Abstract
TSG101 is a recently discovered tumor susceptibility gene whose functional inactivation in mouse fibroblasts results in cell transformation and the ability to form metastatic tumors in nude mice. Although restoration of TSG101 activity reverses tumorigenesis, neoplasia is irreversible in some cells, suggesting that permanent genetic alteration can occur during TSG101 inactivation. Here we describe studies that support this notion. We find that localization of TSG101 is cell cycle-dependent, occurring in the nucleus and Golgi complex during interphase, and in mitotic spindles and centrosomes during mitosis; cells made neoplastic by a deficiency in TSG101 expression show a series of mitosis-related abnormalities, including multiple microtubule organizing centers, aberrant mitotic spindles, abnormal distribution of metaphase chromatin, aneuploidy, and nuclear anomalies. Our findings suggest that TSG101 deficiency may lead to genome instability in addition to previously reported reversible neoplastic transformation.
Tsg101 is a tumor susceptibility gene discovered by using a procedure (random homozygous knockout; RHKO) that allows the identification of unknown genes whose functional inactivation in cells growing in culture produces a phenotype of interest (1). Functional inactivation of TSG101 in mouse NIH 3T3 fibroblasts produced the transformed cell line SL6, which forms colonies on 0.5% agar and metastasizing tumors when injected into athymic (“nude”) mice (1). Restoration of TSG101 function reversed the neoplastic properties of most of the SL6 cell population, implying that tumorigenesis is a direct consequence of TSG101 deficiency; however, some cells remained tumorigenic—suggesting that additional genetic changes may accumulate during TSG101 inactivation.
The mechanism(s) by which interference with TSG101 expression leads to neoplastic transformation and also to phenotypic alterations that persist after restoration of TSG101 function are not known. The occurrence in mouse (1) and human (2) TSG101 proteins of putative DNA-binding and transcriptional activation domains characteristic of transcription factors has suggested that the 43-kDa TSG101 protein may act in the cell nucleus to control gene expression. On the other hand, evidence that a coiled-coil domain within TSG101 can interact with a cytoplasmic phosphoprotein (i.e., stathmin) (3) implicated in microtubule dynamics (4, 5) as well as in cell growth and differentiation (6, 7) implies that TSG101 function may extend beyond the cell nucleus. Supporting this notion is the finding that the N-terminal domain of TSG101 resembles a group of apparently inactive homologues of ubiquitin-conjugating enzymes in which the active site amino acid residue has been replaced (8, 9). This observation has led to the suggestion that TSG101 may have a regulatory role in ubiquitin-mediated protein degradation.
To identify possible sites of action of the TSG101 protein and also to gain information about TSG101 function, we used immunohistochemical and fluorescence methods to investigate the subcellular location of TSG101 during different stages of the cell cycle and also the effects of TSG101 deficiency on key cellular structures. Our results show that the TSG101 protein is present in both the nucleus and cytoplasm of interphase cells, and that it colocalizes with microtubule organizing centers and mitotic spindles during mitosis. These findings, together with observations indicating that TSG101 deficiency is associated with reversible abnormalities in microtubules, mitotic spindles, and nuclei, suggest a mechanism by which reduced TSG101 expression in neoplastically transformed mouse fibroblasts may lead to genetic instability and consequently to the progression of neoplasia.
MATERIALS AND METHODS
Purification of Anti-TSG101 Antibody.
cDNA encoding the full-length murine TSG101 protein was cloned, under the control of the baculovirus polyhedrin promoter, in plasmid pAcC13 (a gift from Leah Conroy, Chiron Corp.) and introduced into insect Sf9 (Spodoptera frugiperda) cells by cotransformation with wild-type viral DNA. Recombinant virus selected by assay for plaques that had lost polyhedrin protein production was used to infect Sf9 cells for production of full-length TSG101 protein. Additionally, the TSG101 peptides indicated in Fig. 1 were synthesized by Research Genetics (Huntsville, AL), and antibodies were raised individually against these peptides and the full-length protein. For affinity purification, peptides were conjugated individually to SulfoLink columns (Pierce) and rabbit antisera were purified as recommended by the column manufacturer.
