Abstract
The mouse retrogene Utp14b is essential for male fertility, and a mutation in its sequence results in the sterile juvenile spermatogonial depletion (jsd) phenotype. It is a retrotransposed copy of the Utp14a gene, which is located on the X chromosome, and is inserted within an intron of the autosomal acyl-CoA synthetase long-chain family member 3 (Acsl3) gene. To elucidate the roles of the Utp14 genes in normal spermatogenic cell development as a basis for understanding the defects that result in the jsd phenotype, we analyzed the various mRNAs produced from the Utp14b retrogene and their expression in different cell types. Two classes of transcripts were identified: variant 1, a transcript driven by the host gene promoter, that is predominantly found in germ cells but is ubiquitously expressed at low levels; and variants 2–5, a group of alternatively spliced transcripts containing some unique untranslated exons that are transcribed from a novel promoter that is germ-cell specific. Utp14b (predominantly variant 1) is expressed at moderately high levels in pachytene spermatocytes, the developmental stage at which the expression of the X-linked Utp14a is suppressed. The levels of both classes of Utp14b transcripts were highest in round spermatids despite the transcription of Utp14a in these cells. We propose that when Utp14b initially inserted into Acsl3, it utilized the Acsl3 promoter to drive expression in pachytene spermatocytes to compensate for inactivation of Utp14a expression. The novel cell-type specific promoter for Utp14b likely evolved later, as the protein may have acquired a germ cell-specific function in spermatid development.
Keywords: Spermatogenesis, retroposons, retrotransposition, Utp14, juvenile spermatogonial depletion, X-chromosome, spermatocytes, spermatogonia, male germ cells
Introduction
Spermatogenesis is a precisely regulated sequence of events during which daughter cells of the stem spermatogonia undergo a series of mitotic divisions to eventually form spermatocytes. In the spermatocytes chromosome pairing and genetic recombination occur, during which the sex chromosomes form the transcriptionally inactive sex body (Ayoub et al., 1997), followed by chromosome desynapsis and the meiotic divisions resulting in haploid spermatids. The spermatids express numerous germ-cell specific genes and, despite the cessation of transcription midway through their development, many messages are stored for later translation, which contribute to the unique morphological and functional characteristics of spermatozoa.
Although retrotransposed copies of genes are widely distributed throughout mammalian genomes, these gene copies, which often have arisen via an mRNA intermediate, generally do not possess promoters, are intronless, carry remnants of 3′ polyadenylation sequences, and therefore are generally not active (Boer et al., 1987). However, a few retrotransposition events have resulted in new functional genes, designated retrogenes. Among the functional retrogenes expressed during spermatogenesis, there is a disproportionately high frequency of ones originating from X-linked progenitors (Emerson et al., 2004; Wang, 2004). It has been proposed that developmental processes during mammalian spermatogenesis are dependent on such autosomal retrogenes to compensate for X-chromosome silencing during meiosis (McCarrey and Thomas, 1987). Alternative hypotheses for the prevalence of testis-biased expression of such retrogenes (Wang, 2004) are that syncytial connections between X- and Y-bearing spermatids do not allow the latter to get sufficient levels of an X-chromosomal gene product, or that these genes have evolved additional functions to meet the special needs of germ cells. The latter alternative, however, does not explain the selectivity for X-chromosome progenitors.
Spontaneous and induced mutations in many genes in mice and humans are known to specifically disrupt spermatogenesis (Matzuk and Lamb, 2002). Mutations in several genes, including Kit, Kitl, Eif2s3y, Sohlh1, and Utp14b (Ballow et al., 2006; Bedell and Mahakali Zama, 2004; Mazeyrat et al., 2001; Rohozinski and Bishop, 2004), cause blocks in spermatogonial differentiation. A frameshift mutation in coding sequence of Utp14b, which introduces a stop codon that truncates the predicted UTP14B protein (Bradley et al., 2004; Rohozinski and Bishop, 2004), results in the juvenile spermatogonial depletion (jsd) phenotype. The only defect in jsd mutant mice is male sterility, characterized by several waves of spermatogenesis in young animals, followed by the progressive failure of type A spermatogonia to differentiate. As a consequence, differentiated germ cells are absent and only type A spermatogonia and Sertoli cells remain in the seminiferous tubules. Reciprocal stem spermatogonial transplantation experiments have shown that the defect is confined to the germ cells themselves rather than the supporting cell lineages (Boettger-Tong et al., 2000; Ohta et al., 2001).
Utp14b is a retrotransposed copy of the X-linked Utp14a gene; whereas the coding region of mouse Utp14a consists of 15 exons, Utp14b is encoded within a single exon. Utp14a and Utp14b are mouse homologs of the yeast UTP14 gene (Bradley et al., 2004; Rohozinski and Bishop, 2004). In yeast, UTP14 is part of the pre-18S-rRNA-processing complex and is required for viability (Dragon et al., 2002). TheUtp14b gene is the first protein-coding retrogene in mammals, to which a phenotype has been ascribed.
Although X-derived retrogenes are important in male reproductive physiology and biology, so far there is only one example, that of Pgk2 in the mouse, of a pathway by which testis-specific expression of these novel genes is acquired. Here the original mRNA transcript of the X-linked Pgk1 is believed to have been retrotransposed onto an autosome, carrying the proximal portion of the progenitor gene’s promoter sequence with it (McCarrey, 1990; McCarrey and Thomas, 1987). Transcription of the retrogene was likely initially regulated like Pgk1, with ubiquitous expression. Later the promoter region of Pgk2 appears to have diverged, losing the CpG-island and subsequently acquiring testis-specific expression (McCarrey, 1990; McCarrey et al., 1992; McCarrey et al., 2005).
A different mechanism for acquisition of testis-specific expression must be involved in the case of Utp14b. There is no evidence that the Utp14b retrogene contains the progenitor gene’s 5′UTR and promoter. Unlike Pgk2, which is located within a gene-less expanse of chromosomal DNA, Utp14b is located within an intron of a host gene, acyl-CoA synthetase long-chain family member 3 (Acsl3) on mouse chromosome 1. It inserted 3′ of the existing promoter element, which drives ubiquitous expression of Acsl3 (Bradley et al., 2004; Rohozinski and Bishop, 2004)
To elucidate the roles of the Utp14 genes in spermatogenic cell development, the selection pressures for high levels of germ cell expression, and to understand their relationship with the jsd phenotype, we analyzed the sequences of the various mRNAs produced from this retrogene and its progenitor and their expression in specific germ and somatic cells of the testis and in other tissues. We report here that the Utp14b gene has multiple transcripts, with the production of one transcript being controlled by the host gene’s (Acsl3) promoter and that of another set of transcripts being regulated by a unique germ cell-specific promoter. Furthermore, we found that Utp14a expression is greatly reduced in pachytene spermatocytes; thus we postulate that Utp14b expression was selected to compensate for sex chromosome inactivation during meiosis. We have also shown that expression of the testis-specific Utp14b transcripts is highest in round spermatids, indicating that UTP14B may have developed a novel germ cell-specific function, and during evolution the second promoter may have been selected for spermatid expression.
Materials and Methods
5′-rapid amplification of cDNA ends (RACE) PCR
RACE-Ready mouse testis cDNA was obtained from Ambion (Austin, TX) and amplified with gene-specific and 5′ RACE-specific primer pairs using a FirstChoice RLM-RACE Kit (Ambion). First-round touchdown PCR was performed with 400 nM 5′ RACE outer primer (supplied in the kit), 400 nM Utp14b- or Acsl3-specific reverse outer primers, 200 mM dNTPs and 0.025 unit/μl of Super Taq™ DNA polymerase (Ambion). The reverse outer primers were Utp14bO, specific for Utp14b exon 3, or Acsl3O for Acsl3 exon 4 (Table 1, Fig. 1). The samples were denatured for 3 min at 94°C, followed by 35 cycles of amplification (94°C 30 sec, 60°C 45 sec, 72°C 1 min) and a final elongation for 7 min at 72°C. From the first-round PCR, 2 μl of product was amplified with the 5′ RACE inner primer (supplied in kit), and one of the following specific reverse inner primers: Utp14bI3 for Utp14b exon 3, Utp14bI2 for Utp14b exon 2, Utp14bI15 for Utp14b exon 1.5, or Acsl3I for Acsl3 exon 3 (Table 1, Fig. 1), using the same PCR conditions as in the first round. Fresh PCR products were run on 3% agarose gels, recovered, cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA) and subsequently sequenced with ABI Big Dye kit (Applied Biosystems, Foster City, CA). The nucleotide sequence data were analyzed by DNAStar software (DNASTAR, Madison, WI).
