a, A12a, A13a, A14a, A15a, A16a, A18a, A20a, A22a, A23a, A24a, A25a, and A37a), Iowa (D28a, D30a, D31a, D32a, D33a, D34a, D37a, D38a, D41a, D42a, D44a, D45a, D48a, and D49a), and Washington state (B13a, B19a, C11a, C18a, C20a, C24a, C25a, G9a, G11a, G12a, G13a, G15a, G17a, G21a, G22a, G26a, H1a, H2a, H3a, H4a, H5a, H8a, H29a, I6a, I7a, I16a, and I22a) or from cecal droppings collected at different broiler houses (2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 14, 15, 16, 19, 21, 22, 23, 24, 25, 26, 33, 34, 35, 36, 39, 41, 42, 43a, 43c, 44, 77, 78, 79, 80, 81, and 83). All the C. jejuni poultry isolates used in this study were passaged less than five times in the laboratory. The primary stock of each of C. jejuni isolate was frozen in citrated bovine blood and stored at −80°C. Escherichia coli INVαF′ (Invitrogen, Carlsbad, CA), S17-1 λ pir, and XL1-Blue MRF′ (Stratagene, La Jolla, CA) were cultured on Luria-Bertani (LB) agar plates at 37°C.
Culture of INT 407 cells.
A culture of INT 407 human epithelial cells (ATCC CCL6) was obtained from the American Type Culture Collection (Manassas, VA). Stock cultures of INT 407 cells were grown in minimal essential medium (MEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT). Cultures were maintained at 37°C in a humidified, 5% CO2 incubator.
C. jejuni-INT 407 cell binding and invasion assay.
For experimental assays, each well of a 24-well tissue culture tray was seeded with 1.4 × 105 INT 407 cells and incubated for 18 h at 37°C in a humidified, 5% CO2 incubator. The cells were rinsed with MEM-1% FBS and inoculated with a suspension of approximately 5 × 107 CFU of bacteria. Tissue culture trays were centrifuged at 600 × g for 5 min and incubated at 37°C in a humidified, 5% CO2 incubator. For binding, the infected monolayers were incubated for 30 min and rinsed three times with phosphate-buffered saline (PBS) and the epithelial cells were lysed with 0.1% (vol/vol) Triton X-100 (Calbiochem, La Jolla, CA). The suspensions were 10-fold serially diluted, and the number of viable, adherent bacteria was determined by counting the resultant colonies on MH-blood agar plates. To measure bacterial internalization, the infected monolayers were incubated for 3 h, rinsed three times with MEM-1% FBS, and incubated for an additional 3 h in MEM-1% FBS containing a bactericidal concentration (250 μg/ml) of gentamicin. The number of internalized bacteria was determined as outlined above. Unless otherwise stated, the reported values represent the mean counts ± standard deviations derived from triplicate wells. All assays were performed a minimum of three times to ensure reproducibility. Significance between samples was determined using Student's t test following log10 transformation of the data. Two-tailed P values were determined for each sample, and a P value <0.01 was considered significant.
Secretion assay.
C. jejuni isolates were harvested from MH-blood agar plates in PBS, pelleted by centrifugation at 6,000 × g, washed twice in MEM, and suspended in MEM lacking methionine (labeling medium; ICN Biomedicals, Inc., Aurora, OH) to an optical density at 540 nm (OD540) of 0.3 (approximately 5 × 108 CFU). Metabolic labeling experiments were performed in 3 ml of labeling medium with the addition of [35S]methionine (PerkinElmer Life Sciences, Inc., Boston, MA) as described elsewhere (37). Following a 3-h labeling period, bacterial cells were pelleted by centrifugation at 6,000 × g and supernatant fluids collected. The supernatant fluids were concentrated fourfold by the addition of 5 volumes of ice-cold 1 mM HCl-acetone followed by centrifugation. The pellets were air dried and resuspended in water. The secreted proteins were resolved in sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gels using the discontinuous buffer system described by Laemmli (41). Gels were treated with Amplify (Amersham, Piscataway, NJ) according to the manufacturer's instructions. Autoradiography was performed with Kodak BioMax MR film at −70°C.
Motility assay.
Motility assays were performed using MH medium supplemented with 0.4% Select agar (motility agar plates; Life Technologies, United Kingdom). A 10-μl suspension of each bacterial isolate was spotted on the surface of the semisolid medium. Motility plates were incubated at 37°C under microaerobic conditions for 48 h.
Macrorestriction enzyme PFGE.
C. jejuni isolates were harvested from MH-blood agar plates in 3 ml PETT IV buffer (1 M NaCl, 10 mM Tris, 10 mM EDTA, pH 8.0) and cell densities adjusted to 0.75 using a Microscan turbidity meter (Dade Behring, West Sacramento, CA) in 12- by 75-mm Falcon round-bottom tubes (Becton Dickinson, Franklin Lakes, NJ). Four hundred microliters of 1.4% (wt/vol) molten (50°C) pulsed-field grade agarose (Bio-Rad, Hercules, CA) was added to an equivalent volume of each bacterial suspension, the suspension was mixed gently, and a 100-μl aliquot was pipetted into agarose plug molds. The agarose plugs were removed from the molds and incubated in 1 ml of ESP buffer (50 mM Tris-HCl [pH 8.0], 50 mM EDTA, 1% [wt/vol] N-lauroylsarcosine, 0.5 mg/ml proteinase K) at 50°C for 1 h. Following cell wall lysis, the agarose blocks were washed three times in sterile water and three times in TE (10 mM Tris, pH 8.0, 1 mM EDTA). Each wash was performed at ambient temperature for 30 min. Individual agarose plugs were incubated with 100 μl of 1× restriction endonuclease buffer containing 20 U of SmaI. The reaction mixtures were incubated at 25°C for a minimum of 4 h. Following incubation, the agarose plugs were loaded into an agarose gel. Restricted genomic DNA was separated in 1% (wt/vol) pulsed-field grade agarose prepared with 0.5× TBE (0.089 M Tris base, 0.089 M boric acid, 0.002 M EDTA [pH 8.0]). Run parameters consisted of a reorientation angle of 120° with a constant voltage of 120 V and a constant temperature of 14°C. An electrophoresis run time of 19 h and a ramped pulse time of 6.8 to 35.4 s were used. For plugs digested with SalI, a run time of 18 h and a ramped pulse time of 5.2 to 42.3 s were used. Gels were stained for 20 min in 3 μg/ml ethidium bromide and destained for 20 min in water. Images were captured using a Bio-Rad FluorS system and processed using Adobe Photoshop.
MLST.
MLST was performed as described by Dingle et al. (15). Portions of the housekeeping genes encoding aspartase A (aspA), glutamine synthetase (glnA), citrate synthase (gltA), serine hydroxymethyl transferase (glyA), phosphoglucomutase (pgm), transketolase (tkt), and the ATP synthase alpha subunit (uncA) were amplified and sequenced. Genomic DNA was recovered from C. jejuni isolates harvested from MH-blood agar plates. Amplification of each gene fragment was performed using approximately 10 ng of genomic DNA and 1.25 U of Taq DNA polymerase (Invitrogen) in a 50-μl reaction volume containing 1 μM forward and reverse primer, 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, and 0.8 mM deoxynucleoside triphosphates (dNTPs). Amplification reactions consisted of the following cycling conditions: an initial denaturation step (94°C, 2 min) followed by annealing (50°C, 1 min) and extension (72°C, 1 min) steps, which were repeated for 35 cycles. PCR amplicons were assessed for quantity and quality by agarose gel electrophoresis. Primers and unincorporated dNTPs were removed by passing amplicons through a commercially available batch column purification system (Qiaquick PCR kit; QIAGEN, Valencia, CA). The nucleotide sequences of PCR amplicons were determined using a modification of Sanger dideoxynucleotide sequencing, incorporating fluorescence dye technology, at the DNA sequencing facility of Washington State University. Sequencing was performed on both strands for a segment of each allele from each isolate. Nucleotide sequences for each allele were aligned using the program Clustal X, and the nucleotide sequences were assigned alleles from the MLST website (http://mlst.zoo.ox.ac.uk). Sequence types, based on the combination of assigned alleles to the seven housekeeping genes, were determined using the MLST website (http://mlst.zoo.ox.ac.uk).
Inoculation of piglets.
Newborn piglets were obtained from sows at the time of farrowing. Feces from each sow were plated on selective Abeyta Hunt Bark agar plates containing sodium cefoperazone (32 mg/liter), rifampin (10 mg/liter), amphotericin B (2 mg/liter), and 4 ml/liter of FBP (62.5 g/liter sodium pyruvate, 62.5 g/liter ferrous sulfate, and 62.5 g/liter sodium metabisulfite). Suspect colonies were tested for C. jejuni as described by Nogva et al. (56). Only piglets obtained from C. jejuni-free sows were used for in vivo studies.
Each piglet was fed five times daily with 50 ml of Similac formula (Ross Laboratories). Overnight-grown cultures of C. jejuni were resuspended in Similac at a concentration of approximately 5 × 109 CFU/ml. Piglets were orally inoculated with approximately 20 ml of either the C. jejuni Turkey or CS isolate. Actual numbers of viable bacteria inoculated were determined by serial dilution and plating of the bacterial suspension.
Piglets were observed for diarrhea, and rectal swabs were plated on Abeyta Hunt Bark medium prior to inoculation and daily thereafter to determine the presence of C. jejuni shedding. Piglets were euthanized when morbidity was observed or at 6 days postinoculation with 2 ml of Beuthanasia-D (Schering Corp.). Gross examinations of the small intestine, colon, and cecum were recorded, and tissue samples were fixed in 10% buffered formalin and examined for histological lesions at the University of Arizona Veterinary Diagnostic Laboratory (Tucson, AZ).
Preparation of whole-cell lysates, outer membrane proteins (OMP), and flagellin.
C. jejuni whole-cell lysates were generated by harvesting bacteria grown overnight on MH-blood agar plates in PBS and subjecting the bacterial suspension to sonication on ice (five 30-s pulses).
OMP were prepared as described by deMelo and Pechère (13) with minor modifications. Briefly, bacterial sonicates were centrifuged at 8,000 × g to remove whole bacterial cells. Supernatant fluids were centrifuged at 100,000 × g for 2 h. The resulting pellets were resuspended in a solution of 1% Sarkosyl (Sigma, St. Louis, MO) in 10 mM Tris-HCl (pH 7.5) and allowed to incubate at ambient temperature for 30 min on a platform rocker. The solutions were centrifuged at 100,000 × g for 2 h, and the supernatants were removed. The pellets were washed in 10 mM Tris-HCl (pH 7.5) and centrifuged at 100,000 × g for 2 h. Finally, the pellets were resuspended in 50 mM Tris-HCl (pH 7.5), and the protein concentration was determined by the BCA protein assay kit according to the manufacturer's instructions (Pierce, Rockford, IL).
