Abstract
Deep-sea vents are the light-independent, highly productive ecosystems driven primarily by chemolithoautotrophic microorganisms, in particular by ε-Proteobacteria phylogenetically related to important pathogens. We analyzed genomes of two deep-sea vent ε-Proteobacteria strains, Sulfurovum sp. NBC37-1 and Nitratiruptor sp. SB155-2, which provide insights not only into their unusual niche on the seafloor, but also into the origins of virulence in their pathogenic relatives, Helicobacter and Campylobacter species. The deep-sea vent ε-proteobacterial genomes encode for multiple systems for respiration, sensing and responding to environment, and detoxifying heavy metals, reflecting their adaptation to the deep-sea vent environment. Although they are nonpathogenic, both deep-sea vent ε-Proteobacteria share many virulence genes with pathogenic ε-Proteobacteria, including genes for virulence factor MviN, hemolysin, invasion antigen CiaB, and the N-linked glycosylation gene cluster. In addition, some virulence determinants (such as the H2-uptake hydrogenase) and genomic plasticity of the pathogenic descendants appear to have roots in deep-sea vent ε-Proteobacteria. These provide ecological advantages for hydrothermal vent ε-Proteobacteria who thrive in their deep-sea habitat and are essential for both the efficient colonization and persistent infections of their pathogenic relatives. Our comparative genomic analysis suggests that there are previously unrecognized evolutionary links between important human/animal pathogens and their nonpathogenic, symbiotic, chemolithoautotrophic deep-sea relatives.
Keywords: ε-Proteobacteria, comparative microbial genomics, deep-sea hydrothermal vent, pathogenesis, symbiosis
Deep-sea hydrothermal vents are areas on the sea floor of high biological productivity fueled primarily by chemosynthesis. Most of the invertebrates thrive in this hostile environment through their relationship with chemolithoautotrophic proteobacterial symbionts. It has become evident that by far the most prevalent microorganisms at deep-sea vents belong to the ε-Proteobacteria (1, 2). They often account for >90% of total rRNA in various hydrothermal habitats as both epi- or endosymbionts of hydrothermal vent invertebrates (3–6) and free-living organisms associated with actively venting sulfide deposits and in areas where vent fluids and seawater mix [supporting information (SI) Fig. 5] (7–9). Until recently, the metabolic role of these key players in the deep-sea vent ecosystem has remained unknown because of their resistance to cultivation. However, in a recent breakthrough, pure cultures of deep-sea vent ε-Proteobacteria provided evidence that the majority of these microbes were mesophilic to thermophilic (capable of growth at 4°C to 70°C) chemolithoautotrophs capable of oxidation of hydrogen and sulfur compounds coupled with the reduction of oxygen, nitrate, and sulfur compounds (9–11).
These deep-sea vent ε-Proteobacteria diverge before their pathogenic relatives in small-subunit rRNA gene trees, and thus comparative genomics can provide insights into the origins and evolution of pathogenesis in the ε-Proteobacteria such as Helicobacter and Campylobacter species. It is estimated that >50% of the human population is infected by Helicobacter pylori, which causes gastric ulcer and cancer (12). In addition, Campylobacter jejuni is the leading cause of food-borne diarrhea (13). Complete genome sequences have been determined for three Helicobacter species (five different strains in total) (12, 14–17), three strains of Campylobacter jejuni (13, 18) and an additional five Campylobacter strains (18). Complete genome sequences and comparative genomic analysis of these pathogenic ε-Proteobacteria have provided important insights into their virulence factors, habitat specificities, and mechanisms of initial colonization, as well as into the significant genome plasticity (15–20). Wolinella succinogenes DSM1740T is the only nonpathogenic, host (cattle)-adapted ε-proteobacterium for which a complete genome sequence is available (21).