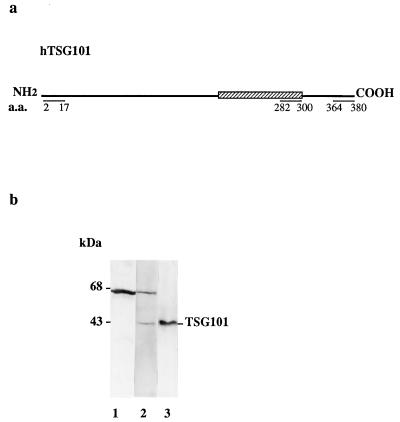
Figure 1.
Generation and analysis of anti-TSG101 antibody. (a) Map of TSG101 protein-coding region showing the location of the N-terminal (amino acids 2–17), coiled-coil domain (amino acids 282–300), and C-terminal peptides (amino acids 364–380), which are identical or highly conserved in human and mouse and were used as antigens. Peptide locations are designated according to the TSG101 sequence shown in ref. 2. (b) Polyclonal antibody detecting a band of 43 kDa in mammalian extract (lane 2); analysis using prebleed serum from the same rabbit (lane 1). Lane 3 shows affinity-purified antibody against the TSG101 coiled-coil domain detecting a single band, migrating at the same location as the mammalian signal, in an extract of Sf9 cells overexpressing TSG101. Purified antibodies generated against the N-terminal or C-terminal domains of TSG101 detected the same band.
Cell Culture and Synchronization.
Mouse fibroblast cell lines and the human HR5 cell line were grown in DMEM supplemented with penicillin G (200 units/ml), streptomycin (0.017%), and either 10% calf serum or 10% fetal bovine serum (FBS) (Life Technologies, Gaithersburg, MD). Mouse cell lines were synchronized in G0 by serum starvation for 48 hr and then allowed to progress into early, mid-, or late G1 by transfer to serum-supplemented (“complete”) growth medium for 4, 8, or 12 hr, respectively. G1/S or S phase synchronization was achieved by culturing in complete growth medium for 16 hr postserum starvation in 200 μM mimosine (Sigma) or 2 μg/ml aphidicolin (Sigma), respectively. Aphidicholin arrest was released in some plates by medium replacement, and cells entering G2/M or M phases were harvested 4 or 8 hr later, respectively (10). Cells were stained with chromomycin A3 as previous described (11) and analyzed by using fluorescence-activated cell sorting (FACS).
Immunofluorescence Staining and Analysis of Golgi Localization.
Cells grown on plates or coverslips were incubated in medium containing brefeldin A (Boehringer Mannheim) (5 μg/ml) for 90 min. Medium was removed by aspiration and cells were washed once with PBS, fixed in 3.7% formaldehyde (Sigma) for 20 min, permeabilized with 0.1% Nonidet P-40 (Sigma) for 20 min, and blocked by incubation in 3% FBS for 30 min. The cells were then incubated with primary antibody for 1 hr, washed in PBS, incubated with secondary antibody conjugated with the fluorescent dyes Oregon Green 514 or Texas Red X (Molecular Probes) for 1 hr, and washed extensively in PBS. Propidium iodide was added with the secondary antibody to stain the DNA for some analyses. TSG101 was stained by using the affinity-purified antibody. Tubulin was stained by using DM1α antibody (a gift from Tim Stearns, Stanford University), and the Golgi complex was stained by using anti-β-COP antibody (a gift from Suzanne Pfeffer, Stanford University) or Bodipy FL C5-ceramide (Molecular Probes). For competition studies, purified anti-TSG101 antibody was preincubated for 10 min at room temperature with 50-fold excess (wt/wt) of purified full-length TSG101 protein or TSG101 peptides, as indicated. The mixtures were then centrifuged at 14,000 rpm for 3 min, and the supernatants were used as primary antibodies.
Construction of GFP-TSG101 Fusion Construct.
Murine tsg101 cDNA was amplified by PCR by using 5′ and 3′ specific primers flanked by BamHI and EcoRI cleavage sites. The PCR product was treated with these two enzymes, and the resulting DNA was cloned into the BglII and EcoRI sites of pEGFP-C1 (CLONTECH). Positive clones were confirmed by restriction enzyme digestion.
Northern Blot Analysis.
Total RNA was extracted by using RNA STAT-60 (Tel-Test, Friendswood, TX) from cells synchronized at various stages of the cell cycle, as described above. Twenty-five micrograms of RNA per lane was electrophoresed on an 0.8% agarose gel overnight and transferred onto Hybond-N membrane (Amersham). After fixing the RNA onto the membrane by using UV crosslinking and oven-baking, the blot was prehybridized in Church’s buffer (12) for 1 hr. The 32P-labeled probe for TSG101 synthesized using single-strand PCR (13) was then allowed to hybridize to the target overnight. The blot was washed in Church’s wash buffer, exposed to x-ray film, stripped, and probed in the same fashion with a β-actin probe generated by using random hexamer priming.
