Abstract
Intracellular transport is best understood for how proteins are shuttled among different compartments of the secretory pathway by membrane-bound transport carriers. However, it remains unclear whether regulation of this transport is modulated by the transported (cargo) proteins in the lumen of transport pathways. In the early secretory pathways that connect the endoplasmic reticulum (ER) and the Golgi complex, the small GTPase ADP-ribosylation factor 1 (ARF1) recruits a cytosolic coat protein complex named COPI onto membranes as a key step in the formation of transport vesicles. Transport of newly synthesized proteins that leave the ER includes a class of cargo proteins with a sequence motif of KDEL. When these KDEL proteins leave the ER to reach the Golgi complex, they are recognized by their receptor and transported retrograde in COPI-coated vesicles back to the ER. We now demonstrate that stimulation of the KDEL receptor by a KDEL protein enhances an interaction between the KDEL receptor and a GTPase-activating protein for ARF1. As a result, more cytosolic GTPase-activating protein is recruited to membranes to inactivate ARF1. Thus, the KDEL proteins are examples of luminal cargo proteins that regulate transport by activating their receptor. Most likely, this regulation affects retrograde transport from the Golgi complex to the ER, as activated KDEL receptor appears to reside only in retrograde COPI-coated vesicles.
Guanine nucleotide exchange factors catalyze the exchange of GDP for GTP on ADP-ribosylation factor 1 (ARF1) (1–4). This activation of ARF1 recruits the cytosolic coat protein complex COPI to membranes to regulate the formation of COPI-coated transport vesicles (5, 6). The GTPase cycle of ARF1 is completed when a GTPase-activating protein (GAP) (7, 8) catalyzes the hydrolysis of GTP to GDP on ARF1. This inactivation of ARF1 results in membrane-bound COPI being released to the cytosol so that transport vesicles can fuse with their target compartment (9). Little is known about whether the regulators of ARF1 drive the GTPase cycle of ARF1 in an autonomous fashion or whether their catalytic activities might be subject to regulation.
In the early secretory pathways, an intracellular, multispanning membrane receptor recognizes a carboxy terminal motif of lysine-aspartate-glutamate-leucine (KDEL) on luminal soluble proteins in the endoplasmic reticulum (ER) and retrieves those KDEL proteins that have leaked to the Golgi complex back to the ER (10). The KDEL proteins function as chaperones for protein folding and assembly in the ER (11). The receptor for the KDEL proteins originally was identified through mutant yeasts that failed to retrieve KDEL proteins, and thus was named ER retention defective complementation group 2 (ERD2) (12). A clue that the KDEL receptor not only retrieves KDEL proteins, but also may regulate intracellular transport came from observations of mutant yeasts that had the KDEL receptor deleted. These yeast cells had profoundly enlarged and disorganized Golgi complexes with dysregulated transport in the early secretory system (12). Overexpression of the KDEL receptor in mammalian cells resulted in perturbations of transport similar to those induced by inhibiting ARF1 guanine nucleotide exchange factor activity (13), suggesting that the KDEL receptor might regulate intracellular transport through ARF1.
Identifying proteins that affect the catalytic activity of either the guanine nucleotide exchange factors or GAP of ARF1 has been difficult, because the direct approach of assessing the activation state of ARF1 is unfeasible, as GTP binding to ARF1 is unstable (14, 15). Studies on the regulation of other small GTPases have circumvented similar obstacles by taking advantage of perturbations in effector functions that can be attributed specifically to exaggerating either the activated (GTP-bound) or inactivated (GDP-bound) state of a small GTPase (16, 17). ARF1 inactivation results in COPI being released from Golgi membranes to the cytosol (9). The uncoating of Golgi membranes causes the Golgi complex to fuse with the ER, which results in a transport block out of the fused compartment (18). By using this phenotype of ARF1 inactivation, we recently demonstrated that the overexpression of the KDEL receptor inactivated ARF1 by enhancing the recruitment of cytosolic ARF1 GAP to membranes, and thus, enhancing the ability of GAP to hydrolyze the membrane-restricted, GTP-bound form of ARF1 (19).
