Abstract
With the completion of the Human Genome Project, over 2 million sequence-verified single nucleotide polymorphisms (SNPs) have been deposited in public databases. The challenge has shifted from SNP identification to high-throughput SNP genotyping. Although this has had little impact on molecular diagnostics, it provides the potential for future molecular diagnostics of complex traits to include SNP profiling. Accordingly, efficient, accurate, and flexible SNP genotyping are needed. In addition, the drive for low cost has pushed genotyping reactions toward multiplexing capability. We compared two SNP genotyping techniques: Affymetrix GenFlex Tag array and Pyrosequencing. The reference method was a well-established, solid-phase, single nucleotide extension reaction technique based on tritium detection. Fourteen SNPs were selected from the fine mapping project of a multiple sclerosis locus on chromosome 17q. Using all three techniques and the reference method, the SNPs were analyzed in 96 related individuals. Without extensive optimization, we successfully genotyped 11 of 14 SNPs with both GenFlex and Pyrosequencing. Our study suggests that the Pyrosequencing technique provides higher accuracy between the two systems which is most likely due to the single-stranded template in the extension reaction. Thus, Pyrosequencing has potential for diagnostic applications. Pyrosequencing, however, is not optimal for large SNP profiling analyses wherein multiplexing potential is an advantage.
The attempts to identify sequence variations that predispose to complex traits, have rarely resulted in identifying mutations that are functionally self evident; such as truncating or modifying the amino acid composition of the translated protein. A more common pattern seems to emerge from situations where a haplotype, comprising of a number of single nucleotide polymorphisms (SNPs), is associated with the trait. If this pattern holds in the future, it will result in complicated diagnostic settings that include both analytical and interpretational challenges. Most of the current diagnostic point mutation detection strategies are based on settings where one, or at most a few, single nucleotide alterations are analyzed in one sample. Currently, over 2 million sequence-verified SNPs are deposited in the public databases. 1 These sources have stimulated scientists to perform linkage analyses and candidate gene association studies in complex diseases with potential to subsequent molecular diagnostics. Accordingly, efficient, flexible, and low-cost SNP genotyping becomes increasingly pertinent to the success of experiments and subsequent diagnostics. If these assays would be translated into clinical or screening settings, the demand of accuracy would become imperative. Genotyping techniques for single nucleotide variations primarily rely on principles, such as the difference in the hybridization dynamic, 2, 3 enzymatic discrimination using ligation, 4, 5 cleavase assay, 6 and polymerase incorporation of allele-specific nucleotides to discriminate allele differences 7 (for review see 8 ). Some of these genotyping systems can be used in multiplexing platforms, such as microarray and flow cytometry. 9 Array and microtiter plate platforms result in major differences in their experimental design. For the array methods, all SNPs are analyzed in one experiment for each sample. The microtiter plate methods, however, typically assay one genotype at a time. The microtiter plate format provides more flexibility in designing the project since the cost per SNP genotype is not so much determined by the size of the sample and the number of SNPs (Figure 1) . On the other hand, most of the microtiter platforms do not provide a capacity needed for high-throughput assays, but represent systems for small to medium size needs. Because of the difference in flexibility and cost for the SNP genotyping systems that are available, we have tested three SNP genotyping techniques. Very few reports exist where the performance of different platforms have been compared with each other. 2, 10, 11 In this study, we have compared the accuracy and reproducibility of single nucleotide variant genotyping with a reference method, solid-phase minisequencing. 7, 12 All of the three techniques share the same basic reaction mechanisms of primer extension. The major difference was in the assay platform; the GenFlex Tag 13 array technique uses arrays, whereas Pyrosequencing 14 and the reference method were performed in microtiter plates. Table 1 illustrates the similarity and the differences between the three systems.
Figure 1.