Table 1.
Primer sequences
| Primer | Sequence | Location and reference sequence | Exon location (see Fig. 1) |
|---|---|---|---|
| Utp14bO | 5′-cagtggatagttttctggcaaatc-3′ | Acsl3 genomic sequence 6200–6223 | Utp14b exon 3 |
| Acsl3O | 5′-atgttggcctttggtttctg-3′ | Acsl3 genomic sequence 30005–30024 | Acsl3 exon 4 |
| Utp14bI3 | 5′-gtagcagcctccttcttctcaca-3′ | Acsl3 genomic sequence 6116–6138 | Utp14b exon 3 |
| Utp14bI2 | 5′-agttggtgatatgccactcag-3′ | Acsl3 genomic sequence 4899–4919 | Utp14b/Acsl3 exon 2 |
| Utp14bI15 | 5′-attcatgcatggttttcttgc-3′ | Acsl3 genomic sequence 2643–2663 | Utp14b/Acsl3 exon 1.5 |
| Acsl3I | 5′-gccatccacactgttgatagatc-3′ | Acsl3 genomic sequence 23519–23541 | Acsl3 exon 3 |
| Acsl3EX1 | 5′-gtcagggtcctgaggaggtggcgct-3′ | Acsl3 genomic sequence 145–169 | Acsl3 exon 1, Utp14b exon 1a |
| Utp14bEX1midF | 5′-tgagatgcatacataggcagc-3′ | Acsl3 genomic sequence 677–697 | Utp14b exon 1b |
| Utp14bEX15F | 5′-catgaattgggatatagatatc-3′ | Acsl3 genomic sequence 2657–2678 | Utp14b exon 1.5 |
| Utp14bEX2F | 5′-ctccttgaacttgctgtgagg-3′ | Acsl3 genomic sequence 4836–4856 | Utp14b/Acsl3 exon 2 |
|
| |||
| Utp14aF | 5′-caagcaactaccccttgagtgcc-3′ | Utp14a genomic sequence 1472–1494 | Utp14a exon 2 |
| Utp14aR | 5′-gtgaagaggtaattccagagtc-3′ | Utp14a genomic sequence 6089–6110 | Utp14a exon 5 |
Note: The numbering system is based on the genomic sequences of Acsl3 (ENSMUSG00000032883) and Utp14a (ENSMUSG00000063785) genomic sequences from Ensembl (v31 May 2005) without any 5′ flanking sequence
Fig. 1.
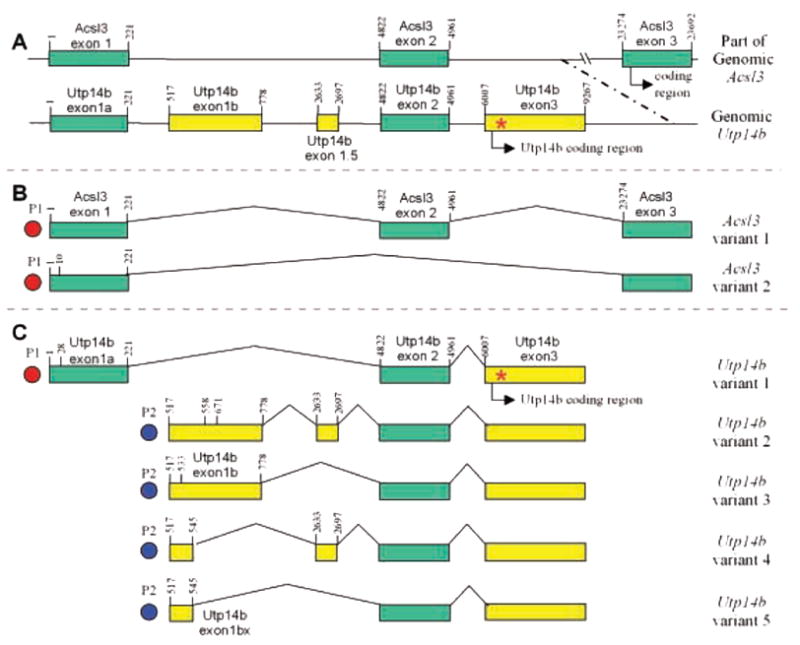
(A) Genomic maps of Acsl3 and Utp14b (not drawn to scale) showing the exons that can be included in transcripts. (B) Transcripts of Acsl3 observed by 5′-RACE. (C) Transcripts of Utp14b observed by 5′-RACE. The nucleotide numbers of the exon boundaries, relative to the major transcriptional start site of Acsl3 and Utp14b variant 1, are indicated vertically above the exons. Downstream alternative transcription start sites, differing by more than a few base pairs, are indicated as vertical lines with nucleotide number within the first exon of the transcript. P1, P2: putative promoters. *Utp14b mutation site for jsd phenotype.
Preparation of spermatogenic cells
Male C57BL/Law mice, bred-in house, and W/Wv mice, whose testes only contain somatic cells and a very few undifferentiated spermatogonia (Ohta et al., 2003), purchased from the Jackson Laboratory (Bar Harbor, ME), were maintained and the M. D. Anderson Cancer Center. Oct4-EGFP transgenic mice (Szabo et al., 2002) were bred and maintained at the University of Washington. All mice were housed in animal facilities approved by the American Association for Accreditation of Laboratory Animal Care and all procedures were approved by the respective Institutional Animal Care and Use Committees.
For each experiment, cell suspensions were prepared from 25 adult C57BL6 mice by a modification of previously published methods (Romrell et al., 1976; Zhang et al., 2003). Seminiferous tubules were isolated by incubating the decapsulated testes with collagenase (0.5 mg/ml) and DNase I (200 μg/ml) in enriched DMEM/F12 (GIBCO, Carlsbad, CA), to which 0.1 mM non-essential amino acids (GIBCO), 1 mM L-glutamine (GIBCO), 1 mM sodium pyruvate (GIBCO), and 5 mM sodium lactate (Sigma-Aldrich, St. Louis, MO) were added. This decapsulated testis tissue was shaken for 15 min at 35°C in a water bath, until it was mostly dispersed into tubules. The dispersed tubules were allowed to settle and, after removal of the supernatant, were resuspended in 32 ml of DMEM/F12 solution. In each of four 50-ml tubes, 8 ml of this suspension was layered onto 40 ml of 5% Percoll solution, and the tubules were allowed to settle until most of the larger tubules and clumps were at the bottom. The supernatants were removed and the settled tubules were washed with DMEM/F12 solution and then further digested with trypsin (1 mg/ml) and DNase I (200 μg/ml) in enriched DMEM/F12 for 20 min at 35°C with shaking. Fetal bovine serum was added to 10%, and the cells were dispersed by pipetting. Total cell suspensions were separated by centrifugal elutriation (JE-6B rotor, Beckman Instruments, Fullerton, CA) to obtain fractions enriched in elongating spermatids (flow rate interval: 12.6 to 18 ml/min, rotor speed: 3000 rpm), round spermatids (9.5 to 14 ml/min, 2000 rpm) and pachytene primary spermatocytes (25 to 37 ml/min, 2250 rpm) (Meistrich et al., 1977). After elution of the pachytene spermatocytes from the chamber, the remaining cells, which were highly enriched in Sertoli cells, were collected. The late spermatid fraction obtained by elutriation was directly used for transcript analysis. Round spermatids and pachytene spermatocytes were further purified on linear, 26%–38%, Percoll gradients (Meistrich et al., 1981). The purified pachytene fraction was obtained by plating the Percoll-enriched fraction on DSA (lectin from Datura stramonium, Sigma) coated dishes, to which Sertoli cells bind strongly, and the pachytene spermatocytes were recovered in the unbound cell fraction (Scarpino et al., 1998). The Sertoli cells recovered from the elutriator were purified by plating them on the DSA-coated dishes, removing the unbound and loosely bound cells, and directly extracting the bound Sertoli cells with RLT lysis buffer (QIAGEN). The purity of each fraction was initially determined by cell smears stained with periodic acid Schiff-hematoxylin.