RESULTS
Specificity of Anti-TSG101 Antisera.
Rabbit polyclonal antisera raised against the full-length TSG101 protein or against peptides comprising the N-terminal, C-terminal, and coiled-coil domains of TSG101, which are identical or highly conserved in mouse and human (Fig. 1a), recognized a 43-kDa band in extracts of NIH 3T3 (American Type Culture Collection) cells (Fig. 1b, lane 2) and also in extracts of Sf9 cells infected with baculovirus carrying a TSG101 overexpression construct (data not shown). This band, which was not detected by preimmune sera from the same rabbits (Fig. 1b, lane 1), corresponds to a protein of the size predicted from the TSG101 cDNA sequence; it was also detected in extracts of Sf9 cells overexpressing the full-length TSG101 protein by antibodies to the full-length protein, as well as by purified antibodies to all three peptides (Fig. 1b, lane 3). Antibodies generated against TSG101 sequences additionally detected an ≈67-kDa band, which reacted also with preimmune sera from these rabbits. All three antipeptide antibodies purified by peptide-affinity chromatography immunoprecipitated an 35S-labeled band of 43 kDa from NIH 3T3 cell extracts. Purified antibody generated against the coiled-coil domain was used for the immunofluorescence studies reported here, except where indicated.
Cell Cycle-Dependent Localization of the TSG101 Protein.
NIH 3T3 cells and Sense 8 cells [a clone of 3T3 cells that overexpresses the TSG101 protein (1)] were synchronized at G0, G1, G1/S, S, G2/M, and M stages of the cell cycle by using serum starvation, mimosine, or aphidocholin (10) as described in Materials and Methods. To confirm synchronization, cells collected at different times were stained with chromomycin A3 for DNA content (11) and examined by FACS. When stained with anti-TSG101 antibody, serum-starved cells showed limited amounts of TSG101 protein, which was present predominantly in the nucleus (Fig. 2a). In cycling cells, TSG101 protein was detected prominently both in nuclei, where it occurred diffusely as well as in discrete foci, and in the cytoplasm, where it was present asymmetrically in a perinuclear location during G1 (Fig. 2b). A fusion protein containing amino acid residues of both TSG101 and jellyfish green fluorescent protein (14, 15) also showed nuclear as well as cytoplasmic and asymmetric perinuclear localization in cycling cells (Fig. 2c).
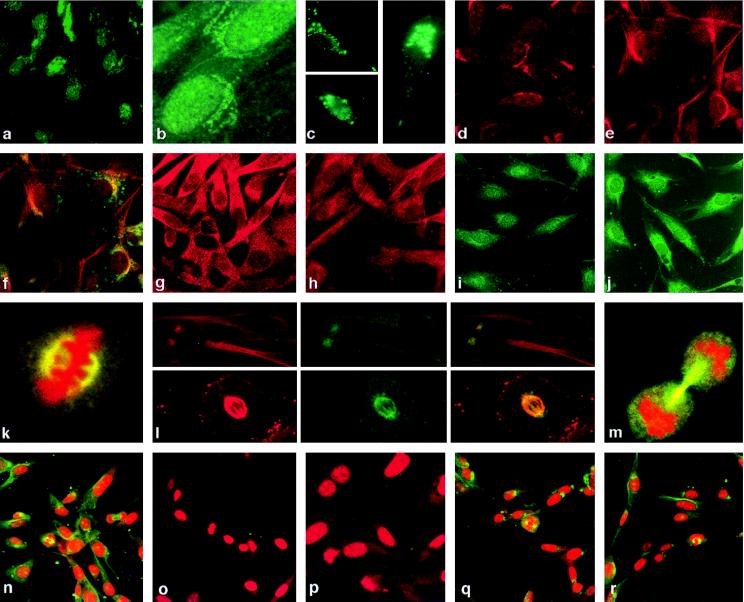
Figure 2.
