Abstract
Since its discovery in 1994, KSHV (also called human herpesvirus-8 or HHV8) has been implicated in a variety of disorders. Although the association of KSHV with Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease has been well established, its presence in some other diseases, such as multiple myeloma, remains controversial. Because most KSHV studies are based on polymerase chain reaction (PCR) analysis, the conflicting data may be attributable to variations in the methods, primer sets, and target sequences selected. To establish an efficient and reliable PCR approach for KSHV detection we designed eight sets of primers to six regions (ORFK1, ORFK2, ORFK9, ORK26, ORF72, and ORF74) of the KSHV genome using appropriate database and software. The detection sensitivity of these primers was carefully assessed and their reliability was strictly validated in a series of positive (15 KS and PEL samples) and negative (16 lymphoid tissues) controls. We found that primer sets to the ORFK9 region showed the highest sensitivity, whereas primer sets to ORFK1 and ORF74 showed the lowest sensitivity. Primer sets to ORFK9, ORF26 and ORF72 regions detected all of the positive cases, whereas other primer sets showed varying detection rates or nonspecific bands. All 16 negative controls were negative with all primer sets. However, six of 16 negative controls became positive when we used nested PCR targeting ORF26. Therefore, multiple target KSHV sequences increase the detection efficiency, while nested PCR protocols are likely to introduce false positivity. Using ORFK9, ORF26 and ORF72 primer sets, we screened bone marrow biopsies from 18 cases of multiple myeloma, and failed to detect any KSHV sequences. This finding supports the conclusion that KSHV is not associated with multiple myeloma. Indeed, our results further confirm that although KSHV is universally present in Kaposi’s sarcoma and primary effusion lymphoma, it is not ubiquitous.
Currently, detection of many pathogens that are not easily cultivated depends heavily on molecular techniques, especially those based on polymerase chain reaction (PCR). Detection of a recently discovered herpesvirus, the Kaposi’s sarcoma-associated herpesvirus (KSHV), is a typical example. Using various PCR protocols, KSHV has been consistently identified in more than 95% of all clinical-epidemiological forms of Kaposi’s sarcoma (KS), 1, 2, 3 essentially all cases of primary effusion lymphoma (PEL), 4 and a significant proportion of cases of multicentric Castleman’s disease. 5, 6 KSHV DNA sequences have also been reported to be identifiable by PCR in many other conditions, including hyperplastic lymph nodes, 7 normal semen and prostate tissue, 8 HIV-associated lymphomas, 9 angiosarcomas, various cutaneous lesions, 10 and, more recently, multiple myeloma. 11 Although these occasional reports indicate that KSHV may be ubiquitous, most KSHV studies have suggested that the virus is not widely disseminated. It is possible that the conflicting data reported in the literature are attributable to technical artifacts or variations in PCR protocols.
In this study, using appropriate database and software, we analyzed the KSHV genome and designed eight sets of primers targeting six regions that either have been previously used for KSHV detection (ORF26, ORF72, and ORF74, encoding a capsid antigen, vCyclin and the KSHV G protein-coupled receptor, respectively) or are known to be specific to the KSHV lineage (ORFK1, ORFK2, ORFK9, encoding an ITAM motif containing protein, viral interleukin 6, and the viral interferon regulatory factor 1, respectively). 12, 13
By optimizing PCR conditions, evaluating primer sensitivity, and performing validation tests on these primers in a series of known KSHV-positive and -negative controls, we have established an efficient and reliable PCR approach to KSHV detection. We have also investigated the controversial association of KSHV with multiple myeloma using this approach.
Materials and Methods
Specimens
For validation tests, 31 DNA samples were retrieved from the DNA bank of the Molecular Pathology Laboratory, Department of Pathology, Weill Medical College of Cornell University. Six samples were prepared from formalin-fixed, paraffin-embedded tissue sections using proteinase K digestion, 14 and the remaining 25 were extracted from fresh tissues or cell lines using a salting-out procedure. 15 Fifteen samples shown to contain KSHV in previous studies 1, 16 were used as positive controls. These positive controls consisted of 11 cases of Kaposi’s sarcoma and 4 cases (including two cell lines, BC1 and BC3) of PEL. For negative controls, two cell lines (HL60 and Raji) and 14 lymphoproliferative diseases without Southern blot-detectable KSHV were included. Nine of the 16 KSHV-negative samples and 3 of the positive controls (PEL) previously had been shown to contain EBV DNA. For analysis of the association of KSHV with multiple myeloma, high molecular weight DNA samples extracted directly from fresh bone marrow core biopsies of 18 untreated multiple myeloma patients were screened with the desired KSHV primer sets. All of the core biopsies were confirmed by histological examination to be involved by myeloma cells.