Undifferentiated spermatogonia were isolated from 8-day-old Oct4-EGFP transgenic mice. Briefly, total cell suspensions were prepared following a previously described procedure (Buaas et al., 2004); cells were sorted using a FACStar cell sorter equipped with CELL QUEST software (Becton-Dickinson, Franklin Lakes, NJ).
The purity of each cell fraction was further tested by real-time PCR for cell-specific marker expression. The markers and their stage-specific expression are as follows: Fthl17 (ferritin, heavy polypeptide-like 17) is mainly expressed in spermatogonia but there is weak expression up to the zygotene spermatocyte stage, Sycp3 (synaptonemal complex protein 3) is strongly expressed in spermatocytes but is also expressed at a low level in spermatogonia and round spermatids (Wang et al., 2005), Acrv1 (acrosomal vesicle protein 1, formerly known as SP10) is specific for round spermatids (Reddi et al., 1999), Prm1 (protamine 1) mRNA is present only in late step 7 through step 14 spermatids (Mali et al., 1989), and Wt1 (Wilms tumor homolog) is specific for Sertoli cells in testis (Sharpe et al., 2003).
Real-time reverse transcription PCR
Total RNA from different tissue and cell samples was extracted using RNeasy Mini or Midi kits (QIAGEN, Valencia, CA). Genomic DNA was removed using RNase-free DNase (QIAGEN). Total RNA (3 μg) was reverse transcribed (RT) using a Superscript first-strand synthesis kit (Invitrogen) and oligo-dT priming according to the manufacturer’s instructions. The RT product was diluted 1:20 and amplified with SYBR Green JumpStart Taq Ready Mix (Sigma) using gene specific primers. Amplification was measured on a Rotor-Gene 3000 real-time thermocycler (Corbett Life Science, Sydney, Australia). Cycle conditions were 94°C for 2 min, followed by 40 cycles of amplification (94°C 15 sec, 60°C 60 sec, 72°C 60 sec). Each experiment was replicated 3 times, and the results were normalized to the amount of Rps2 (ribosomal protein S2) mRNA present in the sample. The absolute amounts of Rps2 mRNA, for the same amounts of RNA used in the RT reaction, in the spermatogonia, pachytene, round spermatid, and Sertoli cell fractions were similar, 79%, 108%, 103%, and 123%, respectively, of the levels in total testis homogenates. Only the late spermatid fraction showed a reduced level of 42% of that in the total testis. RNA samples without reverse transcription and the products from the reverse transcriptions without RNA were used as negative controls. The amplification products of the controls were analyzed by gel electrophoresis, and no transcript-specific PCR products were observed.
For analysis of Utp14b transcripts, a common downstream primer (Utp14bO) that is within the Utp14b coding region (exon 3) was used in combination with one of the following upstream primers: Acsl3EXI (exon 1a), Utp14bEX1midF (exon 1b), Utp14bEX15F (exon 1.5) and Utp14bEX2F (exon 2) (Table 1), which specifically amplify Utp14b mRNA transcript variants 1, 2+3, 2+4, and total Utp14b, respectively (Fig. 1). For Acsl3, a common downstream primer Acsl3I (exon3) was used in combination with upstream primers Acsl3EX1 or Utp14bEX2F to detect Acsl3 transcript variants 1+2 and transcript variant 1, respectively. For Utp14a, the forward, Utp14aF, and reverse, Utp14aR, primers were used.
Results
5′-RACE identification of 5′-UTR and start sites of Utp14b transcripts
Examination of the genomic sequence and predicted exons from Ensembl (ENSMUSG00000032883, version 31, May 2005), showed that the single coding exon of Utp14b is located within an intron of the autosomal gene Acsl3 and is preceded by four possible 5′-non-coding exons (Fig. 1A). Acsl3 itself forms two transcripts with different 5′-UTRs: variant 1 (GenBank Accession NM_028817), the minor transcript, contains exons 1 and 2, whereas variant 2 (NM_001033606), the major transcript, contains only exon 1 and skips exon 2 (Fig. 1A). The coding region of Utp14b is within the second intron of Acsl3 between exons 2 and 3. Exon 1b and exon 1.5 of Utp14b are located in the first intron of Acsl3. Both of them were not found in Acsl3 transcripts as RT-PCR reactions using primer pairs that spanned exons 1 to 3, and exon1b (or exon 1.5) to exon 3 of Acsl3 yielded two bands and no bands, respectively (data not shown).
To identify and compare the transcriptional start sites and 5′-UTR sequences of Utp14b and Acsl3, 5′ RACE was performed with downstream primers in the unique coding regions of Utp14b exon 3, and Acsl3 exon 3. Utp14b produced at least 5 transcripts with different 5′-UTRs, which were formed by initiation at two transcriptional start sites and by alternative splicing (Fig. 1C). Utp14b transcript variant 1 had a 5′-UTR similar to that of Acsl3 variant 1 in that these transcripts shared the first two exons. Furthermore, it had the same transcription start site as variants 1 and 2 of Acsl3 (Fig. 1B, C), indicating that Utp14b variant 1 and Acsl3 share a common promoter. RT-PCR reactions using primer pairs that spanned exons 1a to 2 of Utp14b yielded a single product of the predicted size (data not shown), thus excluding the possibilities of any other Utp14b splice variants that contain Utp14b exons 1b or 1.5 between the exons indicated in variant 1 of Utp14b. Utp14b variant 1 represents a 5′ extension of the sequence reported by Bradley et al (GenBank AY316161) (Bradley et al., 2004), which started at nt 156.
Transcription of Utp14b variants 2, 3, 4, and 5 started at exon 1b, located within the first intron of Acsl3. All variants were observed with the Utp14b exon-3 specific reverse inner primer (Utp14bI3) (Table 1), but additional data regarding start sites for exon 1b were also obtained with the Utp14b exon-2 specific (Utp14bI2) and the Utp14b exon-1.5 specific (Utp14bI15) reverse inner primers. Although Utp14b and Acsl3 variant 1 share exon 2, the 5′ RACE data on the exon 1b start sites from the primer Utp14bI2 within exon 2, must only represent Utp14b and not Acsl3 since the possibility that an Acsl3 splice variant contains Utp14b exon 1b was excluded. Most transcripts started at or within a few bases of nt 517, although there were also transcripts that started at nt 496, 507, 533, 558, and 671. The variant 2 transcript is similar to the sequence of that reported for GenBank accession number AK029972 (Rohozinski and Bishop, 2004), except that the most frequent start site for variant 2 occurred near nt 517. Variant 2 clones with the same start site as AK029972, at nt 558, were also found by 5′-RACE. Both variants 4 and 5 contained only the initial GC-rich 23–29 bp of exon 1b; these variants used an alternative splice donor site at nt 545 to connect to exons 1.5 or 2, respectively. Similarly variants 2 and 3 involved splicing of the full exon 1b, ending at nt 778, to exons 1.5 or 2, respectively. The different transcription start sites of Utp14b variant 1 and Utp14b variants 2, 3, 4, and 5 indicated that they were driven by at least 2 putative promoters: P1 and P2 (Fig. 1C). P1 is either completely or partially shared with the Acsl3 promoter, whereas P2 is a unique promoter within the first intron of Acsl3.