As an effort to address (i) the mechanistic reasons for their prevalence in deep-sea vents and (ii) whether deep-sea vent chemolithoautotrophs could be an evolutionary source of human/animal pathogens, we sequenced and analyzed complete genomes of two representative deep-sea vent ε-Proteobacteria, Sulfurovum sp. NBC37-1 and Nitratiruptor sp. SB155-2. Both strains were isolated from in situ samplers deployed on the actively venting sulfide mound in the Iheya North hydrothermal field, Japan (9).
Results and Discussion
Bacterial Strains.
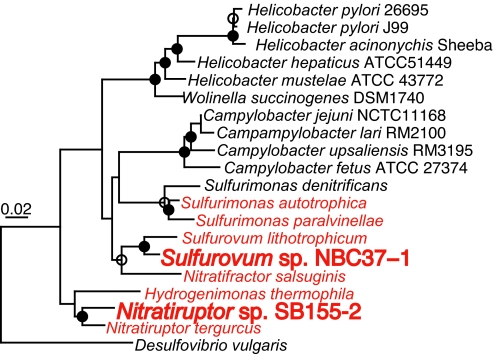
Both ε-proteobacterial strains are Gramnegative chemolithoautotrophs, capable of growth via hydrogen and sulfur-compound oxidation under microaerobic and anaerobic conditions (Table 1) (9). The 16S rRNA gene sequences of strains NBC37-1 and SB155-2 are 99.1% and 95.3% similar with those of Sulfurovum lithotrophicum 42BKT (a strict sulfur-oxidizer) (22) and Nitratiruptor tergarcus MI55–1 (a strict hydrogen-oxidizer) (23), respectively (Fig. 1). DNA–DNA hybridization value between strain NBC37-1 and S. lithotrophicum 42BKT was ≈42%. These results indicated that strains NBC37-1 and SB155-2 are new species within the genera Sulfurovum and Nitratiruptor, respectively. Although, given our sample collection, it is unclear whether these isolates are epibiotic symbionts or any other variation of symbionts, both strains have many close relatives colonizing to hydrothermal vent animals such as polychaetes and shrimps (3, 4, 9). In addition, Sulfurovum sp. NBC37-1 is closely related to the endosymbiont of a hydrothermal vent gastropod Alviniconcha (97.1% similarity of 16S rRNA gene sequence) (6). Therefore, the ε-Proteobacteria sequenced in this study probably have some symbiotic relationship with vent animals. Many genomic features, such as small genome size and high coding density (described below), strongly support their symbiotic lifestyle.
Table 1.
General features of the Sulfurovum sp. NBC37-1 and Nitratiruptor sp. SB155-2
| Characteristics | Sulfurovum sp. NBC37-1 | Nitratiruptor sp. SB155-2 |
|---|---|---|
| Energy source | H2, S2−, S0, S2O32− | H2, S2−, S0, S2O32− |
| Electron acceptor | O2, NO3− | O2, NO3− |
| Temperature range for growth, °C (optimal) | 10–37 (33) | 37–65 (55) |
| Chromosome | ||
| Size, bp | 2,562,277 | 1,877,931 |
| G+C content, mol % | 43.8 | 39.7 |
| Coding sequence (CDS) | ||
| Total no. | 2,466 | 1,857 |
| CDSs with putative function, % | 49.4 | 60.2 |
| Percent of coding region | 90.1 | 95.1 |
| No. of rrn operons | 3 | 3 |
| No. of tRNA genes | 44 | 45 |
Fig. 1.
Phylogenetic tree of 16S rRNA gene sequences. The deep-sea vent ε-Proteobacteria are shown in red. Branch points conserved with bootstrap values of >75% (filled circles) and bootstrap value of 50 to 74% (open circles) are indicated. Scale bar represents the expected number of changes per position.
Genome Features.