Selection of PCR Target Sequences and Primer Design
Six representative regions of the KSHV genome were selected as targets for PCR amplification. These included ORF26, ORF72, and ORF74, which have been frequently used as targets for KSHV PCR detection, and ORFK1 (conserved region), ORFK2, and ORFK9, which have been shown to be specific to KSHV. 12, 13 The complete genomic sequence of KSHV (accession number U74698) was retrieved from GenBank at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Except for primer ORF26out (K330233), which was adapted from previous studies, 1 all primers to the selected KSHV regions were newly designed using Primer 3 software provided by Steve Rozen (steve@genome.wi.mit.edu) and Whitehead Institute/MIT Center for Genome Research (Cambridge, MA). To achieve unified conditions for PCR amplification of the targets, all primers designed contained similar nucleotide length (∼20n), GC ratio (∼50%), and Tm (∼60°C). Multiple primers were generated by the Primer 3 program and were further analyzed for KSHV specificity using the web-based Blast program provided by the National Center for Biotechnology Information. Eight primer sets were selected (Table 1) . Except for the primer set to ORFK2, all of the primer sets selected were highly specific to the targets intended with no or minimal homologies with other known gene sequences. The primer sequences for ORFK2, encoding vIL6, still contained some homology with the human IL6 gene and other loci.
Table 1.
Primer Information
| Name | Gene/region | Sequence | Position* | Product size |
|---|---|---|---|---|
| ORFK1 | ITAM motif | Forward-AACGCCTTACACGTTGACCT | 203–222 | 214 bp |
| Reverse-TGATGGTTGTCCACACAAGG | 416–397 | |||
| ORFK2 | vIL-6 | Forward-AGAAGCTCCATGACGTCCAC | 17493–17512 | 196 bp |
| Reverse-TTCTAGAGCCCGCTGCTATT | 17688–17669 | |||
| ORF26out | Capsid antigen | Forward-AGCCGAAAGGATTCCACCAT | 47287–47306 | 233 bp |
| Reverse-TCCGTGTTGTCTACGTCCAG | 47519–47500 | |||
| ORF26in | Capsid antigen | Forward-TATTCTGCAGCAGCTGTTGG | 47373–47392 | 138 bp |
| Reverse-TCTACGTCCAGACGATATGTGC | 47510–47489 | |||
| ORFK9-1 | vIRF-1 | Forward-GTCTCTGCGCCATTCAAAAC | 84805–84824 | 184 bp |
| Reverse-CCGGACACGACAACTAAGAA | 84988–84969 | |||
| ORFK9-3 | vIRF-1 | Forward-CCCTTTCGCGGATATACACA | 84771–84790 | |
| Reverse-AGTGAGGGGAAAGCGTCAAT | 84935–84954 | |||
| ORF72 | vCyclin | Forward-CGCCTGTAGAACGGAAACAT | 122857–122876 | 138 bp |
| Reverse-TTGCCCGCCTCTATTATCAG | 122994–122975 | |||
| ORF74 | vGPCR | Forward-TTCAGTGTTGTGTGCGTCAG | 129771–129790 | 207 bp |
| Reverse-GTTTCCCGCGTTCTCATAAC | 129977–129958 | |||
| EBV W | EBV BamHI W repeat | Forward-GATTTGGACCCGAAATCTGAT | 17077–17096 | 201 bp |
| Reverse-TCTGGGGGCTTATTCCTCTT | 17277–17258 | |||
| β-globin | β-globin | Forward-ACACAACTGTGTTCACTAGC | 180–199 | 251 bp |
| Reverse-GGAAAATAGACCAATAGGCTG | 430–410 |
Positions are based on the KSHV genome (accession vs. U75698), EBV B95-8 strain genome (accession no. V01555) and the β-globin gene (accession no. L48219) in Genbank at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov).
Control primers included those to β-globin to examine DNA integrity and exclude the presence of PCR inhibitors, and those to EBV W fragment to exclude potential cross-reactivity of the KSHV primers with EBV DNA. 17 The same conditions of the standard PCR described below were used for these primers.