Tissue and cellular distribution of Utp14b variants and Acsl3
The tissue distributions of Utp14b variants and Acsl3 were determined by real-time RT-PCR (Fig. 2). Acsl3 was transcribed ubiquitously, the highest level occurring in the brain, the next highest in testis, and the lowest in lung, kidney, liver, heart, and spleen (Fig. 2A, B). Variant 2, lacking exon 2, was the predominant form. However both variants had nearly identical tissue distributions.
Fig. 2.
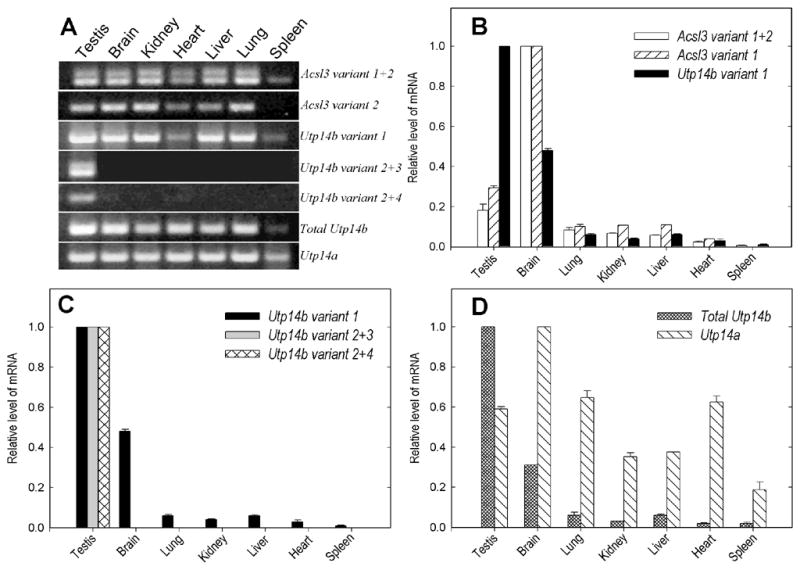
Tissue-specific expression of Acsl3, Utp14b, and Utp14a. (A) Agarose gel electrophoresis analysis of transcript-specific RT-PCR products taken from products of real-time PCR (40 cycles). (B–D) Tissue-specific transcript levels determined by real time-PCR. Expression of (B) Acsl3 (variants 1 and 2) and Utp14b variant 1, (C) Utp14b variant 1, variants 2+3 and variants 2+4, and (D) total Utp14b and Utp14a. Expression levels were first normalized to ribosomal protein Rps2 mRNA levels, and then the values for the tissues with the most abundant expression were set at 1.0.
Utp14b variant 1, which appears to share the promoter P1 with Acsl3, was also transcribed ubiquitously (Fig. 2A, B). Comparison of the tissue distributions showed that the ratios of Acsl3 to Utp14b variant 1 were essentially identical in all somatic tissues tested, supporting the idea that they use a common promoter and demonstrating that the mechanisms for preferentially selecting alternative splice acceptors in these somatic tissues were similar. However, Utp14b expression was higher in the testis than in brain, whereas the reverse was true for Acsl3, suggesting a testis-specific difference in the selection of splice sites or mRNA stability.
In contrast to the ubiquitous expression of Utp14b variant 1, variants 2, 3, and 4 were only detected in testis (Fig. 2A, C) and their transcription was driven by a different shared putative promoter P2. Because we were not able to design forward primers within the truncated 29-bp GC-rich region of exon 1b of variant 5, we were unable to determine its contribution to the total Utp14b transcript population. We estimated the relative contributions of the other transcripts to the total Utp14b transcript present in the testis, using forward primers Acsl3I, Utp14bEX1midF, Utp14bEX15F, and EX2F, which are located in exons 1a, 1b, 1.5, and 2, respectively, and a reverse primer Utp14bO in exon 3. Real-time RT-PCR indicated that there were roughly equal amounts of mRNA of variants 1, 2+3 and 2+4 in total testis (not shown). However, examination of the bands on the gel after real-time PCR indicated that variant 3 was more strongly expressed than variant 2 (Fig. 2A). Conclusions about relative amounts must be made cautiously, however, because quantification may be affected by different upstream primers used and variations in transcript length,
To determine how Utp14b expression is related to the jsd phenotype, we compared the distributions of Utp14b variants in testes with different testicular cell composition using mice of different ages and W/Wv mutants, and in highly enriched populations of Sertoli cells and germ cells at different stages of spermatogenesis. In mice under 10 days of age and W/Wv mutants, low levels of Utp14b variant 1 were expressed, indicating that this transcript is expressed in either gonocytes/primitive spermatogonia or in the somatic cells, or perhaps both (Fig. 3A). There was a significant increase in variant 1 at 10 to 20 days of age, which is when spermatocytes first appear and increase in numbers. Although there was no expression of variants 2+3 in testes of mice under 10 days of age and W/Wv mutants, there were very low but detectible levels of variant 2+4 in these tissues. At 10 and 15 days, there were still extremely low levels of variants 2+3 and continued low levels of variants 2+4. The levels of variant 1 (promoter P1) and variants 2+3 and 2+4 (promoter P2) increased markedly at 20 and 25 days of age, reaching essentially adult levels by day 25. This increase in expression coincides with the increase in the round spermatid population.
Fig. 3.
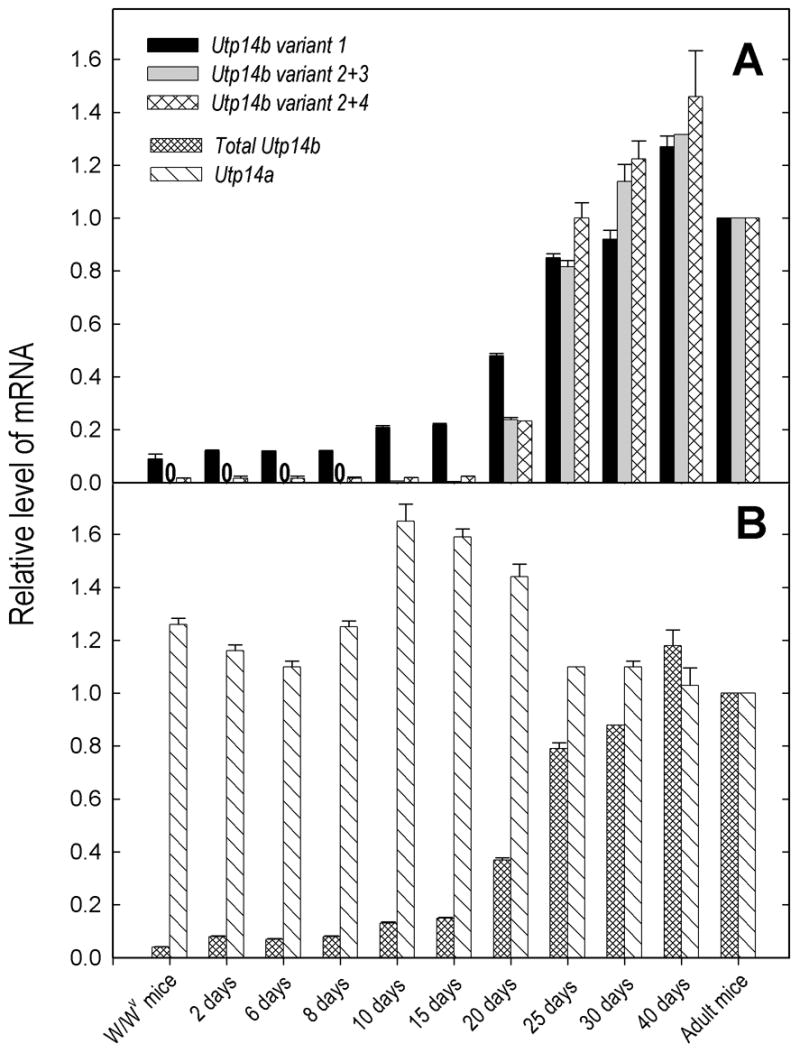
Age-dependent expression of Utp14a and of different Utp14b transcript variants in wild-type mouse testes and expression in adult, germ-cell deficient W/Wv mutants. (A) Comparison of Utp14b variant 1 expression with that of variants 2+3 and 2+4. (B) Comparison of total Utp14b with Utp14a expression. Expression levels were first normalized to ribosomal protein Rps2 mRNA levels, and then the values for the adult wild-type testes were set at 1.0. Zero’s in place of bars, indicate that no transcript-specific RT-PCR product was produced.