Strains NBC37-1 and SB155-2 each have a single circular chromosome (2,562,277 and 1,877,931 bp, respectively) with the GC content of 43.8% and 39.7% and represent the largest and smallest genomes among nonpathogenic, sequenced ε-Proteobacteria (2.1–2.2 Mb (21) (A. Copeland, S. Lucas, A. Lapidus, K. Barry, J. C. Detter, T. Glavina, N. Hammon, S. Israni, S. Pitluck, P. Chain et al., http://genome.jgi-psf.org) (SI Fig. 6 and Table 1). One of the keys to surviving and thriving in the deep-sea vents is having multiple mechanisms to overcome broad fluctuations in the availabilities (both kinds and concentration) of energy sources, electron acceptors, and carbon sources, which might explain the larger genome of strain NBC37-1. The small genome size of strain SB155-2 is comparable to host-adapted ε-proteobacterial pathogens, e.g., Helicobacter hepaticus ATCC51449 (1.8 Mb) (14).
Carbon and Energy Metabolism.
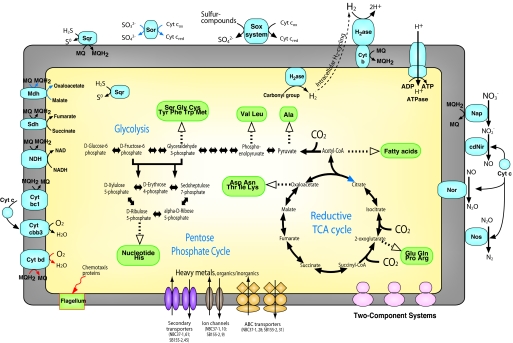
Whereas strain SB155-2 has the small genome, both deep-sea vent ε-proteobacterial genomes do encode for all of the genes of the reductive tricarboxylic acid cycle for CO2 fixation (24) and provide the genetic basis for their versatile respiration, including gene clusters for hydrogenases (described below), sulfur-compounds oxidation system (sox) (25), and denitrification (sequential reduction of nitrate to dinitrogen) pathway (Fig. 2). In contrast to those of facultatively autotrophic sulfur-oxidizing bacteria, the sox genes of deep-sea vent ε-Proteobacteria are not organized in a single region. The scattered organization of sox genes was reported for a strictly autotrophic γ-proteobacterium, Thiomicrospira crunogena (SI Fig. 7) (26). In addition to sox, the strains have both cytoplasmic and periplasmic sulfide-quinone oxidoreductases that catalyze the oxidation of sulfide to elemental sulfur, probably contributing to filamentous sulfur formation by deep-sea vent ε-Proteobacteria (27). Furthermore, consistent with our previous enzyme assay (28), strain NBC37-1 has two copies of sulfite:cytochrome c oxidoreductase (Sor). However, no predicted gene of strain SB155-2 showed significant similarity to any sor in the nonredundant protein database from the National Center for Biotechnology Information, suggesting that this strain has an unique Sor or a different system for sulfite oxidation. Both deep-sea vent ε-Proteobacteria have no dissimilatory sulfite reductase, adenylylsulfate reductase or adenylylsulfate:phosphate adenyltransferase.
Fig. 2.
Central metabolism and solute transport in the deep-sea vent ε-Proteobacteria. Pathways for which no predictable enzymes were found in strains SB155-2 and NBC37-1 genomes are shown in blue and red arrows, respectively. Numbers of transport machineries are shown for both strains. The KEGG database was used for the reconstruction of metabolic pathways. Cyt, cytochrome; H2ase, hydrogenase; Sqr, sulfide-quinone oxidoreductases; Nap, periplasmic nitrate reductase; cdNir, cytochrome cd1 nitrite reductase; Nor, nitric oxide reductase; Nos, nitrous oxide reductase; Mdh, malate dehydrogenase; Sdh, succinate dehydrogenase.
Response to Environment.