Cell cycle-dependent localization of TSG101. Cells were synchronized at G0 (a), G1 (b), G1/S (i), and S (j) stages of the cell cycle and stained with affinity-purified anti-TSG101 antibody. GFP-TSG101 fusion protein localization in cycling cells: c Lower, nuclear staining; c Upper, diffuse cytoplasmic staining; c Right, asymmetric cytoplasmic staining. Cells stained with the Golgi-specific marker β-COP (d) and TSG101 (e). Cells costained with anti-TSG101 antibody (red) and Bodipy FL C5-ceramide (green) showing colocalization (yellow) (f) Brefeldin A-treated cells stained for β-COP (g) and TSG101 (h) showing cytoplasmic dispersion. Mitotic cells (k) showing staining of TSG101 (green) in mitotic spindle and propidium iodide-stained metaphase chromatin (red). (l Upper × 3) Localization of tubulin (red), TSG101 (green), and both (yellow) in centrosomes. (l Lower × 3) The staining of a mitotic spindle, as above. (m) Staining of TSG101 in midbody (green); condensed chromatin is stained in red by propidium iodide. Cells stained with propidium iodide (red) and purified anti-TSG101 antibody (green) (n), and with the same antibody preincubated with the antigen peptide (o) or two irrelevant TSG101 peptides (q and r), are compared with cells stained with prebleed from the same rabbit purified in the same fashion as the immune sera (p). All photographs except for GFP-fusion study were made by using confocal microscopy.
As seen in Fig. 2 d–f, the cellular location of TSG101 and the Golgi-specific markers β-COP and Bodipy FL C5-ceramide was identical, and costaining with anti-TSG101 antibody and Bodipy FL C5-ceramide showed colocalization. Treatment of cells, before fixation, with brefeldin A—which disrupts the Golgi complex by interference with protein transport from the endoplasmic reticulum and produces dispersed staining of Golgi-specific markers (16)—resulted in dispersion of staining for both β-COP (Fig. 2g) and TSG101 (Fig. 2h).
In G1/S (Fig. 2i), S phase (Fig. 2j), and some cells in G2/M populations, anti-TSG101 staining was observed to occur more diffusely in the cytoplasm than during G1. During mitosis, TSG101 was seen prominently in centrosomes and mitotic spindles, where it colocalized with tubulin (Fig. 2 k and l). Strong staining of the TSG101 protein in the midbody region, which contains the postmitotic remains of polar microtubules and exists as a tether between the two daughter cells (17), was observed during cytokinesis (Fig. 2m). Experiments using antibodies raised against TSG101 N-terminal or C-terminal domain peptides or against the full-length TSG101 protein purified from baculovirus-infected Sf9 cells confirmed the nuclear and cytoplasmic localization of TSG101 during interphase, which is shown in Fig. 2 using antibody against the coiled-coil domain. However, although microtubular structures were stained by each of two independently derived antisera raised against the N-terminal peptide, the coiled-coil domain peptide, or the full-length protein, antibodies against the C-terminal domain failed to detect these structures—raising the possibility that epitopes located in the C-terminal domain peptide are masked in microtubules, perhaps by interaction with other proteins. Preimmune serum from rabbits generating the anti-TSG101 antibodies used in these experiments detected only minimal and diffuse cytoplasmic staining during all stages of the cell cycle; no staining of specific cellular structures was seen, demonstrating that the observed staining of nuclei, Golgi complex, and microtubules by immune sera was TSG101-specific. Additionally, staining of these organelles (Fig. 2n), which was not observed in cells stained by the prebleed serum (Fig. 2p), was eliminated by preincubation of immune sera with a 50-fold excess of either purified full-length TSG101 protein or the peptide used to produce the immune serum (Fig. 2o). Two irrelevant TSG101 peptides had no effect (Fig. 2 q and r), further demonstrating the specificity of the reactions detected. Subcellular localization of TSG101 was similar in naive NIH 3T3 cells, NIH 3T3 cells overexpressing TSG101, and human HR5 (HeLa) cells.
TSG101-Specific RNA Remains Constant Through the Cell Cycle.
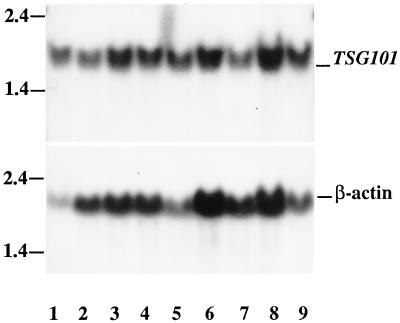
Analysis of RNA extracted from cells synchronized at various stages of the cell cycle showed that the steady-state level of TSG101-specific RNA remained constant throughout the cell cycle (Fig. 3).
