Abstract
Morphological analysis of cytologic samples obtained by fine-needle aspirate (FNA) or bronchoscopy is an important method for diagnosing bronchogenic carcinoma. However, this approach has only about 65 to 80% diagnostic sensitivity. Based on previous studies, the c-myc x E2F-1/p21WAF1/CIP1 (p21 hereafter) gene expression index is highly sensitive and specific for distinguishing normal from malignant bronchial epithelial tissues. In an effort to improve sensitivity of diagnosing lung cancer in cytologic specimens, we used Standardized Reverse Transcriptase Polymerase Chain Reaction (StaRT-PCR) to measure the c-myc x E2F-1/p21 index in cDNA samples from 14 normal lung samples (6 normal lung parenchyma and 8 normal bronchial epithelial cell [NBEC] biopsies), and 16 FNA biopsies from 14 suspected tumors. Based on cytomorphologic criteria, 11 of the 14 suspected tumors were diagnosed as bronchogenic carcinoma and three specimens were non-diagnostic. Subsequent biopsy samples confirmed that the three non-diagnostic samples were derived from lung carcinomas. The index value for each bronchogenic carcinoma was above a cut-off value of 7000 and the index value of all but one normal sample was below 7000. Thus the c-myc x E2F-1/p21 index may augment cytomorphologic diagnosis of bronchogenic carcinoma biopsy samples, particularly those considered non-diagnostic by cytomorphologic criteria.
Most lung cancers are diagnosed by cytomorphologic analysis of bronchial brushings or washings (obtained by bronchoscopy) or transthoracic fine-needle aspirate (FNA) biopsy tissues. 1, 2, 3, 4, 5, 6 Criteria used in selecting the diagnostic method include lesion location, size, and radiographical visibility. Small, peripheral lesions primarily are assessed with transthoracic FNA methods and larger, centrally located masses primarily are evaluated using bronchoscopic techniques. 1, 3, 4, 5, 6 Cytologic samples obtained by bronchoscopy or FNA biopsy have lost their tissue architecture and diagnosis is not as reliable as histomorphologic analysis of surgical or biopsy (core needle or forceps) specimens. In multiple studies, the sensitivity of transthoracic FNA biopsies for diagnosing bronchogenic carcinoma is about 80%. 1, 2, 3 Similarly, the diagnostic yield of brushings and washings is only 60 to 70%. 4, 5, 6 The experience at the Medical College of Ohio is comparable.
Molecular classification of cancer based on expression measurement of many genes and their association to specific phenotypes will have important clinical consequences including improvement of patient care. Specifically, recent studies have shown that molecular classification of lung cancer may increase the ability to distinguish normal from malignant tissues, or metastatic from primary lung tumors and also to predict which tumors will have positive or negative outcome. 7, 8, 9, 10, 11, 12, 13, 14 This potentially will lead to more sensitive and specific diagnostic tests, as well as reveal information not available from morphological criteria regarding cancer causation and best treatment method.
Thus, gene expression analysis has potential to improve sensitivity in diagnosing malignant tissues in cytologic samples from bronchial brushings, washings, and FNA specimens. However, these small, non-renewable tissue samples are challenging to use in gene expression studies. For example, microarray methods are appropriate for screening thousands of genes potentially involved in numerous cancer phenotypes, however they are difficult to use for clinical diagnostic analysis of small cytologic specimens. 15, 16
Standardized Reverse Transcriptase Polymerase Chain Reaction (StaRT-PCR) is optimized for analysis of small clinical samples. It may be used to measure hundreds of genes simultaneously, is standardized, sensitive, and uses cDNA equivalent to 0.01 to 10 ng RNA for each gene expression measurement. 17 This method also is advantageous because it enables quantification at the plateau phase of PCR so that real-time measurement is not necessary. 17, 18, 19, 20, 21 StaRT-PCR products may be separated and quantified using multiple high throughput forms of electrophoresis or mass spectrometry. 19, 21
It is likely that malignant, chemoresistant, and metastatic phenotypes result from the interactive effects of many genes. Because the data are numerical in StaRT-PCR studies, and it is possible to measure hundreds of genes simultaneously, phenotypes can be represented by interactive gene expression indices (IGEI). We reported previously that the c-myc x E2F-1/p21WAF1/CIP1 (p21 hereafter) IGEI predicted malignancy in human bronchial epithelial cells better than any individual gene measured. 22 The association between the c-myc x E2F-1/p21 index and malignant phenotype in BEC was discovered empirically through analysis of 27 cell cycle genes in a group of cultured normal compared to a group of cultured malignant bronchial epithelial cell lines. 22 This empirical association then was validated through analysis of additional groups of normal cultured and primary lung cells and malignant cultured and primary lung cells and tissues. 22 In previous studies thus far, the c-myc x E2F-1/p21 index has been evaluated in 16 malignant specimens (12 carcinoma cell lines and 4 primary lung cancers) and 13 normal lung samples (8 cultured normal bronchial epithelial cells [NBEC] and 5 primary lung parenchyma). 22 While no single gene individually distinguished all malignant from normal samples the IGEI was 100% accurate.
Although discovered empirically, the strong association between this index and malignant phenotype likely has a mechanistic basis since c-myc and E2F-1 both promote cell cycling while p21 inhibits it. Studies are underway to directly test the mechanistic basis for this association.