PCR Amplification
A standard PCR protocol was used for all of the primer sets. The PCR reaction mixture consisted of 100 ng DNA, 10 mmol/L Tris (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.01% Triton X-100, 0.2 mmol/L of each dNTP, 0.2 μmol/L of each primer, 0.001% gelatin, and 0.25 U Taq polymerase (Boehringer Mannheim, Indianapolis, IN) in a total volume of 25 μl. Amplification was performed on an oil-free thermal cycler (Model GeneAmp 2400, Perkin Elmer, Foster City, CA) using a hot-start procedure, 18 in which the PCR mixture was denatured for 3 minutes and the Taq enzyme was added at 58°C. The hot-start procedure was followed by 35 cycles of 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 45 seconds. Series of Mg concentrations (0.5, 1, 1.5, 2, and 2.5 mmol/L) and annealing temperatures (52, 54, 56, 58, and 60°C) were tested for each set of primers to achieve optimal PCR conditions, ie, to obtain single-band PCR products with expected sizes but without nonspecific bands in control BC1 and BC3 cell lines. In addition to standard PCR, a nested PCR procedure was also used for amplification of ORF26. Briefly, 1 μl of the products from the above reaction with the ORF26out primer set was amplified with the ORF26in primer set for an additional 30 cycles, using the same PCR conditions as the first round. PCR products were run on 10% polyacrylamide gels, stained with ethidium bromide, and viewed under ultraviolet light.
Sensitivity Tests
To examine the sensitivity of the primer sets, various amounts (10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001, and 0.000001 ng) of DNA from the KSHV-infected BC3 cell line (10–20 copies of the virus per cell) 19 were mixed with 100 ng of HL60 cell line DNA. Each primer set was tested on these mixed DNA templates using the standard PCR protocol.
Contamination Control
To reduce the risk of cross-contamination, the laboratory was thoroughly cleaned before this study. Preparation of PCR mast mix, template addition, PCR amplification, and PCR product analysis were performed in four physically separated rooms. To monitor contamination, a water control was included in all PCR reactions. All samples were analyzed in duplicate for standard PCR.
Results
Optimization of PCR
All primer sets showed the same optimal Mg concentration at 1.5 mmol/L and annealing temperature at 56°C for PCR. These conditions were included in the standard PCR reaction for subsequent analysis.
Sensitivity of Individual Primer Sets
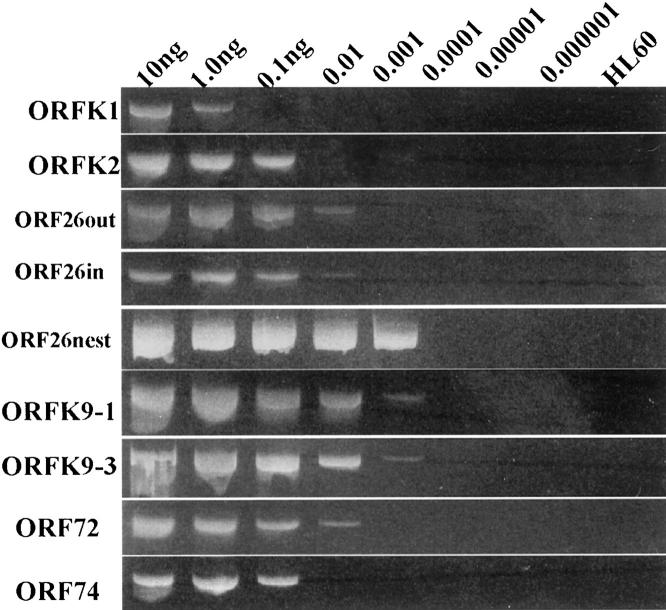
All primer sets generated PCR products with expected sizes when they were tested on the positive control, ie, BC3 (PEL) cell line (10–20 copies of KSHV per cell). As shown in Figure 1 , the primer set targeting the ORFK1 region of KSHV showed the lowest sensitivity (down to 1 ng), primer sets to the ORFK2 and ORF74 regions showed moderate sensitivity (down to 0.1 ng), and primer sets to the ORF26out, ORF26in and ORF72 regions showed high sensitivity (down to 0.01 ng). Primer sets to the ORFK9 region (ORFK9–1 and ORFK9–3) had the highest sensitivity (down to 0.001 ng; approximately 1 to 2 copies per PCR reaction). The nested method with outer and inner ORF26 primer sets had the same sensitivity as primer sets to the ORFK9 region, but showed no obvious quantitative differences within the detection range of different concentrations of BC3 template DNA (Figure 1) .
Figure 1.
Sensitivity tests of the KSHV primer sets. Various amounts (10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001, and 0.000001 ng) of DNA from the KSHV-infected BC3 cell line (10–20 copies of the virus per cell) were mixed with 100 ng of HL60 cell line DNA. Each primer set was separately tested on these mixed DNA samples. ORF26nest, nested PCR with ORF26out as outer primer set and ORF26in as inner primer set.