Deep-sea vent ε-Proteobacteria do not tolerate high oxygen concentrations as other ε-Proteobacteria do (9). Although both strains have no superoxide dismutase (SI Table 2), strain NBC37-1 has catalase. In addition, there are several genes in both genomes that encode for peroxidases and alkyl hydroperoxide reductases (ahp) that provide these microaerobic bacteria with resistance to oxidative stress (SI Table 2). It was recently demonstrated that the Ahp was the major antioxidant enzyme in the endosymbiont of tubeworm Riftia pachyptila inhabiting deep-sea vents (29). Furthermore, fitting to their metal-rich niche, both strains contain a wide array of mineral transport systems including detoxification mechanisms of heavy metals such as arsenate, cadmium, and copper (SI Table 2). Both strains have a large number of two-component signal transduction systems and PAS-GGDEF systems that are used commonly by prokaryotes to sense and respond to environmental signals (SI Table 2) (30). There are almost no flagellar or chemotaxis genes in the strain NBC37-1 genome, whereas the flagellated strain SB155-2 has 35 flagellar genes, 12 chemotaxis (che) genes, and 8 methyl-accepting chemotaxis (mcp) genes (SI Fig. 8), which are organized mostly in a single region with atypical G+C content (SI Fig. 6 and SI Table 3). These genes exhibit the highest similarities to those of phylogenetically distant Bacteria, e.g., Aquifex aeolicus, suggesting that this cluster is a genomic island possibly acquired via horizontal gene transfer. In addition to the potential chemotaxis island in strain SB155-2, we identified several functionally unknown genomic islands (up to 37,176 bp) in both deep-sea vent ε-Proteobacteria genomes (SI Figs. 6 and 9).
Virulence Gene Homologs.
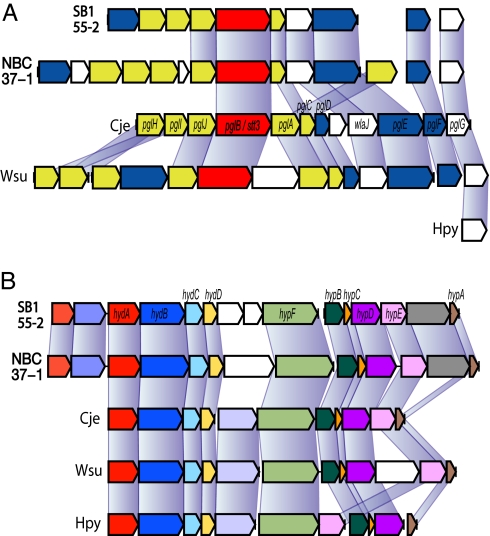
It has been shown that pathogenic ε-Proteobacteria have acquired virulence genes horizontally from evolutionarily distant sources (12, 13). Both deep-sea vent ε-Proteobacteria genomes lack orthologs of virulence genes of pathogenic ε-Proteobacteria, such as type IV secretion pathway and cag pathogenicity island genes. However, both deep-sea vent ε-Proteobacteria share many virulence genes that were identified in pathogenic ε-Proteobacteria, including genes for virulence factor MviN, hemolysin, invasion antigen CiaB, and lytic murein transglycosylase (SI Table 2). One of the most remarkable virulence genes in deep-sea vent ε-Proteobacteria is the N-linked glycosylation (NLG) gene cluster. This cluster includes a pglB/stt3 ortholog encoding for an essential oligosaccharyltransferase with a conserved catalytic motif (WWDYG) (Fig. 3A) (18, 31). The glycosylation by PglB/Stt3 is the most frequent protein-modification system of secretory and membrane protein in Eukarya but was reported exclusively in Campylobacterales in Bacteria (31). Although little is known about the origin of bacterial NLG, glycosylated proteins play important roles in microbial escape from the host immune system in both symbiotic and pathogenic host–bacterial interaction (32). A Campylobacter NLG mutant has reduced ability in adhesion and invasion into host tissue (33). In the PglB/Stt3 phylogenetic tree, deep-sea vent ε-Proteobacteria represent the deepest branching Bacteria (SI Fig. 10), therefore, bacterial NLG probably has arisen in deep-sea ε-Proteobacteria to maintain a symbiotic relationship with hydrothermal vent invertebrates. Establishing a symbiotic relationship (whether endosymbiotic or episymbiotic) with an animal should provide more opportunity in interacting with other microbes, including pathogens to the host, thus leading to acquiring virulence genes in pathogenic descendants.