The inclusion of standardized, competitive templates in every StaRT-PCR reaction allows direct intra-laboratory and inter-laboratory data comparison. 17, 23, 24 The generation of standardized, numerical data will be vital for establishing a common, multi-institutional database. A recent modification of StaRT-PCR, termed multiplex StaRT-PCR, allows further reduction in the amount of starting material needed for gene expression studies. 18 Using multiplex StaRT-PCR, at least 96 genes may simultaneously be evaluated using the amount of cDNA that normally is used to measure one gene. For example, this method was used to simultaneously measure 18 genes putatively associated with chemoresistance in a bronchogenic carcinoma sample obtained by FNA. 18
The purpose of this study was to determine whether a high c-myc x E2F-1/p21 gene expression index would augment cytopathologic diagnosis of bronchogenic carcinoma. Standardized gene expression values for c-myc, E2F-1, and p21 and the IGEI comprising these genes were determined for samples from 14 normal lung samples (from 8 NBEC bronchial brush biopsies and 6 normal lung parenchyma surgical specimens) and compared to 16 transthoracic FNA samples from 14 suspected lung tumors.
Materials and Methods
Acquisition of Bronchogenic Carcinoma, Parenchyma, and Bronchial Epithelial Samples
Transthoracic FNA biopsies of lesions suspicious for primary lung cancer were obtained from 14 patients at the Medical College of Ohio. An informed, signed consent for research use of biopsy material not necessary for diagnosis was obtained from patients according to National Institutes of Health (NIH) and institutional guidelines before each procedure. Normal parenchyma samples were obtained from patients at the time of surgery or from the Cooperative Human Tissue Network (CHTN). NBEC samples were obtained by bronchial brush biopsy as previously described. 23
A sequential order was assigned to each patient at the time of each procedure and tissue collection to protect patient identity and confidentiality, respectively. It is important to note that this research lab collects many different lung tissues (parenchyma, tumors, pleural fluid, brushings, etc) so the numbers representing the patients in this study were not listed in direct order.
Cytomorphologic Preparation of FNA Biopsy Specimens
Most of each biopsy sample was placed directly on slides for diagnostic purposes. The cells were fixed in 95% alcohol, Papanicolaou-stained, and photographed at ×320 magnification. Cell number and viability were determined through morphological analysis by a pathologist. Cells not needed for diagnostic purposes were collected in Preservcyt® Solution (CYTYC, Boxborough, MA).
RNA Preservation, Extraction, and Reverse Transcription
After final cytomorphologic diagnosis, remaining cells from the FNA biopsies stored in Preservcyt at 4°C were transported on ice to the laboratory. Total RNA was extracted from the cell pellet with Tri-Reagent (Molecular Research Center, Cincinnati, OH) according to manufacturer’s protocol. After extraction, RNA quality was evaluated on an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) for detection of 28s and 18s ribosomal RNA bands (data not shown). mRNA was reverse-transcribed using M-MLV reverse transcriptase (Gibco BRL, Gaithersburg, MD) or Sensiscript reverse transcriptase (Qiagen, Valencia, CA), and oligo (dT) primer (Promega, Madison, WI) as previously described according to manufacturers’ instructions. 17, 22, 23
StaRT-PCR
StaRT-PCR was performed using previously published protocols with Gene Express, Inc. (Toledo, OH) system I expression kit. 25 Six internal standard competitive template (CT) mixtures (A-F) and primers for actin, c-myc, E2F-1, and p21 were included in System 1 kit. The concentration of ‘target gene‘ CTs was 10-fold serially diluted in mix A-F compared to the concentration of β-actin CT. In each of the CT mixtures the β-actin CT concentration was 10−13 M so that 1 μl contained 6 × 104 molecules. Some of the cDNA samples had low concentrations and were evaluated using CT mixes diluted 10-fold so that 1 μl contained 6 × 103 molecules of β-actin CT. After reverse transcription, each cDNA sample was diluted so that the native template (NT)/CT β-actin ratio was close to 1.0 when 6 × 104 or 6 × 103 β-actin CT molecules were included in the PCR reaction.
A master mix was prepared containing Rnase-free water, MgCl2 buffer, dNTPs, the amount of cDNA in balance with 6 × 103 or 6 × 104 β-actin molecules, the amount of CT mixture D that contained 6 × 103 or 6 × 104 β-actin CT molecules, and Taq polymerase. The master mix was placed into tubes containing primers for individual genes, and cycled in a Rapidcycler (Idaho Technology, Inc., Idaho Falls, ID). The denaturing temperature was 94°C, annealing temperature was 58°C, and elongation temperature was 72°C for each cycle. After amplification, each PCR product was analyzed by microfluidic capillary electrophoresis on an Agilent 2100 Bioanalyzer machine. The area under the curve for the PCR product of each (NT) was compared to that of its respective internal standard (CT) to determine gene expression values. The unit for each expression value was molecules/per 106 β-actin molecules. If the NT/CT ratio of the PCR products was >10 or <1/10, CT mix C, or E, respectively, was used in the repeat experiment.
Statistical Analysis
A χ2 test was used to determine significant (P < 0.05) differences between primary normal tissue and transthoracic FNA tissue index values. Statistical analysis was conducted using SAS version 6.1 (SAS Institute, Cary, NC).