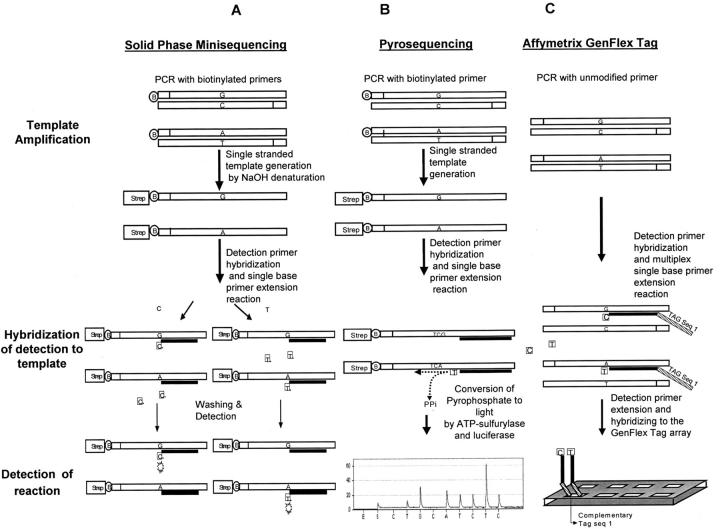
Reaction chemistry of the 3 SNP genotyping systems tested. A: In solid-phase minisequencing, PCR is performed using biotinylated primers. The biotinylated PCR product is captured to a streptavidin (Strep)-coated, solid-phase (eg, microtitration plate), and denatured. The unbound strand is washed away from the reaction solution. A single nucleotide extension reaction using 3H deoxynucleotides is performed on the captured, single-stranded template. B: In Pyrosequencing, the PCR is performed using biotinylated primers and captured to a solid-phase (beads), similar to minisequencing. Yet the beads provide a much greater binding capacity than streptavidin-coated microtitration plates. The unbound strand is washed away. A four enzyme sequencing reaction, for a stretch of 4 to 5 consecutive nucleotides, is performed. Each nucleotide extension releases pyrophosphate. C: In the Tag-Array format, the PCR is performed using regular primers, but the denatured PCR product is captured by hybridization to oligonucleoties which have a Tag sequence as a tail. This tail sequence is complementary to a specific oligonucleotide Tag’s on the array. The primer extension reaction is performed using differently labeled fluorescent dideoxynucleotides.
Table 1.
Comparison of Features of the Three SNP Genotyping Systems
| Solid-phase minisequencing | Pyrosequencing | Affymetrix Tag array | |
|---|---|---|---|
| Template preparation | PCR | PCR | PCR |
| DNA preparation post-PCR | No clean-up needed | Clean-up PCR reaction | Clean-up PCR reaction |
| Template in the detection reaction | Single-stranded template generation (strep primer) | Single-stranded template generation (strep primer) | Double-stranded |
| SNP analysis reaction | Primer extension | Primer extension | Primer extension in multiplex reaction |
| Assaying format | Microtiter | Microtiter | Microarray |
| Allelic signal discrimination | 2 Wells (1 allele/well) | Time-resolving | 2 Fluorphores |
| Allele calling | Ratio of 2 signals | Intensity peak | Ratio of 2 signals |
| Allele coding | MS Excel | Manufacturer’s software | MS Excel |
| Low no. of SNPs, high no. of samples | $$$ | $$$ | $$$$ |
| High no. of SNPs, low no. of samples | $$$ | $$$ | $ |
| High no. of SNPs, high no. of samples | $$$ | $$$ | $–$$ |
Materials and Methods
Study Sample
The study material was a smaller subsample of previously collected Finnish multiple sclerosis families. The study protocol has been described in detail elsewhere. 15, 16 A total of 96 related individuals were selected randomly from the multiple sclerosis pedigrees. Father, mother, and offspring family structures were preferred in our selection of individuals for genotyping because this allows for Mendelian inheritance checking. Extraction of genomic DNA from peripheral blood samples was performed in accordance with standard procedures. 17
Polymerase Chain Reaction
Polymerase chain reaction (PCR) was performed using 40 ng of patient DNA, 2.5 mmol/L MgCl2, and 1 pmol/primer, in a touchdown 66° to 57° (−0.5C/cycle 35 cycles) protocol. All PCR-primers were designed using an online primer design software (www.williamstone.com) to facilitate potential multiplexing. All primers had a Tm between 58°C and 60°C. For Pyrosequencing and minisequencing, one primer was biotinylated to allow solid-phase capturing using streptavidin. In the Affymetrix GenFlex Tag array, Pyrosequencing, and solid-phase minisequencing (the reference method), the DNA template of each SNP was amplified individually and the products were run on EtBr-stained agarose gels to verify the success of amplification.
Selection of SNPs
SNPs were selected from a Multiple Sclerosis (MS) positional cloning project. Putative candidate SNPs were picked by their proximity to the coding regions of the genes in the MS putative locus in chromosome 17q22–24 by querying the LeeLab cSNP database at UCLA. Putative SNPs were verified for heterozygosity by sequencing 12 different individuals. The dbSNP ID numbers were obtained by electronically mapping the cSNP sequence to the database at SNP Consortium (http://snp.cshl.org/).
Affymetrix Tag Array
Primer Design and Template Preparation
The detection primers were designed such that the 3′ end terminates one bp before the SNP site. For each SNP, two potential detection primer sequences from opposite strands were selected. Preference was given to the pair with lower overall potential for primer dimer/hairpin formation as determined by the online primer selection program (www.williamstone.com). All 14 detection primer sequences were chosen with melting temperatures between 58°C and 61°C. Range of primer length ranged from 15 bp to 28 bp. After the selection of detection primer sequences, 10 Affymetrix tag tails (Affymetrix, Santa Clara, CA) were added to each detection primer sequence. This resulted in 10 sets of detection primer sequences with each set capable of interrogating 14 SNP templates obtained from a single individual. A total of 2000 Affymetrix tag tail sequences binned into three levels of hybridization temperature are provided along with the GenFlex Tag array.