Flagella were purified as described by Alm et al. (3). Bacteria were resuspended in PBS and homogenized twice for 3 min. The bacterial suspensions were centrifuged at 8,000 × g to remove whole cells. The supernatant fluids were centrifuged at 100,000 × g for 1 h. The resulting pellets were resuspended in 1% SDS and allowed to incubate at ambient temperature. The solutions were centrifuged at 100,000 × g for 1 h, and the resulting pellets were washed twice in distilled water and centrifuged as above. The final pellets were resuspended in 10 mM Tris-HCl (pH 7.0), and the protein concentration was determined as described above. All protein extracts were stored at −20°C.
Gel electrophoresis.
Two-dimensional gel electrophoresis was performed using the parameters described elsewhere (37). One-dimensional gel electrophoresis was performed by mixing an equal volume of a bacterial suspension (an equivalent of 0.1 OD600 unit) with double-strength electrophoresis sample buffer (37). The samples were placed in boiling water for 5 min and allowed to cool to ambient temperature. Proteins were resolved by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) using the discontinuous buffer system described by Laemmli (41).
Immunoblot analysis.
Proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore Corp., Bedford, MA). The membranes were washed three times in PBS and incubated for 18 h at 4°C with a 1:500 dilution of a rabbit anti-C. jejuni flagellin antibody in PBS (pH 7.4)-0.01% Tween 20 containing 9% dried milk. Bound antibodies were detected using peroxidase-conjugated rabbit anti-goat immunoglobulin G (Sigma) at a 1:1,000 dilution and 4-chloro-1-naphthol (Sigma) as the chromogenic substrate.
RNA isolation.
Total cellular RNA was isolated from the C. jejuni Turkey and CS isolates grown to mid-log phase using the hot-phenol method (74). Briefly, 5 ml of RNA degradation stop solution was added (10% phenol solution, buffered saturated with 0.1 M citrate buffer, pH 4.3 + 0.2, in 100% ethanol) to 50 ml of each bacterial culture. The bacterial suspension was then pelleted and resuspended in 3 ml of 1% buffer-saturated phenol solution. The culture was pipetted into 1.5 ml of boiling lysis buffer in a water bath until cell lysis occurred. This was followed by two extractions with phenol at 60°C and one extraction with phenol-chloroform, and the aqueous phase was precipitated with ethanol and resuspended in diethyl pyrocarbonate-treated water. Contaminating genomic DNA was removed by two consecutive treatments with RQ1-DNase (Promega, Rockford, IL). The absence of genomic DNA was confirmed via PCR using C. jejuni ciaB gene sequence-specific primers (CiaB-F, CTATGCTAGCCATACTTAGGC; CiaB-R, GCCCGCCTTAGAACTTAC). The PCR conditions used were 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, with a final extension time of 5 min (35 cycles in total).
Construction of the C. jejuni DNA microarray.
DNA fragments of individual open reading frames (ORFs) were amplified using primers specific for ORFs present in strain NCTC 11168 (Sigma Genosys, The Woodlands, TX). Each PCR (total reaction volume, 100 μl) consisted of 1× MasterAmp Taq PCR buffer, 1× MasterAmp Taq enhancer, 2.5 mM MgCl2, 200 μM each dNTP, forward and reverse primers at 0.2 μM each, 0.5 U of MasterAmp Taq DNA polymerase (Epicenter, Madison, WI), and approximately 50 ng of genomic DNA from strain NCTC 11168. Thermal cycling was performed using a Tetrad thermal cycler (MJ Research, Waltham, MA) with the following amplification parameters: 30 cycles of 25 s at 94°C, 25 s at 52°C, and 2 min at 72°C and a final extension at 72°C for 5 min. PCR products were analyzed by gel electrophoresis in a 1% (wt/vol) agarose gel (containing 0.5 μg of ethidium bromide/ml) in 1× Tris-acetate-EDTA buffer. DNA bands were examined under UV illumination. A total of 1,530 PCR products were successfully amplified and then purified on a QIAGEN 8000 robot using a Qiaquick 96-well Biorobot kit (QIAGEN), dried, and resuspended to an average concentration of 0.1 to 0.2 μg/μl in 20 μl of 50% dimethyl sulfoxide containing 0.3× saline sodium citrate (SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). All of the PCR probes were then spotted in duplicate on GAPSII slides (Corning, Acton, MA) using an OmniGrid Accent (GeneMachines, Ann Arbor, MI) producing a final array that contained a total of 3,060 features.
Fluorescent labeling of genomic DNA and cDNA.
For each comparative genomic microarray hybridization reaction, genomic DNAs from the reference strain NCTC 11168 and a test strain were fluorescently labeled with Cy5 and Cy3, respectively. Approximately 2 μg of DNA was mixed with 5 μl 10× NEBlot labeling buffer containing random sequence octamer oligonucleotides (New England Biolabs, Beverly, MA) and water to a final volume of 41 μl. This mixture was heated to 95°C for 10 min and then stored for 5 min at 4°C. After this incubation, the remainder of the labeling reaction components were added: 5 μl of 10× dNTP labeling mixture (1.2 mM each dATP, dGTP, dCTP; 0.5 mM dTTP in 10 mM Tris, pH 8.0; 1 mM EDTA), 3 μl of Cy3-dUTP or Cy5-dUTP (Amersham Biosciences, Piscataway, NJ), and 1 μl of Klenow fragment. The labeling reaction mixtures were incubated overnight at 37°C. Fluorescently labeled DNA was purified using Qiaquick PCR purification columns (QIAGEN, Valencia, CA) according to manufacturer's directions.
For the expression profiling arrays, an indirect comparison of gene expression levels was performed (83), where the levels of expression from the various C. jejuni isolates were measured separately on different slides. In this microarray experimental design, each labeled cDNA is combined with a fixed reference source (83), allowing the comparison of different experiments in which a common reference has been used, as in previous studies (17, 45). Sixteen micrograms of total RNA from each C. jejuni isolate was labeled during reverse transcription to cDNA with Cy5-dUTP using Stratascript (Stratagene, Palo Alto, CA). Following 16 h of labeling, RNA was degraded by the addition of NaOH to 0.3 M and incubation at 70°C for 10 min, followed by neutralization with an equimolar amount of HCl. The labeled cDNA was purified using Qiaquick PCR purification columns (QIAGEN) according to the manufacturer's directions. In all expression microarray hybridizations, genomic DNA from C. jejuni strain NCTC 11168 was labeled with Cy3-dUTP as described above. The labeled genomic DNA also served as a quality control for all the spots in the array. In this particular indirect comparison for gene expression, an analysis of repeatable residual color bias from Cy dye swap experiments was removed since the between-slide differences were taken into account (see below) (83).
Microarray hybridization.
For each genomic indexing hybridization, Cy5-labeled reference DNA from C. jejuni strain NCTC 11168 was mixed with Cy3-labeled test DNA or cDNA in 45 μl of Corning hybridization buffer (Corning, Acton, MA) and heated to 95°C for 5 min. Then 15 μl of the hybridization mixture was put onto the microarray slide and sealed with a coverslip in a GeneMachine hybridization chamber (Genomic Solutions, Ann Arbor, MA) and incubated at 42°C for 18 h. This method, known as differential labeling, allows the hybridization of fluorescently labeled control and test DNA or cDNA to be measured for each probe on the microarray. As the genetic composition of the sequenced strain C. jejuni NCTC 11168 is known, it served as a control fluorescence signal for each probe and was used for comparison with the test DNA signal during the statistical analysis (see below). Following hybridization, microarray slides were washed for 2 min in 2× SSC and 0.1% SDS at 42°C to remove the coverslip and then washed twice for 5 min in each of the following buffers: (i) 2× SSC and 0.1% SDS at 42°C, (ii) 0.2× SSC, and finally (iii) 0.01× SSC. Microarray slides were dried by centrifugation at 300 × g for 15 min before scanning.
Microarray data analysis.
DNA microarrays were scanned using a GenePix 4000B microarray laser scanner (Axon Instruments, Union City, CA), and the data for spot and background intensities were processed using GenePix 4.0 software. Poor features were excluded from analysis if they contained abnormalities or a reference signal lower than background plus 3 standard deviations. As described by Eriksson et al. (17), fluorescence ratios were calculated after local background was subtracted from spot signals. To compensate for any effect of the amount of template and uneven Cy-dye incorporation, data normalization was performed as previously described (4, 17, 45) by bringing the median natural logarithm of the ratios for each group of spots printed by the same pin to zero using the following equation: ln(Ti) = ln(Ri/Gi) − c, where T is the centered ratio, i is the gene index, R and G are the red and green intensities, respectively, and c is the 50th percentile of all red/green ratios. Normalized data that passed the quality controls were analyzed using GENESPRING 6.2 software (Silicon Genetics, Palo Alto, CA). For comparison of levels of C. jejuni Turkey and CS gene expression, at least four hybridization measurements were generated per biological experiment (two technical replicate arrays and two replicate features per array) and the experiment was repeated two times (biological replicate). The significance of the centered data at P values >0.05 was determined using a parameter-based statistical t test adjusting the individual P value with the Benjamini and Hochberg false-discovery rate multiple test correction within the GeneSpring analysis package.
Sequencing genes involved in flagellar biosynthesis.
The genes of interest (flhA, flhB, flhF, fliA, fliF, fliG, fliH, fliI, fliM, fliN, fliP, fliQ, fliR, fliY, flgR, flgS, rpoN, Cj0667, Cj0668, and Cj0669) were PCR amplified using KOD DNA polymerase (Novagen, San Diego, CA) and sequenced using gene-specific primers. Both strands of every gene above from both the C. jejuni Turkey and CS isolates were sequenced. All sequencing primers were designed based on C. jejuni NCTC 11168 genomic sequences (60). Sequencing was performed using the Big Dye Terminator kit (Applied Biosystems, Foster City, CA) and the DNA sequencer ABI373 (Applied Biosystems, Foster City, CA). The sequences were analyzed using the MultAlin multiple sequence alignment program.
Real-time RT-PCR.
The relative expression of 26 genes (clpB, cmeA, flaA, flaB, flaC, flgB, flgC, flgD, flgE, flgG, flgH, flgK, fliF, fliG, fliH, fliI, fliM, fliY, flhA, flhB, metA, rpoN, Cj1514, Cj0667, Cj0668, and Cj0669) was determined by real-time quantitative reverse transcription-PCR (RT-PCR). cDNA was synthesized using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's directions. Real-time RT-PCR amplification of 0.5 μl of cDNA was performed in a reaction mixture containing Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA), a 300 nM concentration of forward and reverse primers, and diethyl pyrocarbonate-treated water. Real-time RT-PCR analysis was performed using a Gene Amp 7000 thermocycler (Applied Biosystems, Foster City, CA). PCR conditions included 1 cycle of 2 min at 50°C, followed by 40 cycles of denaturation at 95°C for 15 s and an annealing at 55°C for 1 min. Cycle threshold (CT) values were determined using Prism SDS software version 1.0 (Applied Biosystems). The comparative threshold cycle (ΔΔCT) method was used to calculate change (n-fold) where samples were normalized to glnA, since glnA is a housekeeping gene and was not found to be differentially expressed by microarray analysis. Reactions were performed in duplicate, and two biological replicates were performed for each sample.
Construction of reporter vectors.