Results
Cytomorphologic Analysis of FNA Biopsies
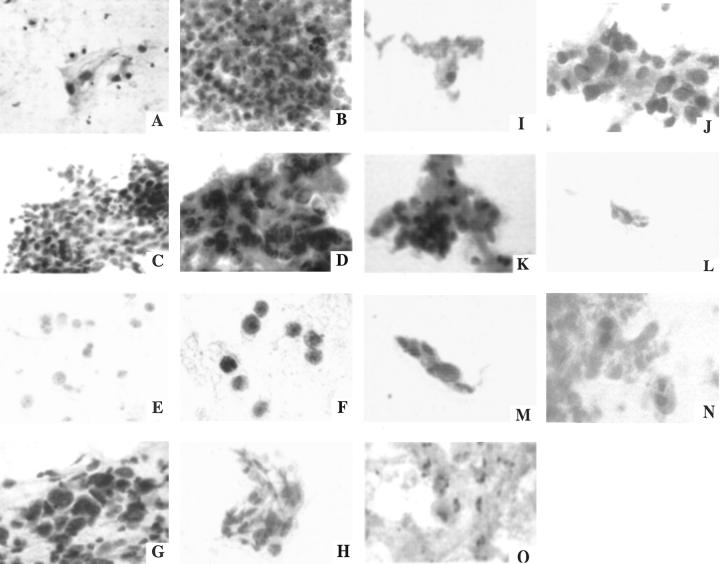
Sixteen FNA biopsies were obtained from 14 suspected tumors. Biographical data and smoking history are provided for patients from whom NBEC brush biopsies (Table 1) or transthoracic FNA biopsies (Table 2) were obtained. The cytomorphologic diagnosis is provided in Table 2 . Representative Papanicolaou stained photomicroscopic images of the cytologic preparation for an FNA from each patient are provided in Figure 1 . These representative photomicrographs were blindly reviewed by an experienced cytopathologist at another institution and the three non-diagnostic specimens were identified as such by the independent reviewer.
Table 1.
Biographic Data for Subjects from Whom Normal Bronchial Epithelial Cells (NBEC) Were Obtained
| Subject | Age | Race | Gender | Smoking History | Reason for bronchoscopy |
|---|---|---|---|---|---|
| 90 | 69 | C* | M | 1/2 ppd† for 20 years | Rule out cancer |
| 115 | 34 | C | M | NA‡ | NA |
| 118 | 72 | C | M | 1/2 ppd for 57 years | Rule out cancer |
| 136 | 38 | AA§ | M | 1 ppd for 22 years | Pneumonia |
| 150 | 70 | C | F | smoker | Rule out cancer |
| 182 | 53 | C | M | 1 1/2 ppd for 35 years | Rule out cancer |
| 191 | 75 | C | M | 1–2 ppd | Pneumonia |
| 212 | 65 | C | M | 1–2 ppd | Rule out cancer |
, Caucasian;
, pack(s) per day;
, not available;
, African-American.
Table 2.
Cytologic, Histologic, and Biographical Data for Subjects from Whom FNA Samples Were Obtained
| Subject | Gender | Age | Race | Smoking history | Cellularity | % Tumor | Viability | FNA diagnosis | Final diagnosis | RNA |
|---|---|---|---|---|---|---|---|---|---|---|
| 110 | F | 69 | C* | 1 ppd†/50 years | I‡ | 90 | H | NSCLC | Squamous | +−§ |
| 123 | M | 75 | C | 1 ppd/62 years | L¶ | 90 | L | NSCLC | Squamous | ++∥ |
| 125 | M | 67 | C | 1 ppd/50 years | H** | 90 | H | NSCLC | NSCLC | ++ |
| 138 | M | 65 | C | NA†† | H | 90 | I | NSCLC | NSCLC | NE‡‡ |
| 148a§§ | M | 77 | C | 1 ppd/65 years | L | - | L | Atypical (80%)¶¶ | SCLC | ++ |
| 148b | L | - | L | Negative | SCLC | −− | ||||
| 165 | F | 45 | C | 1 ppd/26 years | L | - | L | Atypical (20%)¶¶ | SCLC | +− |
| 172 | M | 79 | C | NA | L | 20 | L | NSCLC | NSCLC | −− |
| 200 | M | 69 | AA∥∥ | NA | H | 70 | H | NSCLC | Poor diff. care | ++ |
| 204 | M | 55 | C | Smoker | L | - | L | Suspicious (10%)¶¶ | Poor diff. care | ++ |
| 205 | F | 48 | AA | Smoker | L | 40 | H | NSCLC | NSCLC | +− |
| 219 | F | 64 | C | 1/2 ppd | L | 20 | L | NSCLC | NSCLC | ++ |
| 236a§§ | M | 62 | C | Smoker | L | 40 | H | NSCLC | NSCLC | ++ |
| 236b | I | 90 | H | NSCLC | NSCLC | ++ | ||||
| 242 | F | 74 | C | NA | L | 10 | L | NSCLC | NSCLC | +− |
| 244 | M | 63 | C | 1.5 ppd/40 years | H | 20 | L | NSCLC | Squamous | +− |
Caucasian;
pack(s) per day;
intermediate;
one ribosomal RNA band detected;
Low;
Both 28s and 18s ribosomal RNA bands detected;
High;
not available;
not evaluated;
two different FNA biopsies obtained from subject 148 and 236;
percent cells atypical or suspicious for malignancy in non-diagnostic samples;
African-American.
Figure 1.
Papanicolaou stained photomicroscopic images of transthoracic fine-needle biopsy tissues. A: 110. B: 123. C: 125. D: 138. E: 148a. F: 165. G: 172. H: 200 (Magnification, ×320). I: 204. J: 253. K: 219. L: 236a. M: 236b. N: 242. O: 244 (Magnification, ×320).