Figure 3.
TSG101-specific RNA (Upper) at different stages of the cell cycle, with β-actin RNA control (Lower). Lanes 1–8, RNA extracted from cells synchronized in G0, early G1, mid-G1, late G1, G1/S, S, G2/M, and M stages of the cell cycle. Lane 9, RNA extracted from nonsynchronized cycling cells.
SL6 Cells Show Abnormal Mitotic Spindles, Aneuploidy, and Nuclear Anomalies.
SL6 cells are 3T3 derivatives that have undergone functional inactivation of the TSG101 gene by antisense RNA by using the RHKO procedure (1). To ensure activity of the transactivator, which is encoded by a fusion transcript specifying hygromycin resistance (1), we reselected and maintained cells in hygromycin (500 μg/ml and 200 μg/ml, respectively) before examination. Immunofluorescence analysis of these cells showed sharply reduced, but not totally absent, staining by anti-TSG101 antibody (Fig. 4a Lower), compared with NIH 3T3 cells (Fig. 4a Upper). Staining of SL6 cells with anti-tubulin antibody indicated that organization of the mitotic structures that formed after chromosome condensation was abnormal in this cell line. Multiple mitotic spindles/microtubule organizing centers were seen in 12 of 46 cells examined in metaphase, with some cells containing up to five distinct microtubule organizing centers (Fig. 4 b and c). By comparison, a single instance of an apparent mitotic defect was detected in 350 metaphase NIH 3T3 cells (Table 1). Two lines of NIH 3T3 cells made tumorigenic (K. Liu, L.L., and S.N.C., unpublished data) by functional inactivation of other genes showed no mitotic spindle abnormalities, indicating that the spindle abnormalities observed in SL6 cells are not a general property of transformed cells derived from NIH 3T3.
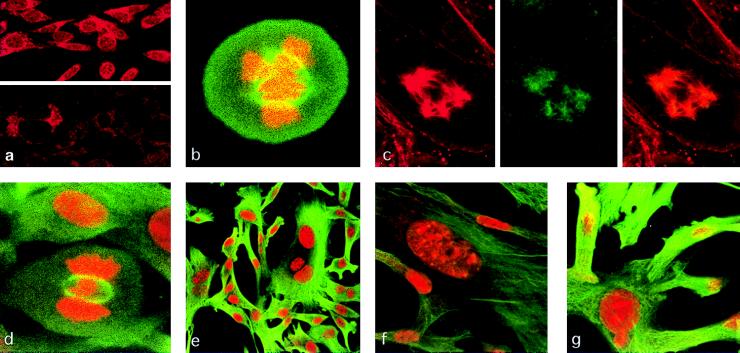
Figure 4.
Mitotic spindles, microtubule organizing centers, and nuclear morphology of SL6 cells. (a) NIH 3T3 (Upper) and SL6 (Lower) cells stained by anti-TSG101 antibody. (b) Cell stained by anti-tubulin antibody (green) and propidium iodide to detect DNA (red). (c) Tubulin (Left), TSG101 (Center), or both (Right) staining in an SL6 cell containing five centrosomes; other metaphase cells in this preparation showed similar abnormalities. (d) “H”-shaped metaphase DNA (red) and tubulin-stained mitotic spindle (green) in SL6 cell. (e–g) Nuclear abnormalities in SL6 cells, stained for tubulin (green) and DNA (red). All photographs were made by using confocal microscopy.
Table 1.
Frequency of mitotic spindle and nuclear abnormalities in cell lines examined
| Cell line | Abnormal spindle | Large nuclei | Multinucleated cells |
|---|---|---|---|
| NIH3T3 | 1/350 | 2/900 | 1/800 |
| SL6 | 12/46 | 15/100 | 20/100 |
| SL6ΔT | 3/200 | 10/400 | 11/400 |
| SL6T2 clones | |||
| 3-1 | 7/100 | 33/400 | 22/400 |
| 3-2 | 9/100 | 22/400 | 25/400 |
| 3-3 | 12/100 | 40/400 | 64/400 |
| 4-2 | 11/100 | 43/400 | 36/400 |
| 4-3 | 16/100 | 48/400 | 35/400 |
| 4-4 | 5/100 | 46/400 | 20/400 |
| 5-1 | 5/100 | 32/400 | 43/400 |
| 5-2 | 10/100 | 28/400 | 16/400 |
| 5-3 | 8/100 | 23/400 | 28/400 |
| 5-5 | 16/100 | 65/400 | 35/400 |
| 6-1 | 10/100 | 30/400 | 24/400 |
| 6-2 | 15/100 | 16/400 | 4/400 |
| 6-4 | 15/100 | 40/400 | 25/400 |
| 6-5 | 13/100 | 26/400 | 26/400 |
The transactivator vector pLLTX(1) was introduced by transfection into SL6ΔT cells, and selection was carried out for 10 days in the presence of hygromycin (500 μg/ml). Cells were stained with anti-tubulin antibody and propidium iodide and visualized by fluorescence microscopy. Mitotic spindle defects were scored based on the number of metaphase cells examined, and the nuclear abnormalities were scored based on the number of interphase cells examined.