The promoter regions of the flaB and metK genes were PCR amplified using the following primer sets: primer set 1, (PmetK-F, ATTTGGATCCCCTTGTGCTCCTGTTTGTGC, and PmetK-R, ATATGGATCCAAAAAGTCCTTTCATTTAAAATGAACC) and, primer set 2 (PflaB-F, AAGGATCCACACTTAAAGGCGCTATGGCTGTGATG, and PflaB-R, AAGGATCCGATGTTGGTGTTTATCCTAAAACC). Primers were designed to include a BamHI site at the 5′ end. The PCR products were cloned into pCR 2.1 using the TOPO TA cloning kit (Invitrogen). The ligated vector was electroporated into E. coli INVαF′, and the transformants were selected on LB agar supplemented with kanamycin (KAN; 50 μg/ml). PflaB-pCR2.1 and PmetK-pCR2.1 were digested with BamHI to give 0.8-kb and 0.78-kb fragments, respectively. Each fragment was gel purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). The pMW10 shuttle vector (80) was digested with BamHI, gel purified, and ligated with the BamHI-PflaB and BamHI-PmetK fragments for 18 h. The ligation mixture was ethanol precipitated and electroporated into E. coli INVαF′. The transformants were selected on LB-KAN agar and tested for insertion by blue-white screening. The sequences of the constructs were verified using a primer that anneals to the aphA3′ gene encoding KAN resistance (TACACTCAAATGGTTCGCTG) as described previously (80).
Electroporation of promoter shuttle vectors in the C. jejuni F38011 and CS isolates.
The vectors (pMW10, PmetK-pMW10, and PflaB-pMW10) were introduced into the C. jejuni F38011 and CS strains by electroporation. The resultant colonies were selected on MH-blood agar plates supplemented with KAN (200 μg/ml).
β-Galactosidase assay.
β-Galactosidase activity was determined as a measure of conversion of o-nitrophenyl-d-galactopyranoside (Sigma) to nitrophenol. C. jejuni transformants were grown for 16 h in MH broth. Harvested bacteria were diluted in PBS until the OD600 was between 0.4 and 0.5. The assay was carried out in triplicate as previously described (80).
Construction of the complementation vectors.
To construct the flgR pRY107 (KAN resistance) complementation vector, the coding region of the flgR gene along with its native promoter (350 bp upstream of the start codon) was PCR amplified using primers containing 5′ BamHI restriction sites (flgRcompF, TATATAGGATCCAATGGGTATGTGAAATTTCTTATAGTG; flgRcompR, TATATAGGATCCCCCAACCGCACCAGTAGC). The PCR product was cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen). The ligated vector was electroporated into E. coli INVαF′, and the transformants were selected on LB-KAN plates (50 μg/ml). The pCR2.1 construct containing the BamHI flgR fragment was digested with BamHI, gel purified, and cloned into the BamHI site of pRY107. The ligated vector was electroporated into E. coli INVαF′, and transformants were selected on LB-KAN plates (50 μg/ml).
To construct the rpoN pRY111 (chloramphenicol [CHL] resistance) complementation vector, the promoter region of Cj0667 was fused with the rpoN ORF. Noteworthy is that the rpoN gene is the fourth gene within the operon (5′-Cj0667-Cj0668-Cj0669-rpoN). The promoter region of Cj0667 was PCR amplified using primers containing 5′ SstI and NdeI restriction sites (Cj0667-SstI-F, TATATAGAGCTCTCAAGGCGTCCTATACCGTG; Cj0667-NdeI-R2, ATATATACATATGTTCAGAAATAGCTCTTC). The rpoN ORF was PCR amplified using primers containing 5′ NdeI and SstI restriction sites (rpoN-NdeI-F, TATAATCATATGTTAAAGCAAAAAATCACCCAAG; rpoN-SstI-R, TATAATCCGCGGAATATTTAAAAACAGTTATTATTGTATCAC). The PCR amplicons were cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen). The ligated vector was electroporated into E. coli INVαF′, and transformants were selected on LB-KAN plates (50 μg/ml). The rpoN-pCR2.1 construct was digested with NdeI and SstI, and the gel-purified fragment was ligated into the NdeI- and SstI-restricted PCj0667-pCR2.1 construct. The ligated vector was electroporated into E. coli INVαF′, and transformants were selected on LB-KAN plates (50 μg/ml). The PCj0776-rpoN-pCR2.1 fusion vector was digested with SstI, and the SstI-PCj0667-rpoN fragment was gel purified and cloned into the SstI site of pRY111. The resulting vector was electroporated into E. coli INVαF′, and transformants were selected on LB agar plates supplemented with CHL (LB-CHL).
Conjugation.
The complementation plasmids were electroporated into E. coli S17-1 λ pir. E. coli S17-1 λ pir harboring the complementation plasmid flgR-pRY107 was then conjugated with the C. jejuni CS isolate. The transconjugants were selected on MH-blood agar plates supplemented with KAN (200 μg/ml) and tetracycline (TET, 50 μg/ml). The C. jejuni CS isolate colonies resistant to both KAN and TET were screened for motility on motility agar plates (Life Technologies, United Kingdom).
E. coli S17-1 λ pir harboring the PCj0667-rpoN-pRY111 complementation plasmid was conjugated with the C. jejuni S2B isolate. The transconjugants were selected on MH-blood agar plates supplemented with CHL (20 μg/ml) and TET (50 μg/ml). The C. jejuni S2B isolate colonies resistant to both KAN and TET were screened for motility on motility agar plates.
Generation of the C. jejuni Turkey flgR mutant.
A disrupted copy of the flgR gene was amplified from the C. jejuni F38011 flgR-cat mutant using flgR gene-specific primers (66), and the resultant product was cloned into pCR2.1. The flgR-cat fragment was digested using EcoRI, gel purified, and cloned into the EcoRI site of the pBluescriptII SK(+) vector. The ligated vector was electroporated into E. coli INVαF′ and selected on LB-CHL. The mutagenesis vector was introduced into the C. jejuni Turkey isolate by electroporation and selected on MH-blood agar plates supplemented with CHL. The putative mutants were confirmed by PCR amplification of flgR, followed by digestion of the PCR product by NheI to release the cat cassette.
Growth curves.
Overnight cultures of C. jejuni Turkey and CS isolates were used to inoculate 250 ml of MH broth at an OD540 of 0.01. The flasks were incubated at 37°C under microaerobic conditions and shaken at 150 rpm. At different time points, 1 ml of bacterial culture was used to determine the OD540 until both cultures reached stationary phase.
Sensitivity to deoxycholate.
Overnight cultures of C. jejuni Turkey, the C. jejuni Turkey flgR mutant, and CS isolates were used to inoculate tubes at an OD540 of 0.05. Different concentrations of sodium deoxycholate were added to each tube in triplicate, and tubes were incubated for 48 h at 37°C under microaerobic conditions and shaken at 150 rpm. At the end of the incubation period, the OD540 for each tube was measured.
Other analytical procedures.
Protein concentrations were determined by the bicinchoninic acid method, with bovine serum albumin as the standard, as outlined by the supplier (Pierce, Rockford, IL). To determine the identity of the protein present in the flagellar extract prepared from the C. jejuni Turkey isolate, the extract was mixed with an equal volume of double-strength electrophoresis sample buffer sample and separated by SDS-PAGE. The proteins were transferred to PVDF membranes and stained with Coomassie brilliant blue G-250 (CBB-G250). The amino-terminal sequence of the 62-kDa protein was determined at the University of British Columbia Biotechnology Laboratory (Protein Sequencing and Peptide Mapping, NAPS Unit, Vancouver, B.C., Canada).
RESULTS
C. jejuni isolates display marked differences in virulence phenotypes.
While there are several reports documenting the variation in the efficiency of C. jejuni isolates to invade cultured cells (18, 38, 54), the molecular mechanisms for these variations have seldom been elucidated. To identify C. jejuni isolates with apparent differences in virulence potential, we performed INT 407 cell invasion assays and secretion assays with six C. jejuni poultry isolates. C. jejuni invasion of INT 407 cells was assessed using the gentamicin protection assay. C. jejuni-INT 407 host cell contact was promoted by a low-speed centrifugation step immediately following inoculation. The levels of invasiveness of the six C. jejuni poultry isolates were markedly different (Table 1). Relative to the C. jejuni 81-176 clinical strain, the C. jejuni Turkey strain was the most invasive environmental isolate tested and the C. jejuni S2B strain was the least invasive. Because C. jejuni binding is a prerequisite for host cell invasion, the data were transformed and presented as the ratio of intracellular to cell-associated bacteria, expressed as a percentage, to more directly assess the invasive potential of each isolate. Three of the six isolates (CS, S2B, and S3) were readily discernible from the other isolates tested in that they displayed lower invasive potential.
TABLE 1.
Binding and internalization of C. jejuni poultry isolates
| Isolate | Origin | Aa | Ib | I/A (%)c |
|---|---|---|---|---|
| 81-176 | Human | (3.0 ± 0.8) × 105 | (2.2 ± 0.3) × 104 | 7.3 |
| CS | Chicken | (5.8 ± 1.0) × 105 | (5.0 ± 1.8) × 102 | 0.09 |
| S1 | Chicken | (3.1 ± 0.7) × 104 | (9.3 ± 1.5) × 102 | 3.0 |
| S2B | Chicken | (3.5 ± 0.4) × 105 | (1.3 ± 0.5) × 102 | 0.04 |
| S3 | Chicken | (3.9 ± 0.4) × 105 | (9.0 ± 2.0) × 101 | 0.02 |
| SC | Chicken | (1.6 ± 0.2) × 105 | (1.5 ± 0.4) × 103 | 0.9 |
| Turkey | Turkey | (1.2 ± 0.2) × 105 | (4.3 ± 0.5) × 103 | 3.6 |
| E. coli XL1-Blue MRF′ | Stratagene | (3.3 ± 1.0) × 105 | (2.1 ± 0.4) × 102 | 0.07 |
Number of cell-associated bacteria per well of C. jejuni-inoculated monolayers.
Number of internalized bacteria per well of C. jejuni-inoculated monolayers treated with gentamicin for 3 h.
Percentage of cell-associated bacteria that are internalized.
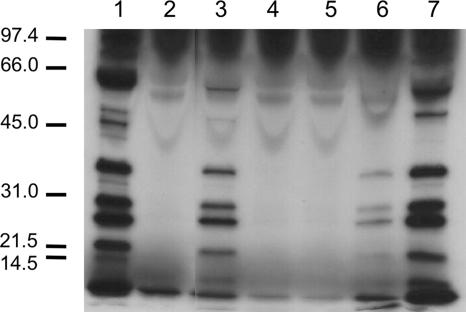
C. jejuni synthesizes and secretes a set of proteins upon coculturing with epithelial cells, which aid in the invasion of the host cells. The secreted proteins are referred to, collectively, as Cia proteins. Since three of the six C. jejuni poultry isolates showed low invasive potential, secretion assays were performed to determine the ability of the isolates to secrete Cia proteins. The assays were performed in the presence of FBS, which serves as an artificial signal to stimulate the synthesis and secretion of the Cia proteins (68). The Cia proteins were readily identifiable in the supernatant fluids of C. jejuni 81-176, S1, SC, and Turkey isolates but not detectable in the supernatant fluids of C. jejuni CS, S2B, and S3 isolates (Fig. 1). The Mrs of the Cia proteins exported from the C. jejuni poultry isolates were consistent with those of the C. jejuni 81-176 human clinical isolate (Fig. 1) and with previous work with other C. jejuni clinical isolates (39, 68). Noteworthy is that the C. jejuni CS, S2B, and S3 isolates displayed a lower invasive potential than the C. jejuni poultry isolates (S1, SC, and Turkey) that secreted the Cia proteins.