Gene Expression Analysis
Among the normal specimens the range of malignancy IGEI values was 3.9 × 102 to 6.6 × 103 for parenchyma and 0.4 × 100 to 1.1 × 104 for BEC (Table 3) . In contrast, the twelve FNA samples (from 11 suspected tumors) diagnosed as bronchogenic carcinoma by cytomorphology had very high index values that ranged from 1.1 × 104 to 3.5 × 106 (Table 4) .
Table 3.
Malignancy Index Gene Expression in mRNA from Normal Lung Parenchyma and Normal Bronchial Epithelial Cells (NBEC)
| Parenchyma | c-myc | E2F-1 | p21 | Index* | |
|---|---|---|---|---|---|
| 53† | Mean‡ | 6.5 × 103 | 3.2 × 102 | 1.1 × 103 | 1.9 × 103 |
| SD§ | 2.3 × 103 | 1.8 × 102 | 3.2 × 102 | ||
| 69 | Mean | 9.2 × 103 | 6.0 × 102 | 4.2 × 103 | 1.3 × 103 |
| SD | 2.8 × 103 | 1.7 × 102 | 8.7 × 102 | ||
| 105 | Mean | 9.4 × 103 | 1.1 × 103 | 5.2 × 103 | 2.0 × 103 |
| SD | 1.2 × 103 | 2.1 × 102 | 2.6 × 103 | ||
| 114 | Mean | 1.9 × 104 | 1.5 × 103 | 7.3 × 104 | 3.9 × 102 |
| SD | 5.5 × 103 | 3.3 × 102 | 3.2 × 104 | ||
| 144 | Mean | 8.2 × 103 | 2.6 × 103 | 7.5 × 103 | 2.8 × 103 |
| SD | 6.2 × 103 | 2.2 × 103 | 4.6 × 103 | ||
| 163 | Mean | 3.3 × 104 | 2.0 × 104 | 1.0 × 105 | 6.6 × 103 |
| SD | 2.5 × 104 | 3.3 × 103 | 5.4 × 104 |
| Normal BEC | c-myc | E2F-1 | p21 | Index | |
|---|---|---|---|---|---|
| 90 | Mean | 5.1 × 103 | 6.0 × 102 | 9.8 × 103 | 3.1 × 102 |
| SD | 6.5 × 102 | 2.8 × 102 | 4.6 × 102 | ||
| 115 | Mean | 5.7 × 104 | 6.5 × 102 | 5.1 × 104 | 7.3 × 102 |
| SD | 3.4 × 103 | 1.6 × 102 | 3.6 × 104 | ||
| 118 | Mean | 7.5 × 103 | 1.0¶ | 1.9 × 104 | 0.4 × 100 |
| SD | 1.7 × 103 | 3.8 × 103 | |||
| 136 | Mean | 3.2 × 103 | 5.0 × 102 | 2.3 × 104 | 7.0 × 101 |
| SD | 1.2 × 103 | 8.2 × 101 | 9.5 × 103 | ||
| 150 | Mean | 5.5 × 103 | 2.5 × 102 | 1.2 × 104 | 1.1 × 102 |
| SD | 1.4 × 103 | 2.4 × 101 | 3.3 × 103 | ||
| 191 | Mean | 6.1 × 104 | 1.0¶ | 1.2 × 104 | 5.0 × 100 |
| SD | 3.2 × 104 | 6.6 × 103 | |||
| 182 | Mean | 6.1 × 104 | 2.6 × 103 | 1.4 × 104 | 1.1 × 104 |
| SD | 3.0 × 104 | 1.1 × 103 | 2.4 × 104 | ||
| 212 | Mean | 1.5 × 104 | 6.4 × 102 | 7.0 × 104 | 1.4 × 102 |
| SD | 3.8 × 103 | 1.2 × 102 | 1.6 × 104 |
,Arbitrary units;
,subject number;
,data are reported as mean number of mRNA molecules per ×106 β-actin mRNA molecules;
,indicates the standard deviation from triplicate expression values;
,E2F-1 expression value too low to measure. Although the internal standard (CT) was detectable.
Native E2F-1 was not detected, so 1.0 was substituted to determine index.
Table 4.