In contrast to the microtubules of 3T3 and Sense 8 cells, both of which showed prominent staining with anti-TSG101 antibody (Fig. 2l), the aberrant microtubules and mitotic spindles of SL6 cells showed little staining; moreover, the microtubule organizing centers of SL6 cells that were treated with antibodies against tubulin (red) and TSG101 (green) showed only small patches of yellow costaining (Fig. 4c), unlike the microtubules of 3T3 cells (Fig. 2l). Interestingly, the condensed metaphase chromatin of some SL6 cells was H-shaped, with the arms of the “H” extending laterally to the mitotic spindles (Fig. 4d). Both mitotic spindles and chromatin appeared to be less compact in SL6 cells vs. the parental NIH 3T3 cell line.
SL6ΔT (1), an SL6-derived cell population that contains one insertionally inactivated tsg101 allele in each cell but lacks the LAP348 transactivator that turns off expression of the second tsg101 allele in SL6 (1), showed defects in microtubules or mitotic spindles in only 3 of 200 metaphase cells (c.f. mitotic spindle defects in >25% of SL6 metaphase cells). Thus, the mitotic abnormalities associated with TSG101 deficiency are largely reversible by restoration of tsg101 function—as is the tumorigenesis associated with TSG101 deficiency (1). To confirm that the high incidence of mitotic defects we observed in SL6 cells is not stochastic, we reintroduced the LAP 348 transactivator gene (1) into SL6ΔT, producing a series of SL6-like derivative clones (i.e., SL6T2 strains) that, because they are the products of separate transformation events, contain the transactivator gene at different chromosomal locations. As seen in Table 1, coincident with reintroduction of the transactivator, the incidence of multiple microtubule organizing centers or tripolar mitotic spindles rose from 1.5% in SL6ΔT to an average of 10% in 14 independently selected SL6T2 clones; 4 SL6T2 clones showed mitotic abnormalities in 15% or more of metaphase cells, approximating the frequency of such abnormalities in SL6.
In addition to showing frequent abnormalities in mitotic structures, SL6 cells also showed extensive aneuploidy, as indicated by the presence of multiple nuclei in more than 20% of cells, as determined by microscopy (Fig. 4e), and an increased fraction of cells having a chromosome number of >4N (1.1% in SL6 vs. 0.3% in 3T3 cells) by FACS analysis. Macronuclei (Fig. 4f), rare, large cells containing 3–5 nuclei with various sizes and DNA content, and cells showing nuclear bulges (Fig. 4g) were observed in an additional 15% of the cell population (Table 1). Whereas these abnormalities were sharply reduced in frequency when transactivation of the antisense promoter was reversed, a high frequency of nuclear abnormalities was again observed after reintroduction of the transactivator (Table 1): various SL6T2 clones showed either variably sized large nuclei and/or multiple nuclei in 10–25% of interphase cells. The parental NIH 3T3 cell line showed a large nucleus in 2 of 900 cells, and 1 cell in 800 contained what appeared to be two nuclei. Nuclear anomalies were similarly rare (less than 0.5% of 1600 interphase cells) in hygromycin-selected NIH3T3 cells that contain the pLLTX (1) transactivator vector only. The nuclear abnormalities and aneuploidy observed in SL6 and SL6T2 cells resemble the effects reported for cells treated with colchicine and other microtubule-poisoning drugs and chemicals (18–20). These findings, together with the high incidence of microtubule and mitotic spindle abnormalities seen in SL6 and SL6T2 cells, lead us to conclude that the observed anomalies are specific effects of reduced TSG101 expression secondary to transactivation of the promoter generating antisense RNA to tsg101 transcripts.