FIG. 1.
Some C. jejuni isolates recovered from poultry do not secrete the Cia proteins. C. jejuni isolates 81-176 (lane 1), CS (lane 2), S1 (lane 3), S2B (lane 4), S3 (lane 5), SC (lane 6), and Turkey (lane 7) were incubated in MEM lacking methionine and containing 1% FBS and labeled with [35S]methionine for 3 h. The supernatant fluids were analyzed by SDS-PAGE and autoradiography as outlined in Materials and Methods. Molecular mass standards, in kDa, are indicated on the left.
Previous work indicates that the C. jejuni Cia proteins are secreted from the flagellar apparatus (39). As a screen to determine whether each isolate possessed a functional flagellar export apparatus, motility assays were performed with each of the six C. jejuni poultry isolates (not shown). In contrast to C. jejuni SC, S1, and Turkey, the C. jejuni CS, S2B, and S3 isolates were nonmotile. These findings raised the possibility that the C. jejuni CS, S2B, and S3 isolates were weakly invasive and did not secrete the Cia proteins because they did not possess a functional secretion apparatus/flagellum.
C. jejuni isolates Turkey and CS are genotypically indistinguishable.
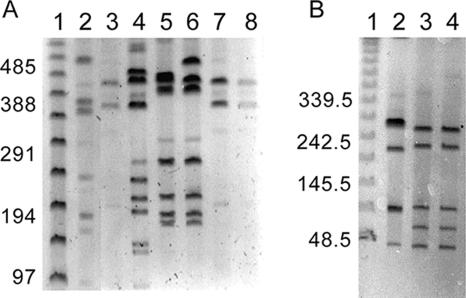
To unravel the molecular basis of the differences in pathogenesis exhibited by the isolates, we wanted to determine their genetic relatedness. Since macrorestriction enzyme PFGE (MRP-PFGE) profiling is the currently accepted method to determine the relatedness of Campylobacter isolates (12, 14, 21, 22, 25, 28, 29, 67, 72, 73, 76), we used it to assess the genomic diversity of the C. jejuni isolates used in this study. Initially, C. jejuni isolates were analyzed by PFGE following digestion of chromosomal DNA with the restriction enzyme SmaI. A representative gel is presented in Fig. 2A. PFGE of SmaI-restricted C. jejuni chromosomal DNA yielded 4 to 10 fragments ranging in size from <97 kb to approximately 485 kb. The C. jejuni CS, SC, and Turkey isolates yielded indistinguishable macrorestriction profiles using the SmaI restriction enzyme. We chose to narrow the remainder of this study to the Turkey and CS isolates because the Turkey isolate was found to be highly invasive for INT 407 cells and was Cia secretion positive whereas the CS isolate displayed a noninvasive phenotype for INT 407 cells and was Cia secretion negative. Noteworthy is that macrorestriction profiles of the C. jejuni Turkey and CS isolates were also indistinguishable using a second restriction enzyme, SalI (Fig. 2B).
FIG. 2.
MRP-PFGE profiles of C. jejuni isolates. Lanes 1 (A and B), PFGE lambda ladder (New England Biolabs, Beverly, MA). The sizes of the molecular mass markers are shown on the left of each panel in kilobase pairs. (A) MRP-PFGE of SmaI-digested DNA from 81-176 (lane 2), CS (lane 3), S1 (lane 4), S2B (lane 5), S3 (lane 6), SC (lane 7), and Turkey (lane 8). (B) MRP-PFGE of SalI-digested DNA from 81-176 (lane 2), Turkey (lane 3), and CS (lane 4).
MLST is a technique that characterizes isolates based upon combinations of alleles at seven “unlinked” housekeeping loci (15). Of interest was that the C. jejuni Turkey and CS isolates were found to belong to the same sequence type (ST-48).
We also examined the gene content of the C. jejuni Turkey and CS isolates by a comparative genomic hybridization analysis (genomotyping) using a C. jejuni NCTC 11168 DNA microarray. It should be noted that only genes that have a feature represented on the C. jejuni NCTC 11168 DNA microarray can be detected using this method. Moreover, some mutations, including frameshift and point mutations that might be present in one isolate and not the other, would not be identified by this method. With these caveats in mind, we found no observable differences between the signal intensities for the C. jejuni Turkey and CS isolates, suggesting that, among those genes conserved with C. jejuni strain NCTC 11168, all three isolates have an identical gene content (not shown). However, both the C. jejuni Turkey and CS isolates exhibited divergence from the NCTC 11168 strain for genes that are contained within the recognized hypervariable regions, including the capsular biosynthesis locus, LOS biosynthesis locus, and restriction modification locus. To confirm divergence in the LOS locus among the isolates, we used a recently described PCR method (59). Both C. jejuni isolates CS and Turkey possess the class B LOS biosynthetic locus (not shown), while strain NCTC 11168 possesses the class C LOS locus.
The C. jejuni Turkey isolate is more pathogenic for piglets.
Because the C. jejuni Turkey and CS isolates were genetically indistinguishable as judged by MRP-PFGE, MLST, and comparative genomic hybridization analysis but differed significantly in their in vitro virulence capabilities, we focused our efforts on these two isolates to decipher the molecular basis for C. jejuni pathogenicity. However, we decided to perform in vivo experiments to assess the pathogenicity of the C. jejuni Turkey and CS isolates prior to comparing the protein and gene expression profiles. Noteworthy is that the infection of piglets with C. jejuni has been used previously as a model for Campylobacter-mediated enteritis, as infected piglets exhibit overt symptoms and histopathology that are characteristic of human infections (4). The C. jejuni Turkey isolate was more pathogenic than the CS isolate for piglets (Table 2). Diarrhea was noted in two of the three piglets inoculated with the C. jejuni Turkey isolate and none of the piglets inoculated with the C. jejuni CS isolate. A single Turkey-inoculated piglet developed hemorrhage in the colon, which was noted upon gross examination. Histological examination of intestinal samples was performed on piglets inoculated with the C. jejuni Turkey and CS isolates. Two of the three piglets inoculated with the C. jejuni Turkey isolate developed significant degenerative lesions in the small intestine. Also apparent in the small intestines of these two piglets were areas of congestion and cell necrosis. In the colon, the crypts were filled with eosinophilic debris and mucosal erosion was apparent, accompanied by a fibrinous exudate. One of the three C. jejuni CS-inoculated piglets exhibited minor pathological changes. This C. jejuni CS-inoculated piglet had congested vessels and slight villus degeneration.
TABLE 2.
The C. jejuni Turkey isolate is more virulent than the CS isolate in the neonatal piglet model
| C. jejuni strain | Fraction of piglets with diarrhea | Onset of diarrhea | Fraction of piglets that experienced:
|
|
|---|---|---|---|---|
| Death (prenecropsy) | Hemorrhage | |||
| Turkey | 2/3 | 24 h | 0/3 | 1/3 |
| CS | 0/3 | Not detected | 0/3 | 0/3 |
The C. jejuni Turkey and CS isolates display differences in protein profiles.
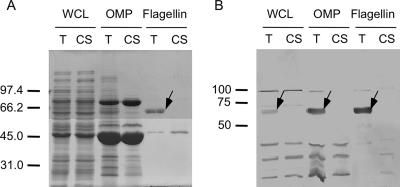
To determine if there were differences in the proteins synthesized by the C. jejuni Turkey and CS isolates in vitro, bacterial whole-cell lysates were analyzed by one- and two-dimensional gel electrophoresis and the proteins stained with CBB-R250 and silver stain (not shown). A minimum of 21 proteins varied in their relative amounts in the whole-cell lysate of the C. jejuni Turkey isolate versus the whole-cell lysate of the CS isolate as judged by two-dimensional electrophoresis coupled with silver staining (not shown). Further, CBB-R250 staining of the two-dimensional gels revealed a predominant band of 62 kDa in the whole-cell lysate of the C. jejuni Turkey isolate that was not observed in the CS isolate. As C. jejuni FlaA monomers are known to have a molecular mass of 62 kDa (30), this difference was further investigated. To confirm whether FlaA was indeed synthesized by the C. jejuni Turkey isolate and not by the C. jejuni CS isolate, bacterial extracts (e.g., whole-cell lysate, outer membrane protein, and flagellar protein) were prepared from both isolates using SDS-PAGE coupled with immunoblot analysis with the flagellin polyclonal antiserum. Again, a 62-kDa immunoreactive band was clearly visible in the various bacterial extracts prepared from the C. jejuni Turkey isolate and absent in the extracts prepared using the C. jejuni CS isolate (Fig. 3). The identity of the 62-kDa protein was confirmed to be FlaA filament protein by N-terminal sequence analysis. More specifically, the amino-terminal amino acid sequence of the mature protein was determined after SDS-PAGE and electrophoretic transfer to PVDF membranes to be GFRINTNVAAL. This sequence corresponds to residues 2 through 12 of the deduced amino acid sequence of the FlaA protein. Collectively, these data indicate that the FlaA protein is synthesized in the C. jejuni Turkey isolate but not in the C. jejuni CS isolate.
FIG. 3.
Electrophoretic and immunoblot analysis of C. jejuni Turkey (T) and CS whole-cell lysates (WCL), OMP, and flagellin preparations. Proteins were separated in SDS-12.5% polyacrylamide gels and either stained with CBB-R250 (A) or transferred to PVDF membranes and reacted a 1:250 dilution of a C. jejuni antiflagellin serum that contains antibodies reactive with both the FlaA and FlaB proteins (B). The arrows indicate the FlaA flagellar protein. Molecular mass standards, in kDa, are indicated on the left.
Decreased expression of flagellum-related transcripts from the C. jejuni CS isolate.
Based on the differences in the phenotypic properties and protein profiles of the C. jejuni Turkey and CS isolates, we anticipated that there would be differences in the gene expression profiles of the two isolates. From MH broth cultures, total RNA was isolated from the C. jejuni Turkey and CS isolates and fluorescently labeled as it was reverse transcribed to cDNA. The labeled cDNA was used to probe the C. jejuni NCTC 11168 DNA microarrays. The expression profiles for the isolates were distinct, showing that 47 genes had significantly reduced transcript levels of twofold or more in the C. jejuni CS isolate versus the Turkey isolate (Table 3). Most strikingly, 20 of the 47 genes are related to flagellar biosynthesis and motility, and this correlates with the decreased motility of the C. jejuni CS isolate. There were also 24 genes for which the transcript levels were increased twofold or more in the C. jejuni CS isolate versus the Turkey isolate (Table 4). Eight of these genes have annotated functions involving purine or amino acid biosynthesis. However, the role that the upregulated genes play in the phenotype displayed by the C. jejuni CS isolate is not known. To assess the accuracy of the microarray data, real-time RT-PCR was performed on four genes that were upregulated (flaB, flgD, flgE, and flgK) and three genes that were downregulated (clpB, metA, and Cj0414) in the array experiments. The expression profile of C. jejuni Turkey and CS isolates using real-time RT-PCR mirrored the microarray results.