Single Base Extension Reaction
PCR of the template was performed as described in the sample preparation section. In the Affymetrix GenFlex Tag array format, PCR products of all 14 SNPs per person were pooled and analyzed together. For each reaction tube, 0.02 pmol of templates for each of the 14 SNPs, 0.2 pmol of each detection primer, 3.2 U of thermosequenase (Amersham Bioscience, Piscataway, NJ), 0.8 nmol of fluorescence-ddGTP, 0.8 nmol fluorescence-ddCTP as well as 0.125 nanomole of biotinylated ddATP (Perkin Elmer) were added to the reaction mixture to a final volume of 32 μl. Single base extension reactions were carried out in a MJ Research (Waltham, MA) thermocycler (95°C for 5 minutes, and 50 cycles for 30 seconds at 94°C, and 12 seconds at 52°C). A total of 10 reaction tubes containing 140 detection primers were pooled together and precipitated by adding 32 μl of sodium acetate (3 mol/L, pH 4.7), 128 μl of MgCl2 (25 mmol/L), 160 μl of water, and 800 μl of 100% ethanol at −20°C for 30 minutes. The reaction tubes were then spun at 13,000 rpm for 15 minutes, decanted, and dried for 30 minutes at 50°C on a heat block.
Affymetrix GenFlex Tag Array Hybridization and Washing
A protocol for hybridization and washing was provided by Affymetix. Briefly, the chip was first prehybridized with 1X MES [2-(N-morpholinoethanesulfonic acid) sodium salt] (pH 6.7) buffer for 10 minutes at 42°C. The precipitated detection primers were mixed in a solution containing 50 μl of 2X MES hybridization buffer, 2 μl of 50X Denhardt’s solution, 5 μl of Affymetrix hybridization control solution, and 43 μl of water. After denaturation, the detection primer solution was then injected into the array and hybridized for 3 hours at 42°C. After washing, the chip was stained with phycoerythrin conjugated streptavidin (SAPE) (Molecular Probes, Eugene, OR): 100 μl 6X SSPE-T, 2.5 μl BSA, 0.3 μl SAPE (1 mg/μl), and 0.3 μl cold streptavidin. Staining was performed in a carousel at room temperature for 15 minutes. After washing the arrays were scanned on a confocal scanner (Affymetrix) using fluorescence at 530 nm (fluorescein) and 560 nm (phycoerythrin) wavelengths.
Calling of Genotypes
For a given marker position, the fluorescence intensity of each of the two signals (fluorescein and phycoerythrin) was corrected for background and nonspecific hybridization by subtracting the intensity of the mismatch cell (MM) from that of the perfect match cell (PM). Each tag position on the chip contains both PM and MM cells which differ only by one nucleotide position in the middle of the tag sequence. SNPs that have negative corrected values were treated as no genotyping call. Metric P, which calculates the relative amount of each allele in the target mixture, was computed. The logarithm of intensity, which is the log of the sum of corrected signals from both channels, was also computed for each of the SNPs. A cluster diagram was generated for each SNP using Metric P and Log intensity as X and Y axes, respectively. Genotypes were determined based on the clustering of the scattered plot. Cluster boundaries were assigned manually.
Solid-Phase Minisequencing
The minisequencing reaction was performed using scintillation microtitration plates as described previously. 7 The detection primers were designed such that the 3′ end terminates one bp before the SNP site. All 14 detection primers have melting temperatures ranging from 58°C to 61°C. The sequence of the detection primer was the same as those used in the Affymetrix GenFlex Tag array with the exception of the “tag tail” sequence which is only used in the GenFlex array platform.
Pyrosequencing
Single Base Extension Reaction
Pyrosequencing reactions were performed according to the manufacturer’s instructions with minor modifications (Pyrosequencing, Uppsala, Sweden). Seven μl (10 μg/μl) of streptavidin-coated Dynabeads (Dynal, Oslo, Norway) were briefly added to each PCR 25 μl product. Then 2X binding-washing buffer (10 mmol/L Trizma base, 2 mol/L NaCl, 1 mmol/L EDTA, and 0.1% Tween 20, pH 7.6), which was equivalent to the combined volume of beads and PCR product, was added to each well. The plates were then sealed and shook at 65°C for 30 minutes. Solid-phase (bound to beads) samples were transferred consecutively to Pyro-sequencing plates containing 0.5 mol/L NaOH (for 1 minute), 1X annealing buffer (20 mmol/L Trizma acetate and 5 mmol/L magnesium acetate, pH 7.6), and 1X annealing buffer containing 15 pmol sequencing primer. Following the last step, samples were heated at 80°C for 2 minutes and then cooled for approximately 15 minutes before PSQ analysis. Pyro-sequencing was performed at room temperature on an automated PSQ 96 instrument (Pyrosequencing) according to the manufacturer’s instructions.