Next we analyzed the levels of different transcripts in purified cells from adult or, in the case of the spermatogonia, from 8-day-old mice. The purities of the fractions of pachytene spermatocytes and round spermatids, assessed on smears, were 97% and 93%, respectively. Of the cells or cell fragments in the late spermatid fraction, 33% were spermatids in steps 9–16 with some residual cytoplasm and 60% were cytoplasmic droplets, of which over 90% originate from elongating and elongated spermatids (Meistrich et al., 1981). Before plating, the Sertoli cell population consisted of 36% Sertoli cells, 56% spermatocytes or spermatids, and 1.6% spermatogonia and early spermatocytes, some of which were still attached to the Sertoli cells; differential cell counts could not be performed after plating since the adherent cells were directly lysed on the dish. The expression of cell-type specific markers (Fig. 4) confirms the cytological assessment of purities of the fractions.
Fig. 4.
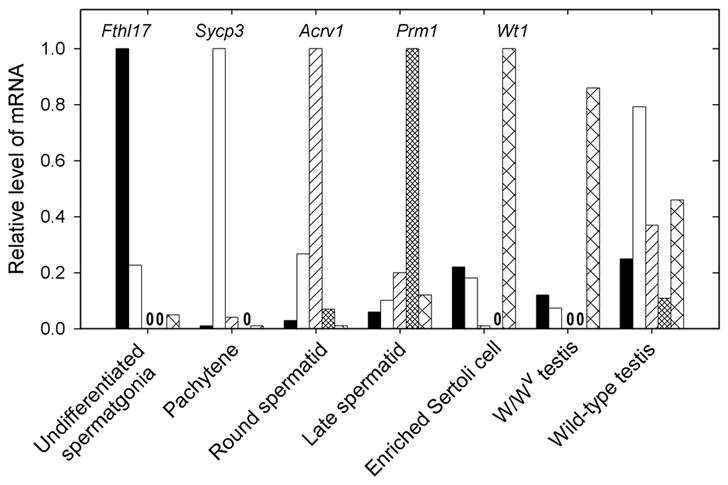
Analysis of purities of cellular fractions with real-time PCR for markers specific for particular cell types. Fthl17, Sycp3, Acrv1, Prm1 and Wt1 were used as markers of spermatogonia, pachytene spermatocytes, round spermatids, late spermatids, and Sertoli cells, respectively. Expression levels were first normalized to ribosomal protein Rps2 mRNA levels and then the values for each gene in the cell type most enriched in that gene were set at 1.0. Zero’s in place of bars, indicate that no transcript-specific RT-PCR product was produced.
In the enriched Sertoli cell population, the levels of Utp14b variant 1 were low, but variants 2, 3, and 4 were even lower, (Fig. 5A, B). These results are consistent with those from the testes of 2- to 10-day old wild-type mice and adult W/Wv mice, in which the homogenates contain primarily somatic cells. The higher levels of these Utp14b variants in the enriched Sertoli cells than in the W/Wv testes may have been caused by contamination of the preparations by some spermatocytes, as indicated by the presence of Sycp3 mRNA in this fraction; the testes of W/Wv mice lack spermatocytes. The absence of Utp14b variants 2+3 in the W/Wv testes demonstrates that these variants are not present in the somatic cells. The low levels of variants 2+4 in W/Wv mice or enriched Sertoli cells could have been contributed by a few spermatogonia or spermatocytes in the preparations. Thus Utp14b variants 2, 3, and 4 appear to be not only testis specific, but also germ-cell specific.
Fig. 5.
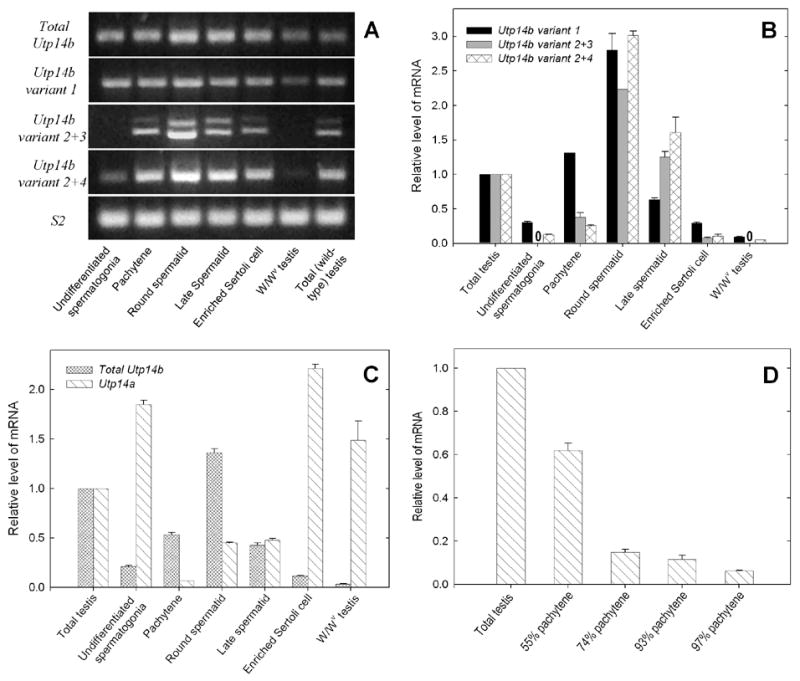
Expression of Utp14a and of different Utp14b variants in testes of wild-type and germ-cell deficient W/Wv mice and in purified cell fractions from wild-type mice testis. (A) Agarose gel analysis of real-time PCR products. (B–D) Cell-type specific transcript levels determined by real time-PCR. Comparison of expression of (B) Utp14b variant 1 with variant 2+3 and variant 2+4, and (C) total Utp14b with Utp14a. (D) Effect of further purification of pachytene spermatocytes on levels of Utp14a expression in fractions. Purities of 55%, 74%, and 93% were obtained with cell suspensions prepared by trypsinization (Meistrich, 1977) and elutriation alone; elutriation plus Percoll gradients; and a combination of elutriation, Percoll gradients, and plating on DSA, respectively. The procedure for obtaining 97% purity is described in the Materials and Methods. Normalization was done as in Fig. 3. Zero’s in place of bars, indicate that no transcript-specific RT-PCR product was produced.
Low levels of Utp14b variant 1 and 2+4 were detected in undifferentiated spermatogonia from 8-day-old mice (Fig. 5A, B). The limited Sertoli cell contamination of the undifferentiated spermatogonia was not enough to account for all of the Utp14b observed. In the undifferentiated spermatogonia fraction, the level of the Sertoli cell-specific marker Wt1 was 6% of that in W/Wv mice, but the levels of Utp14b variants 1 and 2+4 were, respectively, 3.3 and 2.4 times those in W/Wv mice (Fig. 4, 5B). Since we did not detect Utp14b variant 2+3 in the undifferentiated spermatogonia fraction, Utp14b variants 1 and 4 must be the predominant transcripts of Utp14b that are present in undifferentiated spermatogonia.
In the highly purified pachytene spermatocytes, Utp14b variant 1 was expressed at high levels; variants 2, 3 and/or 4 were also expressed but at much lower levels (Fig. 5B). The levels of variant 1 were consistent with increases at 10 and 15 days of age in the developmental study (Fig. 3). The levels of variant 2+3 and 2+4 transcripts in pachytene spermatocytes from mature mice were somewhat higher than expected from the extremely low levels at days 10 and 15 in testes of juvenile mice. Nevertheless, we conclude that there was expression of Utp14b from both promoters in pachytene spermatocytes, but the expression driven by promoter P1 was predominant.