Fig. 3.
Virulence genes in ε Proteobacteria. (A) Organization of N-linked glycosylation gene clusters. Cje, C. jejuni NCTC11168; Hpy, H. pylori 26695; Wsu, W. succinogenes DSM1740. Genes are color-coded by the function of encoded protein (essential oligosaccharyltransferase PglB, red; glycosyltransferase, yellow; enzymes involved in sugar biosynthesis, blue). Connecting lines indicate corresponding orthologs. (B) Organization of H2-uptake and -sensing hydrogenase gene clusters. Orthologs are shown in the same colors. hydA, small subunit gene; hydB, large subunit gene; hydC, cytochrome b subunit gene; others, maturation or accessory genes necessary for the assembly of holoenzyme.
Hydrogenase.
It is increasingly recognized that pathogenic ε-Proteobacteria have virulence determinants that are not classified as virulence genes in general but do play important roles in virulence. For example, Helicobacter species have a H2-uptake hydrogenase encoded outside the pathogenicity island, which is essential for its efficient initial colonization (34, 35). Interestingly, these pathogenic ε-Proteobacteria have only a single H2-uptake hydrogenase (36) and do not have the suite of hydrogenases that the deep-sea vent ε-Proteobacteria have. Strain NBC37-1 has four different hydrogenases (two of H2-uptake type, one H2-sensing type, and one H2-evolving type), and strain SB155-2 has three (one each of H2-uptake type, H2-sensing type, and H2-evolving type) (SI Fig. 11 and SI Table 2). Despite their genomic plasticity (described below), the ε-Proteobacteria have well-conserved H2-uptake hydrogenase gene clusters (Fig. 3B), offering an explanation for the presence of high-affinity hydrogenase in heterotrophic pathogens (35). Because the concentration of H2 in stomachs (≈100 μM) (34) is much higher than in habitats of the deep-sea vent ε-Proteobacteria, which are generally less than 10 μM, it is likely that pathogenic ε-Proteobacteria lost H2-sensing and -evolving hydrogenases in the course of adaptation to constantly H2-rich environments. Likewise, the presence of several genes specific to the reductive tricarboxylic acid cycle in Helicobacter, Campylobacter, and Wolinella (12, 13, 18, 19, 21) further suggests that these heterotrophic relatives of chemolithoautotrophic deep-sea vent ε-Proteobacteria have adapted to their pathogenic lifestyle and no longer need to fix CO2.
Genomic Plasticity.
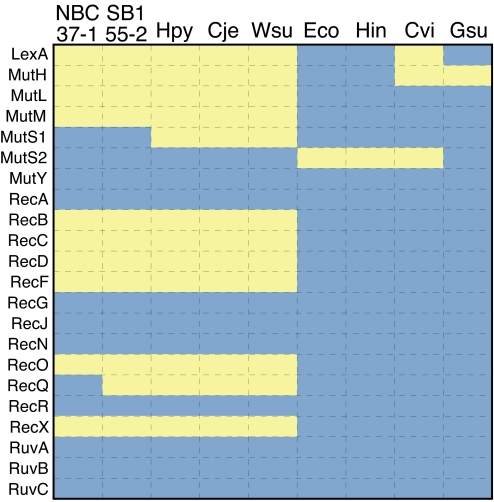
Both deep-sea vent ε-Proteobacteria and their pathogenic relatives lack many DNA-repair genes (Fig. 4), leading to frequent recombination, mutation, gene loss, and horizontal gene transfer (19, 37–39). Such genomic plasticity is not advantageous for organisms living in stable environments (40). Nevertheless, genomic plasticity increases microdiversity and enables H. pylori to persist in infections (37, 39, 41). Similarly, it is likely that the high microdiversity reported for deep-sea vent ε-Proteobacteria (9) results from the genomic plasticity of ε-Proteobacteria and confers a competitive advantage enabling this lineage to thrive in ever changing steep physical–chemical gradients. The genomic plasticity may explain not only the dominance but also the endemism of many ε-Proteobacteria to very specific habitats.