Calling of Genotypes
Genotyping calls were determined automatically using the PSQ HS 96 Software Version 1.0 provided by the manufacturer.
Results
For the three genotyping systems, we tested fourteen sequence-verified SNPs (Table 2) and compared the results to the reference method to determine the success rate and accuracy. The SNPs were selected from our positional cloning project to represent a true research scenario. DNA samples were collected from families that enable us to test for Mendelian inheritance of the genotypes, allowing us to provide an additional level of quality control.
Table 2.
List of SNPs Assayed
| SNP allele ID* | Genes | Position (Dec 2001) UCSC Human Genome Map) | 100 bp 5′ upstream sequence | Alleles | 100 bp 3′ upstream sequence | |
|---|---|---|---|---|---|---|
| 1 | 274579 | no | chr17:59600255-59600456 | ataatatatggtcctcggttggggaaagatacttatgatg | g/a | tagaagtacaactcaatagatggcattaaaacatattgta |
| aaggatattttttaatttaacttttttttaaatattggtaat | gtgtggatatatattttttcttttttaaaatgtgatattgac | |||||
| aggtcggcaacagcaact | gttttattaatattttt | |||||
| 2 | 884927 | no | chr17:66516874-66517074 | gtgattcttagccgaaaaaaaagcgtgtgctcttaaagta | g/a | atattcctttagctgctttgcatatttaacccagtcatcaa |
| tgttcagtttttctgtctacatatgctttgtgtgagttaaa | aaggcataatgacttcaatattttatatatcttggttttc | |||||
| aagggggtggtggtgta | aaggcataaatgacttcaatattttatatatcttggttttc | |||||
| aagtatcattcacattc | ||||||
| 3 | 104364 | RNAHP | chr17:66053470-66053671 | ccagaagccacaggattgaagggaaaggtgatcctg | g/a | acccaacaacgcttttaaagtgtcttctatttcattgtat |
| gtaactgttccaggattgctccaggttgagatggtattg | ttttttttaacttgccccaatgatagaaaagtcttttgctga | |||||
| ctaaattaaaattaaacaaga | aatgattttgatgatttt | |||||
| 4 | 92618 | KIAA0709 | chr17:64712464-64712665 | ccgggcgggggagggctctgcccctggaagagtcccct | g/a | ggttgccagagtcctgggggccccagaggagcaggag |
| gtggggaccaaaataagttccctaacatctcagctcct | tctgggagggcccagagttcaccctctagtggatccagg | |||||
| ggctctggtttggagcaaggggga | aggagcagcacccgagccctgga | |||||
| 5 | 559745 | SLC16A6 | chr17:69396069-69396270 | tgttttaagcttttttttttttgcttgtttttaaagccaaac | g/a | agatatgtagaaagctctttggttcacattccgatatta |
| aaaaacaaccaagcactcttccatatataaatctggct | aaatagtgacatgaactggcaaagtggttttaaagcttt | |||||
| gtattcagtagcaatacaa | cacgtgggataaatgatttt | |||||
| 6 | 641082 | no | chr17:60418570-60418771 | atgttgagttggtgactccagcctctttctcctggaggtc | g/a | catatgtataaccaaactccaagtgataaccagacccat |
| acaagatgatgattgcgtagatgttgcctggtgcaaagtg | ctctcctccaccttgacaaagcagattatagtatacaa | |||||
| ccccaaacagcaatagaaag | ggtaggaattcctgtcctattt | |||||
| 7 | 804731 | TOB1 | chr17:51903762-51903963 | ccaatggaatgttcccaggtgacagcccccttaacctca | g/a | tcttttgtagatggcttgaattttagcttaatacatgca |
| gtcctctccagtacagtaatgcctttgatgtgtttgcagc | gtattctaaccagcaattccagcctgttatggctaactaa | |||||
| ctatggaggcctcaatgagaa | aaaaagaaatgtatcgt | |||||
| 8 | 409259 | AKAP1 | chr17:59089735-59089936 | tgagagtcttttttgcactgttgaatgggcttggcact | c/a | tccatactgtagtcctattgagagacatttcgtctctga |
| caagtcaagatgaactcggaataacaaacatgtcct | ctccagaagtcctttcttt | |||||
| gaaaaaggatggactatgggttctcttcgcaaagcca | aaggatagtgtttaacaagcc | |||||
| 9 | 1565 | NAT2 | chr8:20634137-20642974 | gtcagttaatgtttccaagtccaattgttcctagagttct | g/a | aactaggctgaatgcaatcctcttgcttccagtctgta |
| tatagccaattctttcaaatatgcttcaatgtccatgat | ataaagtgctctcccttccaaactgtgcaagggaagtga | |||||
| ccctttggccagcaacag | tctca | |||||
| 10 | 419952 | MMD | chr17:57212307-57212508 | tgtagtccatgagttatatcctggctcagtggagtgatat | g/a | ttgctttgttgattaatctctcttgttggtgttttaataaat |
| ttatgtattatttttacttttctctcaggtcttatattaag | gaaataggcttgcctttagatcgggtgctgatattgcctg | |||||
| attaacatgttgttaata | tttcctagtaatgggctg | |||||
| 11 | 314335 | no | chr17:54535263-54535464 | tctttttgtctttaatgtttgcgcctctccgaatcagagaa | g/a | cttttgtgcatctaccgctatgtaaaggaagctgatgtca |
| gaagctgccaggattccagtacataccaaaacatgatga | gtagactggggggaacagtaaggcatgtttgtgaccgaa | |||||