The levels of all of the Utp14b transcript variants were highest in the round spermatids (Fig. 5B). The 6-fold increase in levels of the testis-specific Utp14b variants 2, 3 and/or 4, compared to those in the pachytene stage, was particularly striking. This increase is consistent with their dramatic increases at days 20 and 25 in the developmental study (Fig. 3A). The retention of mRNA for variants 2, 3 and/or 4 in late spermatids (Fig. 5B), in which transcription declines to zero (Kierszenbaum and Tres, 1975), suggests that the mRNA is stable and that there might be some function for UTP14B in these cells.
Utp14a is inactivated in pachytene spermatocytes
To test the hypothesis that compensation for X-chromosome silencing during male meiosis was a major factor in the positive selection for the functional Utp14b retrogene that evolved from the X-linked Utp14a progenitor, we determined the cell- and tissue-specific distribution of Utp14a transcripts with particular emphasis on the levels in pachytene spermatocytes. Utp14a was expressed in all tissues, the highest levels occurring in brain, testis, lung, and heart (Fig. 2D). In contrast to the Utp14b transcripts, whose expression levels varied 50-fold between tissues, there was at most a 5-fold variation in the expression of Utp14a. Furthermore, there was only a 1.6-fold variation in Utp14a mRNA levels during testicular development or between wild-type and W/Wv mice (Fig. 3B).
Among the different testicular cell fractions, the highest levels of Utp14a expression were detected in Sertoli cells (Fig. 5C). Enriched undifferentiated spermatogonia from 8-day-old mice also contained high levels of Utp14a. Since the Utp14a level in these spermatogonia was about 84% of the levels found in Sertoli cells, it could not have been accounted for by the small amount of Sertoli cell contamination of this fraction (Fig. 4). Among the differentiated germ cells, moderate levels of Utp14a were detected in both round and late spermatids (Fig. 5C). Since the round spermatid fraction showed 20% of the level of Utp14a that was in Sertoli cell preparations but only 1% of the Wt1 level that was in the Sertoli cells and also showed low levels of spermatogonia contamination (Fig. 4), the Utp14a mRNA must indeed be present in the round spermatids. Although the levels of Utp14a, normalized to Rps2, appear to be similar in round and late spermatids, the actual levels of Utp14a in late spermatids may be lower, because the Rps2 mRNA was also reduced in the late spermatid fraction. However, the maintenance of Utp14a mRNA levels in testes of mice from 20 days of age to adulthood (Fig. 3) further supports the idea that Utp14a mRNA is present in both round and late spermatids, as these constitute the majority of cells in the testis of mature mice.
In pachytene spermatocytes, very low levels of Utp14a transcript (about 7% of that observed in total testis) were detected in the most highly enriched (97% by differential counts in smears) pachytene spermatocyte fraction (Fig. 5C). Calculations based on mRNA analysis of this fraction, which showed 1% of the Wt1 transcript level observed in the Sertoli cell preparations and 4% of the level of Acrv1 detected in round spermatid preparations (Fig. 4), indicated that most of the Utp14a mRNA was due to contamination by these cells. Further support for the notion that the residual level of Utp14a in the pachytene fraction originated from contamination is provided by the demonstration that the levels of Utp14a transcript steadily decreased with the increase of pachytene purity (Fig. 5D). Thus we conclude that Utp14a is either absent in pachytene spermatocytes or present at extremely low levels. These data support the proposal that there is transcriptional inactivation of the X-linked Utp14a in pachytene cells and that the expression of the autosomal retroposon, Utp14b (Fig. 5C), is under selective pressure to compensate for inactivation of the X-linked homolog.
Discussion
In this paper we describe the developmental consequences of an unusual retrotransposition event wherein Utp14a, encoded on the X chromosome, duplicated via an mRNA intermediate and inserted within an intron of an autosomal gene to form Utp14b, and of a mutation in the coding sequence of this retrogene. This duplicate copy survived selection and has acquired tissue-specific expression. Comparison of published sequences indicated that Utp14b has two distinct transcript variants with different first exons and hence has acquired different promoters (Bradley et al., 2004; Rohozinski and Bishop, 2004).
Our results indicate that, after retrotransposition into an intron of Acsl3, expression of Utp14b was most likely driven by the host gene’s promoter. Evidence for this comes from the fact that transcript variant 1 of Utp14b shares the first two exons or just the first exon with variants 1 and 2 of Acsl3, respectively (Fig. 1), and has an expression pattern very similar to that of Acsl3. Thus the transcripts of Acsl3 and Utp14b variant 1 share a promoter and some other transcriptional regulatory elements. Analysis of the sequence upstream from the transcriptional start site of Acsl3 using promoter scan software (http://bimas.dcrt.nih.gov/molbio/proscan/) identified a typical transcription regulatory unit (Fig. 6A). The region of the core promoter does not contain a TATA box (Smale, 1997); however, it does contain a consensus CCAAT (cat) box (Mantovani, 1999) and an oct-B1A binding site (Rosales et al., 1987). Several upstream enhancer elements were also identified within the distal enhancer region, including a c-fos.5 site (Fisch et al., 1987), and multiple Sp1 (Jones and Tjian, 1985) and AP-2 binding sites (Imagawa et al., 1987), which are characteristic of many mammalian promoters.
Fig. 6.
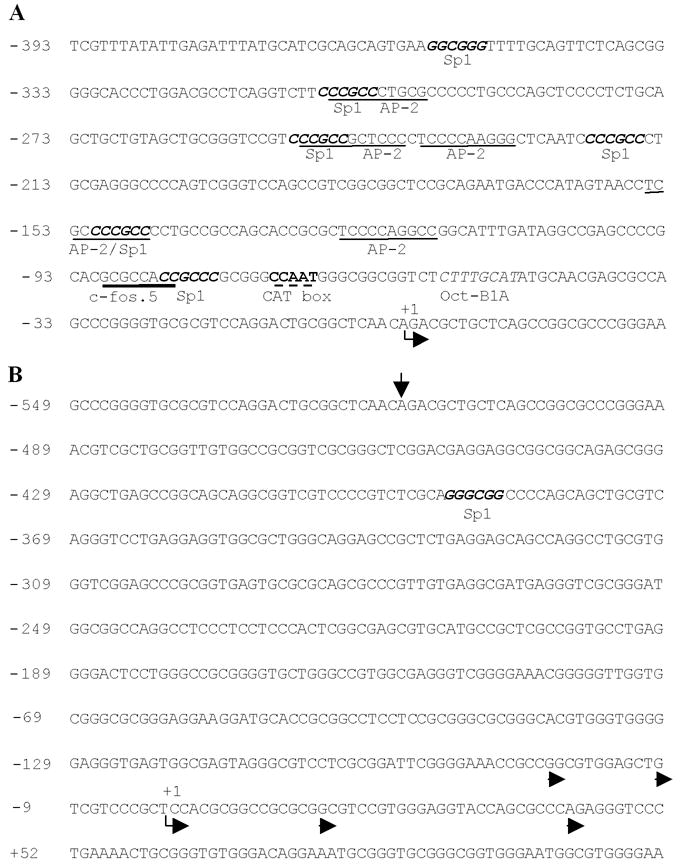
Identification of potential promoter sequences and the transcriptional start sites of Acsl3 and Utp14b. (A) The core promoter region upstream of the Acsl3/Utp14b variant 1 start site contained a single CCAAT (cat) box (bold letters and dashed underline) and an oct-B1A binding site (italics). Upstream there were six Sp1 binding sites (bold and italics), five AP-2 sites (underline), and a single c-fos.5 site (bold underline), some of which were overlapping. An arrow below the text indicates the transcriptional start site. (B) The region upstream of the Utp14b testis-specific transcript start site does not contain any known basal promoter elements or enhancers. There is a single Sp1 site (bold and italics). An arrow above the text indicts the transcriptional start site of Acsl3/Utp14b variant 1. The major consensus start site (start site at that nucleotide or with 3 bp) is indicated by an arrow below the text and other observed start sites are indicated by arrowheads.