Malignancy Index Gene Expression in mRNA from FNA Biopsy Samples
| Malignant cytomorphology | c-myc | E2F-1 | p21 | Index* | |
|---|---|---|---|---|---|
| 110† | Mean‡ | 2.5 × 105 | 4.9 × 103 | 1.1 × 105 | 1.1 × 104 |
| SD§ | 3.8 × 105 | 4.6 × 103 | 3.9 × 104 | ||
| 123 | Mean | 3.3 × 105 | 5.4 × 104 | 9.0 × 104 | 2.0 × 105 |
| SD | 4.5 × 104 | 2.3 × 104 | 4.1 × 104 | ||
| 125 | Mean | 1.2 × 105 | 4.1 × 104 | 1.1 × 105 | 4.5 × 104 |
| SD | 1.4 × 105 | 1.8 × 104 | 3.2 × 104 | ||
| 138 | Mean | 8.7 × 104 | 2.3 × 104 | 2.1 × 104 | 9.5 × 104 |
| SD | 4.0 × 104 | 3.7 × 103 | 6.0 × 103 | ||
| 172 | Mean | 2.9 × 106 | 1.7 × 105 | 1.4 × 105 | 3.5 × 106 |
| SD | 7.5 × 105 | 3.3 × 104 | 1.1 × 105 | ||
| 200 | Mean | 8.6 × 105 | 1.9 × 104 | 8.9 × 103 | 1.8 × 106 |
| SD | 1.8 × 105 | 3.4 × 103 | 5.6 × 103 | ||
| 205 | Mean | 1.6 × 105 | 5.2 × 103 | 5.4 × 104 | 1.5 × 104 |
| SD | 1.4 × 105 | 4.1 × 103 | 8.3 × 103 | ||
| 219 | Mean | 3.8 × 105 | 5.6 × 104 | 1.3 × 104 | 1.6 × 106 |
| SD | 1.7 × 105 | 2.5 × 104 | 5.2 × 103 | ||
| 236a¶ | Mean | 8.1 × 105 | 7.7 × 104 | 1.4 × 105 | 4.4 × 105 |
| SD | 3.7 × 105 | 1.3 × 104 | 7.3 × 104 | ||
| 236b | Mean | 1.1 × 106 | 8.8 × 104 | 2.3 × 105 | 4.2 × 105 |
| SD | 9.8 × 105 | 6.5 × 104 | 8.8 × 104 | ||
| 242 | Mean | 4.5 × 105 | 7.8 × 104 | 3.8 × 104 | 9.2 × 105 |
| SD | 2.1 × 105 | 4.5 × 104 | 1.6 × 104 | ||
| 244 | Mean | 4.3 × 105 | 1.6 × 105 | 4.7 × 105 | 1.5 × 105 |
| SD | 2.5 × 105 | 1.3 × 105 | 1.8 × 105 |
| Non-diagnostic cytomorphology | c-myc | E2F-1 | p21 | Index | |
|---|---|---|---|---|---|
| 148a¶ | Mean | 1.9 × 103 | 1.8 × 105 | 4.8 × 104 | 7.1 × 103 |
| SD | 1.1 × 103 | 2.5 × 104 | 2.4 × 104 | ||
| 148b | Mean | 1.3 × 103 | 3.5 × 106 | 7.3 × 104 | 6.2 × 104 |
| SD | 1.9 × 102 | 4.6 × 105 | 1.1 × 104 | ||
| 165 | Mean | 9.0 × 103 | 3.6 × 104 | 1.6 × 104 | 2.0 × 104 |
| SD | 9.5 × 102 | 1.9 × 104 | 6.0 × 103 | ||
| 204 | Mean | 1.3 × 105 | 1.6 × 105 | 2.3 × 105 | 9.0 × 104 |
| SD | 8.3 × 104 | 6.8 × 105 | 1.3 × 105 |
,Arbitrary units;
,subject number;
,data are reported as mean number of mRNA molecules per 106 β-actin mRNA molecules;
,indicates the standard deviation for triplicate expression values;
,two different FNAs obtained from subjects 236 and 148.
The four FNA biopsies that were non-diagnostic (from three suspected tumors) also exhibited high gene expression index values. For example, for subject 148, two separate FNA biopsies were obtained. The cytomorphologic interpretation was atypical in 148a and negative for malignancy in 148b, yet the index values were high at 7.1 × 103 and 6.2 × 104, respectively (Table 4) . Based on subsequent surgical biopsy samples, the tissues from case 148 were later diagnosed as small cell lung cancer (SCLC).
Similarly, the cytomorphologic interpretation for sample 165 was non-diagnostic (suspicious for SCLC) yet the index value was high at 2.0 × 104 (Table 4) . In addition, a bronchial brush sample of the suspected tumor was obtained by bronchoscopy and again, the cytomorphologic diagnosis was non-diagnostic (suspicious for SCLC) yet the index value was high at 1.1 × 105 (data not shown). For case 165, the correct diagnosis of SCLC was made in a sputum sample.
Furthermore, sample 204 was suspicious but not diagnostic for NSCLC yet had a high index value of 9.0 × 104 indicating the presence of malignant cells. Immunohistochemical evaluation of a second FNA biopsy (no material was provided for research) from this tissue confirmed a diagnosis of NSCLC.
In summary, when comparing these groups of 14 normal lung samples and 16 FNA samples from 14 suspected lung cancers, a cut-off value of 7000 for the c-myc x E2F-1/p21 index had 100% sensitivity and 94% specificity for diagnosing malignancy.
RNA Quality and Morphological Characteristics for FNA Biopsies
RNA quality was evaluated in 15 of 16 FNA specimens (Table 2) . Thirteen samples had high quality (++) or only partially degraded (+−) RNA. As expected, all 13 samples effectively served as substrates for StaRT-PCR amplification. In addition, two samples exhibited poor quality RNA (−−), yet it was possible to complete StaRT-PCR analysis in these as well. Although these two samples exhibited low cellularity and viability (Table 2) , there was not an association between cellularity, viability, and RNA quality. Six of eight samples with low cellularity and viability yielded good quality RNA. One sample was not evaluated (NE) before reverse transcription.
Discussion
Through simultaneous measurement of c-myc, E2F-1, and p21 and the IGEI comprising these genes we obtained data that augment FNA cytologic diagnosis of bronchogenic carcinoma. Similar to our previous study of surgically resected lung cancer specimens, 22 it was possible to identify a cut-off value that discriminated malignant cells from normal parenchyma and bronchial epithelial cells. In this study, the index value for each bronchogenic carcinoma was above a cut-off value of 7000, while the value for all but one normal lung sample was below 7000 for a sensitivity of 100% and specificity of 94%. When the data in this study are combined with data from previous studies, 22 the total is 30 normal primary or cultured lung samples and 27 malignant primary or cultured lung samples. The sensitivity and specificity of the malignancy index among all of these samples is 100% and 96%, respectively.