Fig. 4.
DNA-repair genes in representative Proteobacteria. Blue indicates presence, and yellow indicates absence. Hpy, H. pylori 26695; Cje, C. jejuni NCTC11168; Wsu, W. succinogenes DSM1740; Eco, Escherichia coli K12 (γ-proteobacterium); Hin, Haemophilus influenzae Rd KW20 (γ-proteobacterium); Cvi, Chromobacterium violaceum ATCC12472 (β-proteobacterium); Gsu, Geobacter sulfurreducens PCA (δ-proteobacterium).
Conclusions
Genomes of deep-sea vent ε-Proteobacteria give insights into their versatile energy metabolism, and into an enhanced ability to sense and respond to conditions outside cells. These reflect their adaptation to the deep-sea vent environment by responding to geochemical gradients and through associations with deep-sea invertebrates. Our comparative genomic analysis shows that deep-sea vent chemoautotrophy has provided the core of virulence for important human/animal pathogens. As additional genome sequences become available, the complex forces that have molded this physiologically diverse group of Bacteria will be further resolved. That autotrophy at deep-sea vents and pathogenesis in the acid gut have common genomic roots is evidence for the role genomic plasticity has played in the diversification of microbial life.
Materials and Methods
Bacterial Strains.
The deep-sea vent ε-Proteobacteria strains were isolated from a 30-m-tall sulfide mound in the Iheya North field, Japan (water depth, 1,000 m) (SI Fig. 5) (9). Strains NBC37-1 and SB155-2 were grown without shaking in MMJHS medium at 33 and 55°C, respectively. MMJHS medium contained per liter of synthetic seawater: 1 g of NaHCO3, Na2S2O3·H2O and NaNO3, 30g of S0 and 10 ml of vitamin solution (10, 11). The medium was prepared under a H2/CO2 (80:20) gas phase (300 kPa). The pH of the medium was adjusted to 6.7. Genomic DNA was obtained from late-exponential growth-phase cultures. To clarify the phylogenetic relationship between strain NBC37-1 and S. lithotrophicum 42BKT, we performed DNA–DNA hybridization following the method by Ezaki et al. (42). Briefly, RNase-treated and heat-denatured DNA was adsorbed to microplates (Maxisorb; Nalge Nunc, Rochester, NY). Probe DNA was labeled with photobiotin (Sigma, St. Louis, MO). Salmon sperm DNA (Nacalai Tesque, Kyoto, Japan) was used as a control. After sonication and denaturation of probe DNA, DNA–DNA hybridization was performed at 30°C for 3 h. Streptavidin-conjugated β-d galactosidase (GIBCO BRL, Gaithersburg, MD) and 4-methylumbelliferyl-β-d-galactopyranoside (Sigma) were used as an enzyme and a fluorogenic substrate, respectively. The reaction product was quantified with a microplate reader (Wallac 1420 ARVO sx; PerkinElmer, Boston, MA) at wavelengths of 360 nm for excitation and 450 nm for emission.
Sequencing and Assembly.
The complete genomes were sequenced by using the whole-genome shotgun method as described (43), from libraries of 1.5–2.2 kb and 3–5 kb insert size. Sequences were assembled by using Phred/Phrap (P. Green and B. Ewing, University of Washington, Seattle, WA), producing an average of 8-fold coverage across each genome. Gaps were closed by editing, walking library clones, and linking assemblies by PCR. The sequence assemblies were confirmed by pulsed-field gel electrophoresis and Southern blotting analysis. The low sequence quality region was searched with Phrap and improved by sequencing. Genome sequences of Sulfurovum sp. NBC37-1 and Nitratiruptor sp. SB155-2 are deposited under DDBJ/EMBL/GenBank accession nos. AP009179 and AP009178, respectively.