In this study, we show that overexpression of a KDEL protein enhances an interaction between the KDEL receptor and ARF1 GAP that results in ARF1 inactivation. Thus, the KDEL proteins are examples of luminal cargo proteins that provide a signal to regulate retrograde transport from the Golgi complex to the ER, as activated KDEL receptor has been implicated to reside only in retrograde COPI-coated vesicles (20).
MATERIALS AND METHODS
Cells and Antibodies.
HeLa cells were grown as previously described (19). The following antibodies were used and have been previously described (19): mouse mAb against the myc epitope, mouse mAb against the hemagglutinin (HA) epitope, mouse mAb against the Tac antigen, and rabbit polyclonal anti-ARF1 GAP antiserum. A mouse mAb (F10.6.6) against lysozyme was kindly provided by R. Poljak (Institut Pasteur, Paris, France). Fluorescein-conjugated donkey antibody against mouse IgG and indocarbocyanine-conjugated donkey antibody against rabbit IgG were obtained from Jackson ImmunoResearch.
Plasmids and Transfection.
The following cDNAs were used and have been previously described: HA- and myc-tagged human KDEL receptor (19), chimeric Tac-E19 (13), and myc-tagged human KDEL receptor defective in ligand binding (R169N) (21) (kindly provided by H.R.B. Pelham, Medical Research Council, Cambridge, UK). The tetrapeptides, AARL and KDEL, were appended to the carboxyl terminus of lysozyme by mutagenizing the lysozyme cDNA and using the PCR as described (13). The amplified cDNAs were subcloned into a modified mammalian expression vector pCDLSRα, pXS (13), and verified by DNA sequencing (Sequenase, United States Biochemical). For inducible expression, the cDNA encoding lysozyme-KDEL was subcloned into a modified expression vector, pVAC, that had been generated by replacing the Rous sarcoma virus promoter in the mammalian episomal expression vector pREP7 (Invitrogen) with a synthetic promoter composed of five tandem repeats of the glucocorticoid response element (GRE5) (22).
Transient transfection with the calcium-phosphate method was performed as previously described (13). For stable transfections, constructs were transfected into HeLa cells by the calcium-phosphate method and selected in complete medium containing 250 μg/ml of hygromycin. Inducible constructs were expressed by adding 100 nM of dexamethasone for 24 hr. Transfectants were screened by immunofluorescence microscopy.
Microscopy and Biochemistry Studies.
Immunofluorescence microscopy, immunogold electron microscopy, coprecipitation of KDEL receptor with GAP, and Golgi-specific glycosylation of Tac-E19 were performed as previously described (19).
RESULTS
As KDEL proteins bind the KDEL receptor at the Golgi complex (10), we sought to enhance the activation of the KDEL receptor by overexpressing a chimeric KDEL protein with the KDEL motif appended to the carboxyl terminus of lysozyme. This chimera previously has been shown to deliver high levels of KDEL proteins to the Golgi complex, because lysozyme lacks an ER retention domain that is responsible for retaining much of the endogenous KDEL proteins in the ER (23). The lysozyme-KDEL cDNA was subcloned into an inducible expression vector (22), and then stably integrated into a previously generated cell line that expressed myc-tagged KDEL receptor constitutively (15). As epitope tagging had been demonstrated previously not to affect the function of the KDEL receptor (10, 13), including its interaction with GAP (19), we used this epitope tagging approach, because it facilitated subsequent experiments that required the detection of two different forms of the receptor within the same cell.
KDEL Proteins Modulate an Interaction Between the KDEL Receptor and ARF1 GAP.