Gene Expression Malignancy Index in Non-Diagnostic FNA Biopsies
Measuring the malignancy index using StaRT-PCR may be particularly useful in cases when morphological analysis of cytology specimens from bronchoscopic and/or FNA biopsies are non-diagnostic, for example, when the sample is interpreted as ‘atypical‘ or ‘suspicious for malignancy.‘ Due to high diagnostic accuracy, the malignancy index value may reduce the number of biopsies and of additional invasive procedures required to make a diagnosis and also reduce the time between FNA biopsy and diagnosis. Fewer biopsies performed will reduce patient risk for developing pneumothorax or hemothorax and also reduce discomfort and cost.
Effect of Tumor Cell Fraction on Diagnostic Sensitivity of Index
For each sample the sensitivity of the malignancy index will depend on the tumor cell fraction and the magnitude of the index value. Thus, for sample 172 which had 20% tumor cells by cytomorphology, the index value was 3.5 × 106. By extrapolation, a sample containing only 0.1% (1/1000) tumor cells would have had an index value of 17,500 which is greater than the cut-off value of 7000. In contrast, for sample 148a the index value was 7.1 × 103 in a sample with 80% tumor cells. This sample could not have been diagnosed with a lower fraction of tumor cells. Thus, in a larger study, the sensitivity of the index may be, in part, dependent on the fraction of normal or non-diagnostic cells in the analyzed specimen. However, one advantage of FNA specimens is that malignant cells are preferentially aspirated, resulting in relatively high tumor cell fractions generally.
Significance of High Index in NBEC Sample
Case 182 was diagnosed with NSCLC based on analysis of a FNA sample (none was provided for research purposes). Among NBEC samples, the value for 182 was 15-fold higher than the next highest value (sample 115). One hypothesis to explain this high value is that a small group of malignant cells was present, perhaps a carcinoma in situ. Thus, this patient will be carefully observed for the development of a second tumor. In addition, in the future, all brush biopsies of normal bronchial epithelial cells from patients in this study will be observed by cytomorphologic as well as gene expression criteria.
Comparison of Malignancy Index Gene Expression Data Between Different Studies
One difference in the data reported here compared to our previous studies is that the highest index value reported in malignant tissues in our previous study was 12,000 and ranged as low as 710 mRNA molecules/106 molecules β-actin. 22 In this study, the highest expression value for malignant tissues was 3.5 × 106 and ranged as low as 7.1 × 103 mRNA molecules/per 106 molecules β-actin. The explanation for this difference is the use of different CT mixes in each study. In this study, a commercial standardized mixture of internal standard CTs was used. Because each of these standards was cloned there was sufficient material to quantify by fluorimetry, which is the most accurate method. A sufficient amount of this standardized mixture to last more than 100 years was prepared. In the previous study a mix prepared in our laboratory was used. These standards were not cloned. Quantification was done by comparison of intercalator dye (ethidium bromide)-stained bands to a quantified, stained-size marker. Efficiency of intercalator dye binding is partially dependent on DNA sequence, which can lead to inaccurate quantification. It is important to note, despite differences in actual index values between the two studies, that the relative differences in expression levels between the different tissue types is the same.
Optimal Methods for Clinical Diagnostic Gene Expression Measurement in Small Cytologic Specimens
For institutions currently using the Thin Prep System (CYTYC Corporation) for disease diagnosis, Preservcyt is a suitable tissue collection agent to use in molecular studies. It preserves cell morphology, cellular antigens, and other cell markers for diagnostic purposes yet also preserves nucleic acids for molecular studies. 26, 27, 28 The methods for collecting, preserving, and analyzing cytologic specimens described in this study should be useful for collection and molecular analysis of FNA biopsy material from other tissues (eg, breast, prostate).
Cytologic methods typically are the means by which advanced lung cancers are diagnosed. Diagnosis is required before treatment methods may be implemented. Gene expression analysis at this initial stage may positively affect patient treatment and improve prognosis. Two other recent studies have described. Methods for gene expression analysis using core biopsy samples and microarray gene expression analyses. For each study, it was concluded that microarray analysis was not suitable for the clinical setting. 29, 30 In one study, to address the problem of low RNA concentration, an RNA amplification method was used and multiple RNA core biopsy specimens were combined. 29 In another study, an additional 1 to 4 core biopsies were obtained strictly for research purposes. 30 These approaches are problematic because obtaining additional lung biopsy specimens may increase patient risk for developing pneumothorax/hemothorax and hospitalization. 1, 3, 6
In contrast, StaRT-PCR has many properties ideal for analysis of small clinical specimens. Quality control is facilitated due to the presence of internal standards in every gene expression measurement and the method is sensitive enough to use on small amounts of material remaining after diagnosis. Further, there is no need for RNA amplification or extra needle passes, it is reproducible and it provides standardized data as number of molecules. The suitability of StaRT-PCR in clinical diagnostics has been validated independently in numerous studies. 31, 32, 33, 34 As further validation of StaRT-PCR, the method was recently re-discovered independently from the studies that have taken place in this laboratory over the last 10 years. 21
Future Directions
We have established a high throughput Standardized Expression Measurement (SEM) Center funded by a shared National Cancer Institute (NCI) resource grant that has increased our throughput to 800 to 1,000 gene expression assays per day. The SEM Center incorporates a robotic liquid handler and a Caliper AMS 90 SE for automated microfluidic capillary electrophoresis.