The spermatogenesis-specific transcripts, Utp14b variants 2–5, share a tissue-specific promoter presumably located between the transcriptional start site of exon 1 of Acsl3 and exon 1b of Utp14b, a distance of 517 nucleotides (Fig. 1). Based on currently accepted models of eukaryotic promoter elements, it is presumed that the core promoter, which contains the RNA polymerase binding site, is within -35 nucleotides of the transcriptional start site and that the proximal promoter, containing specific transcriptional factor binding sites, is within -250 nucleotides. Analysis of the region 5′ to the transcriptional start site using publicly available promoter-searching software failed to identify any motifs currently recognized to be associated with promoter activity in vivo. There was a single Sp1 binding site about 396 bases upstream of the transcriptional initiation site (Fig. 6B), which alone is insufficient for promoter activity (Jones and Tjian, 1985). Because this set of transcripts is male germ-cell specific, standard strategies using cultured somatic cell lines to further identify the promoters could not be used.
In addition, the spermatogenesis-specific transcripts, Utp14b variants 2–5, show an array of splice variants. Exon 1b contains an internal splice junction that is sometimes used and generates splice variants 4 and 5 (Fig. 1). The small 66-bp exon 1.5 is sometimes included and generates variants 2 and 4. This complexity makes identification of the individual transcripts by RT-PCR difficult due to an inability to design transcript-unique primer pairs. We therefore used primer pairs that amplified variants 2+3 or 2+4. In some cases, such as the analysis of the undifferentiated spermatogonia from 8-day-old mice, we were able to deduce the levels of specific transcripts from the differences in the levels with the different primer pairs. Nevertheless, the absence of transcripts with either of these primer pairs in somatic cells showed that transcript variants 2, 3, and 4 were only expressed in the spermatogenic cells; we assume the same to be true for variant 5 since it shares the same promoter with these variants.
To determine the developmental stages that would be affected by the jsd mutation in Utp14b, we also analyzed the expression pattern of Utp14a, since Utp14a and Utp14b initially had functional overlap and likely still do so based on sequence comparisons and studies of jsd mutants. Alignment of the mouse Utp14 gene products revealed 66% amino acid identity and 76% homology (Fig. 7). The UTP14 proteins lack functional motifs but alignment of sixty-four UTP14 family members available on the database revealed the presence of two conserved structural domains common to UTP14A and UTP14B that are present in these proteins from all phyla. This indicates that the mouse UTP14 proteins likely have overlapping function. However, extensive amino acid substitution between the mouse UTP14 proteins in other regions indicates that they are in the process of functional divergence. This is further supported by addition of amino acids at the amino terminus and the presence of four deletions and two insertions within core region of the UTP14B peptide. Our observation that jsd mutant mice, which lack UTP14B, show a defect in 18S rRNA processing in spermatocytes (M. Zhao & M. L. Meistrich, unpublished observations) supports the supposition that the pre-18S-rRNA processing function of the yeast homolog has been conserved in Utp14a and Utp14b through mammalian evolution.
Fig. 7.

Alignment of UTP14A and UTP14B proteins showing identities, similarities, and conserved structural motifs. Two regions of containing a large percentage of highly conserved amino acids are shown between the bars; the ones shown in bold italics are conserved (absolutely or as a conserved substitution) across phyla. There is spatial conservation (i.e. the number of amino acids between those marked as conserved) across phyla between the first and last conserved amino acid within these regions. Amino acid deletions and insertions within the core of the UTP14B peptide are indicated by dashes and boxes around the amino acids. At the amino terminus of UTP14B there are an additional 24 amino acids. An asterisk (*) indicates amino acid identity, two dots (:) indicates conserved substitution, one dot (.) indicates similarity, and no symbol indicates other random substitutions. The two peptides share 66% identity and 76% homology.
Utp14a was highly expressed in all somatic cells examined, as would be expected for a gene encoding an essential gene product and consistent with the absence of any somatic abnormalities in jsd mutants. Utp14a was also highly expressed in undifferentiated spermatogonia from 8-day-old mice and at moderate levels in round spermatids, but not in pachytene spermatocytes. The low spermatocyte and high spermatid expression is consistent with in situ hybridization studies (Rohozinski and Bishop, 2004); the failure of in situ hybridization to detect Sertoli cell and spermatogonial expression can be attributed to the close contact of the Sertoli cells with the differentiating germ cells and the low numbers of undifferentiated spermatogonia in the testis, respectively.
The high levels of Utp14a in undifferentiated spermatogonia from 8-day-old mice are consistent with the normal development of spermatogonia during juvenile initiation of spermatogenesis in jsd mice. Utp14b is not necessary in these cells because either Utp14a is present and complements its function or the essential function of the Utp14 genes occur only at stages past this developmental time point. It is not known if Utp14a remains elevated in stem spermatogonia of adult mice. If it does remain elevated, it would explain maintenance of this population of cells in adult jsd mice (Ohta et al., 2001; Shetty and Weng, 2004). The levels of Utp14a or Utp14b in differentiated spermatogonia have not yet been determined because of the difficulties in purifying these cells from adult mice, so at present we cannot test whether the needs for survival and differentiation of these cells contribute to the selective pressure to maintain Utp14b expression and function.
However, although Utp14a is absent or present at extremely low levels in pachytene spermatocytes, Utp14b is expressed in these cells, consistent with previous in situ observations (Rohozinski and Bishop, 2004). Of the Utp14b transcripts, variant 1 was most highly expressed in these cells. This result, along with the failure of spermatocyte development in jsd mutants, strongly suggests that the initial selective advantage of Utp14b was generated by Utp14b variant 1’s transcription from the Acsl3 promoter in the testis to compensate for the absence of Utp14a in spermatocytes. This would have only required the formation of a splice acceptor site near the start of the retrogene. However, the higher levels of Utp14b variant 1 in the testis than of Acsl3 suggests that either the Utp14b message, relative to the Acsl3 message, is more stable in the male germ cells than in other tissues or that the testicular splicing machinery preferentially utilizes the splice acceptor site in exon 3 of Utp14b rather than the one in exon 3 of Acsl3. The increase of Utp14b variant 1 levels in testis, relative to those of Acsl3, may have also been under selective pressure to provide more of its protein during meiosis.
The high levels of expression of Utp14b in spermatids were surprising. Although higher levels of expression in spermatids than in spermatocytes were not observed with in situ hybridization (Rohozinski and Bishop, 2004), we believe our results are valid because real-time PCR is more quantitative than in situ hybridization, the high levels were confirmed with three different primer sets, and the result was not due to changes in levels of the Rps2 mRNA used for normalization. The high level of expression of this retrogene in spermatids suggests that a secondary evolutionary selection process occurred either to compensate for the possibility that some spermatids (Y-bearing) might have low levels of Utp14a if there were unequal sharing across intercellular bridges, or that Utp14b evolved a spermatid-specific function. The latter case may involve mutations in the coding sequence, resulting in a novel or enhanced function of Utp14b in spermatids compared to Utp14a. The increase in Utp14b mRNA levels in spermatids is largely a result of increases in transcript variants 2–5, which are under the control of the germ-cell specific promoter P2 that likely has arisen by the accumulation of multiple mutations due to selective pressure to modify the initial selection for meiotic expression of Utp14b variant 1 from the Acsl3 promoter. However, the role of Utp14b in spermatid development is less clear. In young (3- to 7-week-old) jsd mice, the numbers of round spermatids were reduced from those observed in wild-type (Kojima et al., 1997). The reduction may either be a direct effect of the absence of Utp14b in spermatids or an indirect result of effects on spermatocytes. However, some spermatids did develop to apparently normal sperm even in the absence of Utp14b.