Validation Test
The results of PCR detection are listed in Table 2 . All of the DNA samples were PCR-amplifiable with the β-globin primer set. All DNA negative control samples were negative with all KSHV primer sets in standard PCR tests (Table 2a) . There were obvious variations among PCR tests with different KSHV primer sets in the positive control DNA samples (Table 2b) . The ORFK9–3 and ORF72 primer sets showed the best results; both generated strong, clear and specific products in all 15 KS and PEL samples. All of the KS and PEL cases were also positive with the ORF26out, ORF26in, and ORFK2 primer sets. The ORFK1 primer set missed 5 of the KS and PEL cases, and the OFR74 primer set missed 2, whereas the ORFK9–1 primer set missed 1 of the 15 cases. In comparison with the ORFK9–3 and ORF72 primer sets, all these primers, especially the ORFK1 and ORF74 primer sets, produced slightly weaker products. In addition to specific PCR products, primer sets to ORFK2 also produced one or two weak to moderate nonspecific bands (with unexpected sizes) in some cases even under very stringent conditions (Figure 2 , P1, P2). Very weak bands of unexpected size were also noted in two positive control cases amplified with the ORF74 primer set. Nested PCR for the ORF26 fragment generated strong products in all 15 KS- and PEL-positive controls.
Table 2A.
Results of PCR Validation Tests on KSHV-Negative Cases
| Cases | β-globin | EBV | K1 | K2 | ORF26out | ORF26in | 26nested | K9–1 | K9–3 | ORF72 | ORF74 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 (HL60) | + | − | − | − | − | − | − | − | − | − | − |
| C2 (TN) | ++++ | − | − | −* | − | − | − | − | − | − | − |
| C3 (Ri) | ++++ | ++++ | − | −* | − | − | +++ | − | − | − | − |
| C4 (R) | +++ | − | − | − | − | − | − | − | − | − | − |
| C5 (R) | ++++ | ++++ | − | − | − | − | − | − | − | − | − |
| FL1 | ++++ | +++ | − | − | − | − | − | − | − | − | − |
| FL2 | ++++ | − | − | − | − | − | − | − | − | − | − |
| FL3 | ++++ | ++ | − | − | − | − | − | − | − | − | − |
| FL4 | ++++ | − | − | − | − | − | − | − | − | − | − |
| FL5 | ++++ | ++++ | − | − | − | − | +++ | − | − | − | − |
| B-DLC1 | ++++ | − | − | − | − | − | − | − | − | − | − |
| B-DLC2 | +++ | ++ | − | − | − | − | +++ | − | − | − | − |
| HD-NS | ++++ | ++++ | − | − | − | − | +++ | − | − | − | − |
| PTLPD | ++ | ++++ | − | − | − | − | − | − | − | − | − |
| T-NK1 | + | +++ | − | − | − | − | +++ | − | − | − | − |
| T-NK2 | + | − | − | − | − | − | +++ | − | − | − | − |
C1, HL60 cell line; C2, tonsil; C3, Raji cell line; C4, 5, reactive lymph node; FL, follicular lymphoma; B-DLC, B cell diffuse large cell lymphoma; HD, Hodgkin’s lymphoma; PTLPD, post-transplant lymphoproliferative disorder; T-NK, natural killer cell lymphoma.
Band Intensity: −, negative; +, weak; ++, moderate; +++, strong; ++++, very strong.
Cases showing weak band(s) of unexpected sizes.
Table 2B.
Results of PCR Validation Tests on KSHV-Positive Cases
| Cases | β-globin | EBV | K1 | K2 | ORF26out | ORF26in | 26nested | K9–1 | K9–3 | ORF72 | ORF74 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KS1 | +++ | − | ++ | +++ | +++ | +++ | ++++ | − | ++++ | ++++ | ++ |
| KS2 | ++ | − | + | +++ | +++ | +++ | ++++ | ++++ | +++ | +++ | ++ |
| KS3 | + | − | − | ++* | + | + | ++++ | ++ | ++ | ++ | − |
| KS4 | + | − | +++ | +++* | ++ | +++ | ++++ | ++++ | ++++ | +++ | ++ |
| KS5 | ++++ | − | + | ++ | +++ | ++ | ++++ | +++ | ++++ | +++ | ++ |
| KS6 | ++ | − | +++ | ++++* | ++ | ++ | ++++ | +++ | ++++ | +++ | ++ |
| KS7 | +++ | ++ | − | +++ | ++++ | +++ | ++++ | ++++ | ++++ | +++ | ++ |
| KS8 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| KS9 | +++ | − | − | +++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | +++ |
| KS10 | ++++ | − | − | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| KS11 | + | − | − | +++ | +++ | +++ | ++++ | ++ | +++ | +++ | − |
| PEL1 | ++ | ++++ | ++++ | ++++* | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| PEL2 | ++++ | +++ | +++ | +++* | +++ | +++ | ++++ | ++++ | ++++ | +++ | ++++ |
| PEL3 | + | − | ++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| PEL4 | ++++ | ++++ | +++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ |
KS, Kaposi’s sarcoma; PEL, primary effusion lymphoma.