| caataccctcaactgtgcaa | gctcaatttgccatcacagtg | |||||
| 12 | 572989 | MAP2K6 | chr17:70557575-70660250 | aaaggggaaatgtctcagtcgaaaggcaagaagcgaa | c/a | gagatttagactccaaggcttgcatttctattggaaatca |
| accctggccttaaattccaaagaagcatttgaacaac | gaactttgaggtgaaggcagatgacctggagcctataat | |||||
| ctcagaccagttccacaccacct | ggactgggacgaggtgcgta | |||||
| 13 | 498715 | no | chr17:67227231-67227432 | aaattataacaattttttctttgcagtaaacgatacctc | g/a | taaaataaccccaagccactctctagggtttttattttc |
| atctaagaggctctaatacctaaagagtttatccttaaaa | ttctttctttgtctttatcttttctaactagttttggaatta | |||||
| gtaaagtgactttgtacc | cgttagctactttggtt | |||||
| 14 | 827040 | GK001 | chr17:66126026-66126227 | aaaaggaatgatctatgaaatctgtgtaggttttaaatat | g/a | tgaaatttataggtagataaccagattgttgctttttgttt |
| tttaaaattataatacaaatcatcagtgcttttagtactt | aaaccagacagttgaaatggctataaagactgactctaa | |||||
| cagtgttaaagaaatcc | accaagattctgcaaataat |
Nomenclature according to reference. 1
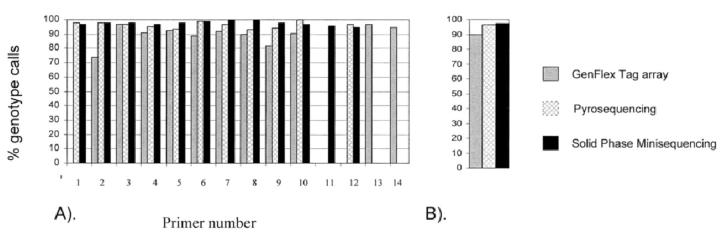
To determine the proportion of successful genotypes for each of the three methods, we determined the percentage of genotypes for each SNP for which a genotype was called from the total number of possible genotypes (Figure 2) . In each of the three systems, the assignment of a SNP genotype to one of the three possible genotypes (homozygote, heterozygote, and homozygote) was determined by the ratio of signal intensities generated by the two alleles. Typically, the SNPs, which fail to generate signals from each of the two alleles, are discarded from genotype assignment. In addition, ambiguous signal intensity ratios resulting in uncertain assignments of genotypes were discarded as well. Of the three SNP genotyping methods tested, none were able to genotype all selected SNPs. No extensive optimization was performed for any individual SNP because that would not be practical in a real high-throughput study. The reference method, solid-phase minisequencing, provided reliable genotyping results for the highest number of the SNPs tested (12 of 14 SNPs) (Figure 2A) . Pyrosequencing and Affymetrix GenFlex Tag arrays were both able to genotype 11 of 14 SNPs. This resulted in an overall genotype call success rate of 96.6% for minisequencing (representing 12 SNPs) and 96.5% for Pyrosequencing (representing 11 SNPs) (Figure 2B) . In the Affymetrix GenFlex Tag array 89.9% of genotype calls were successful. Genotype call success rates for individual SNPs are presented in Figure 2 . Examining the distribution of failed SNP genotypes across the three genotyping methods shows that each of the 14 SNPs could be genotyped by at least one method. This suggests that the designed primers were functioning properly in at least one of the assay systems. Not surprisingly, the two SNPs (SNP 13 and 14), which failed in solid-phase minisequencing, also failed in Pyrosequencing. However, we were able to assign genotypes with the GenFlex Tag array for both of these SNPs. The fact that these two SNPs failed in both minisequencing and Pyrosequencing is likely to be related to the similarity in the preparation of the templates for the primer extension reaction; both Pyrosequencing and Minisequencing use solid-phase capture and single-stranded templates in the reaction. The single-stranded, solid-phase captured format allows efficient hybridization of the detection primers to the target templates. The cause for a lower percentage of genotype calls in the array-based genotyping method might be related to the higher degree of complexity in the primer extension step; for the array-based method the extension reaction is performed on a double-stranded template in a multiplexed format. However, we cannot exclude the possibility that the use of different polymerases and nucleotides in different platforms during the extension reaction can also contribute to the differences in the success of SNP genotyping.