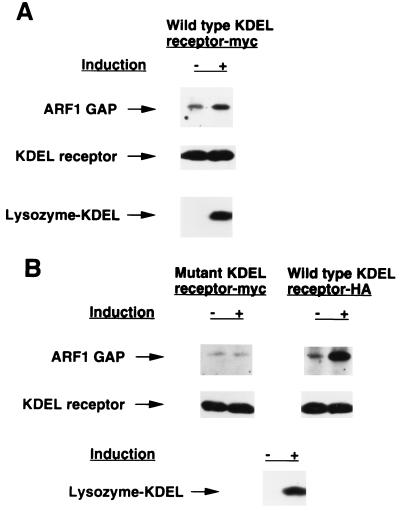
In the newly generated cell line, when expression of lysozyme-KDEL was not induced, an association between the KDEL receptor and GAP was observed by a coprecipitation approach (19), by immunoprecipitating for the myc-tagged receptor followed by immunoblotting for GAP (Fig. 1A). On maximal induction for lysozyme-KDEL, this association was increased, as assessed by equivalent levels of KDEL receptor immunoprecipitated from induced and uninduced cells (Fig. 1A). To assess whether this increased association between the KDEL receptor and GAP required the recognition of the KDEL sequence by the receptor, we generated a cell line with inducible expression for lysozyme-KDEL that stably expressed both an HA-tagged wild-type receptor and a myc-tagged mutant receptor that was defective in ligand binding. Without induction, the association of the mutant receptor with GAP was less than that of the wild-type receptor with GAP, when equivalent levels of mutant and wild-type receptors were assessed (compare uninduced lanes of myc-tagged receptors in Fig. 1 A and B). On induction, the mutant receptor did not associate more with GAP, whereas as a positive control, the wild-type receptor in the same cell line associated more with GAP (Fig. 1B). These results suggested that the increased association between the KDEL receptor and GAP that was induced by KDEL ligand overexpression was mediated specifically through the recognition of the KDEL sequence by the receptor. Moreover, as a ligand-binding defective receptor showed less association with GAP than the wild-type receptor in the uninduced state, this result suggested that the association of the KDEL receptor with GAP normally was modulated by endogenous KDEL proteins that cycled between the ER and the Golgi complex.
Figure 1.
KDEL proteins modulate the association of the KDEL receptor with GAP. (A) The association between the KDEL receptor and GAP is enhanced by the overexpression of lysozyme-KDEL. A HeLa cell line that stably expresses myc-tagged KDEL receptor was either induced or uninduced for lysozyme-KDEL overexpression, and then lysed and immunoprecipitated with an anti-myc antibody followed by immunoblotting with an anti-ARF1 GAP antiserum (Top) or an anti-myc antibody (Middle). The same cell lysate also was immunoprecipitated with an anti-lysozyme antibody followed by Western blotting for lysozyme to assess the level of lysozyme-KDEL expressed (Bottom). (B) The association of KDEL receptor and GAP requires the specific recognition of the KDEL sequence by the receptor. A HeLa cell line that stably expresses both HA-tagged wild-type receptor and myc-tagged mutant receptor was either induced or uninduced for lysozyme-KDEL overexpression, and then immunoprecipitated with either an anti-myc antibody (Left) or an anti-HA antibody (Right) followed by immunoblotting with either an anti-ARF1 GAP antiserum (Top) or the appropriate anti-epitope tag antibody (Middle). The same cell lysate also was immunoprecipitated with an anti-lysozyme antibody followed by Western blotting for lysozyme to assess the level of lysozyme-KDEL expressed (Bottom).
Activation of KDEL Receptor by KDEL Proteins Recruits ARF1 GAP to Membranes.
Because an increased association between the KDEL receptor and GAP caused by receptor overexpression resulted in an enhanced recruitment of cytosolic GAP to membranes (19), we next examined whether receptor stimulation by ligand overexpression also would increase GAP recruitment to membranes. By immunogold electron microscopy, labeling for GAP on membranes of the early secretory system increased on induction of lysozyme-KDEL (Fig. 2). On quantitation, the level of GAP on membranes on induction was 3-fold greater than that detected without induction (Table 1). This degree of enhancement was less than the 8-fold increase in GAP recruitment that was seen in our previous study of GAP on membranes in response to receptor overexpression (19). As KDEL ligand overexpression did not induce a phenotype of ARF1 inactivation (10), whereas KDEL receptor overexpression did (13), these observations were consistent with our previous finding that the level of GAP recruitment to membranes determined whether a phenotype of ARF1 inactivation was achieved (19).
Figure 2.
Overexpression of lysozyme-KDEL increases GAP recruitment to membranes of the early secretory system. A HeLa cell line with inducible expression of lysozyme-KDEL was either uninduced (A) or induced (B), and then examined by immunogold electron microscopy using an anti-ARF1 GAP antiserum (10 nm gold) (bar, 200 nm). g, Golgi; m, mitochondrion; n, nucleus. Quantitation of enhanced GAP recruitment upon induction of lysozyme-KDEL is shown in Table 1.
Table 1.