TABLE 3.
Transcripts detected at lower levels in the C. jejuni CS isolate than in the C. jejuni Turkey isolate
| Gene | Fold change | Description of encoded protein |
|---|---|---|
| Cj0040a,b | 4.30 | Conserved hypothetical protein |
| Cj0041a,b | 3.01 | Conserved hypothetical protein |
| flgDa,b | 6.55 | Flagellar hook assembly protein |
| flgEa,b | 6.52 | Flagellar hook protein |
| Cj0044ca | 2.11 | Conserved hypothetical protein |
| Cj0045ca | 2.60 | Conserved hypothetical protein, putative iron-binding protein |
| aspA | 2.52 | Aspartate ammonia lyase |
| dcuA | 2.57 | C4-dicarboxylate transporter, anaerobic |
| Cj0168c | 3.50 | Hypothetical protein |
| Cj0256 | 2.37 | Conserved hypothetical integral membrane protein, putative sulfatase |
| serB | 2.30 | 3-Phosphoserine phosphatase |
| cheWa | 2.92 | Purine-binding chemotaxis |
| cheAa | 2.75 | Chemotaxis protein |
| cheVa | 2.84 | Chemotaxis protein |
| cmeC | 3.69 | RND efflux system, outer membrane lipoprotein |
| cmeB | 4.40 | RND efflux system, inner membrane transporter |
| cmeA | 4.01 | RND efflux system, membrane fusion protein |
| Cj0391ca,c | 2.77 | Conserved hypothetical protein |
| Cj0420 | 3.52 | Conserved hypothetical protein |
| flgBa | 2.05 | Flagellar basal body rod protein |
| flaGa,b | 2.06 | Possible flagellar protein |
| fliDa,c | 2.48 | Flagellar hook-associated protein 2 |
| Cj0561c | 5.97 | Conserved hypothetical periplasmic protein |
| peb-4 | 2.14 | Cell binding factor 2 precursor, major antigenic peptide |
| fba | 2.78 | Fructose-bisphosphate aldolase |
| peb-2 | 2.25 | Peb2 accessory colonization factor AcfC (acfC) |
| Cj0805 | 2.08 | Conserved hypothetical protein, putative peptidase/protease (M16 family) |
| dapA | 2.98 | Dihydrodipicolinate synthase |
| Cj0807 | 2.18 | Conserved hypothetical protein, putative short chain dehydrogenase |
| flaDa,b | 2.74 | Flagellin family protein |
| Cj0950c | 2.31 | Conserved hypothetical lipoprotein |
| Cj0969 | 2.55 | Unknown |
| Cj0998c | 2.46 | Conserved hypothetical protein |
| Cj1064 | 2.36 | Unknown |
| Cj1159c | 3.40 | Unknown |
| Cj1242 | 3.30 | Conserved hypothetical protein |
| Cj1279c | 2.15 | Conserved hypothetical protein, fibronectin domain-containing lipoprotein |
| flaAa,c | 3.83 | Flagellin |
| flaBa,b | 4.15 | Flagellin |
| Cj1407c | 2.03 | Phosphohexosemutase |
| flgIa,b | 5.63 | Flagellar P-ring protein |
| Cj1463a | 2.94 | Conserved hypothetical protein |
| flgMa,c | 2.00 | Conserved hypothetical protein |
| Cj1465a | 2.07 | Conserved hypothetical protein |
| flgKa,b | 3.02 | Flagellar hook-associated protein |
| Cj1656c | 5.78 | Unknown |
| leuA | 2.36 | 2-Isopropylmalate synthase |
Flagellar biosynthesis-associated gene.
σ54-regulated gene.
σ28-regulated gene.
TABLE 4.
Transcripts detected in greater levels in the C. jejuni CS isolate than in the C. jejuni Turkey isolate
| Gene | Fold change | Description of encoded protein |
|---|---|---|
| Cj0091 | 2.05 | Conserved hypothetical lipoprotein |
| Cj0203 | 2.04 | Unknown |
| Cj0264c | 2.04 | Unknown |
| Cj0265c | 2.19 | Unknown |
| Cj0413 | 2.03 | Conserved hypothetical protein, probable periplasmic protein |
| Cj0414 | 5.34 | Conserved hypothetical protein |
| Cj0415 | 4.50 | Conserved hypothetical protein, putative gluconate dehydrogenase |
| Cj0444 | 2.22 | Unknown |
| thiC | 2.01 | Thiamine biosynthesis protein |
| Cj0501 | 2.61 | Unknown |
| clpB | 2.60 | ATP-dependent Clp protease, ATP-binding subunit |
| hypB | 2.10 | Hydrogenase expression/formation protein |
| Cj0653c | 2.37 | Aminopeptidase (M24 family) |
| Cj0654c | 3.02 | Unknown |
| glnA | 2.49 | Glutamine synthetase |
| putP | 2.04 | Sodium/proline symporter |
| Cj1514c | 2.01 | Conserved hypothetical protein |
| nuoC | 2.30 | NADH-ubiquinone oxidoreductase, subunit C |
| nuoB | 2.32 | NADH-ubiquinone oxidoreductase, subunit B |
| HisG | 2.53 | ATP phosphoribosyltransferase |
| hisD | 2.28 | Histidinol dehydrogenase |
| hisH | 2.35 | Glutamine amidotransferase |
| hisA | 2.19 | N-(5′-Phospho-l-ribosyl-formimino)- 5-amino-1-(5′-phosphoribosyl)-4-imidazolecarboxamide isomerase |
| metA | 2.96 | Homoserine O-succinyltransferase |
| metB | 3.57 | O-Acetylhomoserine sulfhydrylase |
Flagellar class II genes are downregulated in the C. jejuni CS isolate.
Microarray analysis revealed that 20 of the 47 genes expressed at a higher level in the C. jejuni Turkey isolate than in the CS isolate were associated with flagellar biosynthesis. To elucidate the molecular mechanism associated with disruption of flagellar biosynthesis in the C. jejuni CS isolate, we performed real-time RT-PCR on eight class I genes (flhA, flhB, fliF, fliG, fliH, fliI, fliM, and fliY) and eight class II genes (flaB, flaD, flgB, flgD, flgE, flgG, flgH, and flgK) previously shown to be regulated by σ70 and σ54, respectively (10). All class I genes were expressed at a higher level in the C. jejuni CS isolate than in the C. jejuni Turkey isolate. In contrast, the σ54-regulated flagellar genes were expressed at a lower level in the C. jejuni CS isolate than in the C. jejuni Turkey isolate (Fig. 4A). Moreover, flgG and flgH were also found to be expressed at a lower level in the C. jejuni CS isolate than in the C. jejuni Turkey isolate, even though differential expression of these genes was not detected by microarray analysis.
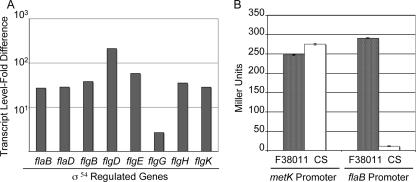
FIG. 4.
(A) Transcript levels of σ54-regulated genes in the C. jejuni Turkey isolate are increased relative to those of the C. jejuni CS isolate. The flaB, flaD, flgB, flgD, flgE, flgG, flgH, and flgK transcripts were measured by real-time RT-PCR using total RNA extracted from the C. jejuni CS and Turkey isolates grown to mid-log phase in MH broth. The change in transcript levels was measured using the comparative CT method, where glnA was used as the internal control and ΔCT of the C. jejuni CS isolate was used as the calibrator. (B) The flaB gene, which is σ54 regulated, is not expressed in the C. jejuni CS isolate. β-Galactosidase activity from the C. jejuni CS and F38011 isolates harboring the PmetK-pMW10 and PflaB-pMW10 constructs was measured. The bacteria were grown in MH broth for 16 h. The values are averages of three independent experiments, with the error bars representing the standard deviations.
The σ54-regulated flaB gene is poorly expressed in the C. jejuni CS isolate.
Real-time RT-PCR indicated that σ54-regulated flagellar genes were downregulated in the C. jejuni CS isolate. To determine if σ54 is active in the C. jejuni CS isolate, we performed reporter assays with the pMW10 lacZ promoterless shuttle vector (80). The promoter regions of the σ54-regulated flaB and σ70-regulated metK genes were cloned upstream of the lacZ gene. C. jejuni F38011 was chosen as a positive control because we were unable to transform the C. jejuni Turkey isolate with a shuttle plasmid. Comparable levels of β-galactosidase activity were noted in the C. jejuni F38011 and CS isolates harboring the PmetK-pMW10 construct. In contrast, the C. jejuni CS isolates harboring the PflaB-pMW10 construct showed almost no β-galactosidase activity (Fig. 4B) compared to the C. jejuni F38011 isolate. This finding further suggests that σ54 (RpoN)-regulated flagellar genes are not expressed in the C. jejuni CS isolate.
To determine why the flaB flagellar gene is not expressed in the C. jejuni CS isolate, the entire rpoN operon (consisting of rpoN, Cj0667, Cj0668, and Cj0669) was sequenced in the C. jejuni Turkey and CS isolates to identify a frameshift or point mutation. No differences were found in the sequences of the genes contained within the rpoN operon of the C. jejuni Turkey and CS isolates. Furthermore, real-time RT-PCR indicated that all four genes in the rpoN operon of the C. jejuni Turkey and CS isolates were expressed at comparable levels.
Identification of point mutations in the flgR gene of the C. jejuni CS isolate.
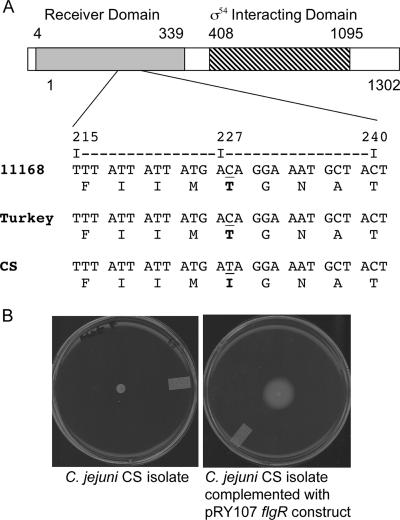
We sequenced the entire ORFs of genes whose products are involved in either flagellar gene regulation or biosynthesis in an attempt to identify the molecular mechanism associated with disruption of flagellar biosynthesis in the C. jejuni CS isolate. The class I genes sequenced in the C. jejuni Turkey and CS isolates were flhA, flhB, flhF, fliF, fliG, fliH, fliI fliM, fliN, fliP, fliQ, and fliR. We also sequenced fliA (σ28) and the genes known to interact or activate σ54 including flgR and flgS. No nucleotide alterations were noted in the flhA, flhB, flhF, fliA, fliF, fliG, fliH, fliI, fliM, fliN, fliP, fliQ, fliR, and flgS genes of the C. jejuni Turkey and CS isolates. However, a difference was noted in a single nucleotide in the flgR gene that resulted in an amino acid change in the C. jejuni CS isolate compared to the Turkey isolate (Fig. 5A). The flgR gene encodes the response regulator of the FlgS/FlgR two-component regulatory system associated with the flagellar regulon. The amino acid change resides within in the receiver domain of the FlgR protein.