Figure 2.
The proportion of genotype calls for each SNP assay platform. A: The height of the bar represents the percentage of successful calls of 96 DNA samples for each SNP. Thus a low bar indicates that a large fraction of samples could not be reliably genotyped. The color coding of the bars indicate the assay platform. A lack of a bar indicates a non-working assay. B: The proportion of successful genotype calls expressed as a mean for all 14 SNPs combined. Color coding as in A.
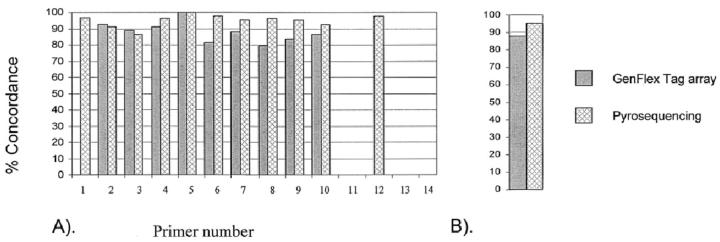
To assess the accuracy and the reliability of the two SNP genotyping methods, we compared the genotype calls with the reference method, solid-phase minisequencing. This choice of the reference method was based on our long experience with the technique. Minisequencing has been routinely used in clinical diagnostics for disease mutation detection because of its robust signal-to-noise ratio. Thus, the concordance of allele calls for each SNP in GenFlex and Pyrosequencing were compared to corresponding calls in the minisequencing assay (Figure 3A) . Pyrosequencing showed a higher average concordance with the reference method (95.1% concordance) than Affymetrix GenFlex Tag array, which had a concordance rate of 88.0% (Figure 3B) . We did not detect any systematic explanation for this difference. The assay failures occurred in different DNA templates excluding the possibility that the differences would be caused by non-working DNA samples.
Figure 3.
Concordance of SNP genotypes from three analysis methods with the reference method. Number of SNPs corresponds to number in Table 2 . A: Height of the bars expresses the percentage of concordance of genotypes for each SNP with the reference method (minisequencing) genotype. The color coding of the bars indicate the assay platform. A lack of a bar indicates a non-working assay. B: The mean concordance of genotypes for all 14 SNPs combined for each platform, compared to the reference method. Color coding as in A.
When checking for Mendelian errors of genotypes, minisequencing produced the highest proportion of genotypes with a correct inheritance pattern (99.5%), which was then followed by Pyrosequencing (97.3%) and Affymetrix GenFlex Tag array (96.5%). The 0.55% Mendelian error rate detected in the minisequencing assay represents a case where homozygote parents had a child with a heterozygote genotype. The minor allele, however, had 3H-counts, which were only one-sixth of that in other samples. This was represented most likely from a leakage in the next well, and thus a miscall. This genotype was discarded in the concordance evaluations. The tests for Mendelian inheritance are parallel with both concordance values, and successfully produced genotype values.
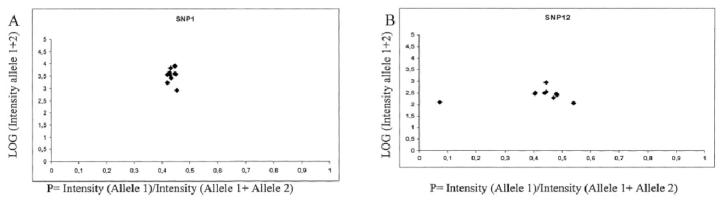
To analyze if individual detection primer sequence differences could explain the variations in SNP genotyping performance (Figure 4.) , we analyzed the primers using the Lasergene PrimerSelect software. 18 All detection primers were analyzed for their GC contents, hairpin formation stability, and dimer formation stability. No significant differences between the working and non-working SNP detection primers were observed in any of the parameters (data not shown)
Figure 4.
Scatter plots highlighting the interference of a tag sequences with the extension primer in the Gene Flex Tag array. A: Represents successful clustering of one SNP (SNP 1) using ten different tags in a sample from one heterozygous individual. B: The same sample as in A analyzed for SNP 12. The outlier observation on the left of the plot indicates cross-reaction between one tag and the extension primer.
In the case of one particular SNP, SLC, we found the genotypes of every individual tested to be heterozygous. The position of the SNP is located at 17q25 MS locus that contains highly duplicated sequences that are shared with the 17q11 region. This position was confirmed by fluorescence in situ hybridization. 16 We have possibly amplified two highly conserved sequences which only differed on the putative SNP site. This results in a heterozygous genotype of every individual in 96 DNA samples. The fact that roughly 5% of the human genome is predicted to be duplicated adds a practical notion to the SNP genotyping. 19 In a diagnostic setting this is rarely a problem as diagnostic mutations are usually extensively studied and well described before being used in clinical applications.