Overexpression of lysozyme-KDEL enhances the recruitment of ARF1 GAP to membranes
| Overexpressed construct | # ARF1-GAP/ unit membrane |
|---|---|
| None | 3.1 ± 0.21 |
| Lysozyme-KDEL | 8.3 ± 0.32 |
Cells were either uninduced or induced for the overexpression of lysozyme-KDEL, and then fixed and labeled for ARF1-GAP. At a calibrated magnification, anti-ARF1-GAP labeling with 10 nm gold along nuclear ER was counted. Statistical difference was P < 1 × 10−4 for the level of ARF1 GAP between uninduced and induced cells, where n = 90 for each set, as assessed by the unpaired t test.
Expression Level of KDEL Receptor Limits the Ability of KDEL Proteins to Inactivate ARF1.
To understand why overexpression of the KDEL receptor, but not its KDEL ligands, resulted in a phenotype of ARF1 inactivation, we examined whether the endogenous level of the receptor was a limiting factor. The phenotype of ARF1 inactivation is characterized by COPI redistribution to the cytosol, Golgi redistribution to the ER, and a secretion block (19). Because Golgi redistribution to the ER was the most specific phenotypic manifestation reflecting ARF1 inactivation, a biochemical assay had been established that quantitatively measured the degree of Golgi redistribution in response to the overexpression of the KDEL receptor (13). Normally, glycoproteins that transit through the medial Golgi complex acquire complex glycosylation that renders them resistant to endoglycosidase H (endo H). However, an ER glycoprotein that does not reach the medial Golgi complex remains sensitive to endo H, unless the Golgi complex (with its resident glycosidases) is redistributed to the ER. Thus, we examined for Golgi redistribution to the ER through the acquisition of endo H resistance by an ER glycoprotein, Tac-E19 (13). In a titration experiment, increasing levels of the KDEL receptor led to proportionally increased degree of Tac-E19 becoming endo H resistant, suggesting that Golgi redistribution to the ER was increased proportionally (Fig. 3A).
Figure 3.
KDEL proteins modulate the ability of the KDEL receptor to induce the phenotype of ARF1 inactivation, as assessed by Golgi redistribution to the ER. (A) Increasing receptor expression increases Golgi redistribution to the ER. HeLa cells were transiently transfected with increasing amounts of a plasmid encoding the KDEL receptor, and then assessed for Golgi-specific glycosylation of Tac-E19 (Upper). Direct immunoblotting of whole-cell lysates with an anti-myc antibody reveals increasing level of receptor expression (Lower). (B) The co-overexpression of lysozyme-KDEL and the KDEL receptor enhances the ability of the receptor to induce Golgi redistribution to the ER. HeLa cells were transiently transfected with either nothing or a plasmid encoding lysozyme-KDEL and increasing amounts of a plasmid encoding the KDEL receptor, and then assessed for Golgi-specific glycosylation of Tac-E19. The ratio of endo H-resistant to endo H-sensitive Tac-E19 is quantified for three separate experiments, and the bar graph represents the mean with standard error. There is a significant difference (P < 10−4) in the effect of co-overexpressing the KDEL receptor with its KDEL ligand (KDEL) versus overexpressing the receptor alone (None), by the two-way factorial ANOVA and contrast test. (C) The co-overexpression of lysozyme-AARL and the KDEL receptor does not enhance the ability of the receptor to induce Golgi redistribution to the ER. HeLa cells were transiently transfected with either nothing or a plasmid encoding lysozyme-AARL and increasing amounts of a plasmid encoding the KDEL receptor, and then assessed for Golgi-specific glycosylation of Tac-E19. The results of three separate experiments are quantified, and their mean with standard error are represented by a bar graph. There is no significant difference (P = 0.96) in comparing the effects of overexpressing the KDEL receptor with a control ligand (AARL) versus overexpressing the receptor alone (None), by the two-way factorial ANOVA and contrast test.