FIG. 5.
(A) Multiple sequence alignment of the flgR gene from C. jejuni 11168, Turkey, and CS isolates. Shown is a segment of the nucleotide sequence of the flgR gene and its predicted translation from C. jejuni strains NCTC 11168 (11168), Turkey (Turkey), and CS (CS). The FlgR receiver and σ54-interacting domains are indicated. Also highlighted (underlined) is the residue that differed in the C. jejuni CS isolate compared to the Turkey isolate. (B) Complementation of the C. jejuni CS isolate with a wild-type copy of flgR gene restores the isolate's motility. Both the C. jejuni CS wild-type and CS flgR pRY107-transformed isolates were spotted onto MH plates supplemented with 0.4% agar, and motility was assessed after 48 h of incubation.
Complementation of the C. jejuni CS isolate with a copy of the flgR gene from C. jejuni NCTC 11168 restores motility.
To determine if FlgR is responsible for the phenotype of the C. jejuni CS isolate, the flgR gene from C. jejuni NCTC 11168 was cloned into the pRY107 shuttle vector and introduced into the C. jejuni CS isolate. The C. jejuni CS isolate transformed with the flgR pRY107 shuttle vector was motile (Fig. 5B). Collectively, these data indicate that a single nucleotide difference in the C. jejuni CS flgR gene, which resulted in an amino acid change, is responsible for the nonmotile phenotype displayed by the isolate. Subsequently, we generated a mutation in the flgR gene of the C. jejuni Turkey isolate. Analysis of this mutant revealed a phenotype that was indistinguishable from that of the C. jejuni CS isolate; the C. jejuni Turkey isolate is nonmotile and incapable of invading INT 407 epithelial cells (data not shown).
The C. jejuni CS isolate displays greater resistance to sodium deoxycholate than the C. jejuni Turkey isolate.
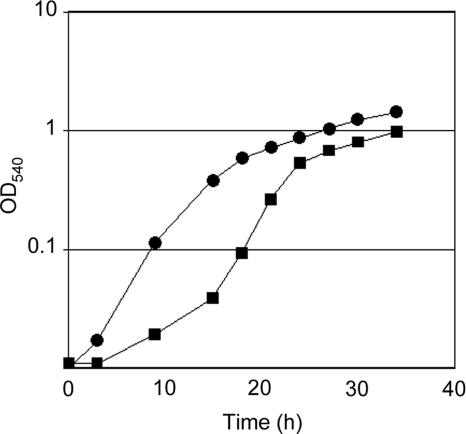
C. jejuni flagellar mutants (i.e., flgS, flgR, and rpoN mutants) have been shown previously to multiply at a rate three times greater than an isogenic wild-type isolate (81). While we did not find any differences in growth rates of the C. jejuni Turkey and CS isolates, the C. jejuni Turkey isolate displayed a longer lag phase than the C. jejuni CS isolate. Also, the C. jejuni CS isolate achieved a greater maximum cell density in MH medium than the C. jejuni Turkey isolate (Fig. 6).
FIG. 6.
Growth of the C. jejuni Turkey and CS isolates in MH broth. MH broth was inoculated with the C. jejuni Turkey and CS isolates to an OD540 of 0.01, and the cultures were incubated at 37°C and 150 rpm, under microaerobic conditions. The points represent the time points at which the optical density of each bacterial culture was determined at a wavelength of 540 nm. Circles, C. jejuni CS isolate; squares, C. jejuni Turkey isolate.
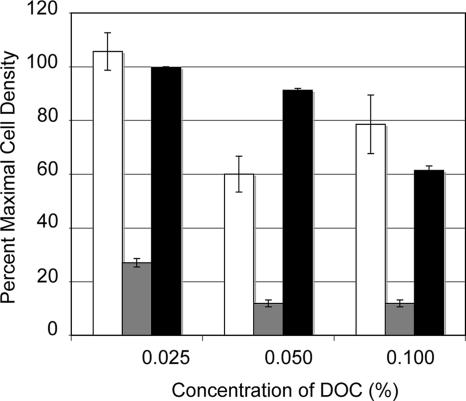
Microarray analysis revealed that some genes encoding membrane-associated proteins were expressed at different levels in the C. jejuni CS isolate compared to the C. jejuni Turkey isolate. Therefore, we studied the ability of the C. jejuni Turkey and CS isolates to resist the antimicrobial activity of the bile salt sodium deoxycholate. Both isolates were exposed to different concentrations of sodium deoxycholate, and the cell density achieved at 48 h was measured. The growth of the C. jejuni Turkey isolate was significantly inhibited by the presence of 0.05% and 0.1% sodium deoxycholate compared to the C. jejuni CS isolate (Fig. 7). This finding was reproduced with the C. jejuni Turkey isolate compared to a C. jejuni Turkey flgR mutant, where the C. jejuni Turkey flgR mutant was found to be more resistant to deoxycholate. Not known is why the C. jejuni Turkey and CS isolates differ in their sensitivities to deoxycholate.
FIG. 7.
The C. jejuni CS isolate and the C. jejuni Turkey flgR mutant display greater resistance to sodium deoxycholate (DOC) than the C. jejuni Turkey isolate. The C. jejuni CS isolate (open bars), C. jejuni Turkey isolate (gray bars), and C. jejuni Turkey flgR mutant (black bars) were incubated in MH broth with 0.1%, 0.05%, 0.025%, and 0% sodium deoxycholate under microaerobic conditions for 48 h at 37°C with shaking. The y axis represents the optical density of bacterial culture relative to the growth achieved without deoxycholate (0% sodium deoxycholate). The error bars represent the standard deviations.
Screening C. jejuni poultry isolates for nonmotile phenotype.
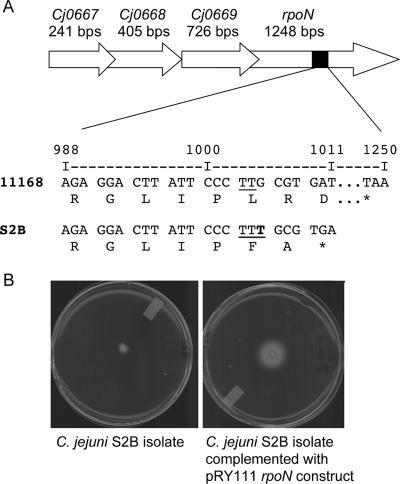
To assess how common are nonmotile isolates within a population of poultry isolates, we screened a total of 99 isolates to determine their motility phenotypes. We identified one additional nonmotile C. jejuni isolate, designated H8a. Counting the initial six C. jejuni isolates used in this study, a total of four nonmotile isolates were identified (CS, S2B, S3, and H8a).
To identify the molecular mechanism for the motility defects, we attempted to transform the C. jejuni S2B, S3, and H8a isolates with the pMW10 flaB promoter construct. Although we were unable to transform the C. jejuni H8a isolate with the pMW10 flaB construct, negligible β-galactosidase activity was detected in the S2B and S3 isolates. Upon sequencing the rpoN genes from each of these isolates, it was found that a thymine base was inserted between bp 1004 and 1005 in the C. jejuni S2B isolate (Fig. 8A). This insertion resulted in an early termination codon at bp 1011. To determine if this frameshift mutation was responsible for the loss in σ54 activity, a wild-type copy of the rpoN from C. jejuni NCTC 11168 was introduced into the C. jejuni S2B isolate using the pRY111 shuttle vector. The C. jejuni S2B isolate harboring the rpoNpRY111 shuttle vector was motile (Fig. 8B). The reason why the C. jejuni S3 and H8a isolates are nonmotile is not known currently.
FIG. 8.
(A) The rpoN operon of the C. jejuni S2B isolate was sequenced, and a single thymine base insertion was identified between bp 1004 and 1005. This base insertion results in a termination codon at bp 1011 as opposed to the normal termination codon at bp 1248. Shown is a segment of the nucleotide sequence of the rpoN gene and its predicted translation from the C. jejuni NCTC 11168 (11168) and S2B (S2B) isolates. Also shown (bottom) is the number of bp from the start codon of rpoN. *, termination codon. (B) Complementation of the C. jejuni S2B strain with a wild-type copy of rpoN restored the motility phenotype. The C. jejuni S2B wild-type and S2B rpoN pRY111-transformed isolates were spotted onto MH plates supplemented with 0.4% agar, and motility was assessed after 48 h of incubation.
DISCUSSION
The clinical presentation of campylobacteriosis varies extensively among individuals. Symptoms range from mild diarrhea to severe abdominal pain associated with diarrhea containing blood and leukocytes. In addition, infections in developing countries are more often reported in children, with symptoms limited to watery diarrhea, while infections in developed countries are associated with more dysentery-like symptoms. Explanations for the variation in the severity of campylobacteriosis include differences in the virulence of C. jejuni strains, repeated exposure to the organism, and host susceptibility.
Genetic variation among strains of Campylobacter spp. is well documented (2, 16, 42, 61, 64, 65, 71, 79). Dingle et al. (15) reported that C. jejuni cells form a weakly clonal population based on the distribution of strains among MLST sequence type complexes. In addition, various groups have shown that strains differ in their genomic contents using microarrays. Dorrell et al. (16) compared 11 C. jejuni strains with NCTC 11168 and found that at least 21% of the NCTC 11168 genome is divergent in one or more of the tested strains. Using a shotgun genomic microarray, Poly et al. (65) showed that there was significant variation between C. jejuni strains ATCC 43431 and NCTC 11168, especially at LOS, capsular biosynthesis, and restriction modification enzyme loci. Comparison of C. jejuni strains 81-176 and ATCC 43431 by the same method showed differences at similar loci (64). Finally, comparison of the whole genome sequences of NCTC 11168 and RM1221 identified the presence of phage insertion elements and polymorphisms at the LOS and capsular biosynthesis loci (23). Noteworthy is that both LOS and capsule biosynthesis have been implicated in the pathogenesis of C. jejuni (6, 27). Changes in the poly(G) tract of cgtA of C. jejuni strain 81-176 results in changes to the LOS core structure (27). A cgtA mutant in which the LOS mimics that of the GM3 ganglioside rather than the GM2 ganglioside demonstrates an increase in invasiveness in cultured epithelial cells. The kpsM gene is involved in the expression of the capsular high-molecular-weight glycan, and a C. jejuni 81-176 kpsM isogenic mutant is reduced in invasiveness and ability to cause diarrhea in the ferret model (6).