Discussion
We have found solid-phase minisequencing to provide the highest number of successful genotype calls and the lowest number of Mendelian errors. This is not surprising when taken into account that we have extensive experience with this technique in a diagnostic setting. The robustness of the technique can be attributed to the large signal-background noise ratio (typically of 60:1) of the systems and the small fraction of no genotyping calls. Also, the process does not require sophisticated conversion of signals to allele calls. However, the laboratory work of the reference method is also the most time consuming, and it uses the most expensive consumables of all techniques tested here. Although the procuring of the cost is less of a concern when individual variations with a high clinical impact are assayed, it becomes a challenge if SNP variation profiles become clinically relevant.
Pyrosequencing, when compared with GenFlex Tag array, had both a higher concordance rate and a higher proportion of successful genotyping calls. Pyrosequencing, however, did not reach the level of accuracy needed in a diagnostic laboratory. As in the case of minisequencing, determining genotyping calls is fairly straightforward. The commercial software included with the equipment allows Pyrosequencing to read a stretch of 5 to 10 nucleotides and makes genotype calling easy. These additional sequence readouts serve as internal controls for monitoring the specificity of the primer extension reaction. The Pyrosequencing equipment can perform over 1000 SNP genotypes in less than 4 hours after the completion of PCR. This performance is about six times the throughput of minisequencing, and the cost of Pyrosequencing is comparable to minisequencing. Therefore, Pyrosequencing has the potential for a clinical diagnostic platform. This would, however, require greater development of the current software, and implementation of more rigorous run-specific quality measurements. Of the three platforms used here, Pyrosequencing was the easiest and needed least technician time. However, also Pyrosequencing is quite laborious and needs substantial in-house development to optimize the work flow.
Affymetrix GenFlex Tag array, which had both a lower concordance rate and a lower percent genotype calls than Pyrosequencing, has the potential to be the highest throughput setup of all of the three systems used. The GenFlex Tag array has 2000 unique “tag” positions which allow simultaneously integrating of 2000 SNPs in a single hybridization. However, in a typical research laboratory, this large platform is both unrealistic, and it poses logistical challenges. Having a large platform, however, could be a relevant scenario in future diagnostic settings, where genetic profiles would be generated based on large number of SNPs and interpreted by sophisticated mathematical algorithms. However, in our particular genotyping setup of 14 SNPs, the cost of the genotype per SNP is far more expensive than in a very large setup where the chips would be used more efficiently. To increase the efficient use of the chip surface, a number of tags can be used for the same SNPs by using the tag to identify the DNA sample in one chip. This strategy was used in our study, where 10 detection primers per SNP were found to be the most economical combination. The cost per SNP genotype is well over $2.00, even though 14 SNP templates are multiplexed in a single reaction tube for primer extension after PCR. Because of the lack of commercially available genotyping software, relatively small signal-to-noise ratio, partial signal cross-talk between two fluorescence dyes and increased reaction complexity, allele-calling from raw data were somewhat challenging.
There are many SNP analysis systems currently available: ABI’s SNaPShot, Perkin Elmer’s fluorescence polarization system, 20 Genometrix’s VastArray, Sequenom’s MassArray, 21 Orchid’s SNP-IT, 22 and the generic tag array. 23 All employ features similar to that of the protocol used by Affymetrix GenFlex Tag array such as using the double-stranded target template, excess use of detection primers to effectively anneal to the target, and the usage of thermocycling in the primer extension step. All these systems have the advantage of implementing multiplexing, which can potentially reduce the cost of SNP genotyping and reduce the consumption of DNA samples. In addition, using thermocycling in the primer extension step improves the reaction kinetics and efficiency compared to constant temperature assays. However, these features and the availability of excess primers can also result in the introduction of noise due to the formation of detection primer-dimers or hairpins. This further emphasizes the importance of careful assay design and quality control of genotypes. Comprehensive evaluations, which compare the accuracy and reproducibility of high-throughput capacity SNP genotyping techniques, are not yet available. For research settings, there is also a great deal of pressure to decrease the current genotyping cost, and thus, within the next few years new, more efficient platforms will be launched. Interestingly enough, none of the techniques tested here seem to reach the level of accuracy needed in diagnostic settings without extensive optimization.
Address reprint requests to Aarno Palotie, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at UCLA, Los Angeles, Gonda Neuroscience and Genetics Research Center, Room 4524, 695 Charles E. Young Drive South, Box 708822, Los Angeles, CA 90095-7088. E-mail: apalotie@mednet.ucla.edu.
Footnotes
Supported in part by National Institutes of Health training grants T32-GM07104 and RO1 NS43559 and a grant from the National Multiple Sclerosis Society.