This ability to titrate the level of KDEL receptor expression and induce a progressively greater degree of ARF1 inactivation, as assessed by Golgi redistribution to the ER, allowed us to test whether the KDEL proteins modulated the ability of their receptor to inactivate ARF1. In the same receptor titration experiment, we overexpressed a fixed level of lysozyme-KDEL with increasing levels of the KDEL receptor, and again assessed for ARF1 inactivation through the acquisition of endo H-resistant Tac-E19. With increasing levels of KDEL receptor expression, the co-overexpression of its ligand increased the proportion of Tac-E19 that became endo H resistant (Fig. 3B). As a control, a lysozyme chimera bearing a carboxy terminal tetrapeptide of AARL that was not recognized by the KDEL receptor (10, 23) also was co-overexpressed with increasing levels of the KDEL receptor. This combination resulted in no significant increase in Tac-E19 becoming endo H resistant over the effect of increasing receptor expression alone (Fig. 3C). Thus, these results suggested that the KDEL proteins modulated the ability of the KDEL receptor to inactivate ARF1.
DISCUSSION
This study suggests that KDEL proteins, as a class of luminal cargo proteins, regulate transport in the early secretory pathways by modulating the ability of the KDEL receptor to inactivate ARF1. Overexpression of a prototypic KDEL protein, lysozyme-KDEL, stimulates more KDEL receptors to associate with ARF1 GAP that results in enhanced GAP recruitment to membranes. However, this degree of GAP recruitment is less than that induced by the overexpression of the KDEL receptor. Moreover, the ability of overexpressed KDEL proteins to induce a phenotype of ARF1 inactivation is dependent on the expression level of KDEL receptor. Thus, our findings suggest that KDEL proteins are capable of affecting the activation status of ARF1 through the KDEL receptor, but the level of the endogenous receptor normally limits this ability. Consistent with this explanation, overexpression of the KDEL receptor induces the phenotype of ARF1 inactivation (13), whereas overexpression of KDEL ligands does not (10).
The mechanism of transport regulation by the KDEL proteins has remarkable similarities to a general mechanism of signal transduction, as exemplified by growth factor receptor signaling (24). Although growth factor receptor signaling and intracellular transport perform two very different cellular functions, they appear to use a fundamentally similar mechanism of regulation. In both cases, ligand-induced activation of a receptor recruits cytosolic effectors to their site of action on membranes. This recruitment controls a class of signal transducing molecules, the small GTPases.
In this context, it seems appropriate to discuss how this signaling process might regulate COPI-mediated transport in the early secretory system. The KDEL receptor with bound KDEL protein has been implicated recently to reside only in retrograde COPI-coated vesicles (20). Thus, as our data suggest that the bound form of the KDEL receptor also recruits GAP, it seems likely that this GAP recruitment regulates COPI-mediated transport in the retrograde direction. This possibility implies that the catalytic activity of GAP must be regulated during the formation of COPI-coated vesicles, as ARF1 activation is required to recruit COPI to form COPI-coated vesicles (5, 6). In this regard, it would seem the most illuminating to examine studies of transport by COPII-coated vesicles, where the mechanistic detail of how an ARF-like GAP regulates vesicular transport is better understood (25).
Activation of an ARF-like small GTPase (Sar1p) recruits COPII to form COPII-coated vesicles (26). In this case, a GAP for Sar1 (Sec23p) is recruited and is required for the initial stage of vesicle formation (27). The exact nature of this requirement during vesicle formation remains unclear, but the catalytic activity of Sec23p would need to be regulated at this initial stage that requires both Sec23p and activated Sar1p (26). After coated vesicle formation, the catalytic activity of Sec23p proceeds to inactivate Sar1p that then allows COPII-coated vesicles to uncoat (28). By analogy, we propose that the KDEL receptor recruits an ARF1 GAP whose catalytic activity inactivates ARF1 so that COPI-coated vesicles that are formed subsequently can uncoat (9). Thus, the catalytic activity of ARF1 GAP also would need to be regulated during the formation of COPI-coated vesicles. How this occurs and whether ARF1 GAP plays a role during vesicle formation remain to be determined.