In the future, we plan to use multiplex StaRT-PCR to measure the c-myc x E2F-1/p21 IGEI as well as a panel of genes recently reported to be associated with poor outcome and a panel of genes associated with cis-platin chemoresistance discovered in this laboratory (Warner K, Crawford E, Zaher A, Roshong-Denk S, Sharief I, Amurao G, Coombs R, El Samaloty H, Yoon Y, Al Astal A, Assaly R, Hernandez D-A, Graves T, Knight C, Harr M, Sheridan T, Demuth J, Zahorchalk R, Hammersly J, Olson D, Durham S, Willey J, submitted). 9, 10, 11, 13 We anticipate that this approach may improve diagnosis of challenging cases as well as treatment outcome of bronchogenic carcinoma.
Acknowledgments
We thank Dr. Andre Moreira for his expert independent review of the cytologic photographs of the FNA samples. We also thank Kelly Goodrich, David Chambers, Tom Miller, Lisa Hojnacki, and Elizabeth Samuelson for their technical assistance in bronchial transthoracic FNA biopsy collection and preparation, and Dr. Sadik Khuder for statistical analysis.
Address reprint requests to James C. Willey, Department of Medicine, Room 0012, Ruppert Building, 3000 Arlington Avenue, Toledo, OH 43699. E-mail: jwilley@mco.edu.
Footnotes
Supported by NIH U01-CA85147, ES07168, R21CA 81126, and the George Isaac Fund for Cancer Research.
K.A.W., E.L.C., R.J.Z., J.P.D., and J.C.W. have significant financial interest in Gene Express, Inc.
References
- 1.Fritscher-Ravens A, Soehendra N, Schirrow L, Sriram PVJ, Meyer A, Hauber H-P, Pforte A: Role of transesophageal endosonography-guided fine-needle aspiration in the diagnosis of lung cancer. Chest 2000, 117:339-345 [DOI] [PubMed] [Google Scholar]
- 2.Delgado PI, Jorda M, Ganjei-Azar P: Small cell carcinoma versus other lung malignancies. Cancer Cytopathol 2000, 90:279-285 [PubMed] [Google Scholar]
- 3.Salazar AM, Westcott JL: The role of transthoracic biopsy for the diagnosis and staging of lung cancer. Clin Chest Med 1993, 14:99-110 [PubMed] [Google Scholar]
- 4.Arroliga AC, Matthay RA: The role of bronchoscopy in lung cancer. Lung Cancer 1993, 14:87-98 [PubMed] [Google Scholar]
- 5.Govert JA, Kopita JM, Matchar D, Kussin PS, Samuelson WM: Cost-effectiveness of collecting routine cytologic specimens during fiberoptic bronchoscopy for endoscopically visible lung tumor. Chest 1996, 109:451-456 [DOI] [PubMed] [Google Scholar]
- 6.Bousamra M, II, Clowry L: Thorascopic fine-needle aspiration of solitary pulmonary nodules. Ann Thorac Surg 1997, 64:1191-1193 [DOI] [PubMed] [Google Scholar]
- 7.Chhieng DC, Cangiarella JF, Zakowski MF, Goswami S, Cohen J-M, Yee HT: Use of thyroid transcription factor 1, PE-10, and cytokeratins 7 and 20 in discriminating between primary lung carcinomas and metastatic lesions in fine-needle aspiration biopsy specimens. Cancer Cytopathol 2001, 93:330-336 [DOI] [PubMed] [Google Scholar]
- 8.Sorensen JB, Hirsch FR, Gazdar A, Olsen JE: Inter-observer variability in histopathologic subtyping and grading of pulmonary adenocarcinoma. Cancer 1993, 71:2971-2976 [DOI] [PubMed] [Google Scholar]
- 9.Wigle DA, Jurisica I, Radulovich N, Pintilie M, Rossant J, Liu N, Lu C, Woodgett J, Seiden I, Johnston M, Keshavjee S, Darling G, Winton T, Breitkreutz B-J, Jorgenson P, Tyers M, Shepherd FA, Tsao MS: Molecular profiling of non-small cell lung cancer and correlation with disease-free survival. Cancer Res 2002, 62:3005-3008 [PubMed] [Google Scholar]
- 10.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Peterson I: Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 2001, 98:13784-13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander E. S., Wong W, Johnson BE, Gloub TR, Sugarbaker DJ, Meyerson M: Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001, 98:13790-13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O-P, Wilfond B, Borg A, Trent J: Gene expression profiles in hereditary breast cancer. N Engl J Med 2001, 344:539-548 [DOI] [PubMed] [Google Scholar]
- 13.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001, 98:10869-10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JCF, Lashkari D, Shalon D, Brown PO, Botstein D: Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA 1999, 96:9212-9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyagi S, Kramer FR: Molecular beacons: probes that fluoresce upon hybridization. Nature Biotech 1996, 14:303-308 [DOI] [PubMed] [Google Scholar]
- 16.DeRisi JL, Iyer VR, Brown PO: Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997, 278:680-686 [DOI] [PubMed] [Google Scholar]