Cases showing weak band(s) of unexpected sizes.
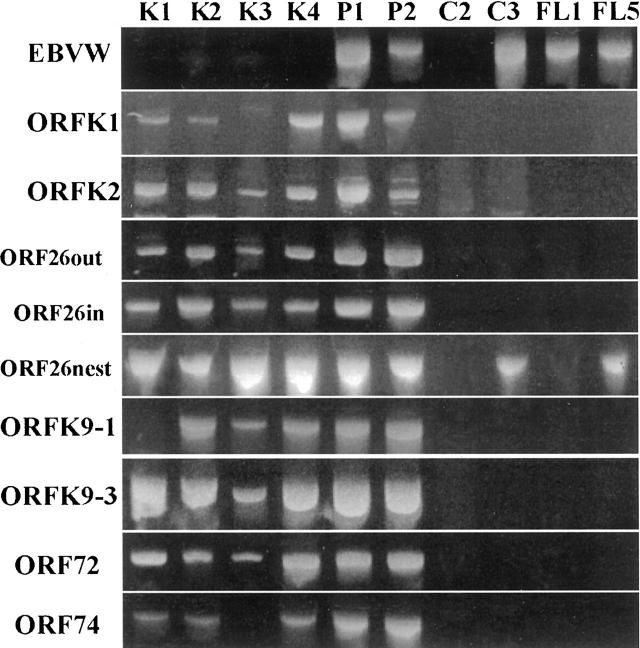
Figure 2.
Selected samples from PCR validation tests with all of the KSHV and EBV-W fragment primer sets. K1–4, Kaposi’s sarcoma cases 1–4; P1,2, Primary effusion lymphoma cases 1 and 2; C2, Negative control tonsil DNA; C3, Negative control Raji cell line DNA; FL1,5, Follicular lymphoma cases 1 and 5.
However, nested PCR also detected KSHV in 6 of the 16 negative controls (Table 2 and Figure 2 ). Using primers to the EBV BamHI W fragment, all 9 KSHV-negative controls and three PEL specimens previously shown to contain EBV generated moderate to strong PCR products with expected size. In addition, two KS cases were also positive for EBV.
Screening for KSHV in Multiple Myeloma Samples with Selected Primer Sets
Based on the results of the validation tests, three KSHV primer sets (ORF26in, ORFK9–3, and ORF72) were selected to form a routine KSHV PCR detection system. This system was tested on DNA samples from 18 cases of multiple myeloma. None of the DNA samples was positive for KSHV, although they were all PCR-amplifiable with β-globin primer set.
Discussion
Through strict sensitivity and validation tests on a series of known KSHV-positive and -negative specimens, we have demonstrated that the efficient and reliable PCR detection of KSHV is dependent on the primer design, target sequence selection, and protocols used. The results of the present study, therefore, provide some explanations for the conflicting data previously reported concerning KSHV distribution in human diseases and allowed us to establish an improved PCR approach for the detection of KSHV.
Since the discovery of KSHV in 1994, 1 the original fragment, KS330 (ORF26), has become the main target for PCR detection of the virus in the majority of KSHV studies. Other regions of KSHV, such as γ2-specific genes encoding vCyclin and vGPCR (ORF72 and ORF74), and KSHV-specific genes encoding an ITAM motif containing protein, vIL-6, and vIRF (ORFK1, ORFK2, and ORFK9), have also been used as targets for KSHV PCR detection. 20 As more and more DNA sequences of various KSHV strains are documented, it has been found that sequence variations exist within these regions of the KSHV genome, ranging from 2% in the ORF26 to 35% in the ORFK1 region. 12, 13, 21, 22, 23 It is theoretically possible that sequence variations in these regions affect the detection efficiency when they are used as targets for PCR amplification. However, so far there have no validation tests reported for comparison of PCR detection efficiencies of these KSHV regions. In the present study, we have demonstrated obvious variations in KSHV detection efficiency among the selected primer sets in our validation tests. Of these primers, the ORFK1 primer sets showed the lowest efficiency in KSHV detection, missing more than 30% of positive control cases, although the primers were designed to target relatively conserved sites of the region. This is in agreement with the observation that ORFK1 contains the highest sequence variability in the KSHV genome. 22 Primer sets to the ORF74 and ORFK9–1 regions also failed to detect some positive cases in the validation tests. The failures, again, are likely to be associated with sequence variations within the primer binding regions. In contrast to ORFK1, ORFK9–1, and ORF74, primer sets to ORFK2, ORF26, ORFK9–3, and ORF72 detected all of the positive cases examined, indicating that sequences of these primer sets are highly conserved among different KSHV strains. These results show that it is necessary to use multiple primer sets targeting different regions of KSHV for reliable detection of the virus in pathological samples.