Regardless of the mechanistic detail, the finding that ARF1 GAP is recruited by activated KDEL receptor and that this recruitment most likely affects retrograde transport in the early secretory system addresses a long-standing question in intracellular transport: whether cargo proteins can regulate their trafficking pathway. This fundamental question has been studied intensely in the peripheral transport compartments where ligand-induced receptor activation at the cell surface results in receptor endocytosis as a mechanism of down-regulating receptor function. In most cases, receptor activation appears to sort a receptor into nascent transport carriers that already are formed by the assembly of clathrin coat proteins on membranes (coated pits) (29, 30). However, it remains unclear whether an activated receptor can ever affect the recruitment of clathrin coat proteins, and thus, suggesting a role for some cargo proteins in regulating their transport pathway. This uncertainty is attributable in part to key factors that would be predicted to regulate the recruitment of clathrin coat proteins, such as an ARF-like small GTPase and its regulatory GAP and guanine nucleotide exchange factors, have yet to be identified (25, 31). In studying the early secretory system where the mechanistic detail of transport regulation is better understood (32, 33), our results suggest that some cargo proteins, such as the KDEL proteins, provide a regulatory signal from the lumen of transport pathways to modulate a critical cytosolic regulator of transport, ARF1. Thus, transport regulation can emanate from transported proteins themselves.
Acknowledgments
We thank Frank Austen, Michael Brenner, and Hans Geuze for support; Dan Cassel and Tom Rapaport for advice; Juan Bonifacino, James Casanova, Jennifer Lippincott-Schwartz, Steven Porcelli, and Marianne Wessling-Resnick for critical reading of the manuscript; Andreas Ambach, Joelle Gaschet, and other members of our respective laboratories for fruitful discussions; and Hugh Pelham for the mutant KDEL receptor. This work is supported in part by the Arthritis Foundation (V.W.H.), the Charles H. Hood Foundation (V.W.H.), and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (T.A.).
ABBREVIATIONS
- ER
endoplasmic reticulum
- ARF1
ADP-ribosylation factor 1
- GAP
GTPase-activating protein
- HA
hemagglutinin
- endo H
endoglycosidase H
References
- 1.Peyroche A, Paris S, Jackson C L. Nature (London) 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 2.Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 3.Morinaga N, Tsai S-C, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meacci E, Tsai S-C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serafini T, Orci L, Amherdt M, Brunner M, Kahn R A, Rothman J E. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson J G, Cassel D, Kahn R A, Klausner R D. Proc Natl Acad Sci USA. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cukierman E, Huber I, Rotman M, Cassel D. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 8.Poon P P, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer R A, Johnston G C. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms J B, Rothman J E. J Cell Biol. 1993;123:1365–1371. doi: 10.1083/jcb.123.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis M J, Pelham H R. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- 11.Pelham H R. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- 12.Semenza J C, Hardwick K G, Dean N, Pelham H R. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 13.Hsu V W, Shah N, Klausner R D. Cell. 1992;69:625–635. doi: 10.1016/0092-8674(92)90226-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss O, Holden J, Rulka C, Kahn R A. J Biol Chem. 1989;264:21066–21072. [PubMed] [Google Scholar]
- 15.Franco M, Chardin P, Chabre M, Paris S. J Biol Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- 16.Lowy D R, Willumsen B M. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 17.Machesky L M, Hall A. Trends Cell Biol. 1996;6:304–310. doi: 10.1016/0962-8924(96)10026-x. [DOI] [PubMed] [Google Scholar]
- 18.Klausner R D, Donaldson J G, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoe T, Cukierman E, Lee A, Cassel D, Peters P J, Hsu V W. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner T H, Rothman J E. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 21.Townsley F M, Wilson D W, Pelham H R. EMBO J. 1993;12:2821–2829. doi: 10.1002/j.1460-2075.1993.tb05943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mader S, White J H. Proc Natl Acad Sci USA. 1993;90:5603–5607. doi: 10.1073/pnas.90.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro S, Pelham H R. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 24.Heldin C H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 25.Schekman R, Orci L. Science. 1996;271:1526–1532. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 26.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 27.Yoshihisa T, Barlowe C, Schekman R. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 28.Oka T, Nakano A. J Cell Biol. 1994;124:425–434. doi: 10.1083/jcb.124.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchhausen T, Bonifacino J S, Riezman H. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 30.Santini F, Keen J H. J Cell Biol. 1996;132:1025–1036. doi: 10.1083/jcb.132.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 32.Lippincott-Schwartz J. Trends Cell Biol. 1993;3:81–88. doi: 10.1016/0962-8924(93)90078-f. [DOI] [PubMed] [Google Scholar]
- 33.Cosson P, Letourneur F. Curr Opin Cell Biol. 1997;9:484–487. doi: 10.1016/s0955-0674(97)80023-3. [DOI] [PubMed] [Google Scholar]