In summary, the absence of Utp14a transcription in pachytene spermatocytes, and the expression of Utp14b from a host genes’ promoter together support the proposal that sex chromosome inactivation is the reason for testis-specific activity of many retrogenes with X-chromosome progenitors. The high levels of Utp14b in spermatids, including the transcript driven by the host genes’ promoter, could also support the model that there was insufficient sharing of X-chromosomal transcripts in spermatids, but it remains to be determined whether that is indeed the case with the Utp14a transcripts. However, it should be noted that unequal distribution across the syncytial connections is the exception rather than the rule (Caldwell and Handel, 1991; Zheng et al., 2001). It appears more likely that under functional selective pressure to increase the efficiency of spermatid development, the Utp14b retrogene acquired additional or enhanced functions and a testis-specific promoter, which drives transcription of Utp14b primarily in spermatids. The possibility of a unique function is suggested by the retention of Utp14b message in late spermatids, after the cessation of transcription, and hence after the need for processing newly synthesized pre-18S rRNA, which is the function of UTP14 in yeast. The exact roles of Utp14b in mouse spermatogenesis still remain to be determined to test the basic parts of these hypotheses.
A significant component of mammalian evolution involves duplication or exchange of genetic material between or within chromosomes by various processes including retrotransposition. Most of the additional copies of genes that arise via retrotransposition are inactive; only a few of them act as functional genes. The present study has further elucidated mechanisms and selection pressures that cause these retrogenes that originate from X-linked progenitors to be preferentially expressed in the testis and to retain function.
Acknowledgments
We thank Kuriakose Abraham for histological preparation, Walter Pagel for editorial advice, and Dr. Ertug Kovanci for critically reading the manuscript. This study is supported by NIH Research Grants HD-40397 (to MLM) and HD-36289 (to CEB) and NIH Cancer Center Support Grant CA-16672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayoub N, Richler C, Wahrman J. Xist RNA is associated with the transcriptionally inactive XY body in mammalian male meiosis. Chromosoma. 1997;106:1–10. doi: 10.1007/s004120050218. [DOI] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Mahakali Zama A. Genetic analysis of Kit ligand functions during mouse spermatogenesis. J Androl. 2004;25:188–199. doi: 10.1002/j.1939-4640.2004.tb02779.x. [DOI] [PubMed] [Google Scholar]
- Boer PH, Adra CN, Lau YF, McBurney MW. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol. 1987;7:3107–3112. doi: 10.1128/mcb.7.9.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger-Tong HL, Johnston DS, Russell LD, Griswold MD, Bishop CE. Juvenile spermatogonial depletion (jsd) mutant seminiferous tubules are capable of supporting transplanted spermatogenesis. Biol Reprod. 2000;63:1185–1191. doi: 10.1095/biolreprod63.4.1185. [DOI] [PubMed] [Google Scholar]
- Bradley J, Baltus A, Skaletsky H, Royce-Tolland M, Dewar K, Page DC. An X-to autosome retrogene is required for spermatogenesis in mice. Nat Genet. 2004;36:872–876. doi: 10.1038/ng1390. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Caldwell KA, Handel MA. Protamine transcript sharing among postmeiotic spermatids. Proc Natl Acad Sci USA. 1991;88:2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Fisch TM, Prywes R, Roeder RG. c-fos sequence necessary for basal expression and induction by epidermal growth factor, 12-O-tetradecanoyl phorbol-13-acetate and the calcium ionophore. Mol Cell Biol. 1987;7:3490–3502. doi: 10.1128/mcb.7.10.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- Jones KA, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. Structural and transcriptional features of the mouse spermatid genome. J Cell Biol. 1975;65:258–270. doi: 10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Kominami K, Dohmae K, Nonomura N, Miki T, Okuyama A, Nishimune Y, Okabe M. Cessation of spermatogenesis in juvenile spermatogonial depletion (jsd/jsd) mice. Int J Urol. 1997;4:500–507. doi: 10.1111/j.1442-2042.1997.tb00293.x. [DOI] [PubMed] [Google Scholar]
- Mali P, Kaipia A, Kangasniemi M, Toppari J, Sandberg M, Hecht NB, Parvinen M. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev. 1989;1:369–382. doi: 10.1071/rd9890369. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–9. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Molecular evolution of the human Pgk-2 retroposon. Nucleic Acids Res. 1990;18:949–55. doi: 10.1093/nar/18.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Geyer CB, Yoshioka H. Epigenetic regulation of testis-specific gene expression. Ann NY Acad Sci. 2005;1061:226–242. doi: 10.1196/annals.1336.025. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- Meistrich ML. Separation of spermatogenic cells and nuclei from rodent testis. In: Prescott DM, editor. Meth Cell Biol. Vol. 15. Academic Press; New York: 1977. pp. 15–54. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Longtin J, Brock WA, Grimes SR, Jr, Mace ML. Purification of rat spermatogenic cells and preliminary biochemical analysis of these cells. Biol Reprod. 1981;25:1065–1077. doi: 10.1095/biolreprod25.5.1065. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Trostle PK, Frapart M, Erickson RP. Biosynthesis and localization of lactate dehydrogenase X in pachytene spermatocytes and spermatids of mouse testes. Dev Biol. 1977;60:428–441. doi: 10.1016/0012-1606(77)90140-3. [DOI] [PubMed] [Google Scholar]
- Ohta H, Tohda A, Nishimune Y. Proliferation and differentiation of spermatogonial stem cells in the W/Wv mutant mouse testis. Biol Reprod. 2003;69:1815–21. doi: 10.1095/biolreprod.103.019323. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yomogida K, Tadokoro Y, Tohda A, Dohmae K, Nishimune Y. Defect in germ cells, not in supporting cells, is the cause of male infertility in the jsd mutant mouse: proliferation of spermatogonial stem cells without differentiation. Int J Androl. 2001;24:15–23. doi: 10.1046/j.1365-2605.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- Reddi PP, Flickinger CJ, Herr JC. Round spermatid-specific transcription of the mouse SP-10 gene is mediated by a 294-base pair proximal promoter. Biol Reprod. 1999;61:1256–1266. doi: 10.1095/biolreprod61.5.1256. [DOI] [PubMed] [Google Scholar]
- Rohozinski J, Bishop C. The mouse juvenile spermatogonial depletion (jsd) phenotype is due to a mutation in the X-derived retrogene, mUtp14b. Proc Natl Acad Sci USA. 2004;101:11695–11700. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romrell LJ, Bellve AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Rosales R, Vigneron M, Macchi M, Davidson I, Xiao JH, Chambon P. In vitro binding of cell-specific and ubiquitous nuclear proteins to the octamer motif of the SV40 enhancer and related motifs present in other promoters and enhancers. EMBO J. 1987;6:3015–3025. doi: 10.1002/j.1460-2075.1987.tb02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpino S, Morena AR, Petersen C, Froysa B, Soder O, Boitani C. A rapid method of Sertoli cell isolation by DSA lectin, allowing mitotic analyses. Mol Cell Endocrinol. 1998;146:121–127. doi: 10.1016/s0303-7207(98)00190-7. [DOI] [PubMed] [Google Scholar]
- Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125:769–784. doi: 10.1530/rep.0.1250769. [DOI] [PubMed] [Google Scholar]
- Shetty G, Weng CCY. Cryptorchidism rescues spermatogonial differentiation in juvenile spermatogonial depletion (jsd) mice. Endocrinology. 2004;145:126–133. doi: 10.1210/en.2003-0928. [DOI] [PubMed] [Google Scholar]
- Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol Metab. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Renfree MB, Short RV. Successful intra- and interspecific male germ cell transplantation in the rat. Biol Reprod. 2003;68:961–967. doi: 10.1095/biolreprod.102.009480. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Deng X, Martin-DeLeon PA. Lack of sharing of Spam1 (Ph-20) among mouse spermatids and transmission ratio distortion. Biol Reprod. 2001;64:1730–1738. doi: 10.1095/biolreprod64.6.1730. [DOI] [PubMed] [Google Scholar]