To achieve uniform properties among multiple primer sets, we used a web-based program, Primer 3, for the primer design. Indeed, all of the primer sets selected were found to react optimally in the same temperature and buffer conditions, which greatly simplified the process for our multiple PCR analyses. However, even under these optimal conditions, there are still obvious variations (up to 1000-fold differences) in the detection sensitivity among the primer sets, with primer sets to ORFK1 and ORF74 showing the lowest sensitivity and primer sets to ORFK9 showing the highest sensitivity. Again, the low sensitivity of ORFK1 and ORF74 primer sets may reflect sequence variations in the region among different KSHV strains. The reasons for the sensitivity differences among other primer sets are not clear, since validation tests indicate that the sequences of these primers are highly conserved among different KSHV-infected samples. Since primer sets with similar properties can result in different sensitivities, it is important to screen sufficient primers to obtain primer sets with desired sensitivity.
Phylogenetic analysis of nucleic acid sequences has placed KSHV in the lymphotropic γ Herpesviridae family, showing significant homologies with herpesvirus saimiri and Epstein-Barr virus. 12 To ensure specificity, we very carefully analyzed all of the primer sequences against documented sequences available in the GenBank through Blast, a Web-based program. All primer sets selected for this study are highly specific to the intended targets with no homologies to other known viral sequences. The specificity of these primer sets is further supported by the lack of evidence for cross-amplification with EBV genome in our validation tests. The high specificity may be the basis for the observation that, except for the ORFK2 and ORF74 primer sets, all primer sets generated a discrete band of expected size with no or minimal background. The primers for the ORFK2 gene still contain some sequences homologous to the human IL6 gene and other loci. Therefore, the nonspecific bands observed with the ORFK2 primer set may reflect coamplification of the relevant human gene fragments in the samples. Since only two KSHV positive control cases amplified with the ORF74 primer set showed weak nonspecific products of unexpected sizes, and none of negative controls exhibited PCR products with the primer set, it is possible that the weak bands observed may be associated with the nature of the samples rather than the nature of the ORF74 primers used.
Sequence variations of KSHV can result in low detection sensitivity of PCR. However, in many studies the high sensitivity of the PCR protocols causes concerns. Most of these studies used PCR methods involving an excessive cycle number (>40) or nested primers, claiming to be able to detect as little as three copies of KSHV genome per 200,000 cells. 8, 24, 25 With these methods, KSHV DNA sequences have been reported in a significant proportion of various diseases and even normal controls by some investigators. 8 It is possible that the enhanced sensitivity of the PCR analysis in these studies might have allowed detection of rare passenger KSHV genomes or minute amounts of randomly contaminated KSHV sequences in the samples examined. Nested PCR assays, based on reamplification of already existing PCR products, carry an increased potential for random contamination compared with standard PCR. However, standard PCR with excessive cycle numbers is also prone to contamination. In comparison with standard PCR in our sensitivity and validation tests, we have noted that nested PCR had the highest sensitivity, but showed no obvious quantitative differences among various concentrations of initial KSHV template DNA, making it impossible to estimate viral loads in the samples. Our nested PCR also detected KSHV sequences in 6 of 16 negative controls. This nested PCR was performed using primers to the ORF26 KS330 region, which has been reported in one study to be widely disseminated. 26 However, our results may be more indicative of the likelihood that with nested PCR, it is extremely easy to get contamination even in strictly controlled environments, as we did not detect these sequences using a direct PCR approach. To avoid contamination and ensure reliable PCR detection of KSHV, we highly recommend the use of standard PCR protocol with more than one set of primers. The ORF26in, ORFK9–3, and ORF72 primer sets described in the current study, which are highly specific and allow detection of one to two copies of DNA in each PCR reaction, should be valuable reagents for routine KSHV PCR detection or viral load analysis using quantitative PCR, such as real-time PCR.