DISCUSSION
The ability of the TSG101 protein to shuttle between different locations during the cell cycle is consistent with the notion that this gene product may act at multiple sites within the cell. As cells progress through interphase, TSG101—which, during G0 and early G1 is observed predominantly in nuclei—appears also in the perinuclear Golgi complex and then becomes dispersed more generally throughout the cytoplasm when cells reach late S phase. Using immunoprecipitation and immunofluorescence methods, Zhong et al. (21) have observed that TSG101 is present mainly in cytoplasmic fractions. Our results indicate that during mitosis, the protein is localized in the mitotic apparatus, which, as we have shown, has a high frequency of abnormalities in cells deficient in TSG101 expression. Transient strong overexpression of TSG101 was observed in these experiments to lead to cell death (W.X. and S.N.C., data not shown). A number of other proteins implicated in the regulation of cell proliferation and/or the progression of cells through the cycle are also known to undergo cycle-dependent translocation, including cyclin-dependent kinases (22), calcium-calmodulin-dependent protein kinase II (23), and human cyclins B1 and B2 (24). One of these, calcium-calmodulin-dependent protein kinase II, is present in the cytoplasm and nucleus during interphase and in the mitotic apparatus during mitosis, as was observed for TSG101.
Although our data do not formally establish that the microtubule, mitotic spindle, and nuclear abnormalities we observed in SL6 cell mitotic structures are a direct consequence of the observed TSG101 deficiency in these structures, the association of these defects with reduced concentration of the TSG101 protein and their dependence on antisense inactivation of the tsg101 gene argue strongly for this conclusion. The high frequency of microtubule and mitotic spindle anomalies seen in SL6 cells was not observed in either the parental cell line or SL6ΔT cells, from which the transactivator regulating the antisense promoter that accomplishes homozygous inactivation of tsg101 has been excised. However, these abnormalities reappeared when the transactivator was reintroduced into SL6ΔT.
Despite the infrequency of microtubule and mitotic spindle abnormalities in SL6ΔT cells, which have one chromosomal copy of TSG101 inactivated by a retroviral insertion (1) and also were derived from a neoplastic cell population (i.e., SL6), SL6ΔT cells were not entirely normal; nuclear aberrations were observed at a somewhat higher frequency than in parental NIH 3T3 cells, and metaphase chromatin and microtubules both appeared to be less compact. Whether these effects are a consequence of loss of function of one tsg101 allele or result from secondary genetic changes that may have occurred during homozygous inactivation of TSG101 in the SL6 parent of SL6ΔT is not known. Although all of the SL6T2 clones we produced by reestablishment of functional homozygous knockout showed an increased incidence of mitotic and nuclear abnormalities, the frequency of these defects differed among the SL6T2 clones, presumably reflecting variable expression of the transactivator at different chromosomal insertion sites. In any case, because frequent mitotic and nuclear anomalies were observed in 14 independent SL6T2 clones generated from a population of SL6ΔT cells that showed a much lower frequency of abnormalities, the observed defects in SL6 and SL6T2 are unlikely to be a result of stochastic events unrelated to TSG101 deficiency.
Genome instability has long been known to be a key feature of neoplasia. Accumulating evidence indicates that such instability is associated not only with defects in DNA repair, but also with cell cycling defects and abnormalities in cell cycle checkpoints (25–27). In transgenic mouse cells that express the simian virus 40 tumor antigen and in S. cerevisiae mutants that uncouple the duplication of mitotic spindle pole bodies from DNA replication, mitotic spindle defects can lead to multipolar mitosis and aneuploidy (28, 29). Whereas studies of neoplastic transformation have focused largely on the role of “gatekeeper genes,” which regulate the growth of tumors directly by inhibiting cell growth or promoting death, it has become increasingly evident that “caretaker” genes, whose inactivation can promote genetic instability and consequently increase the frequency of mutation of all genes (including gatekeepers and caretakers), may have an important role in neoplasia, particularly in tumor progression (25, 26).
Earlier work has shown that TSG101 inactivation in NIH 3T3 fibroblasts is associated with neoplastic transformation (1). The irreversibility of neoplasia in some cells after the restoration of TSG101 activity is consistent with the notion that the TSG101 gene product may also have a “caretaker” function that may lead to other genetic alterations, facilitating cancer progression. Our finding that TSG101 colocalizes with microtubules during mitosis, and the demonstration that defective TSG101 expression in SL6 cells is associated with multiple microtubule organizing centers, abnormal mitotic spindles, aneuploidy, and nuclear anomalies, imply that the postulated caretaker function of TSG101 may be mediated at the level of mitosis.