In addition to strain-to-strain genetic variation, intrastrain variation has been observed in Campylobacter spp. For example, MRP-PFGE profile changes were found in five out of six Campylobacter coli isolates that had been passaged up to 50 times in the laboratory after initial isolation from separate pig herds (58). With respect to C. jejuni, Mixter et al. (50) observed that genomic changes occurred in C. jejuni strain F38011, as determined by SmaI MRP-PFGE, after intraperitoneal inoculation of mice. Carrillo et al. (10) reported that a C. jejuni NCTC 11168 strain that had undergone long-term passage in laboratory colonized chickens poorly and was less motile than the original stock strain (10). Although a specific mutation was not identified in a C. jejuni NCTC 11168 nonmotile variant, microarray analysis revealed an expression profile that was comparable to that for an flhA flagellar biosynthetic mutant. Gaynor et al. (24) also found gene expression differences between the genome-sequenced variant of C. jejuni NCTC 11168 and the original freezer stock; the two strains also showed differences in their abilities to colonize chickens.
Virulence-related phenotypic differences between C. jejuni strains have been described. Multiple investigators have found differences in the invasion of C. jejuni strains into cultured epithelial cells (38, 54). Konkel and Joens (38) showed that the invasion of cultured epithelial cells by C. jejuni clinical isolates varied considerably. Overall, clinical isolates were more invasive than those obtained from culture collections (e.g., American Type Culture Collection, Manassas, VA). Everest et al. (18) found that C. jejuni isolates recovered from individuals with colitis were more invasive than those isolated from individuals with noninflammatory diarrhea. Newell et al. (54) observed that C. jejuni and C. coli isolates recovered from water sources were less invasive than clinical isolates. Variation in invasion efficiency among C. jejuni strains supports the notion that strain-to-strain genetic differences may play a role in the range of clinical symptoms of campylobacteriosis.
In this study, we examined the genotypic and phenotypic characteristics of C. jejuni poultry isolates. These isolates differed in both their abilities to invade INT 407 cells and levels of secretion of virulence proteins. All the isolates found to be nonmotile also showed lack of invasion and secretion potential. Conversely, all isolates that were able to secrete virulence proteins were also able to migrate through semisolid media, which is widely used as an indication of swarming motility. These data are consistent with previously published work demonstrating that the flagellar apparatus is required for the secretion of virulence proteins by C. jejuni (39). In addition, motility has been associated with C. jejuni pathogenesis both in terms of invasion of epithelial cells (77) and colonization of chickens (78).
We found that the C. jejuni Turkey and CS isolates used in this study were indistinguishable by MRP-PFGE and therefore constituted a clonally matched pair. MLST confirmed this finding, with both strains belonging to ST-48. Genomotyping using a microarray consisting of ORFs from NCTC 11168 revealed no differences in genomic content between the C. jejuni CS and Turkey isolates. Although clonally indistinguishable, the Turkey and CS isolates differed in their levels of putative pathogenic potential as judged by in vitro assays. The C. jejuni Turkey isolate secreted virulence proteins, invaded INT 407 epithelial cells at a high level, and was motile, while the C. jejuni CS isolate failed to secrete, invaded INT 407 epithelial cells at a low level, and was nonmotile. Also, the levels of pathogenic potential of C. jejuni isolates CS and Turkey were tested in the neonatal piglet model as described by Babakhani et al. (5). In agreement with our in vitro assays, piglets inoculated with the C. jejuni CS isolate did not develop diarrhea. Taken together, these results indicate a good correlation between the in vitro and in vivo models with respect to determining an isolate's pathogenic potential. Moreover, the C. jejuni Turkey isolate was assessed to be more pathogenic than the CS isolate in all assays.
No detectable differences in the genomic contents of the C. jejuni Turkey and CS isolates were noted by genomotyping or genomic subtractive hybridization (not shown), indicating that a difference in gene expression might result in the observed phenotypes. The protein profiles of the C. jejuni Turkey and CS isolates were examined. Based on the fact that the C. jejuni CS isolate was nonmotile, it is not surprising that the FlaA flagellin protein was not synthesized in the CS isolate, as judged by the absence of a 62-kDa protein in whole-cell extracts, outer membrane protein preparations, and a flagellar extract. The flagellin protein in the C. jejuni Turkey isolate, but not the C. jejuni CS isolate, was confirmed to be FlaA by N-terminal sequence analysis. Total RNA was also extracted from the C. jejuni Turkey and CS isolates and used in an expression microarray to identify genes that differed in their expression. The C. jejuni Turkey isolate had significantly higher expression of genes involved in flagellar biosynthesis than the C. jejuni CS isolate.
Flagellar biosynthesis in C. jejuni is divided into three stages. The expression of class I genes is under the control of σ70 and is critical for the expression of class II genes. Class II genes are under the control of σ54. Class III genes are under the control of σ28. In addition to the participation of different sigma factors, the FlgS/R two-component signal transduction system participates in the regulation of the flagellar regulon. Real-time RT-PCR analysis of the isolates in this study indicated that all class I genes were upregulated in the C. jejuni CS isolate, whereas all the class II genes were downregulated. These findings suggested that a mutation might exist in either a class I flagellar biosynthesis gene (e.g., flhA) or a regulator of flagellar biosynthesis (rpoN, flgR, or flgS). To test this possibility, the ORFs of genes involved in flagellar gene regulation from both isolates were sequenced, including flhA, flhB, flhF, fliA, fliF, fliG, fliH, fliI, fliM, fliN, fliP, fliQ, fliR, rpoN, Cj0667, Cj0668, and Cj0669. Although we were unable to identify a point mutation in any gene contained within the rpoN operon, it was evident from the β-galactosidase reporter assay that the σ54-regulated flagellar genes were not expressed in the C. jejuni CS isolate. Since FlgS and FlgR are required for σ54 activity, we sequenced the flgS and flgR ORFs. Comparison of the ORFs of the flgR genes of C. jejuni Turkey and CS revealed a difference in one nucleotide that altered the amino acid sequence. This change in the deduced amino acid sequence proved to be responsible for the nonmotile phenotype of the C. jejuni CS isolate, as indicated by the restoration of motility via complementation of the isolate with a wild-type copy of the flgR gene on the pRY107 shuttle vector. Hendrixson (31) reported a phase-variable mechanism responsible for modulating motility, which in turn helps the bacteria establish a commensal state. FlgR phase variation was achieved by the gain or loss of a nucleotide in the homopolymeric adenine or thymine tract. Noteworthy is that the region in which the mutation occurred in the flgR gene of the C. jejuni CS isolate was not identified previously as a phase-variable region (31).
To assess the frequency of nonmotile strains in a population of poultry isolates, we screened a total of 105 isolates and found 4 nonmotile strains (CS, S2B, S3, and H8a). Of the four nonmotile isolates recovered from poultry, we identified mutations in two distinct genes involved in flagellar biosynthesis. We were able to restore the motility of both the C. jejuni CS and S2B isolates and found a motile revertant of the S3 isolate (not shown). The latter finding indicates that the S3 isolate likely has a single point mutation that alters its motility phenotype. We have sequenced the rpoN and flgR genes from the C. jejuni S3 isolate and did not identify a nucleotide difference that altered the deduced amino acid sequence from that of the C. jejuni NCTC 11168 isolate. Thus, the S3 isolate possesses a mutation in a gene distinct from rpoN and flgR, indicating that a mutation of a third gene can also lead to a nonmotile phenotype. Based on these findings, it appears that a nonmotile phenotype likely offers a selective advantage to C. jejuni within the host or environment.
One of the differences noted in the phenotypes of C. jejuni Turkey and CS isolates was in their resistance to the bile salt deoxycholate; the C. jejuni CS isolate is more resistant to deoxycholate. The difference in sensitivity appears to be specifically associated with the flgR mutation, as the same observation was noted for C. jejuni Turkey compared to a C. jejuni Turkey flgR mutant. It is presently not known why an flgR mutant is more resistant to deoxycholate than a wild-type isolate. In C. jejuni, the CmeABC multidrug efflux pump plays a major role in the organism's resistance to detergents, antimicrobials including different classes of antibiotics, and heavy-metal-containing salts (43, 44). A C. jejuni cmeB mutant is 64-fold more sensitive to sodium deoxycholate than a wild-type strain (44). In contrast to what one might predict, microarray analysis revealed that the cmeABC genes were downregulated in the C. jejuni CS isolates compared to the C. jejuni Turkey isolate. The reduction in expression of the cmeABC genes in the CS isolate was confirmed by real-time RT-PCR. Clearly additional work is needed to understand this phenomenon.
Microarray analysis of the C. jejuni CS isolate (flgR mutant) and rpoN mutant revealed that many more genes were not expressed in a rpoN mutant compared to its isogenic wild-type isolate, unlike the results of a similar comparison between the C. jejuni CS isolate and the Turkey isolate (data not shown). Of the nine flagellum-related genes whose expression was lower in the C. jejuni F38011 rpoN mutant than in the F38011 wild-type isolate, eight of the same nine genes had lower expression in the C. jejuni CS isolate than in the C. jejuni Turkey isolate. However, a total of 80 genes were downregulated in the defined rpoN mutant versus a total of 47 genes in the C. jejuni CS isolate. Based on this finding, we propose that FlgR is necessary for the expression of σ54-regulated flagellar genes but that σ54 is responsible for the expression of other genes independent of the FlgR response regulator. Additional work is required to determine the genes of the σ54 regulon.
The role of motility in the pathogenesis of C. jejuni has been well documented. Newell et al. (52, 53) reported that, in a murine intestinal colonization model, a nonflagellated C. jejuni isolate colonized poorly and was cleared within 7 days postinfection. Nachamkin et al. (51) found that only C. jejuni isolates with intact and functional flagella colonized the 3-day-old chick model, while Wassenaar et al. (78) found that expression of the FlaA flagellin subunit maximized colonization of 1-day-old chicks by C. jejuni. Consistent with the hypothesis that motility is an important virulence attribute for C. jejuni, only motile variants were recovered from human volunteers challenged with a mixture of motile and nonmotile variants of C. jejuni (8). The results of this study also support the hypothesis that motility is an important C. jejuni virulence determinant as it affects an isolate's pathogenicity in the neonatal piglet model of campylobacteriosis. Our findings further underscore the need to assess whether a C. jejuni isolate is motile prior to assessing its in vitro and in vivo virulence potential.
In summary, we have compared the genetically matched C. jejuni Turkey and CS isolates and found that the differences in the pathogenicities of these two isolates are due to a point mutation in the flgR gene of the C. jejuni CS isolate. The C. jejuni CS isolate, compared to the C. jejuni Turkey isolate, exhibited a shorter lag phase in MH broth and achieved a greater maximum cell density in MH medium (Fig. 6). Inspection of the data published by Wösten et al. (81) also revealed differences in lag phase and maximal cell density of a flgR mutant compared to a wild-type isolate, but the basis of these differences was not analyzed. It is unclear if the increased expression of certain nucleotide and amino acid biosynthesis genes by the C. jejuni CS isolate contributes to its higher maximal cell density. Nevertheless, it seems possible that a mutation in flgR may provide an organism with a selective advantage in a host (e.g., chicken intestinal tract) because it can reach a higher cell density. The resistance of a C. jejuni isolate to the bile salt deoxycholate might provide a selective advantage to the organism in the intestinal tracts of both humans and poultry. Finally, this study provides an approach to identify the molecular basis for C. jejuni pathogenicity. Moreover, we believe that it will be possible to identify novel C. jejuni virulence factors by comparing C. jejuni genetically matched isolates that display differences in virulence potential.