Sequence Analysis and Annotation.
We identified ORFs using Glimmer (44) and GeneMarkS (45). Regions <30 aa were eliminated. Additional ORFs >50 aa were searched from intergenic regions. The ORFs were compared with the nonredundant protein database of the NCBI by using BLASTP (46). The protein domain families were searched against the PFAM database (47). The annotated functions were classified based on Clusters of Orthologous Groups (COG) database (48). The metabolic pathway was reconstructed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (49). Signal peptides in proteins were predicted by using the SIGNALP (50), and membrane-spanning domains were predicted by using SOSUI (51). Transfer RNAs were predicted by tRNAscan-SE (52). By using cumulative GC profile method (53), we detected genomic segments that are different in GC content from the rest of the genome. We identified a segment as horizontally transferred genomic island when it has genomic island-specific features (53), such as biased codon usage, the presence of mobile genes (e.g., integrases and transposases), and the presence of repeat elements and a tRNA locus at junctions.
Phylogenetic Tree Constructions.
A phylogenetic tree of 16S rRNA gene sequences was constructed by using ARB (54). Alignments were manually checked, and unambiguously aligned nucleotide positions were used. Distances were estimated with the Jukes–Cantor correction. Bootstrapping (100 resamplings) was performed with PAUP* 4.0. Desulfovibrio vulgaris (δ-proteobacterium) served as outgroup. Phylogenetic trees based on amino acid sequences of PglB/Stt3 and hydrogenase (SI Figs. 10 and 11) was created by using ClustalX (55).
Supplementary Material
Acknowledgments
We thank the crew of the RV Natsushima and the DSROV Hyper Dolphin team for their assistance and K. Uematsu for technical help with TEM. A.-L.R. was supported by National Science Foundation Grant OCE-0242038.
Abbreviation
- NLG
N-linked glycosylation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome sequences of Sulfurovum sp. NBC37-1 and Nitratiruptor sp. SB155-2 have been deposited in DDBJ/EMBL/GenBank databases under the project accession nos. AP009179 and AP009178, respectively.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700687104/DC1.
References
- 1.Campbell BJ, Engel AS, Porter ML, Takai K. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 2.Takai K, Nakagawa S, Reysenbach A-L, Hoek J. In: Geophysical Monograph Series. Christie D, Fisher C, Lee S-M, Givens S, editors. Washington, DC: American Geophysical Union; 2006. pp. 185–213. [Google Scholar]
- 3.Polz MF, Cavanaugh CM. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cary SC, Cottrell MT, Stein JL, Camacho F, Desbruyères D. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urakawa H, Dubilier N, Fujiwara Y, Cunningham DE, Kojima S, Stahl DA. Environ Microbiol. 2005;7:750–754. doi: 10.1111/j.1462-2920.2005.00753.x. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y, Sasaki T, Suzuki M, Nogi Y, Miwa T, Takai K, Nealson KH, Horikoshi K. Appl Environ Microbiol. 2005;71:5440–5450. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longnecker K, Reysenbach AL. FEMS Microbiol Ecol. 2001;35:287–293. doi: 10.1111/j.1574-6941.2001.tb00814.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa S, Takai K, Inagaki F, Chiba H, Ishibashi J, Kataoka S, Hirayama H, Nunoura T, Horikoshi K, Sako Y. FEMS Microbiol Ecol. 2005;54:141–155. doi: 10.1016/j.femsec.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa S, Takai K, Inagaki F, Hirayama H, Nunoura T, Horikoshi K, Sako Y. Environ Microbiol. 2005;7:1619–1632. doi: 10.1111/j.1462-2920.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- 10.Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y, Nealson KH, Horikoshi K. FEMS Microbiol Lett. 2003;218:167–174. doi: 10.1111/j.1574-6968.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa S, Takai K. Methods Microbiol. 2006;35:55–91. [Google Scholar]