Acknowledgments
We thank Leah Conroy of Chiron Corporation for helping us express TSG101 in baculovirus-infected Sf9 cells and also Tim Stearns, Uta Francke, and Wen-Hwa Lee for helpful discussions and comments on the manuscript. This work was supported in part by the 1993 Helmut Horten Research Award (S.N.C.) and by a gift from Chiron. W.X. is the recipient of a Markey Foundation Predoctoral Fellowship Award.
References
- 1.Li L, Cohen S N. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Li X, Francke U, Cohen S N. Cell. 1997;88:143–154. doi: 10.1016/s0092-8674(00)81866-8. [DOI] [PubMed] [Google Scholar]
- 3.Maucuer A, Camonis J H, Sobel A. Proc Natl Acad Sci USA. 1995;92:3100–3104. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marklund U, Larsson N, Melander H, Brattsand G, Gullberg M. EMBO J. 1996;15:5290–5298. [PMC free article] [PubMed] [Google Scholar]
- 5.Belmont L D, Mitchison T J. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 6.Marklund U, Brattsand G, Osterman O, Ohlsson P I, Gullberg M. J Biol Chem. 1993;268:25671–25680. [PubMed] [Google Scholar]
- 7.Sobel A. Trends Biochem Sci. 1991;16:301–305. doi: 10.1016/0968-0004(91)90123-d. [DOI] [PubMed] [Google Scholar]
- 8.Koonin E, Abagyan R. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 9.Ponting C P, Cai Y-D, Bork P. J Mol Med (Berlin) 1997;75:467–469. [PubMed] [Google Scholar]
- 10.Williams R T, Wu L, Carbonara-Hall D A, Tolo V T, Hall F L. J Biol Chem. 1993;268:8871–8880. [PubMed] [Google Scholar]
- 11.Gray J W, Coffino P. Methods Enzymol. 1979;58:233–248. doi: 10.1016/s0076-6879(79)58140-3. [DOI] [PubMed] [Google Scholar]
- 12.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bednarczuk T A, Wiggins R C, Konat G W. Biotechniques. 1991;10:478. [PubMed] [Google Scholar]
- 14.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 15.Kain S R, Adams M A, Kondepudi A, Yang T T, Ward W W, Kitts P. Biotechniques. 1995;19:650–655. [PubMed] [Google Scholar]
- 16.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara K. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 17.Sellitto C, Kuriyama R. J Cell Biol. 1988;106:431–440. doi: 10.1083/jcb.106.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, Alder I-D. Mutagenesis. 1990;5:371–374. doi: 10.1093/mutage/5.4.371. [DOI] [PubMed] [Google Scholar]
- 19.Miller B M, Alder I-D. Mutagenesis. 1992;7:69–76. doi: 10.1093/mutage/7.1.69. [DOI] [PubMed] [Google Scholar]
- 20.Bonatti S, Cavalieri Z, Viaggi S, Abbondandolo A. Mutagenesis. 1992;7:111–114. doi: 10.1093/mutage/7.2.111. [DOI] [PubMed] [Google Scholar]
- 21.Zhong Q, Chen C-F, Chen Y, Chen P-L, Lee W-H. Cancer Res. 1997;57:4225–4228. [PubMed] [Google Scholar]
- 22.Gallant, P., Fry, A. M. & Nigg, E. A. (1995) J. Cell Sci.19, Suppl., 21–28. [DOI] [PubMed]
- 23.Ohta Y, Ohba T, Miyamoto E. Proc Natl Acad Sci USA. 1990;87:5341–5345. doi: 10.1073/pnas.87.14.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackman M, Firth M, Pines J. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinzler K W, Vogelstein B. Nature (London) 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell L H, Weinert T A, Kadyk L, Garvik B. The Molecular Genetics of Cancer. Cold Spring, NY: Cold Spring Harbor Lab. Press; 1994. pp. 259–263. [DOI] [PubMed] [Google Scholar]
- 27.Hartwell L H, Weinert T A. Science. 1989;246:629–635. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 28.Levine D S, Sanchez C A, Rabinovitch P S, Reid B J. Proc Natl Acad Sci USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baum P, Yip C, Goetsch L, Byers B. Mol Cell Biol. 1988;8:5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]