Acknowledgments
We are grateful to Vanessa Rivera-Amill for performing the secretion assays and Marshall Monteville for performing the binding and internalization assays. We are also thankful to Jeffrey Christensen for technical assistance in constructing the suicide and shuttle vectors and for real-time RT-PCR. We thank Qijing Zhang (Department of Veterinary Microbiology and Preventive Medicine, Iowa State University, Ames, IA) for providing some of the C. jejuni poultry isolates. Finally, we appreciate the time of Mike Kahn in discussing the data.
This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2006-35201-16553, awarded to M.E.K.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Aeschbacher, M., and J.-C. Piffaretti. 1989. Population genetics of human and animal enteric Campylobacter strains. Infect. Immun. 57:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, I. H., G. Manning, T. M. Wassenaar, S. Cawthraw, and D. G. Newell. 2002. Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiology 148:1203-1212. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., P. Guerry, M. E. Power, and T. J. Trust. 1992. Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J. Bacteriol. 174:4230-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anjum, M. F., S. Lucchini, A. Thompson, J. C. Hinton, and M. J. Woodward. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 71:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babakhani, F. K., G. A. Bradley, and L. A. Joens. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 61:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 7.Belbouri, A., and F. Megraud. 1988. Enterotoxin-like activity produced by Campylobacter jejuni and Campylobacter coli isolated from patients and healthy controls in Algeria. FEMS Microbiol. Lett. 28:25-28. [Google Scholar]
- 8.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 9.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16:53-64. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo, C. D., E. Taboada, J. H. Nash, E. P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 11.Daikoku, T., M. Kawaguchi, K. Takama, and S. Suzuki. 1990. Partial purification and characterization of the endotoxin produced by Campylobacter jejuni. Infect. Immun. 58:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer, P., B. Duim, A. Rigter, J. van Der Plas, W. F. Jacobs-Reitsma, and J. A. Wagenaar. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Melo, M. A., and J.-C. Pechère. 1990. Identification of Campylobacter jejuni surface proteins that bind to eucaryotic cells in vitro. Infect. Immun. 58:1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickins, M. A., S. Franklin, R. Stefanova, G. E. Schutze, K. D. Eisenach, I. Wesley, and M. D. Cave. 2002. Diversity of Campylobacter isolates from retail poultry carcasses and from humans as demonstrated by pulsed-field gel electrophoresis. J. Food Prot. 65:957-962. [DOI] [PubMed] [Google Scholar]
- 15.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 18.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 19.Fauchère, J.-L., M. Kervella, A. Rosenau, K. Mohanna, and M. Veron. 1989. Adhesion to HeLa cells of Campylobacter jejuni and C. coli outer membrane components. Res. Microbiol. 140:379-392. [DOI] [PubMed] [Google Scholar]
- 20.Fauchère, J.-L., A. Rosenau, M. Veron, E. N. Moyen, S. Richard, and A. Pfister. 1986. Association with HeLa cells of Campylobacter jejuni and Campylobacter coli isolated from human feces. Infect. Immun. 54:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3:72-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson, J. R., C. Fitzgerald, and R. J. Owen. 1995. Comparison of PFGE, ribotyping and phage-typing in the epidemiological analysis of Campylobacter jejuni serotype HS2 infection. Epidemiol. Infect. 115:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goossens, H., J. P. Butzler, and Y. Takeda. 1985. Demonstration of cholera-like enterotoxin production by Campylobacter jejuni. FEMS Microbiol. Lett. 29:73-76. [Google Scholar]
- 27.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänninen, M.-L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänninen, M.-L., P. Perko-Mäkelä, H. Rautelin, B. Duim, and J. A. Wagenaar. 2001. Genomic relatedness within five common Finnish Campylobacter jejuni pulsed-field gel electrophoresis genotypes studied by amplified fragment length polymorphism analysis, ribotyping, and serotyping. Appl. Environ. Microbiol. 67:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris, L. A., S. M. Logan, P. Guerry, and T. J. Trust. 1987. Antigenic variation of Campylobacter flagella. J. Bacteriol. 169:5066-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 32.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 33.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, W. M., and H. Lior. 1986. Cytotoxic and cytotonic factors produced by Campylobacter jejuni, C. coli, and C. laridis. J. Clin. Microbiol. 24:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kervella, M., J.-M. Pagès, Z. Pei, G. Grollier, M. J. Blaser, and J.-L. Fauchère. 1993. Isolation and characterization of two Campylobacter glycine-extracted proteins that bind to HeLa cell membranes. Infect. Immun. 61:3440-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 37.Konkel, M. E., and W. Cieplak, Jr. 1992. Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect. Immun. 60:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konkel, M. E., and L. A. Joens. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 57:2984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konkel, M. E., Y. Lobet, and W. Cieplak, Jr. 1992. Examination of multiple isolates of Campylobacter jejuni for evidence of cholera toxin-like activity, p. 193-198. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, DC.
- 41.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 42.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 43.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahajan, S., and F. G. Rodgers. 1990. Isolation, characterization, and host-cell-binding properties of a cytotoxin from Campylobacter jejuni. J. Clin. Microbiol. 28:1314-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCardell, B. A., J. M. Madden, and E. C. Lee. 1986. Campylobacter jejuni and Campylobacter coli production of a cytotonic toxin immunologically similar to cholera toxin. J. Food Prot. 47:943-949. [DOI] [PubMed] [Google Scholar]
- 49.McSweegan, E., and R. I. Walker. 1986. Identification and characterization of two Campylobacter jejuni adhesins for cellular and mucous substrates. Infect. Immun. 53:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mixter, P. F., J. D. Klena, G. A. Flom, A. M. Siegesmund, and M. E. Konkel. 2003. In vivo tracking of Campylobacter jejuni using a novel recombinant expressing green fluorescent protein. Appl. Environ. Microbiol. 69:2864-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nachamkin, I., X. H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newell, D. G. 1986. Monoclonal antibodies directed against the flagella of Campylobacter jejuni: production, characterization and lack of effect on the colonization of infant mice. J. Hyg. 96:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newell, D. G., H. McBride, and J. M. Dolby. 1985. Investigations on the role of flagella in the colonization of infant mice with Campylobacter jejuni and attachment of Campylobacter jejuni to human epithelial cell lines. J. Hyg. 95:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newell, D. G., H. McBride, F. Saunders, Y. Dehele, and A. D. Pearson. 1985. The virulence of clinical and environmental isolates of Campylobacter jejuni. J. Hyg. 94:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newell, D. G., and A. Pearson. 1984. The invasion of epithelial cell lines and the intestinal epithelium of infant mice by Campylobacter jejuni/coli. J. Diarrhoeal Dis. Res. 2:19-26. [PubMed] [Google Scholar]
- 56.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oelschlaeger, T. A., P. Guerry, and D. J. Kopecko. 1993. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. USA 90:6884-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.On, S. L. W. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 59.Parker, C. T., S. T. Horn, M. Gilbert, W. G. Miller, D. L. Woodward, and R. E. Mandrell. 2005. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43:2771-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 61.Pearson, B. M., C. Pin, J. Wright, K. l'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 62.Pei, Z., C. Burucoa, B. Grignon, S. Baqar, X.-Z. Huang, D. J. Kopecko, A. L. Bourgeois, J.-L. Fauchère, and M. J. Blaser. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez-Perez, G. I., D. L. Cohn, R. L. Guerrant, C. M. Patton, L. B. Reller, and M. J. Blaser. 1989. Clinical and immunologic significance of cholera-like toxin and cytotoxin production by Campylobacter species in patients with acute inflammatory diarrhea in the USA. J. Infect. Dis. 160:460-468. [DOI] [PubMed] [Google Scholar]
- 64.Poly, F., D. Threadgill, and A. Stintzi. 2005. Genomic diversity in Campylobacter jejuni: identification of C. jejuni 81-176-specific genes. J. Clin. Microbiol. 43:2330-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poly, F., D. Threadgill, and A. Stintzi. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raphael, B. H., S. Pereira, G. A. Flom, Q. Zhang, J. M. Ketley, and M. E. Konkel. 2005. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J. Bacteriol. 187:3662-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivera-Amill, V., B. J. Kim, J. Seshu, and M. E. Konkel. 2001. Secretion of the virulence associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J. Infect. Dis. 183:1607-1616. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz-Palacios, G. M., J. Torres, N. I. Torres, E. Escamilla, B. R. Ruiz-Palacios, and J. Tamayo. 1983. Cholera-like enterotoxin produced by Campylobacter jejuni: characterization and clinical significance. Lancet ii:250-251. [DOI] [PubMed] [Google Scholar]
- 70.Russell, R. G., M. O'Donnoghue, D. C. Blake, Jr., J. Zulty, and L. J. DeTolla. 1993. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168:210-215. [DOI] [PubMed] [Google Scholar]
- 71.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing correlate with strain associations identified by multilocus enzyme electrophoresis. J. Clin. Microbiol. 41:4058-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stephens, C. P., S. L. W. On, and J. A. Gibson. 1998. An outbreak of infectious hepatitis in commercially reared ostriches associated with Campylobacter coli and Campylobacter jejuni. Vet. Microbiol. 61:183-190. [DOI] [PubMed] [Google Scholar]
- 74.Tong, S., A. Porco, T. Isturiz, and T. Conway. 1996. Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J. Bacteriol. 178:3260-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Spreeuwel, J. P., G. C. Duursma, C. J. L. M. Meijer, R. Bax, P. C. M. Rosekrans, and J. Lindeman. 1985. Campylobacter colitis: histological, immunohistochemical and ultrastructural findings. Gut 26:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vargas, A. C., M. M. Costa, M. H. Vainstein, L. C. Kreutz, and J. P. Neves. 2003. Phenotypic and molecular characterization of bovine Campylobacter fetus strains isolated in Brazil. Vet. Microbiol. 93:121-132. [DOI] [PubMed] [Google Scholar]
- 77.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 79.Whitehouse, C. A., P. B. Balbo, E. C. Pesci, D. L. Cottle, P. M. Mirabito, and C. L. Pickett. 1998. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect. Immun. 66:1934-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wösten, M. M. S. M., M. Boeve, M. G. A. Koot, A. C. van Nuene, and B. A. M. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wösten, M. M. S. M., J. A. Wagenaar, and J. P. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 82.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiological application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]
- 83.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
- 84.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and nonmotile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 85.Yeen, W. P., S. D. Puthucheary, and T. Pang. 1983. Demonstration of a cytotoxin from Campylobacter jejuni. J. Clin. Pathol. 36:1237-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]