Using a combination of ORF26in, ORFK9–3, and ORF72 KSHV primer sets, we failed to detect KSHV in multiple myeloma. Initial studies similarly failed to recognize KSHV in bone marrow aspirates from multiple myeloma samples. 9, 27, 28 However, subsequent reports of the presence of this virus in adherent cells obtained from bone marrow aspirates and in bone marrow biopsy samples from patients with myeloma 11, 29 suggested that KSHV-infected cells were too rare to be detected in aspirates, but could be detected in the actual core biopsies. Though some investigators have been able to reproduce the alleged KSHV positivity in multiple myeloma, 30, 31 the vast majority of studies have failed to find such an association. 32 Furthermore, multiple serological studies have indicated that patients with multiple myeloma lack antibodies to KSHV, and a monoclonal antibody specific to the KSHV latent nuclear antigen has failed to detect KSHV in bone marrow biopsies involved with multiple myeloma. 33 Studies in our laboratory have also suggested that patients with myeloma lack antibodies to KSHV, 34 and we have failed to detect KSHV in myeloma bone marrow biopsies by immunohistochemistry using antibodies to latent nuclear antigen and vIL-6 (E Hyjek and E Cesarman, unpublished observations). The studies presented here are strongly supportive of the large body of evidence indicating that KSHV is not associated with multiple myeloma.
Address reprint requests to Dr. Langxing Pan, Department of Pathology, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10021. E-mail: lpan@mail.med.cornell.edu.
References
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS: Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266:1865-1869 [DOI] [PubMed] [Google Scholar]
- 2.Boshoff C, Whitby D, Hatziioannou T, Fisher C, van der Walt J, Hatzakis A, Weiss R, Schulz T: Kaposi’s-sarcoma-associated herpesvirus in HIV-negative Kaposi’s sarcoma (letter). Lancet 1995, 345:1043-1044 [DOI] [PubMed] [Google Scholar]
- 3.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y: Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol 1996, 70:549-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nador RG, Cesarman E, Knowles DM, Said JW: Herpes-like DNA sequences in a body-cavity-based lymphoma in an HIV-negative patient (letter). N Engl J Med 1995, 333:943. [DOI] [PubMed] [Google Scholar]
- 5.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L: Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995, 86:1276-1280 [PubMed] [Google Scholar]
- 6.Dupin N, Diss TL, Kellam P, Tulliez M, Du MQ, Sicard D, Weiss RA, Isaacson PG, Boshoff C: HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 2000, 95:1406-1412 [PubMed] [Google Scholar]
- 7.Luppi M, Barozzi P, Maiorana A, Artusi T, Trovato R, Marasca R, Savarino M, Ceccherini-Nelli L, Torelli G: Human herpesvirus-8 DNA sequences in human immunodeficiency virus-negative angioimmunoblastic lymphadenopathy and benign lymphadenopathy with giant germinal center hyperplasia and increased vascularity. Blood 1996, 87:3903-3909 [PubMed] [Google Scholar]
- 8.Monini P, de Lellis L, Fabris M, Rigolin F, Cassai E: Kaposi’s sarcoma-associated herpesvirus DNA sequences in prostate tissue and human semen. N Engl J Med 1996, 334:1168-1172 [DOI] [PubMed] [Google Scholar]
- 9.Otsuki T, Kumar S, Ensoli B, Kingma DW, Yano T, Stetler-Stevenson M, Jaffe ES, Raffeld M: Detection of HHV-8/KSHV DNA sequences in AIDS-associated extranodal lymphoid malignancies. Leukemia 1996, 10:1358-1362 [PubMed] [Google Scholar]
- 10.Rady PL, Yen A, Rollefson JL, Orengo I, Bruce S, Hughes TK, Tyring SK: Herpesvirus-like DNA sequences in non-Kaposi’s sarcoma skin lesions of transplant patients. Lancet 1995, 345:1339-1340 [DOI] [PubMed] [Google Scholar]
- 11.Rettig MB, Ma HJ, Vescio RA, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said JW, Berenson JR: Kaposi’s sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science 1997, 276:1851-1854 [DOI] [PubMed] [Google Scholar]
- 12.McGeoch DJ, Davison AJ: The descent of human herpesvirus 8. Semin Cancer Biol 1999, 9:201-209 [DOI] [PubMed] [Google Scholar]