References
- 1.Single Nucleotide Polymorphisms for Biomedical Research. The SNP Consortium Ltd. (Deerfield, IL), http://snp.cshl.org/
- 2.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES: Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 1999, 1999, 22:231-238 [DOI] [PubMed] [Google Scholar]
- 3.Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, Chakravarti A: High-throughput variation detection and genotyping using microarrays. Genome Res 2001, 2001, 11:1913-1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Livak KJ, Kwok PY: A homogeneous, ligase-mediated DNA diagnostic test. Genome Res 1998, 1998, 8:549-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvius J, Nilsson M, Landegren U: Oligonucleotide ligation assay. Methods Mol Biol 2003, 2003, 212:215-228 [DOI] [PubMed] [Google Scholar]
- 6.Ryan D, Nuccie B, Arvan D: Non-PCR-dependent detection of the factor V Leiden mutation from genomic DNA using a homogeneous invader microtiter plate assay. Mol Diagn 1999, 1999, 4:135-144 [DOI] [PubMed] [Google Scholar]
- 7.Ihalainen J, Siitari H, Laine S, Syvanen AC, Palotie A: Towards automatic detection of point mutations: use of scintillating microplates in solid-phase minisequencing. Biotechniques 1994, 1994, 16:938-943 [PubMed] [Google Scholar]
- 8.Syvanen AC: Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet 2001, 2001, 2:930-942 [DOI] [PubMed] [Google Scholar]
- 9.Cai H, WP, Torney D, Deshpande A, Wang Z, Keller RA, Marrone B, Nolan JP: Flow cytometry-based minisequencing: a new platform for high-throughput single-nucleotide polymorphism scoring. Genomics 2000, 2000, 66:135-143 [DOI] [PubMed] [Google Scholar]
- 10.Holloway JW, Beghe B, Turner S, Hinks LJ, Day IN, Howell WM: Comparison of three methods for single nucleotide polymorphism typing for DNA bank studies: sequence-specific oligonucleotide probe hybridisation, TaqMan liquid phase hybridisation, and microplate array diagonal gel electrophoresis (MADGE). Hum Mutat 1999, 1999, 14:340-347 [DOI] [PubMed] [Google Scholar]
- 11.Le Hellard S, Ballereau SJ, Visscher PM, Torrance HS, Pinson J, Morris SW, Thomson ML, Semple CA, Muir WJ, Blackwood DH, Porteous DJ, Evans KL: SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi-automated method for data storage and analysis. Nucleic Acids Res 2002, 2002, 30:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Syvänen AC, Aalto-Setala K, Harju L, Kontula K, Soderlund H: A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics 1990, 1990, 8:684-692 [DOI] [PubMed] [Google Scholar]
- 13.Fan JB, CX, Halushka MK, Berno A, Huang X, Ryder T, Lipshutz RJ, Lockhart DJ, Chakravarti A: Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res 2000, 2000, 10:853-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronaghi M, Uhlen M, Nyren P: A sequencing method based on real-time pyrophosphate. Science 1998, 1998, 281:363-365 [DOI] [PubMed] [Google Scholar]
- 15.Kuokkanen S, Gschwend M, Rioux JD, Daly MJ, Terwilliger JD, Tienari PJ, Wikstrom J, Palo J, Stein LD, Hudson TJ, Lander ES, Peltonen L: Genome-wide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet 1997, 1997, 61:1379-1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saarela J, Schoenberg Fejzo M, Chen D, Finnila S, Parkkonen M, Kuokkanen S, Sobel E, Tienari PJ, Sumelahti ML, Wikstrom J, Elovaara I, Koivisto K, Pirttila T, Reunanen M, Palotie A, Peltonen L: Fine mapping of a multiple sclerosis locus to 2.5 Mb on chromosome 17q22–q24. Hum Mol Genet 2002, 2002, 11:2257-2267 [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J: Molecular Cloning: A Laboratory Manual ed 3 2001. Cold Spring Harbor Laboratory Cold Spring Harbor, NY
- 18.DNASTAR, Inc. (Madison, WI), http://www.lasergene.com
- 19.Samonte RV, Eichler EE: Segmental duplications and the evolution of the primate genome. Nat Rev Genet 2002, 2002, 3:65-72 [DOI] [PubMed] [Google Scholar]
- 20.Chen X, LL, Kwok PY: Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 1999, 1999, 9:492-498 [PMC free article] [PubMed] [Google Scholar]
- 21.Jurinke C, van den Boom D, Cantor CR, Koster H: Automated genotyping using the DNA MassArray technology. Methods Mol Biol 2002, 2002, 187:179-192 [DOI] [PubMed] [Google Scholar]
- 22.Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, Kopla R,, Modica F, Pohl M, Varde S, Zhao R, Zhao X, Boyce-Jacino MT: SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques Jun; 2002, (Suppl):70-77 [PubMed] [Google Scholar]
- 23.Hirschhorn JN, Sklar P, Lindblad-Toh K, Lim YM, Ruiz-Gutierrez M, Bolk S, Langhorst B, Schaffner S, Winchester E, Lander ES: SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc Natl Acad Sci USA 2000, 2000, 97:12164-12169 [DOI] [PMC free article] [PubMed] [Google Scholar]