- 12.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 13.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, et al. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 14.Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, Droge M, Fartmann B, Fischer HP, Ge Z, et al. Proc Natl Acad Sci USA. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, et al. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, et al. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 17.Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, Keller H, Morelli G, Gressmann H, Achtman M, Schuster SC. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, et al. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eppinger M, Baar C, Raddatz G, Huson DH, Schuster SC. Nat Rev Microbiol. 2004;2:872–885. doi: 10.1038/nrmicro1024. [DOI] [PubMed] [Google Scholar]
- 20.Marais A, Mendz GL, Hazell SL, Mégraud F. Microbiol Mol Biol Rev. 1999;63:642–674. doi: 10.1128/mmbr.63.3.642-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, Klimmek O, Nandakumar R, Gross R, Rosinus A, Keller H, et al. Proc Natl Acad Sci USA. 2003;100:11690–11695. doi: 10.1073/pnas.1932838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inagaki F, Takai K, Nealson KH, Horikoshi K. Int J Syst Evol Microbiol. 2004;54:1477–1482. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa S, Takai K, Inagaki F, Horikoshi K, Sako Y. Int J Syst Evol Microbiol. 2005;55:925–933. doi: 10.1099/ijs.0.63480-0. [DOI] [PubMed] [Google Scholar]
- 24.Hügler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. J Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, Boller AJ, Chain PS, Clark JA, Davis CR, et al. PLoS Biol. 2006;4:e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor CD, Wirsen CO. Science. 1997;277:1483–1485. [Google Scholar]
- 28.Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, Hirayama H, Nakagawa S, Suzuki Y, Inagaki F, et al. Appl Environ Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hügler M, Albrecht D, Robidart J, Bench S, Feldman RA, et al. Science. 2007;315:247–250. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 30.Galperin MY, Nikolskaya AN, Koonin EV. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 31.Szymanski CM, Wren BW. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 32.Hooper LV, Gordon JI. Glycobiology. 2001;11:1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 33.Szymanski CM, Burr DH, Guerry P. Infect Immun. 2002;70:2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson JW, Maier RJ. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 35.Maier RJ, Olson J, Olczak A. J Bacteriol. 2003;185:2680–2682. doi: 10.1128/JB.185.8.2680-2682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vignais PM, Billoud B, Meyer J. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 37.Monack DM, Mueller A, Falkow S. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 38.Kraft C, Suerbaum S. Int J Med Microbiol. 2005;295:299–305. doi: 10.1016/j.ijmm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Kang J, Blaser MJ. Nat Rev Microbiol. 2006;4:826–836. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 40.Tamames J. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0020. RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson K, Loughlin MF, Potter R, Jenks PJ. J Infect Dis. 2005;191:579–587. doi: 10.1086/427657. [DOI] [PubMed] [Google Scholar]
- 42.Ezaki T, Hashimoto Y, Yabuuchi E. Int J Syst Bacteriol. 1989;39:224–229. doi: 10.1099/00207713-40-3-305. [DOI] [PubMed] [Google Scholar]
- 43.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JA, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 44.Salzberg SL, Delcher AL, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besemer J, Borodovsky M. Nucleic Acids Res. 2005;33:W451–454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatusov RL, Galperin MY, Natale DA, Koonin EV. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S, Kawashima S, Nakaya A. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 51.Hirokawa T, Boon-Chieng S, Mitaku S. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 52.Lowe TM, Eddy SR. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang R, Zhang CT. Bioinformatics. 2004;20:612–622. doi: 10.1093/bioinformatics/btg453. [DOI] [PubMed] [Google Scholar]
- 54.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar B, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.