- 17.Willey JC, Crawford EL, Jackson CM, Weaver DA, Hoban JC, Khuder SA, De Muth JP: Expression measurement of many genes simultaneously by quantitative RT-PCR using standardized mixtures of competitive templates. Am J Respir Cell Mol Biol 1998, 19:6-17 [DOI] [PubMed] [Google Scholar]
- 18.Crawford EL, Warner KA, Khuder SA, Zahorchak RJ, Willey JC: Multiplex standardized RT-PCR for expression analysis of many genes in small clinical samples. Biochem Biophys Res Commun 2002, 293:509-516 [DOI] [PubMed] [Google Scholar]
- 19.Crawford EL, Warner KA, Weaver DA, Willey JC: Quantitative end-point RT-PCR expression measurement using the Agilent 2100 Bioanalyzer and standardized RT-PCR. Agilent Application Sept 2001, 1-8; http://www.chem.agilent.com/temp/rad9D6B0/00029012.pdf
- 20.Lyon E, Millson A, Lowery CM, Woods R, Wittwer CT: Quantification of HER2/neu gene amplification by competetive PCR using fluorescent melting curve analysis. Clin Chem 2001, 47:844-851 [PubMed] [Google Scholar]
- 21.Ding C, Cantor CR: A high-throughput gene expression analysis technique using competetive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci USA 2003, 100:3059-3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeMuth JP, Jackson CM, Weaver DA, Crawford EL, Durzinsky DS, Durham SJ, Zaher A, Phillips ER, Khuder SA, Willey JC: The gene expression index of c-myc x E2F-1/p21 is highly predictive of malignant phenotype in human bronchial epithelial cells. Am J Respir Cell Mol Biol 1998, 19:18-24 [DOI] [PubMed] [Google Scholar]
- 23.Crawford EL, Khuder SA, Durham SJ, Frampton M, Utell M, Thilly WG, Weaver DA, Ferencak WJ, Jennings CA, Hammersley JR, Olson DE, Willey JC: Normal bronchial epithelial cell expression of glutathione transferase P1, glutathione transferase M3, and glutathione peroxidase is low in subjects with bronchogenic carcinoma. Cancer Res 2000, 60:1609-1618 [PubMed] [Google Scholar]
- 24.Crawford EL, Peters GJ, Noordhuis P, Rots MG, Vondracek M, Grafstrom RC, Lieuallen K, Lennon G, Zahorchak RJ, Georgeson MJ, Wali A, Lechner JF, Fan PS, Kahaleh MB, Khuder SA, Warner KA, Weaver DA, Willey JC: Reproducible gene expression measurement among multiple laboratories obtained in a blinded study using standardized RT (StaRT)-PCR. Mol Diagn 2001, 6:217-225 [DOI] [PubMed] [Google Scholar]
- 25.Gene Express System 1 Instruction Manual. Gene Express Inc. 2000, www.genexpressinc.com
- 26.Dimulescu I, Unger ER, Lee DR, Reeves WC, Vernon SD: Characterization of RNA in cytological samples preserved in a methanol-based collection solution. J Mol Diagn 1998, 3:67-72 [DOI] [PubMed] [Google Scholar]
- 27.Tabbara SO, Sidawy MK, Frost AR, Brosky KR, Coles V, Hecth S, Radcliffe G, Shermanm ME: The stability of estrogen and progesterone receptor expression on breast cancer cells stored as Preservcyt suspensions and Thin Prep slides. Cancer 1998, 84:355-360 [PubMed] [Google Scholar]
- 28.Kaplan MA, Segura AM, Wang HH, Schnitt SJ, Upton MP: Evaluation of Cytolyt and Preservcyt as preservatives for immunocytochemistry for cytokeratin in fine-needle aspiration. Appl Immunohisto 1998, 6:23-29 [Google Scholar]
- 29.Sotiriou C, Khanna C, Jazaeri AA, Petersen D, Liu ET: Core biopsies can be used to distinguish differences in expression profiling by cDNA microarrays. J Mol Diagn 2002, 4:30-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis M, Davis N, Coop A, Lie M, Schumaker L, Lee RY, Srikanchana S, Russel CG, Singh B, Miller WR, Stearns V, Pennanen M, Tsangaris T, Gallagher A, Liu A, Zwart A, Hayes DF, Lippman ME, Wang Y, Clarke R: Development and validation of a method for using breast core-needle biopsies for gene expression microarray analyses. Clin Cancer Res 2002, 8:1155-1166 [PubMed] [Google Scholar]
- 31.Blaschke V, Reich K, Blaschke S, Zipprich S, Neumann C: Quantitative RT-PCR: comparing real-time LightCycler technology with quantitative competitive RT-PCR. Biochemica, 2002, 1:6-7; http://www.roche-appliedscience.com/biochemica/no1_02/PDF/p6.pdf
- 32.Allen JT, Knight RA, Bloor CA, Spiteri MA: Enhanced insulin-like growth factor binding protein-related protein 2 (connective tissue growth factor) expression in patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am J Respir Cell Mol Biol 1999, 21:693-700 [DOI] [PubMed] [Google Scholar]
- 33.Loitsch SM, Kippenberger S, Dauletbaev N, Wagner TO, Bargon J: Reverse transcription-competitive multiplex PCR improves quantification of mRNA in clinical samples: application to the low abundance CFTR mRNA. Clin Chem 1999, 45:619-624 [PubMed] [Google Scholar]
- 34.Mollerup S, Ryberg D, Hewer A, Phillips DH, Haugen A: Sex differences in lung CYP1A1 expression and DNA adduct levels among lung cancer patients. Cancer Res 1999, 59:3317-3320 [PubMed] [Google Scholar]