- 13.Poole LJ, Zong JC, Ciufo DM, Alcendor DJ, Cannon JS, Ambinder R, Orenstein JM, Reitz MS, Hayward GS: Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi’s sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol 1999, 73:6646-6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diss TC, Pan LX, Peng HZ, Wotherspoon AC, Isaacson PG: Sources of DNA for detecting B cell monoclonality using PCR. J Clin Pathol 1994, 47:493-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988, 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesarman E, Nador RG, Aozasa K, Delsol G, Said JW, Knowles DM: Kaposi’s sarcoma-associated herpesvirus in non-AIDS related lymphomas occurring in body cavities. Am J Pathol 1996, 149:53-57 [PMC free article] [PubMed] [Google Scholar]
- 17.Diss TC, Wotherspoon AC, Speight PM, Pan LX, Isaacson PG: B cell monoclonality, Epstein Barr virus and t(14;18) in myoepithelial sialadenitis and low grade B cell MALT lymphoma of the parotid gland. Am J Surg Pathol 1995, 19:531-536 [DOI] [PubMed] [Google Scholar]
- 18.D’Aquila RT, Bechtel LJ, Videler JA, Eron JJ, Kaplan JC: Maximizing sensitivity and specificity of PCR by pre-amplification heating. Nucleic Acids Res 1991, 19:3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E: Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 1996, 88:2648-2654 [PubMed] [Google Scholar]
- 20.Berenson JR, Vescio RA: HHV-8 is present in multiple myeloma patients. Blood 1999, 93:3157-3159 [PubMed] [Google Scholar]
- 21.Hayward GS: KSHV strains: the origins and global spread of the virus. Semin Cancer Biol 1999, 9:187-199 [DOI] [PubMed] [Google Scholar]
- 22.Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS: High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol 1999, 73:4156-4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelbrecht S, Treurnicht FK, Schneider JW, Jordaan HF, Steytler JG, Wranz PA, van Rensburg EJ: Detection of human herpes virus 8 DNA and sequence polymorphism in classical, epidemic, and iatrogenic Kaposi’s sarcoma in South Africa. J Med Virol 1997, 52:168-172 [PubMed] [Google Scholar]
- 24.Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O’Leary JJ: Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med 1995, 1:1274-1278 [DOI] [PubMed] [Google Scholar]
- 25.Tasaka T, Said JW, Morosetti R, Park D, Verbeek W, Nagai M, Takahara J, Koeffler HP: Is Kaposi’s sarcoma-associated herpesvirus ubiquitous in urogenital and prostate tissues? Blood 1997, 89:1686-1689 [PubMed] [Google Scholar]
- 26.Tisdale JF, Stewart AK, Dickstein B, Little RF, Dube I, Cappe D, Dunbar CE, Brown KE: Molecular and serological examination of the relationship of human herpesvirus 8 to multiple myeloma: orf 26 sequences in bone marrow stroma are not restricted to myeloma patients and other regions of the genome are not detected. Blood 1998, 92:2681-2687 [PubMed] [Google Scholar]
- 27.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM: Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 1995, 332:1186-1191 [DOI] [PubMed] [Google Scholar]
- 28.Gessain A, Briere J, Angelin-Duclos C, Valensi F, Beral HM, Davi F, Nicola MA, Sudaka A, Fouchard N, Gabarre J, Troussard X, Dulmet E, Audouin J, Diebold J, De The G: Human herpes virus 8 (Kaposi’s sarcoma herpes virus) and malignant lymphoproliferations in France: a molecular study of 250 cases including two AIDS-associated body cavity based lymphomas. Leukemia 1997, 11:266-272 [DOI] [PubMed] [Google Scholar]
- 29.Said JW, Rettig MR, Heppner K, Vescio R, Schiller G, Ma HJ, Belson D, Savage A, Shintaku IP, Koeffler HP, Asou H, Pinkus G, Pinkus J, Schrage M, Green E, Berenson JR: Localization of Kaposi’s sarcoma-associated herpesvirus in bone marrow biopsy samples from patients with multiple myeloma. Blood 1997, 90:4278-4282 [PubMed] [Google Scholar]
- 30.Brousset P, Meggetto F, Attal M, Delsol G: Kaposi’s sarcoma-associated herpesvirus infection and multiple myeloma (letter; comment). Science 1997, 278:1972-1973 [PubMed] [Google Scholar]
- 31.Agbalika F, Mariette X, Marolleau JP, Fermand JP, Brouet JC: Detection of human herpesvirus-8 DNA in bone marrow biopsies from patients with multiple myeloma and Waldenstrom’s macroglobulinemia (letter). Blood 1998, 91:4393-4394 [PubMed] [Google Scholar]
- 32.Tarte K, Chang Y, Klein B: Kaposi’s sarcoma-associated herpesvirus and multiple myeloma: lack of criteria for causality. Blood 1999, 93:3159-3163 [PubMed] [Google Scholar]
- 33.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, Weiss RA, Alitalo K, Boshoff C: Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc Natl Acad Sci USA 1999, 96:4546-4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyjek E, Fata F, Niesvisly R, Michaeli J, Cesarman E: Low prevalence of antibodies to Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in patients with multiple myeloma. Blood 1997, 90:87A (abstr.)